Abstract
Introduction
Previous studies have shown that individuals with human papillomavirus (HPV)‐related cervical lesions have different prognoses according to the HPV genotype. However, these studies failed to account for possible diagnostic misclassification. In this retrospective cohort study, we aimed to clarify the natural course of cervical lesions according to HPV genotype to account for any diagnostic misclassification.
Materials and Methods
Our cohort included 729 patients classified as having cervical intraepithelial neoplasia (CIN). HPV was genotyped in all patients, who were followed up or treated for cervical lesions at the University of Tokyo Hospital from October 1, 2008 to March 31, 2015. Hidden Markov models were applied to estimate the diagnostic misclassification probabilities of the current diagnostic practice (histology and cytology) and the transitions between true states. We then simulated two‐year transition probabilities between true cervical states according to HPV genotype.
Results
Compared with lesions in patients with other HPV genotypes, lesions in HPV 16‐positive patients were estimated to be more likely to increase in severity (i.e., CIN3/cancer); over 2 years, 17.7% (95% confidence interval [CI], 9.3%–29.3%) and 27.8% (95% CI, 16.6%–43.5%) of those with HPV 16 progressed to CIN3/cancer from the true states of CIN1 and CIN2, respectively, whereas 55%–70% of CIN1/2 patients infected with HPV 52/58 remained in the CIN1/2 category. Misclassification was estimated to occur at a rate of 3%–38% in the current diagnostic practice.
Conclusion
This study contributes robust evidence to current literature on cervical lesion prognosis according to HPV genotype and quantifies the diagnostic misclassification of true cervical lesions.
Keywords: cervical cancer, cervical intraepithelial neoplasia (CIN), hidden Markov model, human papillomavirus (HPV)
In this retrospective cohort study, hidden Markov models were applied to clarify the natural course of cervical lesions according to human papillomavirus (HPV) genotype to account for any diagnostic misclassification. We revealed: (i) the current diagnostic practice (i.e., histology and cytology) was subject to diagnostic misclassification, and (ii) HPV 16‐positive patients were more likely to progress to more severe lesions than those with other HPV genotypes.
![]()
1. INTRODUCTION
Cervical cancer is the fourth most common cancer in women worldwide, as approximately 570,000 women developed cervical cancer and 311,000 died of the disease in 2018. 1 Human papillomavirus (HPV) infection is a common cause of cervical cancer and its precancerous lesions as well as cervical intraepithelial neoplasia (CIN). Previous studies have indicated that cervical lesions should be managed according to HPV genotype. 2 Besides HPV genotype, patients with cervical lesions are also managed according to their CIN classification (CIN1, CIN2, and CIN3). The risk stratification of patients with cervical lesions according to HPV genotype and CIN classification is critical to present effective treatment strategies while avoiding obstetric complications. 3 , 4
In this context, the prognosis of CIN lesions according to HPV genotype has been assessed. 5 A continuous‐time multistate Markov model was also applied to accommodate the bidirectional feature of CIN lesions 6 ; for example, CIN2 can regress to CIN1 or to a normal condition, remain as CIN2, or progress to CIN3 or cervical cancer. 7 However, another concern when building structural models of CIN lesion prognosis is diagnostic misclassification. Although CIN diagnosis is based on the combination of cytological and histological examinations aided by colposcopy, the accuracy of CIN diagnosis is limited, resulting in the misclassification of the “true” pathology of the lesion. For example, cytologic diagnosis was shown to have low sensitivity in detecting CIN2 or more severe lesions. 8 Colposcopy‐directed biopsy also tends to underestimate the CIN severity compared with a diagnosis confirmed by surgery. 9 Nevertheless, few studies have evaluated HPV pathogenesis, accounting for the probability of diagnostic misclassification. 10
To accommodate these types of measurement challenges, various latent variable models (e.g., factor models and structural equation models) have been adopted in medical research. 11 Latent variable models can manage unobserved random variables. Recently, another latent variable model, a hidden Markov model, was applied to model (i) the transition between the (unobserved) “true” states and (ii) the probabilities of the “observed” state conditional on the “true” states (misclassification probabilities). 12 , 13 In contrast, a Markov model can model only transitions between observed states. In addition to other latent variable models, hidden Markov models have been applied in clinical settings with measurement challenges, such as frailty, HIV infection, and diabetic retinopathy. 14 , 15 , 16
In this study, we applied a hidden Markov model to our cohort of HPV‐infected patients to clarify the natural course of CIN according to HPV genotype, which accounted for the misclassification probability. We aimed to confirm the robustness of the current literature, including a previous study that used a Markov model, on the CIN characteristics according to HPV genotype. Using this model, we also quantified the misclassification probability in CIN diagnosis.
2. MATERIALS AND METHODS
2.1. Study cohort
Between October 1, 2008 and March 31, 2015, 1427 female patients underwent HPV genotyping at the Obstetrics and Gynecology Department of the University of Tokyo Hospital (Tokyo, Japan). Patients with mild cervical lesions were also followed up at this hospital instead of being referred to other clinics. We reviewed the electronic medical records (EMRs) of those patients and extracted information on pathological (cytological and histological) examinations and treatment of cervical lesions as previously described. 6
To clarify the natural course of cervical lesions according to the CIN classification, we constructed a dataset that included patients (i) diagnosed with normal cervical lesions, CIN1, or CIN2 at the time of entry and (ii) who visited the Obstetrics and Gynecology Department at least twice during the follow‐up period. Patients were excluded if they had HPV 6‐single‐positive lesions with the sole diagnosis of condyloma during the follow‐up period and if they had only glandular lesions. Patients with multiple HPV genotypes were excluded. One patient with malignant lymphoma was also excluded. Patients were followed up until they received treatment, until they were diagnosed with CIN3 or cervical cancer, until they were moved to another hospital, or until March 31, 2018, whichever occurred first. Finally, 729 patients (6082 observations) were included in the dataset (Figure 1).
FIGURE 1.
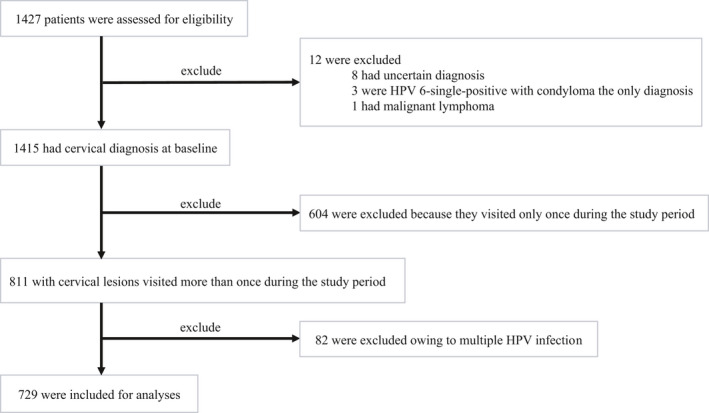
Flowchart illustrating the sample selection
This study was performed under the principles of the Declaration of Helsinki and was approved by the ethics committee of the Graduate School of Medicine, University of Tokyo (nos. 1390–1, G10082‐11, and G0637‐8).
2.2. Variables
2.2.1. HPV genotype
A cervical swab sample was collected from each patient at the time of entry or at the first visit when the HPV DNA genotype was determined. DNA was extracted from the cervical samples with a DNeasy Blood Mini Kit (Qiagen), the procedure for which has been previously described. 6 For each patient, we confirmed the HPV genotype, which was recorded in the EMRs. Since the genotyping was performed only once for each patient, the HPV genotype did not change over time. We classified HPVs into Group 1 or Group 2A (HPVs 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) as “high‐risk HPVs” (hrHPVs) according to the classification of the International Agency for Research on Cancer. 17 We focused on HPVs 16, 18, 52, and 58. 6 hrHPVs other than these four genotypes were defined as “other hrHPVs.” The remaining HPVs were classified as “no hrHPVs.” Patients without HPV infection were placed into this category (i.e., “no hrHPV” patients).
2.2.2. Pathological diagnosis
The dates of visits and the trajectory of cytological and histological results were maintained in the EMRs of each patient. At the patient’s first visit, we collected a histological sample via colposcopy. Thereafter, we performed repeated cytological and/or histological examinations in line with each patient’s visits. The histological examination was performed at the gynecologists’ discretion, especially when colposcopic findings indicated severe lesions compared with the previous diagnosis, the disease had progressed since the previous assessment, or when lesions that tended to exhibit discrepancies in cytological and histological diagnoses were observed.
Based on the cytological and histological results obtained after each visit, we grouped the pathological diagnoses at each visit into one of the following four CIN categories: normal, CIN1, CIN2, and CIN3/cancer. For some controversial records, the investigators (gynecologic oncology experts) discussed and determined the following: (i) CIN1–2 was classified as CIN1, (ii) CIN2–3 was classified as CIN2, (iii) uncertain diagnoses (e.g., atypical squamous cells of uncertain significance, atypical squamous cells that cannot exclude high‐grade squamous intraepithelial lesion [HSIL], and dysplasia without grading) were excluded from diagnostic reliability, and (iv) if the histological and cytological results were not identical, we accepted the more severe result as the diagnosis. We classified CIN1–2 as CIN1 and CIN2–3 as CIN2 in order to mitigate the overdiagnosis from the fourth protocol.
Furthermore, based on the dates of patients’ visits, we calculated the follow‐up time at the time of each examination. The age of the patients at the time of entry was extracted from the EMRs.
2.3. Continuous‐time multistate hidden Markov model
We applied the continuous‐time multistate hidden Markov model to account for the possibility that pathological examinations were subject to misclassification and for the natural bidirectional course of cervical lesions. This statistical model contains two parts: (i) the transition between the “true” pathological states and (ii) the probabilities of the “observed” diagnosis conditional on the “true” pathological states (misclassification probabilities). 13 , 18
We assumed transitions of the “true” pathological states and the misclassification patterns as shown in Figure 2. For each of the underlying true states and the observed states, the state space was {Normal, CIN1, CIN2, and CIN3/cancer} with each element corresponding to state 1, state 2, state 3, and state 4, respectively. Each observed state was determined by the underlying true state and misclassification. All transitions between adjacent true states were allowed in this model, except for the transition from CIN3/cancer to CIN2, as CIN3/cancer was the absorbing state. The transition parameter represents the transition intensity, which can be interpreted as an instantaneous rate of transition from the true pathological state i to the true pathological state j (e.g., denotes the transition intensity from true CIN1 to true CIN2). Based on the clinical assumption that adjacent misclassifications could account for most diagnostic misclassifications, we assumed the following misclassification matrix for the estimation (Figure 2):
where denotes the probability of observing the state s conditionally on the true state r (e.g., denotes the probability of observing CIN3 when the true state is CIN2). In other words, we allowed for just a “one‐step” misclassification adjacent to the true state.
FIGURE 2.
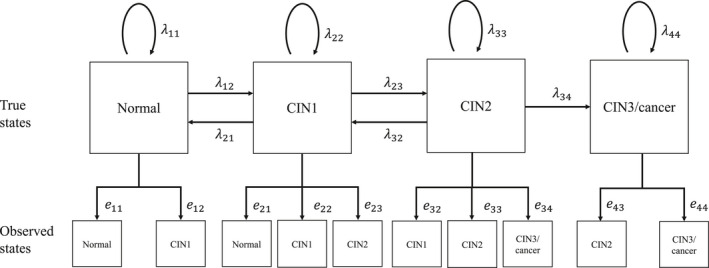
Hidden Markov model for the underlying true pathological states (CIN categorization). We defined four states: normal (state 1), cervical intraepithelial neoplasia 1 (CIN1, state 2), CIN2 (state 3), and CIN3/cancer (state 4). The arrows in this figure specify possible transitions among these states; all transitions between adjacent states were allowed, except for the transition from CIN3/cancer to CIN2 (i.e., CIN3/cancer was the absorbing state). The transition parameter represents an instantaneous rate of transition from the true pathological state i to the true pathological state j. denotes the probability of observing the state s conditionally on the true state r ( = 1−, = 1−−, = 1−, = 1−)
2.4. Statistical analysis
First, we reported the summary statistics of patients according to HPV genotype (HPV 16, 18, 52, 58, other hrHPVs, and no hrHPVs) and observed diagnoses at the time of entry. We also showed the transition of diagnoses according to HPV genotype over two successive examinations.
Next, a maximum likelihood procedure was applied to estimate the parameters and using the msm package in R. 18 The estimates were derived to maximize a likelihood function composed of the transition probabilities among the true states and the misclassification probabilities conditional on the true state. Along with the parameter estimation, the “true” initial distribution was also estimated in the models. The detailed formulation of the likelihood function is described elsewhere. 18 To account for possible differences in the natures of HPV genotypes, we introduced dummy variables representing each HPV genotype (except for “no hrHPVs,” the reference genotype). These dummy variables were included in the model as covariates for the transition parameter . Furthermore, we simulated the probabilities of transitions among the true states for up to 2 years. We selected the span of 2 years for two reasons. First, a two‐year transition prediction is considered a good benchmark for the prognosis of hrHPV‐related cervical lesions. At least half of those lesions have been reported to regress to a normal state and approximately 10% progress to CIN3 within 2 years. 5 Second, the median follow‐up period of our subjects was approximately three years. Hence, a transition prediction beyond this period was unwarranted. Finally, we evaluated our model by visually comparing the prevalence of observed diagnoses and that of simulated observed diagnoses, which was derived by right‐multiplying the simulated “true” prevalence matrix by the estimated misclassification matrix. R 3.6.2 (R Foundation) was used for all analyses. 19
2.5. Sensitivity analysis
To confirm the robustness of our primary analysis, we performed two different sensitivity analyses. First, we assumed another misclassification matrix for the CIN categorization. Specifically, we set in the misclassification matrix to zero. We performed this sensitivity analysis because the estimation of the parameter jointly with in the CIN analysis was supposedly unstable due to the nature of our dataset. Since the analysis of patients in our dataset with the CIN categorization was truncated once they were diagnosed with CIN3/cancer, the available observations to estimate were reasonably scarce. Using this sensitivity analysis, we aimed to stabilize the estimation of to examine whether the estimates in the primary analysis were subject to this instability.
Second, to clarify the natural course of cervical lesions according to the two‐tier classification (World Health Organization 2020), 20 we constructed another dataset based on that classification (i.e., normal, low‐grade squamous intraepithelial lesion [LSIL], and HSIL). The details of this dataset are described in the Supplemental material (eAppendix 1, Figure S1). We applied the same analysis as in the CIN categorization for the two‐tier classification according to the model specified in Figure S2. For these sensitivity analyses, the estimated misclassification matrix was presented. The probabilities of transitions among the true states up to two years were then simulated.
3. RESULTS
3.1. Patient characteristics
Table 1 shows the characteristics of our study subjects. In total, 729 patients in the CIN category were enrolled with a mean follow‐up of 3.3 years (standard deviation [SD]: 2.4 years). The mean age at the time of entry was 39.1 years (SD: 9.8 years), and the mean number of visits was 8.3 (SD: 5.4). At the time of entry, 185 (25.3%), 270 (37.0%), and 274 (37.6%) patients were diagnosed as normal or with CIN1 or CIN2, respectively.
TABLE 1.
Basic characteristics of the study subjects at the time of entry (CIN categorization)
| Diagnosis at the time of entry | HPV 16 | HPV 18 | HPV 52 | HPV 58 | Other hrHPVs | No hrHPVs | All | |
|---|---|---|---|---|---|---|---|---|
| Normal | N | 8 | 7 | 14 | 10 | 24 | 122 | 185 |
| Age at entry (years), mean (SD) | 39.9 (8.2) | 45.3 (15.9) | 38.0 (10.1) | 44.9 (17.4) | 42.7 (16.3) | 41.2 (10.5) | 41.5 (11.9) | |
| Number of visits, mean (SD) | 6.6 (5.8) | 8.4 (2.7) | 8.9 (7.2) | 7.0 (5.0) | 8.0 (4.8) | 6.5 (4.0) | 7.0 (4.5) | |
| Follow‐up interval (years), mean (SD) | 0.47 (0.35) | 0.48 (0.30) | 0.51 (0.37) | 0.49 (0.44) | 0.47 (0.27) | 0.52 (0.39) | 0.50 (0.36) | |
| Follow‐up period (years), mean (SD) | 2.7 (3.0) | 3.4 (1.1) | 4.3 (3.3) | 3.0 (2.4) | 3.4 (2.2) | 3.1 (2.1) | 3.2 (2.3) | |
| CIN1 | N | 20 | 8 | 32 | 23 | 67 | 120 | 270 |
| Age at entry (years), mean (SD) | 35.3 (9.2) | 33.0 (10.6) | 39.1 (8.7) | 36.7 (8.3) | 34.5 (7.1) | 38.8 (10.1) | 37.1 (9.2) | |
| Number of visits, mean (SD) | 8.8 (5.4) | 6.5 (2.8) | 9.4 (5.0) | 11.0 (5.5) | 9.4 (4.4) | 9.3 (5.4) | 9.3 (5.1) | |
| Follow‐up interval (years), mean (SD) | 0.38 (0.19) | 0.51 (0.56) | 0.38 (0.21) | 0.42 (0.28) | 0.38 (0.18) | 0.42 (0.30) | 0.41 (0.27) | |
| Follow‐up period (years), mean (SD) | 3.5 (2.6) | 3.0 (1.8) | 3.6 (2.1) | 4.2 (2.5) | 3.6 (2.0) | 3.9 (2.4) | 3.7 (2.3) | |
| CIN2 | N | 64 | 12 | 52 | 37 | 51 | 58 | 274 |
| Age at entry (years), mean (SD) | 38.4 (7.8) | 42.5 (5.7) | 41.4 (8.0) | 41.1 (8.1) | 39.1 (7.8) | 37.0 (9.3) | 39.4 (8.3) | |
| Number of visits, mean (SD) | 6.4 (5.9) | 6.7 (5.2) | 8.5 (6.3) | 8.4 (5.6) | 8.7 (5.7) | 9.4 (5.7) | 8.1 (5.9) | |
| Follow‐up interval (years), mean (SD) | 0.33 (0.19) | 0.31 (0.11) | 0.35 (0.19) | 0.36 (0.22) | 0.38 (0.41) | 0.37 (0.35) | 0.35 (0.28) | |
| Follow‐up period (years), mean (SD) | 1.9 (2.3) | 1.9 (1.9) | 2.9 (2.5) | 3.1 (2.4) | 3.2 (2.2) | 3.5 (2.4) | 2.8 (2.4) |
Other hrHPVs included HPV 31, 33, 35, 39, 45, 51, 56, 59, and 68. No hrHPVs were HPVs other than HPV 16, 18, 52, 58, or other hrHPVs.
Abbreviations: hrHPV, high‐risk human papillomavirus; SD, standard deviation.
Table 2 shows the visit‐wise transitions of observed lesions according to HPV genotype. For the CIN categorization, 5353 transitions were observed, of which 551, 165, 770, 569, 1130, and 2168 transitions were observed for HPVs 16, 18, 52, 58, other hrHPVs, and no hrHPVs, respectively. The HPV genotype pattern in transitions was similar for the two‐tier classification. Most (78%–90%) patients in the normal state remained in that state over consecutive observations (e.g., 86.7% of HPV 16‐positive patients in the normal state observed at a certain visit were also categorized as normal at the next visit). The progression from CIN2 to CIN3 was dependent on HPV genotype: 11.9%, 7.5%, 5.1%, 3.7%, 5.4%, and 2.0% of those with HPVs 16, 18, 52, 58, other hrHPVs, and no hrHPVs, progressed to CIN3, respectively.
TABLE 2.
Transitions from each diagnosis of cervical epithelial lesions according to HPV genotype (CIN categorization)
| Diagnosis at (t − 1) visit | HPV category | Diagnosis at t visit | ||||
|---|---|---|---|---|---|---|
| Normal | CIN1 | CIN2 | CIN3 | Cancer | ||
| Normal | HPV 16 | 190 (86.7) | 10 (4.5) | 16 (7.3) | 3 (1.3) | 0 (0.0) |
| HPV 18 | 60 (80.0) | 11 (14.6) | 4 (5.3) | 0 (0.0) | 0 (0.0) | |
| HPV 52 | 251 (77.7) | 44 (13.6) | 26 (8.0) | 2 (0.6) | 0 (0.0) | |
| HPV 58 | 196 (84.1) | 21 (9.0) | 14 (6.0) | 2 (0.8) | 0 (0.0) | |
| Other hrHPVs | 573 (86.8) | 71 (10.7) | 14 (2.1) | 2 (0.3) | 0 (0.0) | |
| No hrHPVs | 1348 (89.9) | 119 (7.9) | 28 (1.8) | 3 (0.2) | 1 (0.0) | |
| CIN1 | HPV 16 | 24 (26.9) | 30 (33.7) | 33 (37.0) | 2 (2.2) | 0 (0.0) |
| HPV 18 | 15 (40.5) | 13 (35.1) | 8 (21.6) | 1 (2.7) | 0 (0.0) | |
| HPV 52 | 65 (33.6) | 80 (41.4) | 46 (23.8) | 2 (1.0) | 0 (0.0) | |
| HPV 58 | 39 (26.5) | 69 (46.9) | 36 (24.4) | 3 (2.0) | 0 (0.0) | |
| Other hrHPVs | 122 (45.3) | 113 (42.0) | 33 (12.2) | 1 (0.3) | 0 (0.0) | |
| No hrHPVs | 225 (52.5) | 162 (37.8) | 38 (8.8) | 3 (0.7) | 0 (0.0) | |
| CIN2 | HPV 16 | 26 (10.6) | 34 (13.9) | 153 (62.9) | 29 (11.9) | 1 (0.4) |
| HPV 18 | 7 (13.2) | 6 (11.3) | 36 (67.9) | 4 (7.5) | 0 (0.0) | |
| HPV 52 | 36 (14.1) | 44 (17.3) | 161 (63.3) | 13 (5.1) | 0 (0.0) | |
| HPV 58 | 21 (11.1) | 43 (22.7) | 118 (62.4) | 7 (3.7) | 0 (0.0) | |
| Other hrHPVs | 40 (19.9) | 35 (17.4) | 115 (57.2) | 11 (5.4) | 0 (0.0) | |
| No hrHPVs | 57 (23.6) | 42 (17.4) | 137 (56.8) | 5 (2.0) | 0 (0.0) | |
Values are the number (percentage) of observed transitions from a visit to the next visit.
Other hrHPVs included HPV 31, 33, 35, 39, 45, 51, 56, 59, and 68.
No hrHPVs were HPVs other than HPV 16, 18, 52, 58, or other hrHPVs.
Abbreviations: hrHPV: high‐risk human papillomavirus.
3.2. Primary analysis
Table 3 represents our estimates for the misclassification matrix for the CIN categorization. Of patients with the normal state as their true state, 95.7% (95% confidence interval [CI], 94.2%–96.9%) were estimated to be diagnosed correctly. The model estimated that, of patients with the true state of CIN1, 61.9% (95% CI, 51.6%–71.2%) were diagnosed correctly, while 24.4% (95% CI, 17.3%–33.4%) and 13.5% (95% CI, 9.3%–19.2%) were misclassified as having normal and CIN2, respectively. For those with the true state of CIN2, 88.7% (95% CI, 80.4%–93.8%) were estimated to be diagnosed correctly as having CIN2, while 6.2% (95% CI, 3.8%–10.0%) and 4.9% (95% CI, 3.1%–7.7%) were estimated to be misclassified as having CIN1 and CIN3/cancer, respectively. For those with the true state of CIN3/cancer, 95.5% (95% CI, 24.6%–99.9%) were estimated to be diagnosed correctly. However, this wide 95% CI implied the instability of the estimation.
TABLE 3.
Misclassification probabilities for the CIN categorization based on cytology and histology
| True underlying state | Observed state | |||
|---|---|---|---|---|
| Normal | CIN1 | CIN2 | CIN3/cancer | |
| Normal | 0.957 (0.942–0.969) | 0.042 (0.030–0.057) | 0.000 (0.000–0.000) | 0.000 (0.000–0.000) |
| CIN1 | 0.244 (0.173–0.334) | 0.619 (0.516–0.712) | 0.135 (0.093–0.192) | 0.000 (0.000–0.000) |
| CIN2 | 0.000 (0.000–0.000) | 0.062 (0.038–0.100) | 0.887 (0.804–0.938) | 0.049 (0.031–0.077) |
| CIN3/cancer | 0.000 (0.000–0.000) | 0.000 (0.000–0.000) | 0.044 (0.000–0.753) | 0.955 (0.246–0.999) |
Values are the estimated emission probabilities (95% confidence interval).
Table 4 shows the predicted transition probabilities of true lesions after two years according to HPV genotype. For HPV 16, 2.3% (95% CI, 0.9%–5.5%), 17.7% (95% CI, 9.3%–29.3%), and 27.8% (95% CI, 16.6%–43.5%) of patients progressed to CIN3/cancer from the true state of normal, CIN1, and CIN2, respectively. On the contrary, 44.4% (95% CI, 32.5%–55.5%) and 29.9% (95% CI, 20.3%–39.9%) of HPV 16‐positive patients regressed to a normal state from CIN1 and CIN2, respectively. For HPV 18‐positive patients, the progression to CIN3/cancer was less likely than for HPV 16‐positive patients; 0.7% (95% CI, 0.0%–14.4%) transitioned from a normal state, 2.6% (95% CI, 0.0%–35.2%) transitioned from CIN1, and 10.9% (95% CI, 0.0%–96.5%) transitioned from CIN2. For HPV 52/58‐positive patients, the transitions were more likely to be stable over the 2 years than for HPV 16/18‐positive patients; approximately 55%–70% of CIN1/2 patients remained as CIN1/2 after 2 years. Figure S3 illustrates the observed prevalence of the test‐revealed lesions and the simulated prevalence of observed lesions by HPV genotype.
TABLE 4.
Predicted two‐year transition probabilities according to HPV genotype (CIN categorization)
| Current state | HPV category | State after two years | |||
|---|---|---|---|---|---|
| Normal | CIN1 | CIN2 | CIN3/cancer | ||
| Normal | HPV 16 | 0.832 (0.685–0.913) | 0.064 (0.033–0.126) | 0.079 (0.036–0.155) | 0.023 (0.009–0.055) |
| HPV 18 | 0.736 (0.493–0.897) | 0.159 (0.054–0.310) | 0.096 (0.006–0.214) | 0.007 (0.000–0.144) | |
| HPV 52 | 0.933 (0.168–0.998) | 0.045 (0.001–0.475) | 0.020 (0.000–0.312) | 0.001 (0.000–0.037) | |
| HPV 58 | 0.974 (0.050–0.999) | 0.017 (0.000–0.581) | 0.007 (0.000–0.324) | 0.000 (0.000–0.052) | |
| Other hrHPVs | 0.889 (0.808–0.940) | 0.090 (0.049–0.157) | 0.017 (0.008–0.036) | 0.002 (0.000–0.007) | |
| No hrHPVs | 0.910 (0.768–0.966) | 0.068 (0.025–0.172) | 0.019 (0.006–0.050) | 0.001 (0.000–0.005) | |
| CIN1 | HPV 16 | 0.444 (0.325–0.555) | 0.127 (0.085–0.194) | 0.250 (0.168–0.339) | 0.177 (0.093–0.293) |
| HPV 18 | 0.622 (0.385–0.752) | 0.161 (0.075–0.314) | 0.189 (0.007–0.370) | 0.026 (0.000–0.352) | |
| HPV 52 | 0.392 (0.153–0.485) | 0.317 (0.246–0.479) | 0.256 (0.186–0.356) | 0.033 (0.010–0.105) | |
| HPV 58 | 0.410 (0.050–0.508) | 0.330 (0.241–0.569) | 0.238 (0.145–0.365) | 0.020 (0.002–0.175) | |
| Other hrHPVs | 0.734 (0.648–0.791) | 0.171 (0.124–0.232) | 0.071 (0.038–0.111) | 0.023 (0.009–0.055) | |
| No hrHPVs | 0.673 (0.614–0.712) | 0.189 (0.159–0.239) | 0.117 (0.091–0.145) | 0.020 (0.009–0.041) | |
| CIN2 | HPV 16 | 0.299 (0.203–0.399) | 0.137 (0.091–0.200) | 0.285 (0.189–0.389) | 0.278 (0.166–0.435) |
| HPV 18 | 0.285 (0.024–0.484) | 0.143 (0.006–0.267) | 0.461 (0.000–0.730) | 0.109 (0.000–0.965) | |
| HPV 52 | 0.236 (0.103–0.317) | 0.349 (0.265–0.461) | 0.334 (0.229–0.438) | 0.079 (0.026–0.240) | |
| HPV 58 | 0.252 (0.041–0.345) | 0.372 (0.242–0.514) | 0.323 (0.150–0.443) | 0.052 (0.006–0.407) | |
| Other hrHPVs | 0.437 (0.316–0.546) | 0.219 (0.155–0.271) | 0.195 (0.099–0.313) | 0.146 (0.065–0.308) | |
| No hrHPVs | 0.406 (0.334–0.468) | 0.250 (0.213–0.290) | 0.253 (0.189–0.326) | 0.089 (0.044–0.179) | |
Values are the predicted transition probabilities (95% confidence interval) from the current true lesions over two years.
Other hrHPVs included HPV 31, 33, 35, 39, 45, 51, 56, 59, and 68.
No hrHPVs were HPVs other than HPV 16, 18, 52, 58, or other hrHPVs.
Abbreviation: hrHPV, high‐risk human papillomavirus.
3.3. Sensitivity analysis
Tables S1 and S2 show the misclassification matrix and predicted transition probabilities of true lesions for the sensitivity analysis, respectively. The results of this sensitivity analysis were consistent with those of our primary analysis for CIN categorization. Furthermore, compared with our primary analysis, the sensitivity analysis resulted in narrower confidence intervals for the misclassification matrix (Table 3 and Table S1).
For the two‐tier classification, dataset characteristics and visit‐wise transitions of observed lesions are shown in Tables S3 and S4, respectively. Table S5 contains our estimates for the misclassification matrix in the two‐tier classification. Of patients with a normal state as their true state, 96.6% (95% CI, 95.1%–97.7%) were estimated to be diagnosed correctly. It was estimated that for those with the true state of LSIL, 61.6% (95% CI, 52.8%–69.8%) were diagnosed correctly, while 24.7% (95% CI, 19.3%–31.0%) and 13.5% (95% CI, 9.8%–18.4%) were misclassified as normal and having HSIL, respectively. For those with the true state of HSIL, 95.4% (95% CI, 93.0%–97.1%) were estimated to be diagnosed correctly.
Table S6 shows the predicted transition probabilities of true lesions after two years according to HPV genotype. For HPV 16, 10.7% (95% CI, 5.7%–19.6%) and 41.6% (95% CI, 31.3%–52.0%) of patients progressed to HSIL from the true states of normal and LSIL, respectively. In contrast, 44.8% (95% CI, 33.8%–55.5%) and 22.4% (95% CI, 15.1%–30.5%) of HPV 16‐positive patients regressed to a normal state from LSIL and HSIL, respectively. The progression to HSIL was less likely in HPV 18‐positive patients compared with HPV 16‐positive patients; 4.6% (95% CI, 0.7%–17.8%) from a normal state and 20.3% (95% CI, 7.3%–40.7%) from LSIL. The transitions were more likely to be stable over two years in HPV 52/58‐positive patients than in HPV 16/18‐positive patients; approximately 30%–35% of LSIL patients remained in the same state after 2 years. Figure S4 illustrates the observed prevalence of the test‐revealed lesions and the simulated prevalence of observed lesions by HPV genotype.
4. DISCUSSION
In this study, we applied the continuous‐time multistate hidden Markov model and successfully estimated the transition probabilities of cervical lesions according to HPV genotype, which accommodated the misclassification probabilities of pathological lesions. Adopting a latent variable approach (i.e., hidden Markov model) as in previous medical research, we revealed the following two points: (i) the current diagnostic practice (i.e., histology and cytology) was subject to diagnostic misclassification at a rate of 3%–38% and (ii) even when the diagnostic misclassification was accounted for, HPV 16‐positive patients were more likely to progress to more severe lesions (i.e., CIN3/cancer or HSIL) than those with other HPV genotypes, which was consistent with previous studies. 5 , 6
Our results showed that even when we accounted for the diagnostic misclassification in current practice, the progression rate toward CIN3/cancer was higher among HPV 16‐ positive patients than among others. In contrast, patients with HPV 52/58 tended to remain in the CIN1/2 category. Moreover, the progression and regression rates were similar between patients with other hrHPVs and those with no hrHPVs. These results are consistent with those seen in the literature and corroborate the finding that patients with HPV 16 are more likely to develop more severe lesions than those with HPV 52, HPV 58, other hrHPVs, or no hrHPVs. 6 By applying the hidden Markov model to accommodate the diagnostic misclassification, our study confirmed the robustness of the current literature on CIN characteristics according to HPV genotype. Our results were robust regardless of the cervical lesion categorization. We performed a sensitivity analysis for another dataset based on the two‐tier classification and the results were similar to those of our primary analysis as follows: HPV 16‐positive patients were more likely to progress to severer lesions (i.e., HSIL) than those with other HPV genotypes.
Our prognostic prediction was different from that in a previous study, which used a continuous‐time multistate Markov model. 6 Notably, patients with HPV 52/58‐derived CIN1/2 were more likely to be classified as having CIN1 over two years than in the previous study. Furthermore, compared with the corresponding women in the previous study, those in the initial normal state in this study were more likely to remain in the normal state over two years (e.g., 83.2% vs. 59.8% for HPV 16). This discrepancy could be attributed to the compositional difference of the initial states between the two studies, as the diagnostic misclassification was accounted for in this study; for example, the normal state in the previous study was reasonably composed of patients with true normal and more severe states. This accommodation of diagnostic misclassification also helped us reveal the stable nature of CIN1/2 in HPV 52/58‐positive patients. Although the slightly different CIN definitions between the studies and the (non‐)existence of an absorbing state in those models might explain the different results, our model, which incorporates diagnostic misclassification, could successfully derive a better picture of cervical lesion prognosis according to CIN classification and HPV genotype.
Our results estimated that diagnosis through histological and cytological examinations was subject to misclassification at a rate of 3%–38%. These misclassifications can be explained by two mechanisms. First, histological and cytological examinations may be subject to sampling errors. The diagnostic precision of histological examination has been shown to depend on the quality of specimen processing. 21 , 22 , 23 Furthermore, it is sometimes difficult to distinguish metaplastic epithelial cells from atypical epithelial cells in a cytological examination. 24 , 25 , 26 Second, cervical lesions are representative of a continuous disease spectrum rather than discrete states. Categorizing this continuous spectrum into discrete systems (i.e., CIN classification and the two‐tier classification) can underestimate the extent of variation in the disease spectrum (i.e., loss of information), which may result in diagnostic misclassification. 27 , 28
To the best of our knowledge, this is the first study to apply a hidden Markov model to quantify the misclassification probabilities of the cervical diagnostic procedure as well as the natural history of HPV infections; Kang and Lagakos applied a hidden Markov model only to the natural history of HPV infections under the prespecified misclassification probability. 10 Notably, the misclassification probability in patients with the true state of CIN1 was higher than those with the other true states (37.9% for CIN1 vs. 4.2%–11.1% for the other true states). This result was biologically plausible, as CIN1 is a low‐grade lesion with a high propensity for regression due to immune responses, but this lesion type may also progress to a high‐grade lesion. 29 , 30 , 31 The high misclassification probabilities in CIN1 patients may reflect the biological instability of these lesions due to the smooth transition imposed by the hidden Markov model. In contrast, the misclassification probability of observing CIN2 in patients with the true state of CIN3/cancer was only 4.4% (11% for the sensitivity analysis). To analyze the clinical implications, we applied Bayes’ theorem to the misclassification estimate of observing CIN2 with CIN3/cancer as the true state; we then calculated the probability of the true state of CIN3/cancer when CIN2 was observed. We may assume subjectively that the marginal probabilities of observing CIN2 and the true state of CIN3/cancer are 40% and 10%, respectively. Then, Bayes’ theorem indicates that the probability of the true state of CIN3/cancer is 1.1% (0.044*0.1/0.4 = 0.011 or 1.1%; 2.8% for the sensitivity analysis). Although the “true state” estimated from the hidden Markov model was not the “clinical true state” that was histologically validated by conization or total hysterectomy, this small probability of overlooking true CIN3/cancer with CIN2 observed may justify the current Japanese practice, where patients with (observed) CIN2 are carefully followed up without treatment. To further validate this practice, an attempt to predict a patient's true state more accurately may be a valid direction for future work.
Some limitations of this study should be acknowledged. First, we could not accommodate the effect of concurrent multiple HPV infections on the progression and regression of cervical lesions. Even for interaction terms between two different HPV genotypes, our cohort was too small to account for multiple HPV infections. Compared with single infections, coinfections with multiple α9 species have been shown to be associated with an increased risk of CIN2 or more severe lesions. 32 Second, because the genotyping was performed only once at the baseline and the sample size was not large enough, we could not investigate the temporary transitions of HPV infections, which could affect the progression and regression of cervical lesions. 33 , 34 Third, we could not account for patient heterogeneity in terms of various factors, such as age, sexual activity, menopausal status, contraception, and HPV vaccination. It is possible that these patients’ characteristics can affect the pathogenesis of cervical lesions or the precision of the diagnosis. 35 , 36 , 37 , 38 , 39 , 40 , 41 However, we did not have information on sexual activity, menopausal status, contraception, or HPV vaccination. Preliminary analyses in which we included age as a covariate was unsuccessful in estimation convergence. We could not include the heterogeneity by HPV genotypes for estimating the initial distribution of cervical lesions (i.e., normal, CIN1, and CIN2) while achieving the model convergence. Nevertheless, the impacts of some of these characteristics should be minor in this study for the following reasons: (i) oral contraception was not prevalent in Japan during the study period 42 ; (ii) HPV vaccination coverage was considered to be low in our cohort, because free vaccination against HPV began in 2010, particularly aiming for girls aged 12–16 years, and very few patients in our cohort (median age at the time of entry: 37.9 years [interquartile range: 32.1–44.7 years]) were eligible for it. 43 Further studies with a larger number of participants are expected to accommodate these covariates in a hidden Markov model.
Fourth, our model specification might not be optimal. Our estimation in the analysis of the CIN categorization was unstable due to the nature of our dataset. Specifically, the confidence intervals were wide for the misclassification probability with the true underlying state CIN3/cancer, since the analysis of patients in our dataset with the CIN categorization was truncated once they were diagnosed with CIN3/cancer; this dataset structure increased the difficulty in estimating both the probability of observing CIN2 in patients believed to have CIN3/cancer (i.e., ) and the probability of observing CIN3/cancer in patients with CIN2 (i.e., ) in the same model. Furthermore, only “one‐step” misclassifications were allowed in this study, although other misclassifications were possible. Misclassification heterogeneity may have been present due to different diagnostic procedures (i.e., cytologic and histologic examinations) or physicians in charge of patients. In our preliminary analyses, most of the more generous misclassification patterns did not achieve estimation convergence. Despite these concerns, our analyses should provide reliable estimates: (i) we confirmed that our results were robust to the alternative specification of the misclassification matrix, where the confidence intervals were much smaller, and (ii) our model specification with just “one‐step” misclassifications is an appropriate abstraction of the diagnostic reality if other misclassifications were sufficiently rarer than these misclassifications.
Fifth, our model assumed that the transition intensities were independent of the time spent in each state. Previous studies have proposed time‐inhomogeneous transition model 10 , 44 ; however, our cohort was too small to implement it. Future studies are expected to explore the possible time‐inhomogeneous transition intensities between cervical lesions.
Lastly, this was a single‐institution study. Patients included in this study were restricted to those who previously had abnormal cytology and who had visited the university hospital. Hence, the generalizability of our results to other settings warrants further research.
In conclusion, we applied a continuous‐time multistate hidden Markov model to reveal the different prognosis of cervical lesions according to HPV genotype; HPV 16‐positive patients were more likely to progress to CIN3/cancer than those with other HPV genotypes, and those with HPV 52/58‐derived CIN1/2 tended to remain in the CIN1 category. We believe this study contributes robust evidence to the current literature on cervical lesion prognosis according to HPV genotype and quantifies the diagnostic misclassification of true cervical lesions.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceptualization, Ryo Ikesu, Ayumi Taguchi, Konan Hara, and Kei Kawana; methodology, Ryo Ikesu, Ayumi Taguchi, and Konan Hara; data collection and data curation, Ayumi Taguchi and Tetsushi Tsuruga; supervision, Kei Kawana, Tetsushi Tsuruga, Jun Tomio, and Yutaka Osuga; funding acquisition, Ayumi Taguchi. All authors reviewed the manuscript and edited it for intellectual content and gave final approval for this version.
ETHICAL APPROVAL STATEMENT
The Ethics Committee of the Graduate School of Medicine, University of Tokyo approved this study (nos. 1390–1, G10082‐11, and G0637‐8), which was performed in accordance with the Declaration of Helsinki.
CONSENT FOR PUBLICATION
All the patients included in this study provided consent for the publication of the research findings.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S6
Appendix
ACKNOWLEDGMENTS
We thank all the women included in this study for contributing to our dataset. We also thank Takuya Ishihara and Yuki Ito for their help and advice with the literature review on hidden Markov models in the context of statistics.
Ikesu R, Taguchi A, Hara K, et al. Prognosis of high‐risk human papillomavirus‐related cervical lesions: A hidden Markov model analysis of a single‐center cohort in Japan. Cancer Med. 2022;11:664–675. doi: 10.1002/cam4.4470
Ryo Ikesu and Ayumi Taguchi equally contributed to this work.
Funding information
This study was supported by AMED under Grant Number 21wm0325014h0002 and the Seiichi Imai Memorial Foundation Research Grant. The funders did not have any role in study design, analyses, interpretation of the data, or preparation of the manuscript.
Contributor Information
Konan Hara, harakonan@email.arizona.edu.
Kei Kawana, Email: kkawana-tky@umin.org.
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available upon reasonable request.
REFERENCES
- 1. Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Heal. 2020;8:e191‐e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith JS, Lindsay L, Hoots B, et al. Human papillomavirus type distribution in invasive cervical cancer and high‐grade cervical lesions: a meta‐analysis update. Int J Cancer. 2007;121:621‐632. [DOI] [PubMed] [Google Scholar]
- 3. Kyrgiou M, Koliopoulos G, Martin‐Hirsch P, Arbyn M, Prendiville W, Paraskevaidis E. Obstetric outcomes after conservative treatment for intraepithelial or early invasive cervical lesions: systematic review and meta‐analysis. Lancet. 2006;367:489‐498. [DOI] [PubMed] [Google Scholar]
- 4. Kyrgiou M, Athanasiou A, Paraskevaidi M, et al. Adverse obstetric outcomes after local treatment for cervical preinvasive and early invasive disease according to cone depth: systematic review and meta‐analysis. BMJ. 2016;354;i3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsumoto K, Oki A, Furuta R, et al. Predicting the progression of cervical precursor lesions by human papillomavirus genotyping: a prospective cohort study. Int J Cancer. 2011;128:2898‐2910. [DOI] [PubMed] [Google Scholar]
- 6. Taguchi A, Hara K, Tomio J, et al. Multistate Markov model to predict the prognosis of high‐risk human papillomavirus‐related cervical lesions. Cancers (Basel). 2020;12:1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Insinga RP, Dasbach EJ, Elbasha EH. Epidemiologic natural history and clinical management of human papillomavirus (HPV) disease: a critical and systematic review of the literature in the development of an HPV dynamic transmission model. BMC Infect Dis. 2009;9:1‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright TC, Schiffman M, Solomon D, et al. Interim guidance for the use of human papillomavirus DNA testing as an adjunct to cervical cytology for screening. Obstet Gynecol. 2004;103:304‐309. [DOI] [PubMed] [Google Scholar]
- 9. Stoler MH, Vichnin MD, Ferenczy A, et al. The accuracy of colposcopic biopsy: analyses from the placebo arm of the Gardasil clinical trials. Int J Cancer. 2011;128:1354‐1362. [DOI] [PubMed] [Google Scholar]
- 10. Kang M, Lagakos SW. Statistical methods for panel data from a semi‐Markov process, with application to HPV. Biostatistics. 2007;8:252‐264. [DOI] [PubMed] [Google Scholar]
- 11. Rabe‐Hesketh S, Skrondal A. Classical latent variable models for medical research. Stat Methods Med Res. 2008;17:5‐32. [DOI] [PubMed] [Google Scholar]
- 12. Eddy SR. What is a hidden Markov model? Nat Biotechnol. 2004;22:1315‐1316. Available from http://www.nature.com/naturebiotechnology [DOI] [PubMed] [Google Scholar]
- 13. Bureau A, Shiboski S, Hughes JP. Applications of continuous time hidden Markov models to the study of misclassified disease outcomes. Stat Med. 2003;22:441‐462. [DOI] [PubMed] [Google Scholar]
- 14. Murphy RA, Ip EH, Zhang Q, et al. Transition to sarcopenia and determinants of transitions in older adults: a population‐based study. Journals Gerontol ‐ Ser A Biol Sci Med Sci. 2014;69:751‐758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mangal TD. Joint estimation of CD4+ cell progression and survival in untreated individuals with HIV‐1 infection. AIDS. 2017;31:1073‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Looker HC, Nyangoma SO, Cromie DT, et al. Predicted impact of extending the screening interval for diabetic retinopathy: the Scottish Diabetic Retinopathy Screening programme. Diabetologia. 2013;56:1716‐1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bzhalava D, Guan P, Franceschi S, Dillner J, Clifford G. A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types. Virology. 2013;445:224‐231. Available from: 10.1016/j.virol.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 18. Jackson CH. Multi‐state models for panel data: the msm package for R. J Stat Softw. 2011;38:1‐28. [Google Scholar]
- 19. R Core Team . R: a language and environment for statistical computing. 2020; Available from: https://www.r‐project.org/
- 20. WHO Classification of Tumours Editorial Board , ed. Female Genital Tumours: WHO Classification of Tumours. 5th ed. World Health Organization. 2020. [Google Scholar]
- 21. Fadare O, Rodriguez R. Squamous dysplasia of the uterine cervix: tissue sampling‐related diagnostic considerations in 600 consecutive biopsies. Int J Gynecol Pathol. 2007;26:469‐474. [DOI] [PubMed] [Google Scholar]
- 22. Heatley MK. How many histological levels should be examined from tissue blocks originating in cone biopsy and large loop excision of the transformation zone specimens of cervix? J Clin Pathol. 2001;54:650‐651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Villegas‐Hinojosa E, Terán‐Figueroa Y, Gallegos‐García V, et al. Histopathological diagnosis of cervical biopsies: reduction of sampling errors with the evaluation of a third histologic level. Cancer Manag Res. 2020;12:5099‐5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Regauer S, Reich O. CK17 and p16 expression patterns distinguish (atypical) immature squamous metaplasia from high‐grade cervical intraepithelial neoplasia (CIN III). Histopathology. 2007;50:629‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vooijs GP, van Aspert‐van Erp AJM, Bulten J. Benign proliferative reactions, intraepithelial neoplasia, and invasive cancer of the uterine cervix. In: Bibbo M, Wilbur D, (eds). Comprehensive Sytopathology. 2008;131‐212. [Google Scholar]
- 26. Song T, Seong SJ, Lee SK, et al. Searching for an ideal cervical cancer screening model to reduce false‐negative errors in a country with high prevalence of cervical cancer. J Obstet Gynaecol (Lahore). 2020;40(2):240‐246. Available from: 10.1080/01443615.2019.1621813 [DOI] [PubMed] [Google Scholar]
- 27. Altman DG, Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332:1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moons KGM, Altman DG, Reitsma JB, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1‐W73. [DOI] [PubMed] [Google Scholar]
- 29. Bansal N, Wright JD, Cohen CJ, Herzog TJ. Natural history of established low grade cervical intraepithelial (CIN 1) lesions. Anticancer Res. 2008;28:1763‐1766. [PubMed] [Google Scholar]
- 30. Monnier‐Benoit S, Mauny F, Riethmuller D, et al. Immunohistochemical analysis of CD4+ and CD8+ T‐cell subsets in high risk human papillomavirus‐associated pre‐malignant and malignant lesions of the uterine cervix. Gynecol Oncol. 2006;102:22‐31. [DOI] [PubMed] [Google Scholar]
- 31. De Gruijl TD, Bontkes HJ, Walboomers JMM, et al. Differential T helper cell responses to human papillomavirus type 16 E7 related to viral clearance or persistence in patients with cervical neoplasia: a longitudinal study. Cancer Res. 1998;58:1700‐1706. [PubMed] [Google Scholar]
- 32. Chaturvedi AK, Katki HA, Hildesheim A, et al. Human papillomavirus infection with multiple types: pattern of coinfection and risk of cervical disease. J Infect Dis. 2011;203:910‐920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown DR, Shew ML, Qadadri B, et al. A longitudinal study of genital human papillomavirus infection in a cohort of closely followed adolescent women. J Infect Dis. 2005;191:182‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho HW, So KA, Lee JK, Hong JH. Type‐specific persistence or regression of human papillomavirus genotypes in women with cervical intraepithelial neoplasia 1: a prospective cohort study. Obstet Gynecol Sci. 2015;58:40‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Benard VB, Watson M, Castle PE, Saraiya M. Cervical carcinoma rates among young females in the United States. Obstet Gynecol. 2012;120:1117‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmeink CE, Massuger LFAG, Lenselink CH, et al. Prospective follow‐up of 2,065 young unscreened women to study human papillomavirus incidence and clearance. Int J Cancer. 2013;133:172‐181. [DOI] [PubMed] [Google Scholar]
- 37. Moore KN, Bannon RJ, Lanneau GS, Zuna RE, Walker JL, Gold MA. Cervical dysplasia among women over 35 years of age. Am J Obstet Gynecol. 2008;199(471):e1‐e5. [DOI] [PubMed] [Google Scholar]
- 38. Gilani SM, Mazzara PF. Cytohistologic correlation in premenopausal and postmenopausal women. Acta Cytol. 2013;57:575‐580. [DOI] [PubMed] [Google Scholar]
- 39. De Villiers EM. Relationship between steroid hormone contraceptives and HPV, cervical intraepithelial neoplasia and cervical carcinoma. Int J Cancer. 2003;103:705‐708. [DOI] [PubMed] [Google Scholar]
- 40. Marks M, Gravitt PE, Gupta SB, et al. The association of hormonal contraceptive use and HPV prevalence. Int J Cancer. 2011;128:2962‐2970. [DOI] [PubMed] [Google Scholar]
- 41. Huh WK, Joura EA, Giuliano AR, et al. Final efficacy, immunogenicity, and safety analyses of a nine‐valent human papillomavirus vaccine in women aged 16–26 years: a randomised, double‐blind trial. Lancet. 2017;390(10108):2143‐2159. Available from: http://dx.doi.org/ 10.1016/S0140-6736(17)31821-4 [DOI] [PubMed] [Google Scholar]
- 42. Yoshida H, Sakamoto H, Leslie A, Takahashi O, Tsuboi S, Kitamura K. Contraception in Japan: current trends. Contraception. 2016;93(6):475‐477. Available from: 10.1016/j.contraception.2016.02.006 [DOI] [PubMed] [Google Scholar]
- 43. Hanley SJB, Yoshioka E, Ito Y, Kishi R. HPV vaccination crisis in Japan. Lancet. 2015;385(9987):2571. Available from: 10.1016/S0140-6736(15)61152-7 [DOI] [PubMed] [Google Scholar]
- 44. Titman AC, Sharples LD. Semi‐Markov models with phase‐type sojourn distributions. Biometrics. 2010;66:742‐752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S6
Appendix
Data Availability Statement
The data supporting the findings of this study are available upon reasonable request.


