Abstract
The recovery of blood supply after a period of myocardial ischaemia does not restore the heart function and instead results in a serious dysfunction called myocardial ischaemia‐reperfusion injury (IRI), which involves several complex pathophysiological processes. Mitochondria have a wide range of functions in maintaining the cellular energy supply, cell signalling and programmed cell death. When mitochondrial function is insufficient or disordered, it may have adverse effects on myocardial ischaemia‐reperfusion and therefore mitochondrial dysfunction caused by oxidative stress a core molecular mechanism of IRI. Peroxisome proliferator‐activated receptor gamma co‐activator 1α (PGC‐1α) is an important antioxidant molecule found in mitochondria. However, its role in IRI has not yet been systematically summarized. In this review, we speculate the role of PGC‐1α as a key regulator of mitonuclear communication, which may interacts with nuclear factor, erythroid 2 like ‐1 and ‐2 (NRF‐1/2) to inhibit mitochondrial oxidative stress, promote the clearance of damaged mitochondria, enhance mitochondrial biogenesis, and reduce the burden of IRI.
Keywords: mitochondria; myocardial ischaemia‐reperfusion injury (IRI); nuclear factor, erythroid 2 like 1/2 (NRF‐1/2); nucleus; oxidative stress; peroxisome proliferator‐activated receptor gamma co‐activator‐1α (PGC‐1α)
1. INTRODUCTION
Cardiovascular diseases are key medical issues with frequently fatal outcomes. According to statistics of the World Health Organization, about one sixth of the global mortality annually is caused by myocardial ischaemia‐reperfusion injury (IRI). 1 In the past decades, researchers have found several therapeutic approaches that can prevent and reduce IRI, such as calcium channel blockers, anti‐inflammatory drugs, erythropoietin, atorvastatin, adenosine and anti‐hyperglycaemic agents. 2 , 3 , 4 However, limited by their inherent characteristics, these therapeutics have so far remained largely theoretical and not yet achieved satisfactory clinical results. Therefore, there is an urgent need to find more suitable and clinically actionable treatments for IRI.
Peroxisome proliferator‐activated receptor gamma co‐activator‐1α (PGC‐1α) is one of the most studied nuclear cofactor family members. As an auxiliary regulator, it plays an important role in polyprotein complexes. On one hand, it binds transcription factors binding to DNA sequences in complexes; on the other hand, it can interact with some nuclear factors, such as nuclear factor, erythroid 2 like‐1 (NRF‐1) and −2 (NRF‐2). 5 , 6 , 7 , 8 In addition, previous studies have found that overexpression of PGC‐1α helps to reduce myocardial ischaemia. Therefore, PGC‐1α may be a potential therapeutic target for IRI.
Mitochondria mainly exist in the myocardium, which has a high energy demand. 10 Here, they do not only produce large amounts of adenosine triphosphate via oxidative phosphorylation in order to maintain the normal physiological function of cells, but also participate in the cell matrix metabolism, signal transduction pathways, and other cellular activities. 11 In order to maintain the mitochondrial function stability, mitochondria participate in the anterograde regulation of the nucleus, as well as produce a retrograde response to regulate nuclear gene expression. 12 , 13 , 14 This mitonuclear communication by positive regulation and reverse feedback constitutes a functional interaction which helps to maintain mitochondrial function and can protect cells from external damage. 15
This article reviews the cardioprotective effects of PGC‐1α in IRI. Firstly, the structure, expression, and location of PGC‐1α are described to provide an overview of PGC‐1α. In addition, we summarized the importance of mitonuclear communication in the myocardium. At the same time, the potential protective effects of PGC‐1α on IRI via mitonuclear communication were described in detail. Finally, the therapeutic potential and future research directions involving PGC‐1α in IRI are presented.
2. OVERVIEW OF PGC‐1α
2.1. Structural characteristics of PGC‐1α
The PGC‐1α gene is highly conserved and located on the reverse strand of chromosome 4 (23, 755, 041‐23, 904, 089). 16 There are 18 transcripts (splice variants), 182 lineage homologues and 2 lineage homologues of PGC‐1α, and it shares similar domain structures with other nuclear cofactor family members. The protein contains three functional domains, including an N‐terminal activation domain, a central regulatory domain, and a C‐terminal ribonucleic acid binding domain. 17 The N‐terminal activation domain consists of a transcriptional activation domain and a leucine‐rich LXXLL motif, which is a key motif for nuclear receptor interaction. 18 The C‐terminal ribonucleic acid binding domain consists of a ribonucleic acid recognition and binding site (RRM, ribonucleic acid recognition motif) and a serine‐ and arginine‐rich structural domain (RS, serine/arginine enrichment domain). 19 As the PGC‐1α family lacks enzymatic activity, PGC‐1α acts as an anchoring platform for other proteins with histone acetyltransferase activity, and triggers gene transcription by promoting the assembly of transcription mechanisms, thus playing a regulatory role in gene transcription (Figure 1). PGC‐1 α promoter contains a common sequence (ARE) are binding to NRF. NRF is likely to interact with the N‐terminal activation domain, C‐terminal SR and RNA binding domain of PGC‐1 α.
FIGURE 1.
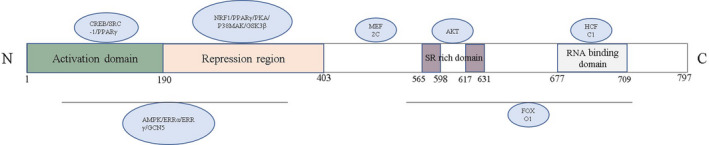
Functional domains of PGC‐1α. A schematic representation of PGC‐1α functional domains involved in the regulation of IRI
2.2. Expression and localization of PGC‐1α
Since PGC‐1α was discovered to contribute to heat production, specifically in brown adipose tissue, in the 1990s, 20 many studies on PGC‐1α have accumulated. PGC‐1α typically is located in the cytosol and nucleus, where it performs its biological functions under physiological and pathological conditions. PGC‐1α is highly expressed in metabolically active tissues, such as brown adipose tissue, heart, kidney, skeletal muscle, and brain. 21 In addition, the expression of PGC‐1α can be further induced under various stress conditions. For example, PGC‐1α is activated by nutritional deprivation, oxidative damage, and chemotherapy. 22 Moreover, high expression levels of PGC‐1α have been found to be a critical protective factor in IRI. 23 , 24
3. ROLE OF MITONUCLEAR COMMUNICATION IN IRI
3.1. Mitochondrial dysfunction during IRI
Mitochondria are the key producers of energy in eukaryotic cells. The number of mitochondria varies in different tissues and cells based on varying amounts of energy demand. 25 Cardiac muscle cells are highly metabolically active and energy‐demanding and therefore have a high number of mitochondria, which account for 40% of the cell volume. 26 During myocardial ischaemia, an increase in reactive oxygen species (ROS) in the myocardial submucosal matrix and an inhibition of mitochondrial permeability transition pore (mPTP) opening are observed. The opening of mPTP and oxidative stress caused by ROS are considered the main mechanisms of mitochondrial and myocardial dysfunction. 27 In addition, mitochondria in myocardial fibrous matrix decrease the activity of aconitase, an enzyme of the tricarboxylic acid cycle, and have no inhibitory effect on the production of ROS and the opening of mPTP. 22 Compared with the cardiac fibrous matrix, the cardiac submucosal matrix has a protective effect on mitochondria, which is related to the reduction of ROS production by inhibition of the activity of the mitochondrial electron transfer chain complex. 28 When rats were deprived of oxygen for a long time, the production of mitochondrial ROS decreased, the opening of mPTP was inhibited, and the apoptosis of subcellular stromal cells decreased. 29 Therefore, cardiomyocytes have been thought to be beneficial to mitochondria in hypoxic environments. However, during reperfusion, tissues receive blood supply again, which greatly increases the number of free radicals, including superoxide anions, hydrogen peroxide and hydroxyl groups, resulting in an aggravated injury. 26 , 30 Free radicals are highly reactive and they can attack mitochondria in myocardial tissue, leading to mitochondrial dysfunction. However, mitochondria are not only a main target of ROS damage, but also an important source of ROS. 31 ROS are predominantly produced by electron transfer chain complexes I and III. Concurrently, ROS production in mitochondria destroys the structure of mitochondrial membranes, resulting in the release of cytochrome C, thus triggering apoptosis via the mitochondrial pathway. It has been reported that mitochondria have a positive feedback regulatory role in the initial ROS production by increasing their own ROS levels, which is called ROS‐induced ROS release (RIRR). 32 ROS produced by the RIRR pathway lead to opening of mPTP, Ca2+ overload, and the collapse of the mitochondrial membrane potential, thus further aggravating IRI. These changes can lead to reperfusion arrhythmia, myocardial stunning, apoptosis and necrosis, resulting in microvascular and macrovascular injuries. 33 Several drugs target mitochondria in order to protect myocardial cells from IRI by reducing the levels of ROS, for example the peptide bendavia. Myocardial cells treated with bendavia exhibit higher survival rates than those of the control group. 34 This protective effect is achieved by reducing ROS‐induced cell death during reperfusion and maintaining the mitochondria transmembrane potential (ΔΨm). Singer et al have found that ammonium tetrathiomolybdate (ATTM) significantly reduced the infarct size after myocardial ischaemia or cerebral ischaemia by reducing mitochondrial ROS, thus improving the survival rate of mice by preventing a severe haemorrhage. 35 Although these drugs have achieved good efficacy in animals, they may have side effects in humans, so clinical experiments must be conducted to test their efficacy (Figure 2).
FIGURE 2.
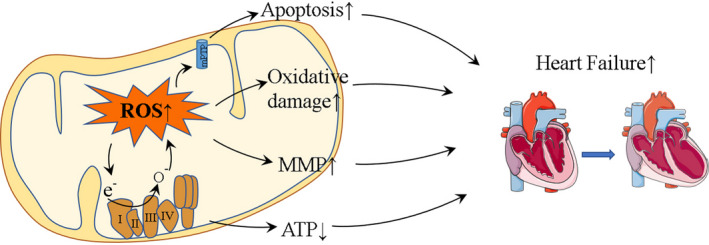
Role of mitochondria in IRI. In IRI, mitochondria attacked by ROS in turn produce increased levels of ROS, which can exacerbate damage. In addition, ROS lead to a decrease of ATP levels, oxidative damage, MMP and apoptosis, which eventually leads to heart failure
3.2. Mitonuclear communication during IRI
Eukaryote cell structures are the result of the co‐evolution of mitochondrial and nuclear genomes. 36 Therefore, it is conceivable that this cooperation resulted in genes which are adapted to environmental challenges via nuclear‐mitochondrial genetic interactions. Among the 1200 proteins in mitochondria, 13 are encoded by mitochondrial genes (mtDNA), 37 which account for a majority of the mitochondrial electron transfer chain complex. 38 As only a small fraction of mitochondrial proteins is encoded by mtDNA genes, the nucleus and mitochondria must continuously coordinate the transcription and translation of mitochondrial proteins, as well as translocations and imports. KJ et al 39 have investigated this by using mitochondrial‐nuclear eXchange mice, in which the nuclear and mitochondrial genomes were exchanged among different murine strains. They showed that the combination of the nuclear‐mitochondrial genetic backgrounds significantly changed metabolic efficiency and body composition. Mendelian genetics and mitochondrial genetics do not unilaterally control gene expression and as a result, when mitochondria are stressed, the nucleus reacts accordingly.
PGC‐1α plays a key role in nuclear‐mitochondrial crosstalk. Mitochondria are not only the core production site of cellular energy, but also the regulator of many cellular functions such as the metabolism and apoptosis. 40 Moreover, the function of mitochondria is tightly controlled by the nucleus, which can reduce or increase the activity of mitochondria and promote the biogenesis of mitochondria, depending on the bioenergetic need of the cell. 41 For example, the nucleus can regulate the transcription of mitochondrial DNA and mitochondrial regulatory genes, such as AMP‐activated catalytic subunit alpha 1 (AMPK), sirtuin 1 (Sirt1), and PGC‐1α. 42 , 43 Conversely, mitochondria can generate a ‘retrograde regulation’ to send signals to the nucleus and change the expression of nuclear genes, thus changing the function of cells and re‐planning cellular metabolism. 44 This response can be triggered by decreases in ATP levels, an increase in ROS or the release of mitochondrial Ca2+. 45 Low levels of ATP activate AMPK and PGC‐1α, which regulate mitochondrial biogenesis and mitochondrial homeostasis. The increase of ROS and Ca2+ also activates AMPK, PGC‐1 α, and c‐Jun N‐terminal kinase (JNK) pathways via anterograde regulation. 46
4. PGC‐1Α PROTECTS FROM IRI THROUGH ANTEROGRADE REGULATION OF MITONUCLEAR COMMUNICATION
During IRI, the function of mitochondria is compromised due to an increase of ROS. Clearing ROS and injured mitochondria, and generating new mitochondria is critical for myocardial cells in order to maintain their normal functions. 47 We speculate that PGC‐1α actively regulates downstream factors through the mitonuclear communication during IRI, and ensures mitochondrial homeostasis by reducing ROS, which activates mitochondrial biogenesis and mitophagy (Figure 3).
FIGURE 3.
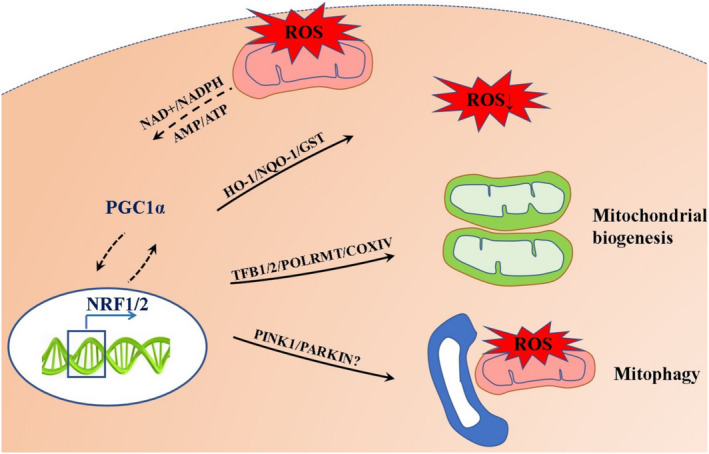
Role of PGC‐1α in IRI. Proposed model for the PGC‐1α‐NRF‐1/NRF‐2 signalling pathway in IRI. Mitochondrial dysfunction caused by myocardial ischaemia‐reperfusion injury leads to cell death. We illustrate how the PGC‐1α‐NRF‐1/NRF‐2 signalling pathway may orchestrate protection from mitochondria damage
4.1. ROS clearance pathway
NRF‐1 is closely associated with mitochondrial oxidative stress. Under oxidative stress, NRF‐1 binds to small Maf (sMaf) to form the NRF1‐MAFG heterodimer, which binds preferentially to the antioxidant response element sequence (ARE). 48 In addition, the knockdown of NRF‐1 inhibits the expression of antioxidant genes encoding glutamate‐cysteine ligase catalytic subunit (GCLC), glutathione peroxidase 1 (GP1) and metallothionein 2 (MT‐2) in MC3T3‐E1 cells treated with lipopolysaccharide. 49 These findings are consistent with the results of targeted NRF‐1 knockout in a severe oxidative stress model of mouse fibroblasts. 50 What' more, the absence of NRF‐1 was not compensated by the presence of NRF‐2, which indicates that NRF‐1 plays a key role in antioxidant stress. 51 , 52 , 53 Therefore, PGC‐1α may play a protective role in IRI by binding and co‐activating NRF‐1 and its downstream antioxidant genes.
NRF‐2 binds to ARE elements and transactivates a group of protective enzymes, such as haem oxygenase 1 (HO‐1), gamma‐glutamyl cysteine synthase (gamma‐GCS), peroxidase 1 (PRDX1), glutathione reductase (GR), and thioredoxin (SRXN). 54 Drug metabolism and detoxification enzymes NADH quinone dehydrogenase 1 (NQO1), glutathione‐S‐transferase (GST), UDP‐glucuronosyltransferase and glucose‐6‐phosphate dehydrogenase have been used to alleviate oxidative stress injury. 55 , 56 , 57 In addition, ARE‐like elements present in the promoter of NRF‐2 enhance its own transcription. 58 Previous studies have shown that NRF‐2 activates downstream antioxidant genes during IRI, which significantly reduce myocardial injury. 51 , 59
Similar to NRF‐1, NRF‐2 also activates ARE elements (sMaF) by binding to sMaf. Under normal conditions, sMaf is a heteromeric chaperone of NRF‐1. However, under oxidative stress, NRF‐2 recruits sMaf proteins and deprives NRF‐1 of sMaf. 60 This indicates that NRF‐2 may play a major role in the clearance of ROS. In addition, the absence of NRF‐1 is not compensated for by the presence of NRF‐2, which indicates that NRF‐1 plays an important role in coping with cellular stress. In both young and old mice, NRF‐2 knockout did not increase the compensatory ability of NRF‐1, but decreased the expression of NRF‐1. 61 It has been found that NRF‐2 knockout reduces both the transcription of NRF‐1 and the downstream mitochondrial transcription factor A (mtTFA) in ROS as well as the nitric oxide (NO)‐induced adaptive responses of mouse skeletal muscle cells to exercise. 62 However, NRF2 knockout mice were used in the experiment, and it is necessary to verify the interaction mechanism between NRF‐1 and NRF‐2 at the cellular level. From these studies, we speculated that NRF‐2 promotes the stability of NRF‐1 in complex networks.
ROS is also the early inducement of metabolic syndrome. Metabolic syndrome refers to the simultaneous existence of risk factors for atherosclerosis in an individual, including hyperglycaemia, dyslipidemia and hypertension. For example, in rats with metabolic syndrome induced by high fat and high sugar diet, excessive production of ROS is accompanied. 63 Rats with large amounts of ROS have earlier insulin resistance, hyperlipidemia, and hypertension. 64 Patients with metabolic syndrome may aggravate myocardial ischaemia‐reperfusion injury. PGC‐1α is the main way to eliminate ROS, which means that targeting PGC‐1α can eliminate ROS, reduce the occurrence of metabolic syndrome and ensure the health of the heart.
4.2. Mitochondrial biogenesis
Mitochondrial biogenesis refers to the formation of new mitochondria and their ability to produce ATP. 65 Mitochondria have relatively independent genetic systems and biosynthetic sites. 66 However, mitochondrial proliferation is a complex process that depends on the regulation of nuclear coding genes, with PGC‐1α being an important regulator of mitochondrial proliferation. PGC‐1α stimulates mitochondrial proliferation by activating transcription factors. It has been proved that NRF can bind to are sequence in promoter ARE region of mitochondrial biogenesis related genes (such as nuclear respiratory factor 1/2, Nrf1/2) to stimulate mitochondrial synthesis. 67
Nrf1 activates the expression of nuclear genes essential for mitochondrial biogenesis and function, including key genes of the mitochondrial respiratory complex subunits, haem biosynthetase and regulatory factors involved in mtDNA replication and transcription. 68 In addition, Nrf1 activates TFAM and transcription factor B1/2 (TFB1M/2M) to promote the transcription of mitochondrial genes. 69 The mitochondrial transcription mechanism, consisting of TFAM, TFB2M and mitochondrial RNA polymerase (POLRMT), initiates the expression of mtDNA. 70 TFAM binds to mtDNA to change its structure. TFB2M is also encoded by nuclear DNA and is transported from the cytoplasm to mitochondria as a transcriptional factor of mitochondrial genes. In addition, TFB2M and POLRMT interact with TFAM to induce nucleus gene expression. 71
Recent studies have shown that activation of the Nrf2 pathway enhances mitochondrial biogenesis. NRF‐2 binds specifically to cis elements in the promoter of cytochrome oxidase subunit IV (COXIV) to induce COXIV transcription. 72 In addition, Nrf2 interacts with many other genes related to respiratory chain expression, including TFAM, TFB1M and TFB2M, to promote mitochondrial biogenesis. 73 , 74
In many cases, Nrf1 and Nrf2 have the same targeted genes. Nrf1 and Nrf2 both bind to the TFB1M and TFB2M promoters, as was detected by chromatin immunoprecipitation. 75 Three of the four succinate dehydrogenase (complex II) subunit genes also have Nrf1/2 binding sites in their promoter. 76 Piantadosi and his colleagues have shown that Nrf2 activation enhances mitochondrial biogenesis in cardiomyocytes via multiple Nrf2 binding sites in the Nrf1 promoter. Therefore, Nrf1 plays a major role in mitochondrial biogenesis. Like Nrf1, Nrf2 is involved in the regulation of the expression of essential respiratory chain proteins and key components of the mitochondrial transcription mechanism. This complex mechanism ensures the coordination among genes and promotes mitochondrial biogenesis.
4.3. Mitophagy pathway
During IRI, damage to mitochondria is further aggravated by oxidative stress. Therefore, it is vital to remove dysfunctional mitochondria. Mitophagy refers to the process of selective removal of redundant or damaged mitochondria by autophagy, and it plays a significant role in mitochondrial control and cell survival. 77
The PTEN induced kinase 1 (PINK1)/ parkin RBR E3 ubiquitin protein ligase (Parkin)‐mediated signalling pathway is the best‐characterized mitophagy pathway. 78 Upon oxidative stress, mitochondrial depolarization and a decrease in membrane potential occurs; PINK1 can no longer enter the mitochondria, and its protein hydrolysis, cleavage and subsequent degradation are inhibited. Furthermore, PINK1 accumulates in the depolarized mitochondrial outer membrane, where it is phosphorylated, and then recruits several other proteins, including Parkin and TANK‐binding kinase 1. After Parkin is activated by phosphorylation, mitophagy is induced by ubiquitination of voltage‐dependent anion channel (VDAC) and recruitment of the receptor protein sequestosome 1 (p62/SQSTM1). 79 Phosphorylation of parkin is one of the key steps of mitophagy. Therefore, other potential phosphorylation sites can be found by bioinformatics, liquid chromatography‐mass spectrometry, and microarray. In addition, mitochondrial dynamics can also affect mitophagy. The size of mammalian autophagy bodies ranged from 500 nm to 1500 nm. Therefore, it can be predicted that mitochondrion division occurs before mitochondrial autophagy. Experimental evidence also confirmed that the expression of mitochondrion fission‐related factors or the degradation of mitochondrial fusion‐related factors is necessary to induce mitophagy. These factors regulate mitophagy by interacting with microtubule associated protein 1 light chain 3 alpha (LC3) adaptor protein or LC3 receptor. For example, dynamin 1 like (Drp1), as an important mitochondrial fission protein, can interact with LC3 receptors FUN14 domain containing 1 and BCL2 like 13 to induce mitophagy. In addition, optic atrophy 1 (OPA1), as a mitochondrial fusion protein, can also be used as an important factor to induce mitophagy.
PGC‐1α has been shown to participate in mitophagy, 80 but the specific mechanism is still unclear. Both PGC‐1α and PINK1 are highly expressed in the brain, 81 however, when the expression of PINK1 is knocked down, the PGC‐1α expression is also reduced. 82 The exact interaction mechanism is unclear, but it is possible that PGC‐1α contributes to transcriptional regulation and activates the promoter region of PINK1. Down‐regulation of PINK1, a protein kinase, may in turn affect the phosphorylation of PGC‐1α and decrease its cytoplasmic stability. In addition, the theoretical downstream target genes of NRF‐1 include PINK1 and Parkin, 83 hence we believed that PGC‐1α may regulate the expression of PINK1 and Parkin indirectly by regulating NRF‐1, and thus may participate in mitophagy. In addition, the p62/SQSTM1 promoter contains ARE elements, allowing NRF‐1 and −2 to bind and mediate transcription, 84 and it has been found that NRF‐1 rapidly activates p62/SQSTM1 and induces autophagy under oxidative stress. 85 NRF‐2 has also been shown to promote the expression of p62/SQSTM1 by interaction with the transcription co‐regulator SPBP. 86 The STGE motif allows p62/SQSTM1 to bind and target kelch‐like ECH‐associated protein 1 (Keap1) for autophagic degradation, eliminating negative regulation, and thus promoting an accumulation of NRF‐2. 87 Ser‐349 in the STGE motif of p62/SQSTM1 can be phosphorylated by mammalian target of rapamycin complex 1, which senses oxidative stress, 88 and phosphorylation of Ser‐349 enhances its affinity for Keap1 binding. 89 Keap1 removal results in an increase in NRF‐2 protein expression. p62/SQSTM1, the autophagic connector proteins CALCOCO‐2/NDP52, serine/threonine kinase ULK1, E3 ubiquitin ligase ATG5 and ubiquitin‐like modifier GABA Type A receptor‐related protein 1 all have been demonstrated to interact with NRF‐2.
5. CONCLUSIONS AND FUTURE PERSPECTIVES
Oxidative stress is the main factor leading to mitochondrial dysfunction. The damaged mitochondria are fed back to the nucleus through ‘retrograde regulation’. The nucleus resists mitochondrial damage by ‘anterograde regulation’. We propose the important role of mitonuclear communication mechanism in IRI. PGC‐1α can resist IRI‐induced mitochondrial damage by up‐regulating a series of mitochondrial related genes. PGC‐1α/NRF‐1/2 reduce the production of ROS caused by IRI via up‐regulation of antioxidant genes and furthermore eliminate damaged mitochondria by enhancing mitophagy. Additionally, PGC‐1α/NRF‐1/2 promotes the biogenesis of mitochondria in the myocardium. In addition, we believe that mitochondrial biogenesis can help mitophagy to produce healthy mitochondria. However, it is still unclear which of the three signalling pathways mediated by PGC‐1α/NRF‐1/2 play a major role in IRI. This may depend on whether the patient has other underlying diseases and the degree of blood supply. In conclusion, PGC‐1α may effectively resist IRI through ‘anterograde regulation’ of mitonuclear communication.
Although a large amount of evidence has elucidated the underlying mechanism of PGC‐1 α associated with heart disease, drug development for successful treatment of PGC‐1 α is still in its infancy. Therefore, we need to continue to explore the mechanism of PGC‐1 α in the heart. In addition, data on the pathological effects of PGC‐1 α on heart disease in rodents and humans are limited and often contradictory. Future research should focus on clarifying the signalling network of PGC‐1 α in IRI. In addition, there are many problems to be solved. For example, how does PGC‐1 α activity change in IRI? How to control PGC‐1 α signalling pathway to play a powerful role in height regulation without side effects? Therefore, it is necessary to have a more comprehensive and detailed understanding of the role of PGC‐1 α in heart disease before studying the therapy for PGC‐1 α signalling pathway.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Yanqing Li: Formal analysis (equal); Project administration (equal); Software (equal); Validation (equal); Writing‐original draft (equal). Yan Jiao: Data curation (equal); Methodology (equal); Visualization (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Ya‐Nan Liu: Supervision (equal); Writing‐review & editing (equal). Jiaying Fu: Validation (equal). Lian‐Kun Sun: Conceptualization (equal); Funding acquisition (equal); Investigation (equal); Resources (equal); Writing‐original draft (equal). Jing Su: Conceptualization (equal); Funding acquisition (equal); Resources (equal); Validation (equal); Writing‐review & editing (equal).
ACKNOWLEDGEMENTS
The authors would like to thank all the reviewers who participated in the review and MJEditor (www.mjeditor.com) for its linguistic assistance during the preparation of this manuscript.
Li Y‐Q, Jiao Y, Liu Y‐N, Fu J‐Y, Sun L‐K, Su J. PGC‐1α protects from myocardial ischaemia‐reperfusion injury by regulating mitonuclear communication. J Cell Mol Med.2022;26:593‐600. 10.1111/jcmm.16236
Yanqing Li and Yan Jiao are the co‐first author.
Contributor Information
Lian‐Kun Sun, Email: sunlk@jlu.edu.cn.
Jing Su, Email: sujing@jlu.edu.cn.
REFERENCES
- 1. Topjian AA, de Caen A, Wainwright MS, et al. Pediatric post‐cardiac arrest care: a scientific statement from the american heart association. Circulation. 2019;140:e194‐e233. [DOI] [PubMed] [Google Scholar]
- 2. Heusch G. Myocardial ischaemia‐reperfusion injury and cardioprotection in perspective. Nat Rev Cardiol. 2020;17(12):773‐789. [DOI] [PubMed] [Google Scholar]
- 3. Han D, Wang Y, Chen J, et al. Activation of melatonin receptor 2 but not melatonin receptor 1 mediates melatonin‐conferred cardioprotection against myocardial ischemia/reperfusion injury. J Pineal Res. 2019;67:e12571. [DOI] [PubMed] [Google Scholar]
- 4. Penna C, Andreadou I, Aragno M, et al. Effect of hyperglycaemia and diabetes on acute myocardial ischaemia‐reperfusion injury and cardioprotection by ischaemic conditioning protocols. Br J Pharmacol. 2020;177:5312‐5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Du X, Shen T, Wang H, et al. Adaptations of hepatic lipid metabolism and mitochondria in dairy cows with mild fatty liver. J Dairy Sci. 2018;101:9544‐9558. [DOI] [PubMed] [Google Scholar]
- 6. Wu H, Liu B, Chen Z, Li G, Zhang Z. MSC‐induced lncRNA HCP5 drove fatty acid oxidation through miR‐3619‐5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo‐resistance of gastric cancer. Cell Death Dis. 2020;11:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen YT, Hu Y, Yang QY, et al. Excessive glucocorticoids during pregnancy impair fetal brown fat development and predispose offspring to metabolic dysfunctions. Diabetes. 2020;69:1662‐1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan HWS, Anjum B, Shen HM, Ghosh S, Yen PM, Sinha RA. Lysosomal inhibition attenuates peroxisomal gene transcription via suppression of PPARA and PPARGC1A levels. Autophagy. 2019;15:1455‐1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tian L, Cao W, Yue R, et al. Pretreatment with tilianin improves mitochondrial energy metabolism and oxidative stress in rats with myocardial ischemia/reperfusion injury via AMPK/SIRT1/PGC‐1 alpha signaling pathway. J Pharmacol Sci. 2019;139:352‐360. [DOI] [PubMed] [Google Scholar]
- 10. Youle RJ. Mitochondria‐striking a balance between host and endosymbiont. Science (New York, NY). 2019;365. [DOI] [PubMed] [Google Scholar]
- 11. Akbari M, Kirkwood TBL, Bohr VA. Mitochondria in the signaling pathways that control longevity and health span. Ageing Res Rev. 2019;54:100940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lisowski P, Kannan P, Mlody B, Prigione A. Mitochondria and the dynamic control of stem cell homeostasis. EMBO Rep. 2018;19.e45432‐e45432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grünewald A, Kumar KR, Sue CM. New insights into the complex role of mitochondria in parkinson's disease. Prog Neurogibol. 2019;177:73‐93. [DOI] [PubMed] [Google Scholar]
- 14. Fernández‐Ruiz I. Imaging: Mitochondria shine light on heart function. Nat Rev Cardiol. 2017;14:633. [DOI] [PubMed] [Google Scholar]
- 15. Quirós PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213‐226. [DOI] [PubMed] [Google Scholar]
- 16. Nierenberg AA, Ghaznavi SA, Sande Mathias I, Ellard KK, Janos JA, Sylvia LG. Peroxisome proliferator‐activated receptor gamma coactivator‐1 alpha as a novel target for bipolar disorder and other neuropsychiatric disorders. Biol Psychiat. 2018;83:761‐769. [DOI] [PubMed] [Google Scholar]
- 17. Lv J, Jiang S, Yang Z, et al. PGC‐1α sparks the fire of neuroprotection against neurodegenerative disorders. Ageing Res Rev. 2018;44:8‐21. [DOI] [PubMed] [Google Scholar]
- 18. Petr M, Maciejewska‐Skrendo A, Zajac A, Chycki J, Stastny P. Association of elite sports status with gene variants of peroxisome proliferator activated receptors and their transcriptional coactivator. Int J Mol Sci. 2020;21(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng Z, Zheng L, Almeida FA. Epigenetic reprogramming in metabolic disorders: nutritional factors and beyond. The Journal of nutritional biochemistry. 2018;54:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold‐inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829‐839. [DOI] [PubMed] [Google Scholar]
- 21. Martínez‐Redondo V, Pettersson AT, Ruas JL. The hitchhiker's guide to PGC‐1α isoform structure and biological functions. Diabetologia. 2015;58:1969‐1977. [DOI] [PubMed] [Google Scholar]
- 22. Kim JY, Park S, Park SH, et al. Overexpression of pigment epithelium‐derived factor in placenta‐derived mesenchymal stem cells promotes mitochondrial biogenesis in retinal cells. Lab Invest. 2020;101: 51‐69. [DOI] [PubMed] [Google Scholar]
- 23. Hu MZ, Zhou B, Mao HY, et al. Exogenous hydrogen sul fi de postconditioning protects isolated rat hearts from ischemia/reperfusion injury through Sirt1/PGC‐1alpha signaling pathway. International heart journal. 2016;57:477‐482. [DOI] [PubMed] [Google Scholar]
- 24. Li Y, Li J, Hou Z, Yu Y, Yu B. KLF5 overexpression attenuates cardiomyocyte inflammation induced by oxygen‐glucose deprivation/reperfusion through the PPARgamma/PGC‐1alpha/TNF‐alpha signaling pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;84:940‐946. [DOI] [PubMed] [Google Scholar]
- 25. Mohanraj K, Nowicka U, Chacinska A. Mitochondrial control of cellular protein homeostasis. Biochem J. 2020;477:3033‐3054. [DOI] [PubMed] [Google Scholar]
- 26. Rahman J, Singh P, Merle NS, Niyonzima N, Kemper C. Complement's favorite organelle ‐ mitochondria? Br J Pharmacol. 2020;25 1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Penna C, Perrelli MG, Pagliaro P. Mitochondrial pathways, permeability transition pore, and redox signaling in cardioprotection: therapeutic implications. Antioxid Redox Signal. 2013;18:556‐599. [DOI] [PubMed] [Google Scholar]
- 28. Guo M, Zhang X, Liu J, Hou L, Liu H, Zhao X. OsProDH negatively regulates thermotolerance in rice by modulating proline metabolism and reactive oxygen species scavenging. Rice (New York, NY). 2020;13:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang B, Qian J, Ma J, et al. Myocardial transfection of hypoxia‐inducible factor‐1α and co‐transplantation of mesenchymal stem cells enhance cardiac repair in rats with experimental myocardial infarction. Stem Cell Res Ther. 2014;5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pagliaro P, Penna C. Redox signalling and cardioprotection: translatability and mechanism. Br J Pharmacol. 2015;172:1974‐1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Picca A, Mankowski RT, Burman JL, et al. Mitochondrial quality control mechanisms as molecular targets in cardiac ageing. Nat Rev Cardiol. 2018;15:543‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim YM, Kim SJ, Tatsunami R, Yamamura H, Fukai T, Ushio‐Fukai M. ROS‐induced ROS release orchestrated by Nox4, Nox2, and mitochondria in VEGF signaling and angiogenesis. Am J Physiol Cell Physiol. 2017;312:C749‐C764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li D, Wang X, Huang Q, Li S, Zhou Y, Li Z. Cardioprotection of CAPE‐oNO(2) against myocardial ischemia/reperfusion induced ROS generation via regulating the SIRT1/eNOS/NF‐κB pathway in vivo and in vitro. Redox Biol. 2018;15:62‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao W, Xu Z, Cao J, et al. Elamipretide (SS‐31) improves mitochondrial dysfunction, synaptic and memory impairment induced by lipopolysaccharide in mice. J Neuroinflammation. 2019;16:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dyson A, Dal‐Pizzol F, Sabbatini G, et al. Ammonium tetrathiomolybdate following ischemia/reperfusion injury: Chemistry, pharmacology, and impact of a new class of sulfide donor in preclinical injury models. PLoS Medicine. 2017;14:e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brunk CF, Martin WF. Archaeal histone contributions to the origin of eukaryotes. Trends Microbiol. 2019;27:703‐714. [DOI] [PubMed] [Google Scholar]
- 37. Bahat A, Gross A. Mitochondrial plasticity in cell fate regulation. The Journal of biological chemistry. 2019;294:13852‐13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benayoun BA, Lee C. MOTS‐c: a mitochondrial‐encoded regulator of the nucleus. BioEssays : news and reviews in molecular, cellular and developmental biology. 2019;41:e1900046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dunham‐Snary KJ, Sandel MW, Sammy MJ, et al. Mitochondrial ‐ nuclear genetic interaction modulates whole body metabolism, adiposity and gene expression in vivo. EBioMedicine. 2018;36:316‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan DC. Mitochondrial dynamics and its involvement in disease. Annual review of pathology. 2020;15:235‐259. [DOI] [PubMed] [Google Scholar]
- 41. Mangalhara KC, Shadel GS. A mitochondrial‐derived peptide exercises the nuclear option. Cell Metab. 2018;28:330‐331. [DOI] [PubMed] [Google Scholar]
- 42. Mello T, Simeone I, Galli A. Mito‐nuclear communication in hepatocellular carcinoma metabolic rewiring. Cells. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eisenberg‐Bord M, Schuldiner M. Ground control to major TOM: mitochondria‐nucleus communication. The FEBS journal. 2017;284:196‐210. [DOI] [PubMed] [Google Scholar]
- 44. Vizioli MG, Liu T, Miller KN, et al. Mitochondria‐to‐nucleus retrograde signaling drives formation of cytoplasmic chromatin and inflammation in senescence. Genes Dev. 2020;34:428‐445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei Y, Chen L, Xu H, Xie C, Zhou Y, Zhou F. Mitochondrial dysfunctions regulated radioresistance through mitochondria‐to‐nucleus retrograde signaling pathway of NF‐κB/PI3K/AKT2/mTOR. Radiat Res. 2018;190:204‐215. [DOI] [PubMed] [Google Scholar]
- 46. Quan N, Wang L, Chen X, et al. Sestrin2 prevents age‐related intolerance to post myocardial infarction via AMPK/PGC‐1α pathway. J Mol Cell Cardiol. 2018;115:170‐178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stanzani G, Duchen MR, Singer M. The role of mitochondria in sepsis‐induced cardiomyopathy. Biochim Biophys Acta. 2019;1865:759‐773. [DOI] [PubMed] [Google Scholar]
- 48. Katsuoka F, Otsuki A, Takahashi M, Ito S, Yamamoto M. Direct and specific functional evaluation of the Nrf2 and MafG heterodimer by introducing a tethered dimer into small maf‐deficient cells. Mol Cell Biol. 2019;39:e00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lou B, Hu Y, Lu X, et al. Long‐isoform NRF1 protects against arsenic cytotoxicity in mouse bone marrow‐derived mesenchymal stem cells by suppressing mitochondrial ROS and facilitating arsenic efflux. Toxicol Appl Pharmacol. 2020;407:115251. [DOI] [PubMed] [Google Scholar]
- 50. Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003;278:48021‐48029. [DOI] [PubMed] [Google Scholar]
- 51. Weng G, Zhou B, Liu T, Huang Z, Yang H. Sitagliptin promotes mitochondrial biogenesis in human SH‐SY5Y cells by increasing the expression of PGC‐1α/NRF1/TFAM. IUBMB Life. 2019;71:1515‐1521. [DOI] [PubMed] [Google Scholar]
- 52. Kunkel GH, Chaturvedi P, Thelian N, Nair R, Tyagi SC. Mechanisms of TFAM‐mediated cardiomyocyte protection. Can J Physiol Pharmacol. 2018;96:173‐181. [DOI] [PubMed] [Google Scholar]
- 53. Suliman HB, Keenan JE, Piantadosi CA. Mitochondrial quality‐control dysregulation in conditional HO‐1(‐/‐) mice. JCI insight. 2017;2:e89676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buendia I, Michalska P, Navarro E, Gameiro I, Egea J, León R. Nrf2‐ARE pathway: An emerging target against oxidative stress and neuroinflammation in neurodegenerative diseases. Pharmacol Ther. 2016;157:84‐104. [DOI] [PubMed] [Google Scholar]
- 55. Matzinger M, Fischhuber K, Heiss EH. Activation of Nrf2 signaling by natural products‐can it alleviate diabetes? Biotechnol Adv. 2018;36:1738‐1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ferrandiz ML, Nacher‐Juan J, Alcaraz MJ. Nrf2 as a therapeutic target for rheumatic diseases. Biochem Pharmacol. 2018;152:338‐346. [DOI] [PubMed] [Google Scholar]
- 57. Ma S, Paiboonrungruan C, Yan T, Williams KP, Major MB, Chen XL. Targeted therapy of esophageal squamous cell carcinoma: the NRF2 signaling pathway as target. Ann N Y Acad Sci. 2018;1434:164‐172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tonelli C, Chio IIC, Tuveson DA. Transcriptional regulation by Nrf2. Antioxid Redox Signal. 2018;29:1727‐1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shen Y, Liu X, Shi J, Wu X. Involvement of Nrf2 in myocardial ischemia and reperfusion injury. Int J Biol Macromol. 2019;125:496‐502. [DOI] [PubMed] [Google Scholar]
- 60. Katsuoka F, Yamazaki H, Yamamoto M. Small maf deficiency recapitulates the liver phenotypes of Nrf1‐ and Nrf2‐deficient mice. Genes cells. 2016;21:1309‐1319. [DOI] [PubMed] [Google Scholar]
- 61. Guan P, Liang Y, Wang N. Fasudil alleviates pressure overload‐induced heart failure by activating Nrf2‐mediated antioxidant responses. J Cell Biochem. 2018;119:6452‐6460. [DOI] [PubMed] [Google Scholar]
- 62. Merry TL, Ristow M. Nuclear factor erythroid‐derived 2‐like 2 (NFE2L2, Nrf2) mediates exercise‐induced mitochondrial biogenesis and the anti‐oxidant response in mice. J Physiol. 2016;594:5195‐5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carrier A. Metabolic syndrome and oxidative stress: a complex relationship. Antioxid Redox Signal. 2017;26:429‐431. [DOI] [PubMed] [Google Scholar]
- 64. López‐Acosta O, de los Angeles Fortis‐Barrera M, Barrios‐Maya MA, Ramírez AR, Aguilar FJA, El‐Hafidi M. Reactive oxygen species from NADPH oxidase and mitochondria participate in the proliferation of aortic smooth muscle cells from a model of metabolic syndrome. Oxid Med Cell Longev. 2018;2018:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Signorile A, Sgaramella G, Bellomo F, De Rasmo D. Prohibitins: a critical role in mitochondrial functions and implication in diseases. Cells. 2019;8:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Priesnitz C, Becker T. Pathways to balance mitochondrial translation and protein import. Genes Dev. 2018;32:1285‐1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase‐1 regulates cardiac mitochondrial biogenesis via Nrf2‐mediated transcriptional control of nuclear respiratory factor‐1. Circulation research. 2008;103:1232‐1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kiyama T, Chen CK, Wang SW, et al. Essential roles of mitochondrial biogenesis regulator Nrf1 in retinal development and homeostasis. Mol Neurodegener. 2018;13:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. King GA, Hashemi Shabestari M, Taris KH, et al. Acetylation and phosphorylation of human TFAM regulate TFAM‐DNA interactions via contrasting mechanisms. Nucleic Acids Res. 2018;46:3633‐3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hillen HS, Temiakov D, Cramer P. Structural basis of mitochondrial transcription. Nat Struct Mol Biol. 2018;25:754‐765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bouda E, Stapon A, Garcia‐Diaz M. Mechanisms of mammalian mitochondrial transcription. Protein science : a publication of the Protein Society. 2019;28:1594‐1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chang CW, Chen YS, Tsay YG, et al. ROS‐independent ER stress‐mediated NRF2 activation promotes warburg effect to maintain stemness‐associated properties of cancer‐initiating cells. Cell Death Dis. 2018;9:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ammal Kaidery N, Ahuja M, Thomas B. Crosstalk between Nrf2 signaling and mitochondrial function in Parkinson's disease. Mol Cell Neurosci. 2019;101:103413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huang DD, Fan SD, Chen XY, et al. Nrf2 deficiency exacerbates frailty and sarcopenia by impairing skeletal muscle mitochondrial biogenesis and dynamics in an age‐dependent manner. Exp Gerontol. 2019;119:61‐73. [DOI] [PubMed] [Google Scholar]
- 75. Hämäläinen M, Teppo HR, Skarp S, et al. NRF1 and NRF2 mRNA and protein expression decrease early during melanoma carcinogenesis: an insight into survival and microRNAs. Oxidative medicine and cellular longevity. 2019;2019:2647068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhu YP, Zheng Z, Hu S, et al. Unification of opposites between two antioxidant transcription factors Nrf1 and Nrf2 in mediating distinct cellular responses to the endoplasmic reticulum stressor tunicamycin. Antioxidants (Basel, Switzerland). 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang J, Culp ML, Craver JG, Darley‐Usmar V. Mitochondrial function and autophagy: Integrating proteotoxic, redox, and metabolic stress in Parkinson's disease. J Neurochem. 2018;144:691‐709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Parousis A, Carter HN, Tran C, et al. Contractile activity attenuates autophagy suppression and reverses mitochondrial defects in skeletal muscle cells. Autophagy. 2018;14:1886‐1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu P, Lin H, Xu Y, et al. Frataxin‐mediated PINK1‐Parkin‐dependent mitophagy in hepatic steatosis: The protective effects of quercetin. Mol Nutr Food Res. 2018;62:e1800164. [DOI] [PubMed] [Google Scholar]
- 80. Kaarniranta K, Kajdanek J, Morawiec J, Pawlowska E, Blasiak J. PGC‐1α protects RPE cells of the aging retina against oxidative stress‐induced degeneration through the regulation of senescence and mitochondrial quality control. the significance for AMD pathogenesis. Int J Mol Sci. 2018;19(8):2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Choi J, Ravipati A, Nimmagadda V, Schubert M, Castellani RJ, Russell JW. Potential roles of PINK1 for increased PGC‐1α‐mediated mitochondrial fatty acid oxidation and their associations with alzheimer disease and diabetes. Mitochondrion. 2014;18:41‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yeo D, Kang C, Gomez‐Cabrera MC, Vina J, Ji LL. Intensified mitophagy in skeletal muscle with aging is downregulated by PGC‐1alpha overexpression in vivo. Free Radic Biol Med. 2019;130:361‐368. [DOI] [PubMed] [Google Scholar]
- 83. Dong YZ, Li L, Espe M, Lu KL, Rahimnejad S. Hydroxytyrosol attenuates hepatic fat accumulation via activating mitochondrial biogenesis and autophagy through the AMPK pathway. Journal of agricultural and food chemistry. 2020;68:9377‐9386. [DOI] [PubMed] [Google Scholar]
- 84. Guilbert SM, Lambert H, Rodrigue MA, Fuchs M, Landry J, Lavoie JN. HSPB8 and BAG3 cooperate to promote spatial sequestration of ubiquitinated proteins and coordinate the cellular adaptive response to proteasome insufficiency. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2018;32:3518‐3535. [DOI] [PubMed] [Google Scholar]
- 85. Sha Z, Schnell HM, Ruoff K, Goldberg A. Rapid induction of p62 and GABARAPL1 upon proteasome inhibition promotes survival before autophagy activation. J Cell Biol. 2018;217:1757‐1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gilardini Montani MS, Santarelli R, Granato M, et al. EBV reduces autophagy, intracellular ROS and mitochondria to impair monocyte survival and differentiation. Autophagy. 2019;15:652‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gatliff J, East D, Crosby J, et al. TSPO interacts with VDAC1 and triggers a ROS‐mediated inhibition of mitochondrial quality control. Autophagy. 2014;10:2279‐2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang J, Duran A, Reina‐Campos M, et al. Adipocyte p62/SQSTM1 suppresses tumorigenesis through opposite regulations of metabolism in adipose tissue and tumor. Cancer Cell. 2018;33(770–84):e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ichimura Y, Komatsu M. Activation of p62/SQSTM1‐Keap1‐nuclear factor erythroid 2‐related factor 2 pathway in cancer. Frontiers in oncology. 2018;8:210. [DOI] [PMC free article] [PubMed] [Google Scholar]


