Abstract
A panel of seven recombinant antigens, derived from Ehrlichia phagocytophila (the agent of human granulocytic ehrlichiosis), was evaluated by class-specific enzyme-linked immunosorbent assays (ELISAs) for utility in the diagnosis of the infection. Fourteen genomic fragments, obtained by serologic expression screening, contained open reading frames (ORFs) encoding 16 immunodominant antigens. Eleven of these antigens were members of the major surface protein (MSP) multigene family. Alignment of their predicted protein sequences revealed a pattern of conserved sequences, which contained short direct repeats, flanking a variable region. In addition, two genomic clones contained two and three MSP ORFs, respectively, indicating that these genes are clustered in tandem copies. The implications for this pattern of both genomic and protein arrangements in antigenic variations of MSPs and in their utilities in a diagnostic assay are discussed. In addition to two MSP recombinant antigens (rHGE-1 and -3) and a fusion protein of these antigens (rErf-1), five further recombinants were evaluated by ELISA. Two of these antigens (rHGE-14 and -15) were novel, while a third (rHGE-2), with no known function, has been described. The final two recombinant antigens (rHGE-9 and -17) represent overlapping segments of the ankyrin gene (ank). The addition of rHGE-9 ELISA data resulted in the detection of 78% (21 of 27) of acute-phase sera. When serologic data for all recombinants are combined, 96.2% (26 of 27) of convalescent-phase patient serum samples and 85.2% (23 of 27) of acute-phase patient serum samples are detected, indicating the potential of these antigens for use in the development of a rapid serologic assay for the detection of E. phagocytophila infection.
Ehrlichia species have become recognized as major tick-borne pathogens of humans in both the United States and Europe (2, 12, 14). Human granulocytic ehrlichiosis (HGE) was first described in the United States in 1994 in patients from Minnesota and Wisconsin (2, 3, 7) and differed from infections due to the previously described Ehrlichia chaffeensis (human monocytic ehrlichiosis [HME]) in infecting only granulocytic hematopoietic cells. HGE appears to result from the attachment of infected tick vectors including the deer tick (Ixodes scapularis [also called Ixodes dammini]), while HME has been associated with the Amblyomma americanum tick (3, 15, 33, 34, 38). Increases in the prevalence of ehrlichiosis cases over time can be attributed to awareness of these pathogens by health care practitioners and to better diagnostic tools. However, human ehrlichiosis might also be occurring more frequently because of the steady encroachment of susceptible humans and pets into habitats occupied by the infected tick vectors (35). Recently, a study conducted in Sweden revealed that 11.4% of 185 individuals tested were seropositive for HGE, while 1.1% were positive for HME (13).
Because these organisms can cause fever or even fatal disease if they are not detected early, the development of rapid, sensitive, and specific diagnostic assays is needed for timely and efficacious therapy (4, 11, 13, 18, 23, 35). However, diagnostic tests for both HGE and HME are labor-intensive, expensive, and time-consuming. Tests include, roughly in order of their availability, examination of peripheral blood smears, serology, PCR, and isolation and culture of the organism (39). Serologic diagnosis of E. chaffeensis infection by using the Ehrlichia canis organism was standard until E. chaffeensis was isolated and cultured and it was determined that E. canis-containing material lacked optimal sensitivity. The use of whole organisms for diagnosis lacks specificity, and thus, confirmation depends upon protein immunoblotting, which is time-consuming and expensive (11). Antibodies from the serum of patients infected with the agent of HGE (referred to here as Ehrlichia phagocytophila) do not react with E. chaffeensis proteins but do react with proteins of approximately 44 kDa from either Ehrlichia equi or E. phagocytophila by Western blot analysis (10, 35). However, tests with these antigens or antigens from different strains of E. phagocytophila derived from either horses or cultured HL-60 cells give variable results, possibly due to the expression of variant forms of the immunodominant proteins (1, 21).
Diagnosis of HGE is further complicated by the potential coinfection of patients with the tick-borne pathogens of human babesiosis and Lyme borreliosis (25, 27). These tick-borne diseases share many of the same symptoms, making it difficult to differentiate between the infections in their early stages (5). Serologic cross-reactivity among members of the genus Ehrlichia is well known, and over one-third of patients with HME have concurrent diagnostic titers of antibodies to agents of other infections including Rocky Mountain spotted fever, murine typhus, Q fever, Lyme disease, babesiosis, and brucellosis (9, 40). Also complicating diagnosis is the possibility for false-positive Lyme disease serology in patients with HGE by using enzyme-linked immunosorbent assay (ELISA) or immunoblot assays (40). False-positive antibody assays for Lyme disease are also documented to occur in patients with a variety of other infections and autoimmune diseases (25). All of these complications can affect the accuracy of the overall diagnosis, particularly in the early stages of disease, when the appropriate therapeutic regimen needs to be determined. The use of a recombinant protein-based ELISA with E. phagocytophila major outer membrane proteins shows potential for use in the development of a rapid serodiagnostic test (21, 37). A rapid test with high degrees of sensitivity and specificity, however, ultimately may depend on the use of multiple recombinant proteins or a fusion protein consisting of several proteins or epitopes (19). In this report we describe a comprehensive effort to identify diagnostically relevant proteins of a human isolate of E. phagocytophila, along with initial efforts to characterize the serologic response to these proteins.
MATERIALS AND METHODS
Culture and isolation of E. phagocytophila genomic DNA.
A strain of E. phagocytophila (isolate WI 1), isolated from a west-central Wisconsin (Spooner, Wis., area) patient with clinical and laboratory-confirmed HGE, was grown in and purified from human promyelocytic leukemia cell line HL-60 (ATCC CCL 240) by a previously described protocol (17, 24). Genomic DNA was isolated from a fifth-passage culture with an IsoQuick Nucleic Acid Extraction kit (Orca Research Inc., Bothell, Wash.).
Genomic expression library construction.
Twenty micrograms of E. phagocytophila genomic DNA was resuspended in 400 μl of TE (Tris-EDTA) buffer and sonicated for 5 s at 50% continuous power with a Labsonic sonicator (B. Braun Biotech, Inc., Allentown, Pa.) to generate fragments of approximately 0.5 to 5.0 kbp. DNA fragments were blunted with T4 DNA polymerase (Gibco BRL) and ligated to EcoRI adapters (Stratagene) with T4 ligase (Stratagene). The adapted inserts were then phosphorylated with T4 polynucleotide kinase (Stratagene) and size selected with a Sephacryl S-400-HR column (Sigma). Approximately 0.25 μg of insert was ligated to a 1.0-μg Lambda ZAP II, EcoRI/calf intestinal alkaline phosphatase-treated vector (Stratagene), and the ligation mixture was packaged with Gigapack II Gold packaging extract (Stratagene) following the manufacturer's instructions.
Expression screening.
Immunoreactive proteins were screened from approximately 4 × 105 PFU. Twenty 150-mm petri dishes were plated with approximately 2 × 104 PFU and incubated at 42°C until plaques formed. Nitrocellulose filters (Schleicher & Schuell, Keene, N.H.) prewetted with 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG) were placed on the plates, and the plates were then incubated overnight at 37°C. The filters were then washed three times with phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBST; Sigma), blocked with 1.0% bovine serum albumin (Sigma) in PBST, and washed three times with PBST. The filters were next incubated overnight with Escherichia coli-adsorbed, E. phagocytophila-infected BALB/c mouse serum (24), washed three times with PBST, and incubated with a goat anti-mouse immunoglobulin G (IgG; heavy and light chains), alkaline phosphatase-conjugated secondary antibody (diluted 1:1,000 with PBST) for 1 h. The filters were finally washed three times with PBST and two times with alkaline phosphatase buffer (pH 9.5) and developed with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate (Gibco BRL). Reactive plaques were then excised, and a second or a third plaque purification was performed. Excision of phagemid followed the Stratagene Lambda ZAP II protocol, and the resulting plasmid DNA was sequenced with an automated sequencer (Perkin-Elmer ABI PRISM 377; Applied Biosystems, Inc.) with M13 forward, reverse, and internal DNA sequencing primers. Initial nucleic acid and protein homology searches were performed with DNA Star (Madison, Wis.) against the EMBL and GenBank (release 99) and the Swiss, Protein Information Resource (PIR), and Translated (release 97) databases. Protein analysis was performed with the PSORT program (National Institute for Basic Biology, Okazaki, Japan) and with the IDENTIFY program of EMOTIF (Department of Biochemistry, Stanford University). Sequence alignments were produced with the Megalign program (Clustal) of DNA Star.
Expression and purification of recombinant protein.
Expression of recombinant E. phagocytophila proteins (HGE-2, HGE-3, HGE-9, full-length HGE-14 [HGE-14fl] and the carboxy terminus of HGE-14 [HGE-14c], HGE-15, and HGE-17) was accomplished by amplifying the cloned plasmid inserts with Pfu polymerase (Stratagene) and the primers HGE-2 HIS (CAATTACATATGCATCACCATCACCATCACTATGGTATAGATATAGAGCTAA GTG) and HGE-2 TERM (CGAGAAAGAATTCCTAATAACTTAGAACATC), HGE-3 HIS (CAATTACATATGCATCACCATCACCATCACTTCTATATTGGTTTGGATTACAGTCCAG) and HGE-3 TERM (CTACGGGATCCGGTATTCAGAGTTAAAGATGG), HGE-9 HIS (CAATTAGCTAGCCATCACCATCACCATCACAAAGGGGCTCCAGCAACGCAG) and HGE-9 TERM (ACTACGGAATTCTAACGAGTAGCTGGAACCTGAGG), HGE-14fl HIS (CAAT TAGC TAGCCATCACCATCACCATCAC TC TGCGGAATATAAAGAAACTG) and HGE-14 TERM (ACTACGAATTCCAAGATCATGCTCTTCGCG), HGE-14c HIS (pGACAAGAAATACGGAAGATATTTCAATGC), HGE-15 HIS (CAATTACATATGCATCACCATCACCATCACAAGTTGTCTAATTCTGGCAACGGAC) and HGE-15 TERM (ACTATTGGATCCTAAATGTATACAGTCTCAGATTC), and HGE-17 HIS (CAATTACATATGCATCACCATCACCATCACAACATTGCAGATAAAGTGTATGGC) and HGE-17 TERM (GAGATAGAATTCTTACTTATATAGCTTACCGTC). Primers contained restriction sites for cloning (boldface) and a 6-histidine tag (italic) for protein purification (amino terminus). The amplification product was digested with the restriction enzymes NdeI or NheI and BamHI or EcoRI, depending on the primer set used, isolated by gel electrophoresis, and ligated to a pET17b plasmid vector (Novagen) previously cut with NdeI or NheI and BamHI or EcoRI and dephosphorylated. HGE-14c was cloned as a blunt-EcoRI fragment into a pET28 plasmid vector (Novagen), modified to include an amino-terminal 6-histidine tag. The ligation mixture was transformed into XL1-Blue competent cells (Stratagene), and plasmid DNA was prepared for sequencing (Qiagen). Recombinant protein was expressed by transforming plasmid DNA into BL21 pLysS competent cells (Novagen) and inducing a single-colony cell culture with 2 mM IPTG (Sigma). Recombinant protein was recovered from cell lysate with Ni-nitrilotriacetic acid agarose beads (Qiagen), following the manufacturer's instructions, and dialyzed in 10 mM Tris (pH 9.0). Recombinant proteins were quality checked for purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by staining with Coomassie blue stain and N-terminal protein sequencing (26), and were quantified by a Micro bicinchoninic acid assay (Pierce, Rockford, Ill.). Recombinants were assayed for endotoxin contamination by the Limulus assay (Bio Whittaker, Walkersville, Md.).
Recombinant HGE-1 protein was expressed in XL1-Blue cells (Stratagene) as a lacZ fusion protein and then isolated by gel electrophoresis with a Whole Gel Eluter (Bio-Rad). A fusion construct composed of the HGE-1- and HGE-3-coding sequences was constructed by amplifying the HGE-1 sequence with primers PDM-208 (GAGCTTGAGATTGGTTACGAGCGCTTC) and PDM-265 (CAATTACTCGAGAATTCATTAAAAAGCGAGCC) and the HGE-3 sequence with primers PDM-263 (CTACATCACGTGTTCTATATTGGTTTGGATTAC) and PDM-264 (GGTTAACTCGAGTACTAAGATGGTTTGTGTAATG) (restriction sites for cloning are indicated in boldface). The HGE-3 product was digested with the restriction enzymes Eco72I and XhoI and cloned into a pET28 plasmid vector (Novagen) modified to include an amino-terminal 6-histidine tag (pPDM). The HGE-1 product was then cloned into the ScaI site in the HGE-3–pPDM construct and then screened for correct orientation.
Study population.
The 43 serum samples from HGE patients used in the present study were positive by IgG-based immunofluorescence assay (IFA; ICN Pharmaceuticals, Inc., Costa Mesa, Calif.) and clinical history at the Mayo Clinic, and the results were verified by IFA at the Centers for Disease Control and Prevention with cultured E. equi and E. chaffeensis IFA substrates. Eleven of these patients provided both acute-phase and convalescent-phase serum samples, giving a total of 54 serum samples: 27 acute-phase serum samples and 27 convalescent-phase serum samples. Acute-phase patients were defined as infected individuals at 1 to 10 days after the onset of symptoms, and convalescent-phase patients were defined as those at 11 days to several weeks or months after the onset of symptoms. Random donor serum samples, used for development of cutoff values and for assay specificity, were purchased from Boston Biomedica (West Bridgewater, Mass.).
ELISA.
Each well of 96-well microtiter plates (Costar; Corning, Cambridge, Mass.) was coated overnight at 4°C with 200 ng of the recombinant proteins HGE-1, -2, -3, -9, -14c, -15, and -17 and Erf-1. The plates were then aspirated and blocked with PBS containing 1% (wt/vol) bovine serum albumin (BSA) for 2 h at room temperature. This was followed by washing in PBST. Serum diluted (1/100) in PBS containing 0.1% BSA was added to the wells, and the plates were incubated for 30 min at room temperature, followed by washing of the plates six times with PBST and then incubation with protein A-horseradish peroxidase conjugate (1/20,000 dilution; Sigma Chemical Co., St. Louis, Mo.) for 30 min. The plates were then washed six times in PBST and then incubated with tetramethylbenzidine substrate (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) for 15 min. The reaction was stopped by the addition of 1 N sulfuric acid, and the plates were read at 450 nm with an ELISA plate reader (EL311; Biotek Instruments, Hyland Park, Va.). The cutoff for the assays was determined from the mean for the negative population plus 3 standard deviations of the mean.
Western blot analysis.
Recombinant antigens (200 ng/lane) were subjected to SDS-PAGE analysis with 15% polyacrylamide minigels. The antigens were transferred to nitrocellulose BA-85 (Schleicher & Schuell) and blocked for 1 h at room temperature with PBS containing 1% Tween 20. The blots were then washed three times for 10 min each time in PBST–0.5 M sodium chloride (wash buffer). Next, the blots were probed for 1 h at room temperature with serum diluted 1:500 in wash buffer, followed by three washes (10 min each time) in wash buffer. The blots were then incubated for 45 min at room temperature with protein A-horseradish peroxidase diluted 1:20,000 in wash buffer and again washed three times for 10 min each time in wash buffer. Finally, the blots were incubated in chemiluminescent substrate (ECL; Amersham Plc, Little Charlton, United Kingdom) for ∼1 min and were then exposed to X-ray film (XAR5) for 10 to 60 s, as required.
Nucleotide sequence accession numbers.
The GenBank accession numbers for clones hge-1, -2, -3, -14, -15, and -17 are AF356507, AF356508, AF356509, AF356510, AF356511, and AF356512, respectively.
RESULTS
Expression cloning and molecular characterization of E. phagocytophila antigens.
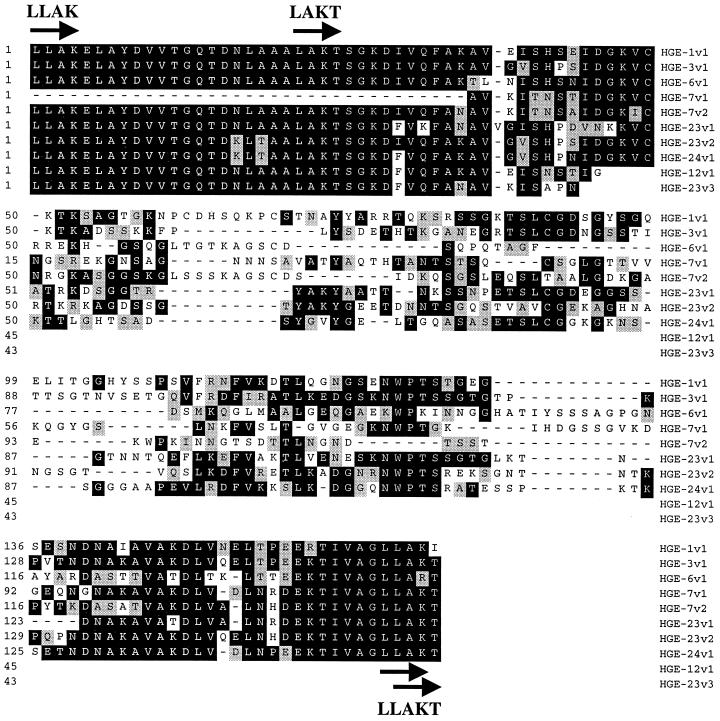
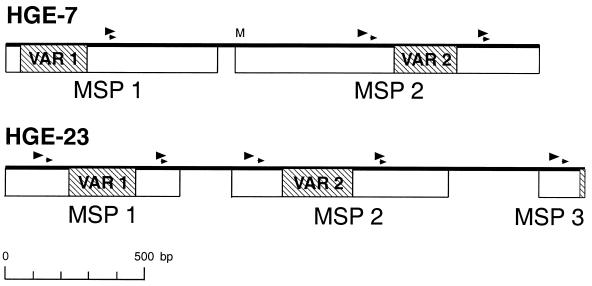
Serologic expression screening resulted in the cloning of 14 genomic DNA fragments containing predicted protein coding regions. Eight clones, hge-1, -3, -6, -7, -8, -12, -23, and -24, contained open reading frames (ORFs) encoding 11 members of the major surface protein (MSP) gene family (hge-7, two copies; hge-23, three copies). The predicted protein sequence for clone hge-3 (HGE-3) showed 98% identity over 323 amino acid residues (aa) with the sequence for GenBank entry MSP-2C (accession number AF029323), while the predicted protein sequences for the other 10 ORFs showed lower degrees of homology due to a variable region within the ORFs. Alignment of the predicted protein sequences for the 11 ORFs showed high degrees of homology at the amino and carboxy regions with central and carboxy-terminal variable regions (Fig. 1). Amino-terminal variations were due primarily to truncations in ORFs that produced MSP copies with no obvious initiation codons (i.e., pseudogenes). Two clones contained multiple copies of the MSP-like genes: hge-7 contained two copies (HGE-7.1 and -7.2) and hge-23 contained three copies (HGE-23.1, -23.2, and -23.3) aligned in tandem (Fig. 2). One ORF, HGE-23.2, contained a predicted full-length protein, while other ORFs were truncated at the amino terminus (HGE-1, -3, -7.1, and -23.1) or the carboxy terminus (HGE-8, -7.2, and -23.3), or both termini (HGE-6, -12, and -24). Predicted ATG initiation sites were obvious only for ORFs HGE-7.2 and HGE-8. ORFs containing amino termini that did not have initiation sites included HGE-23.2 and -23.3. Variable regions, contained in each ORF, were flanked by direct repeats (4 aa, 12 nucleotides [nt]) approximately 49 aa upstream and 25 aa downstream (LLAK). A second set of direct repeats occurred approximately 28 aa upstream and 26 aa downstream of the variable region (LAKT), with the two downstream repeats overlapping (LLAKT; arrows and arrowheads, Fig. 1 and 2, respectively). Direct repeats for each MSP copy (Table 1) show approximately 83% conservation of the consensus nucleotide sequence over 12 nt and 100% conservation of the consensus nucleotide sequence over 10 nt. Conservation of these repeats may have relevance to antigenic variation of these molecules (see below).
FIG. 1.
Alignment of partial ORFs representing the variable regions and flanking conserved regions of 10 MSP antigens. The protein sequence, if available, extends from an upstream direct repeat (LLAK; arrow) to a downstream direct repeat (LLAKT; arrows). A second upstream direct repeat (LAKT) is also indicated by an arrow. Identity (in amino acid residues) is indicated by a black background, and conserved residues are indicated by shading. Gaps in sequences are indicated by dashes. The amino acid residue number is indicated on the left side, and the sequence identity is indicated on the right side. Sequence identity includes the MSP clone number (HGE-N) and MSP copy number (vN), as shown in Fig. 2.
FIG. 2.
Diagram of the orientation of MSP ORFs in two genomic clones, hge-7 and hge-23. The locations of variable regions (VAR) are indicated by shaded boxes, and the locations of direct repeats are indicated by arrowheads. A potential initiation start codon is indicated by an M. The scale for the sizes of the clones (in base pairs) is indicated below.
TABLE 1.
Alignment of direct repeats that flank the MSP variable regionsa
| MSP copy | Amino acid sequence
|
|||
|---|---|---|---|---|
| LLAK DR1 | LAKT DR1 | LLAK DR2 | LAKT DR2 | |
| HGE-1 VAR1 | CTA CTA GCT AAG | TTA CTT GCT AAA | ||
| HGE-3 VAR1 | CTA CTA GCT AAG | TTA CTA GCT AAG | ||
| HGE-6 VAR1 | CTA CTA GCT AAG | TTA CTA GCT AGA | ||
| HGE-7 VAR1 | NAb | TTA CTA GCT AAA | ||
| HGE-7 VAR2 | CTA CTA GCT AAG | TTA CTA GCT AAA | ||
| HGE-8 VAR1 | CTA CTA GCT AAG | NA | ||
| HGE-12 VAR1 | CTA CTA GCT AAG | NA | ||
| HGE-23 VAR1 | CTA CTA GCT AAG | CTA CTA GCT AAA | ||
| HGE-23 VAR2 | CTA CTA GCT AAG | TTA CTA GCT AAA | ||
| HGE-23 VAR3 | CTA CTA GCT AAG | NA | ||
| HGE-24 VAR1 | CTA CTG GCT AAG | TTA CTA GCT AAA | ||
| Consensus | CTA CTA GCT AAG | TTA CTA GCT AAA | ||
| HGE-1 VAR1 | CTT GCC AAA ACC | CTT GCT AAA ATT | ||
| HGE-3 VAR1 | CTT GCT AAG ACC | CTA GCT AAG ACT | ||
| HGE-6 VAR1 | CTT GCC AAG ACT | CTA GCT AGA ACT | ||
| HGE-7 VAR1 | NA | CTA GCT AAA ACT | ||
| HGE-7 VAR2 | CTT GCC AAG ACC | CTA GCT AAA ACT | ||
| HGE-8 VAR1 | NA | NA | ||
| HGE-12 VAR1 | CTT GCT AAA ACC | NA | ||
| HGE-23 VAR1 | CTT GCC AAA ACC | CTA GCT AAA ACT | ||
| HGE-23 VAR2 | CTT GCC AAA ACC | CTA GCT AAA ACT | ||
| HGE-23 VAR3 | CTT GCC AAA ACT | NA | ||
| HGE-24 VAR1 | CTT GCT AAG ACC | CTA GCT AAA ACT | ||
| Consensus | CTT GCC AAA ACC | CTA GCT AAA ACT | ||
The MSP copy number and the amino acid sequences of direct repeats 1 (DR1) and 2 (DR2) as well as the respective nucleotide sequence alignments and consensus sequences are shown. The direct repeats for LLAK and LAKT are indicated in Fig. 2 by large and small arrowheads, respectively. The MSP copy number includes the clone name (HGE-N) and the associated variable region (VARn), as shown in Fig. 2.
NA, the sequence was not available for evaluation.
Clone hge-2 codes for a polypeptide of 578 aa predicted to be 61.4 kDa. Searches of the GenBank protein database resulted in a 99% identity of the sequence with that of an E. phagocytophila 100-kDa antigen (36), a protein of unknown function (GenBank accession number AF020523). The predicted protein sequence for HGE-2 contained three degenerate repeats of 124 aa and an additional repeat of 12 aa (VS VEADAGMQQE) found within two of the three repeats of 124 aa. The HGE-2 sequence contained no predicted signal or transmembrane regions, and the cellular location was predicted to be cytoplasmic.
Clones hge-9 and hge-17 code for overlapping segments of the E. phagocytophila ankyrin gene (36). The predicted protein sequence for both clones is identical to a product of 1,231 aa from a Wisconsin isolate deposited in the GenBank database (GenBank accession number AAF42730). The HGE-17 protein sequence aligns with aa 178 to 1231 of the Wisconsin isolate, while HGE-9 aligns with aa 735 to 1110 of this sequence, suggesting that the antigenic region of this protein lies within the shared sequence.
Clone hge-14 (3,735 bp) encodes a predicted 81.1-kDa polypeptide of 752 aa to which no sequences in the GenBank database are identical. Protein analysis indicates that the polypeptide contains no leader sequence but does contain one or two transmembrane domains (the second one has a lower probability score) and is predicted to be a type II membrane protein. The carboxy half of the polypeptide sequence contains four conserved repeats of 41 aa that are followed by two similar truncated repeats of 7 and 14 aa (Fig. 3). Additional degenerate repeats are seen within the sequence before and after the repeats of 41 aa.
FIG. 3.
Sequence of the HGE-14 protein gene. The nucleotide sequence number is shown on the right, and two potential ATG initiation codons are shown in italics and underlined. The predicted amino acid sequence is translated beneath the nucleotide sequence, and the amino terminus of the recombinant protein (HGE-14c) is indicated by an arrowhead. Potential transmembrane stretches are indicated by double underlining, and four repeats of 41 aa are italicized and indicated by arrowheads. Two truncated repeats that follow the four repeats of 41 aa are also indicated by arrowheads. Additional degenerate repeats are indicated by a dashed underline.
Clones hge-15 (2,322 bp) and hge-25 (5,521 bp) encode overlapping segments of a predicted 66.1-kDa (HGE-15) polypeptide with no identical sequences in the GenBank database. Both ORFs are truncated at the carboxy terminus, with HGE-15 having 590 aa and HGE-25 having 530 aa. Protein analysis indicates a potential cleavable (first ATG codon) or uncleavable (second ATG codon) amino-terminal signal sequence (Fig. 4). A search of the sequences in the GenBank protein database showed similarity with Vibrio cholerae (23% identity and 41% similarity over 530 aa; GenBank accession number AAF95066), Haemophilus influenzae (21% identity and 45% similarity over 357 aa; GenBank accession number P44092), and Rhodabacter sphaeroides (22% identity and 39% similarity over 560 aa [YbaU]; GenBank accession number AAD09115) peptidyl-prolyl cis-trans isomerase D (PpiD). In E. coli, this protein is anchored to the inner membrane, facing the periplasm, and is required for the proper folding of outer membrane proteins (8). Consistent with this potential PpiD homology, motif searches indicate putative chaperonin signatures (chaperonin 10 and chaperonin T-complex polypeptide 1) and two potential peptidyl-prolyl isomerase C peptidyl-prolyl isomerase (PPIase) signatures (Fig. 4).
FIG. 4.
Sequence of the HGE-15 protein gene. The nucleotide sequence number is shown on the right. Two potential ATG initiation codons are shown in italics and underlined, and potential ribosome-binding sites are underlined. The predicted amino acid sequence is translated beneath the nucleotide sequence, and potential transmembrane stretches are underlined. The amino terminus of the recombinant protein (rHGE-15) is indicated by an arrowhead. Putative chaperonin signatures (chaperonin 10 and chaperonin TCP-1, respectively) are shown in italics and underlined. Potential PPIC PPIase signatures are shown in italics and are underlined with a dashed line.
Recombinant protein expression and purification.
Seven clones were chosen for recombinant protein expression in E. coli (proteins HGE-1, -2, -3, -9, -14, -15, and -17). These expression constructs were engineered to include an N-terminal 6-histidine tag for ease of purification with an Ni-nitrilotriacetic acid agarose column. Recombinant expression constructs contained specific E. phagocytophila DNA inserts of 972, 1,731, 969, 1,128, 2,193, 1,049, 1,632, and 1,326 nt that coded for predicted proteins of 34.9 (38.9 as a LacZ fusion), 61.3, 34.5, 39.8, 78.8, 36.3, 61.0, and 46.7 kDa, respectively, for HGE-1, -2, -3, -9, -14fl, -14c, -15, and -17, respectively. Recombinant proteins HGE-2 and HGE-3 correspond to aa 2 to 578 and 42 to 364 of a 100-kDa protein (GenBank accession number AF020523 [36]) and MSP-2C (GenBank accession number AF029323 [28]), respectively. Recombinant HGE-1 is missing 119 aa of the amino-terminal conserved sequence found in MSP-2C. Recombinants HGE-9 and -17 represent two overlapping (overlap of 28 aa) segments of the E. phagocytophila ankyrin-like protein (GenBank accession numbers AF047897 and AF020521). Recombinant HGE-17 (rHGE-17) comprises amino acid residues 322 to 763, while rHGE-9 covers residues 735 to 1111 of the ankyrin-like protein of 1,232 aa. Recombinant HGE-14fl includes aa 22 to 752, and HGE-14c includes aa 410 to 752 of the full-length protein sequence (Fig. 3). Recombinant HGE-15 comprises aa 32 to 575 of the sequence shown in Fig. 4. HGE-1 and -3, two distinct members of the MSP gene family, were also engineered as a fusion polypeptide (Erf-1). This expression construct contained a sequence of 1,950 nt that coded for a predicted 69.7-kDa polypeptide with an additional 6-histidine tag at the amino terminus. All recombinant proteins were tested for purity by N-terminal sequencing and Coomassie blue staining and were determined to be over 90% pure.
Western blot analysis.
Western blot analysis was used to assess the expression levels and qualities of the recombinant proteins and to monitor recombinant protein purification. Most recombinants migrated in an SDS-polyacrylamide gel to positions of the predicted size. However, some recombinants, such as HGE-14c, migrated to positions of a higher relative molecular size, possibly due to a high proline content (13.4%). rHGE-14fl was not expressed at a level sufficient for purification. Western blot analysis, however, revealed significant reactivity of the recombinant protein with a pool of five serum samples from HGE patients (data not shown). Because of this reactivity and difficulty with full-length protein expression and purification, we chose to express and purify the more hydrophilic HGE-14c.
ELISA.
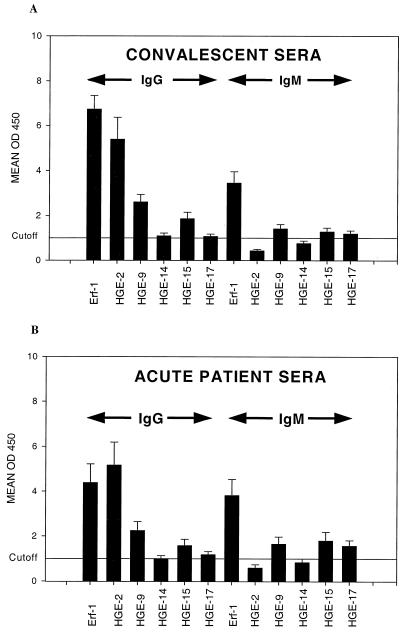
Comparison of ELISA results for recombinant proteins HGE-1, HGE-3, and Erf-1 (HGE-1–HGE-3 fusion protein) by using a panel of IFA-reactive sera from HGE patients demonstrated that rHGE-1 and rHGE-3 were reactive with overlapping subsets of patient sera and that the Erf-1 fusion polyprotein provided the greatest sensitivity. rHGE-1 was reactive with 14 of 27 acute-phase serum samples from HGE patients and 21 of 27 convalescent-phase serum samples from HGE patients, while rHGE-3 was reactive with 14 of 27 acute-phase serum samples and 25 of 27 convalescent-phase serum samples in an IgG-based assay. The reactivity of recombinant fusion polyprotein Erf-1 (rErf-1) was similar to the reactivities of rHGE-1 and rHGE-3, as determined from combined ELISA values, and thus, we chose to develop rErf-1 in subsequent experiments. The ELISA reactivities for recombinant proteins Erf-1, HGE-2, HGE-9, HGE-14c, HGE-15, and HGE-17 in both IgG- and IgM-based assays are summarized in Table 2 and Fig. 5. rErf-1 was reactive with 55.6% of acute-phase serum samples (15 of 27 in an IgM-based assay) from HGE patients and 92.6% of convalescent-phase serum samples (25 of 27 in an IgG-based assay) and gave relatively high mean assay values (Fig. 5). The other recombinant E. phagocytophila proteins showed variable sensitivities relative to that of rErf-1; however, most of the recombinant proteins added theoretically to the sensitivity of rErf-1. Although rHGE-2 was reactive with a relatively high percentage of acute-phase (55.6%) and convalescent-phase (77.8%) patient sera and gave a high mean optical density value by IgG-based ELISA, it adds little (one acute-phase serum sample from an HGE patient) to the sensitivity of rErf-1. However, the theoretical addition of rHGE-9 to the rErf-1 assay increased its sensitivity by adding five additional acute-phase patient samples (Table 2). Recombinant HGE-14c, while being reactive with three additional acute-phase patient samples in an IgG-based assay, shows an overall weak reactivity, as seen in Fig. 5. Recombinants HGE-15 and -17 each provided one additional patient sample to the theoretical rErf-1 combination IgG-based assay. The five recombinant proteins add little to the reactivity of rErf-1in IgM-based assays. Recombinants HGE-2, -9, -14c, -15, and -17 gave sensitivities of 55.6, 74.1, 51.8, 48.1, and 55.6%, respectively, for acute-phase serum samples in theoretically combined IgM- and IgG-based assays, and they gave sensitivities of 77.8, 74.1, 48.1, 70.4, and 70.4%, respectively, for convalescent-phase serum samples. When the data are combined, the six recombinant antigens are reactive with 85.2 and 96.2% of acute- and convalescent-phase serum samples, respectively, and 94.1 and 96% of acute- and convalescent-phase serum samples, respectively, with an IgG-based IFA titer equal to or greater than 64. The five acute-phase serum samples not detected by ELISAs with the six recombinants had IFA titers of 64 or less (64, 32, 16, <16, and <16, respectively). The one convalescent-phase serum sample missed had an IFA titer of 64. Conversely, of 12 samples that were negative by IFA (IFA titer, ≤32), 8 were detected with these recombinant antigens. Three serum samples that were not detected in the acute phase were detected in the convalescent phase. The specificities for the recombinants ranged from 94 to 100%, depending on the antigen and the secondary antibody used (Table 2).
TABLE 2.
ELISA results for sera from patients and random donors
| ELISA and immunoglobulina | Sensitivityb
|
Specificityc | |
|---|---|---|---|
| Acute-phase sera | Convalescent-phase sera | ||
| Erf-1 | |||
| IgG | 14/27 (51.8) | 25/27 (92.6) | 97.2 (1/36) |
| IgM | 15/27 (55.6) | 23/27 (85.2) | 100 (0/36) |
| IgG + IgM | 15/27 (55.6) | 25/27 (92.6) | 97.2 (1/36) |
| HGE-9 | |||
| IgG | 18/27 (66.7) | 19/27 (70.4) | 97.3 (1/37) |
| IgM | 12/27 (44.4) | 18/27 (66.7) | 100 (0/37) |
| IgG + IgM | 20/27 (74.1) | 20/27 (74.1) | 97.3 (1/37) |
| Erf-1 + HGE-9 | |||
| IgG | 19/27 (70.4) | 25/27 (92.6) | |
| IgM | 16/27 (59.2) | 23/27 (85.2) | |
| IgG + IgM | 21/27 (77.8) | 25/27 (92.6) | |
| HGE-2 | |||
| IgG | 15/27 (55.6) | 21/27 (77.8) | 97.3 (1/37) |
| IgM | 4/27 (14.8) | 3/27 (11.1) | 94.6 (2/37) |
| IgG + IgM | 15/27 (55.6) | 21/27 (77.8) | 91.9 (3/37) |
| HGE-14 | |||
| IgG | 13/27 (48.1) | 13/27 (48.1) | 96.8 (1/31) |
| IgM | 8/27 (29.6) | 7/27 (25.9) | 93.5 (2/31) |
| IgG + IgM | 14/27 (51.8) | 13/27 (48.1) | 93.5 (2/31) |
| HGE-15 | |||
| IgG | 12/27 (44.4) | 17/27 (63.0) | 97.3 (1/37) |
| IgM | 12/27 (44.4) | 14/27 (51.8) | 97.3 (1/37) |
| IgG + IgM | 13/27 (48.1) | 19/27 (70.4) | 94.6 (2/37) |
| HGE-17 | |||
| IgG | 12/27 (44.4) | 14/27 (51.8) | 94.6 (2/37) |
| IgM | 14/27 (51.8) | 15/27 (55.6) | 100 (0/37) |
| IgG + IgM | 15/27 (55.6) | 19/27 (70.4) | 94.6 (2/37) |
| Total (all antigens) | |||
| IgG | 21/27 (77.8) | 26/27 (96.3) | |
| IgM | 16/27 (59.2) | 22/27 (81.5) | |
| IgG + IgM | 23/27 (85.2) | 26/27 (96.2) | |
| IFA (≥64) (IgG + IgM) | |||
| Erf-1 + HGE-9 | 15/17 (88.2) | 23/25 (92.0) | |
| All antigens | 16/17 (94.1) | 24/25 (96.0) | |
The recombinant protein identity and the secondary antibody used in the assay are indicated.
Results for sensitivity are number of samples with positive reactions/total number of samples tested (percent positive results). Cutoff values were determined from the mean value plus 3 standard deviations of the mean for samples from random donors from areas of nonendemicity. Mean assay values with standard errors of the mean for each patient group tested are shown in Figure 5.
Results for specificity are percent positive results (number of samples with positive reactions/total number of samples tested).
FIG. 5.
Mean values of the optical density at 450 nm (OD450) obtained by ELISA are provided for convalescent-phase sera from HGE patients (A) and acute-phase sera from HGE patients (B) for the six recombinant E. phagocytophila proteins. Arrows indicate values for IgG- and IgM-based assays, and the vertical lines on each bar give the respective standard error of the mean for each mean value. Recombinant protein identities are shown below, and mean values of the optical density at 450 nm are shown to the left. These data were normalized to a cutoff value of 1.0 (optical density at 450 nm values/cutoff value), as indicated by the horizontal line. The original cutoff values (mean for the random donor population plus 3 standard deviations of the mean) for each assay are 0.271, 0.218, 0.485, 0.314, 0.532, and 0.918, respectively, for IgG-based assays and 0.160, 0.538, 0.364, 0.245, 0.312, and 0.554, respectively, for IgM-based assays.
DISCUSSION
The MSPs of E. phagocytophila have been demonstrated to be the immunodominant antigens in HGE infections (20, 21, 22, 28, 37, 42, 43, 44). Although these antigens are reported to have high sensitivities (87% [21] and 89.7% [37]) and specificities (98%) in ELISAs, additional antigens might be necessary to achieve optimal sensitivity, especially for acute-phase patient sera. In the present study we identified, by serologic expression screening, 14 genomic clones containing one or more ORFs encoding immunoreactive proteins. Eight of these clones encoded 11 unique members of the MSP multigene family; one clone contained two MSP copies and a second clone contained three MSP copies. The occurrence of multiple, tandem copies of the MSP ORFs has been described previously for E. phagocytophila (28, 43) and for Ehrlichia chaffeensis (30) and E. canis (29). Also previously reported is the observation that while some ORFs contain an ATG initiation codon, other ORFs do not (42). Of the MSP ORFs reported here that have complete amino termini, two contained initiation codons and two had no obvious start codon. Alignment of the predicted polypeptides from the 11 ORFs resulted in a pattern of conserved and variable regions. The predicted amino termini are well conserved except for variability at the extreme amino end due to variations in ORF size (i.e., amino-terminal truncations). This conserved region is followed by a variable region of approximately 71 to 91 aa and then a second conserved region near the carboxy termini. The extreme carboxy termini are variable in sequence and in length. The conserved regions have been suggested to be involved in rearrangements, through genomic recombination, that might result in antigenic variation (16). Of interest is the finding of direct repeats of 12 bp (4 aa) that flank the central variable region (Fig. 1 and 2 and Table 1). A similar genomic arrangement is seen in Borrelia burgdorferi, which is probably involved in antigenic variation of the surface-exposed lipoprotein VlsE (41). In this system, a vls expression site is closely associated with 15 additional, tandemly arranged, silent vls cassettes. Conserved sequences, on either side of the variable region, are thought to facilitate recombination between the expressed and silent vls sequences, and conserved 17-bp direct repeats that flank all variable regions may be involved in alignment or in binding of a proposed site-specific recombinase (41). In E. phagocytophila one finds (i) tandemly arranged silent and expressed MSP copies, (ii) a variable region flanked by conserved regions in each copy, and (iii) in-frame, direct repeats of 12 nt (4 aa) that flank the variable regions. This system of antigenic variation through recombination of variable regions could produce variability in the MSPs and thus aid in immune evasion. In the closely related organism Anaplasma marginale, investigators (16, 31, 32) demonstrate that true MSP2 structural antigenic variants emerge during each cycle of persistent rickettsemia. They also show that the MSP central hydrophilic variable region is the sole site of MSP2 structural polymorphism among expressed variants and that the structure of the MSP2 genes (i.e., variable region flanked by conserved regions) provides the basis for homologous recombination by gene conversion. From a practical standpoint, inclusion of the most common variants initially encountered in human infection may be an important consideration in serologic test design.
Three clones, hge-2, hge-9, and hge-17, contain ORFs encoding antigens that have been described previously (6, 36). HGE-2, a predicted 61.4-kDa protein of unknown function with three repeats of 124 aa, differs by 1 aa from the 100-kDa antigen described by Storey et al. (36). They show that the 100-kDa antigen shares similarities with a 120-kDa antigen of E. chaffeensis and might be surface associated. Clones hge-9 and hge-17 encode partial ORFs for the E. phagocytophila ankyrin gene (6). The HGE-17 sequence is identical to 86% of the sequence of the carboxy end of the E. phagocytophila ankyrin protein, and the HGE-9 sequence is identical to 30% of the central region of the E. phagocytophila ankyrin protein (GenBank accession number AAF42730).
Three additional clones, hge-14, hge-15, and hge-25, contained ORFs with no identities in the GenBank database. ORFs encoded by hge-15 and hge-25 are identical, and both are truncated at the carboxy terminus. Predicted protein HGE-14 is an 81.1-kDa polypeptide with four repeats of 41 aa that are bordered by degenerate repeats. It is predicted to be a type II transmembrane protein, with the carboxy 600 aa potentially being extracellular. HGE-15 also has an amino-terminal leader sequence or transmembrane domain that is predicted to be cleavable or uncleaved, depending on the initiation codon used. Interestingly, this antigen has homology with bacterial PpiD at both the amino acid sequence level and at the structural level and contains both chaperonin and potential PPIC PPIase signatures (Fig. 4). E. coli ppiD encodes a periplasmic peptidyl-prolyl isomerase that is required for proper folding of the outer membrane proteins (8). E. coli ppiD is under control of the sigma factor regulon transcribing cytosolic heat shock proteins and participates in the folding of noncytoplasmic proteins. In E. coli, elevated temperatures (37°C or greater) lead to the aggregation of proteins in general and also to changes in the composition of the outer membrane and in the abundance of outer membrane proteins. Thus, E. coli uses this important catalyst of folding, needed for membrane biogenesis, which is regulated by a classical heat shock sigma factor (ς32) (8). One might speculate that in Ehrlichia spp. potential host-related changes in membrane composition are partly due to the different temperatures of vertebrate and invertebrate hosts and are regulated by similar stress regulons and PPIase activities, possibly including HGE-15.
To assay for immunogenicity and the potential use of E. phagocytophila recombinant proteins in a serum-based immunoassay, seven ORFs were reengineered for expression in E. coli. Two of the MSP ORFs, HGE-1 and HGE-3, were reconstructed for recombinant protein expression. These proteins were also expressed as a fusion protein, rErf-1. ORFs HGE-9 and -17 were expressed as overlapping portions of the ankyrin gene. HGE-2, HGE-14, and HGE-15 were also expressed as recombinant proteins. Initial ELISA data indicated that the rErf-1 fusion protein was as reactive as rHGE-1 and -3 combined were with sera from patients with HGE. rErf-1 detected 92.6% of convalescent-phase sera from HGE patients and only 55.6% of acute-phase sera from HGE patients in an IgG- and IgM-based assay. However, rHGE-9 detected 74% of acute-phase sera from HGE patients and detected 77.8% of acute-phase sera from these patients when it was theoretically combined with rErf-1. rHGE-2, rHGE-14c, and rHGE-15 detected 78, 48, and 70% of convalescent-phase sera, respectively, and 56, 52, and 48% of acute-phase sera, respectively. When the data for both the IgM- and the IgG-based assays were combined, the antigens detected 96.2% of convalescent-phase sera from HGE patients and 85.2% of acute-phase sera from HGE patients. The percentage of acute-phase patient sera detected rose to 94.1% when an IFA titer of ≥64 in serum was used as a cutoff for use in the assay. The five patient serum samples not detected with these recombinant antigens had IFA titers of 64 or less, and ELISA values for these sera were just under the cutoff values, suggesting that greater sensitivity might be obtained with an increase in recombinant protein purity. Our data indicate that additional, non-MSP recombinant proteins can add to the sensitivity of a serum-based assay for the detection of HGE without compromising specificity. Additional efforts are under way to optimize assay conditions and recombinant protein purity and to select the appropriate cocktail of recombinant antigens for a fusion polyprotein that would be most useful for the detection of both convalescent-phase and acute-phase E. phagocytophila infection.
ACKNOWLEDGMENTS
We thank Thomas Vedvick and Darrick Carter for protein sequence data and Dan Hoppe and Joe Parsons for assistance with DNA sequencing. We also thank Jonathan Clapper and Peter Phan for performing lipopolysaccharide assays with purified recombinant protein.
M.J.L. and R.L.H. contributed equally to the data presented here.
This work was supported by NIH grants AI42416 (to M.J.L.) and AI32403 (to D.H.P.), as well as cooperative agreement U50/CCU-510343 from the Centers for Disease Control and Prevention (to D.H.P.).
REFERENCES
- 1.Aguero-Rosenfeld M E, Kalantarpour F, Baluch M, Horowitz H W, McKenna D F, Raffalli J T, Hsieh T C, Wu J, Dumler J S, Wormser G P. Serology of culture-confirmed cases of human granulocytic ehrlichiosis. J Clin Microbiol. 2000;38:635–638. doi: 10.1128/jcm.38.2.635-638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakken J S. The discovery of human granulocytotropic ehrlichiosis. J Lab Clin Med. 1998;132:175–180. doi: 10.1016/s0022-2143(98)90165-2. [DOI] [PubMed] [Google Scholar]
- 3.Bakken J S, Dumler J S, Chen S-M, Eckman M R, Van Etta L L, Walker D H. Human granulocytic ehrlichiosis in the upper Midwest United States. JAMA. 1994;272:212–218. [PubMed] [Google Scholar]
- 4.Bakken J S, Krueth J, Wilson-Nordskog C, Tilden R L, Asanovich K, Dumler J S. Clinical and laboratory characteristics of human granulocytic ehrlichiosis. JAMA. 1996;275:199–205. [PubMed] [Google Scholar]
- 5.Belongia E A, Reed K D, Mitchell P D, Chyou P H, Mueller-Rizner N, Finkel M F, Schriefer M E. Clinical and epidemiological features of early Lyme disease and human granulocytic ehrlichiosis in Wisconsin. Clin Infect Dis. 1999;29:1472–1477. doi: 10.1086/313532. [DOI] [PubMed] [Google Scholar]
- 6.Caturegli P, Asanovich K M, Walls J J, Bakken J S, Madigan J E, Popov V L, Dumler J S. AnkA: an Ehrlichia phagocytophilagroup gene encoding a cytoplasmic protein antigen with ankyrin repeats. Infect Immun. 2000;68:5277–5283. doi: 10.1128/iai.68.9.5277-5283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen S-M, Dumler J S, Bakken J S, Walker D H. Identification of a granulocytotropic Ehrlichiaspecies as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–595. doi: 10.1128/jcm.32.3.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dartigalongu C, Raina S. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawson J E, Rikihisa Y, Ewing S A, Fishbein D B. Serologic diagnosis of human ehrlichiosis using two Ehrlichia canisisolates. J Infect Dis. 1991;163:564–567. doi: 10.1093/infdis/163.3.564. [DOI] [PubMed] [Google Scholar]
- 10.Dumler J S, Asanovich K M, Bakken J S, Richter P, Kimsey R, Madigan J E. Serologic cross-reactions among Ehrlichia equi. Ehrlichia phagocytophila, and human granulocytic ehrlichia. J Clin Microbiol. 1995;33:1098–1103. doi: 10.1128/jcm.33.5.1098-1103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumler J S, Bakken J S. Ehrlichial diseases of humans: emerging tick-borne infections. Clin Infect Dis. 1995;20:1102–1110. doi: 10.1093/clinids/20.5.1102. [DOI] [PubMed] [Google Scholar]
- 12.Dumler J S, Bakken J S. Human granulocytic ehrlichiosis in Wisconsin and Minnesota: a frequent infection with the potential for persistence. J Infect Dis. 1996;173:1027–1030. doi: 10.1093/infdis/173.4.1027. [DOI] [PubMed] [Google Scholar]
- 13.Dumler J S, Dotevall L, Gustafson R, Granström M. A. population-based seroepidemiologic study of human granulocytic ehrlichiosis and Lyme borreliosis on the west coast of Sweden. J Infect Dis. 1997;175:720–722. doi: 10.1093/infdis/175.3.720. [DOI] [PubMed] [Google Scholar]
- 14.Eng T R, Harkess J R, Fishbein D B, Dawson J E, Greene C N, Redus M A, Satalowich F T. Epidemiologic, clinical, and laboratory findings of human ehrlichiosis in the United States, 1988. JAMA. 1990;264:2251–2258. [PubMed] [Google Scholar]
- 15.Fishbein D B, Dawson J E, Robinson L E. Human ehrlichiosis in the United States. Ann Intern Med. 1994;120:736–743. doi: 10.7326/0003-4819-120-9-199405010-00003. [DOI] [PubMed] [Google Scholar]
- 16.French D M, Brown W C, Palmer G H. Emergence of Anaplasma marginaleantigenic variants during persistent rickettsemia. Infect Immun. 1999;67:5834–5840. doi: 10.1128/iai.67.11.5834-5840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman J L, Nelson C, Vitale B, Madigan J E, Dumler J S, Kurtti T J, Munderloh U G. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med. 1996;334:209–215. doi: 10.1056/NEJM199601253340401. [DOI] [PubMed] [Google Scholar]
- 18.Hardalo C J, Quagliarello V, Dumler J S. Human granulocytic ehrlichiosis in Connecticut: report of a fatal case. Clin Infect Dis. 1995;21:910–914. doi: 10.1093/clinids/21.4.910. [DOI] [PubMed] [Google Scholar]
- 19.Houghton R L, Benson D R, Reynolds L D, McNeill P D, Sleath P, Lodes M J, Skeiky Y A, Leiby D, Badaro R, Reed S G. A. multiepitope synthetic peptide and recombinant protein for the detection of antibodies to Trypanosoma cruziin radioimmunoprecipitation-confirmed and consensus-positive sera. J Infect Dis. 1999;179:1226–1234. doi: 10.1086/314723. [DOI] [PubMed] [Google Scholar]
- 20.IJdo J W, Sun W, Zhang Y, Magnarelli L A, Fikrig E. Cloning of the gene encoding the 44-kilodalton antigen of the agent of human granulocytic ehrlichiosis and characterization of the humoral response. Infect Immun. 1998;66:3264–3269. doi: 10.1128/iai.66.7.3264-3269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.IJdo J W, Wu C, Magnarelli L A, Fikrig E. Serodiagnosis of human granulocytic ehrlichiosis by a recombinant HGE-44-based enzyme-linked immunosorbent assay. J Clin Microbiol. 1999;37:3540–3544. doi: 10.1128/jcm.37.11.3540-3544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.IJdo J W, Zhang Y, Hodzic E, Magnarelli L A, Wilson M L, Telford III S R, Barthold S W, Fikrig E. The early humoral response in human granulocytic ehrlichiosis. J Infect Dis. 1997;176:687–692. doi: 10.1086/514091. [DOI] [PubMed] [Google Scholar]
- 23.Jahangir A, Kolbert C, Edwards W, Mitchell P, Dumler J S, Persing D H. Fatal pancarditis associated with human granulocytic ehrlichiosis in a 44-year-old man. Clin Infect Dis. 1998;27:1424–1427. doi: 10.1086/515014. [DOI] [PubMed] [Google Scholar]
- 24.Kolbert C P, Bruinsma E S, Abdulkarim A S, Hofmeister E K, Tompkins R B, Telford III S R, Mitchell P D, Adams-Stich J, Persing D H. Characterization of an immunoreactive protein from the agent of human granulocytic ehrlichiosis. J Clin Microbiol. 1997;35:1172–1178. doi: 10.1128/jcm.35.5.1172-1178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnarelli L A, Dumler J S, Anderson J F, Johnson R C, Fikrig E. Coexistence of antibodies to tick-borne pathogens of babesiosis, ehrlichiosis, and Lyme borreliosis in human sera. J Clin Microbiol. 1995;33:3054–3057. doi: 10.1128/jcm.33.11.3054-3057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 27.Mitchell P D, Reed K D, Hofkes J M. Immunoserologic evidence of coinfection with Borrelia burgdorferi, Babesia microti, and human granulocytic Ehrlichiaspecies in residents of Wisconsin and Minnesota. J Clin Microbiol. 1996;34:724–727. doi: 10.1128/jcm.34.3.724-727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy C I, Storey J R, Recchia J, Doros-Richert L A, Gingrich-Baker C, Munroe K, Bakken J S, Coughlin R T, Beltz G A. Major antigenic proteins of the agent of human granulocytic ehrlichiosis are encoded by members of a multigene family. Infect Immun. 1998;66:3711–3718. doi: 10.1128/iai.66.8.3711-3718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohashi N, Unver A, Zhi N, Rikihisa Y. Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canisand application of the recombinant protein for serodiagnosis. J Clin Microbiol. 1998;36:2671–2680. doi: 10.1128/jcm.36.9.2671-2680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohashi N, Zhi N, Zhang Y, Rikihisa Y. Immunodominant major outer membrane proteins of Ehrlichia chaffeensisare encoded by a polymorphic multigene family. Infect Immun. 1998;66:132–139. doi: 10.1128/iai.66.1.132-139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer G H, Brown W C, Rurangirwa F R. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. 2000;2:167–176. doi: 10.1016/s1286-4579(00)00271-9. [DOI] [PubMed] [Google Scholar]
- 32.Palmer G H, Eid G, Barbet A F, McGuire T C, McElwain T F. The immunoprotective Anaplasma marginalemajor surface protein 2 is encoded by a polymorphic multigene family. Infect Immun. 1994;62:3808–3816. doi: 10.1128/iai.62.9.3808-3816.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pancholi P, Kolbert C P, Mitchell P D, Reed K D, Dumler J S, Bakken J S, Telford S R, Persing D H. Ixodes damminias a potential vector of human granulocytic ehrlichiosis. J Infect Dis. 1995;172:1007–1012. doi: 10.1093/infdis/172.4.1007. [DOI] [PubMed] [Google Scholar]
- 34.Piesman J, Mather T N, Telford S R, Spielman A. Concurrent Borrelia burgdorferi and Babesia microti infection in nymphal Ixodes dammini. J Clin Microbiol. 1986;24:446–447. doi: 10.1128/jcm.24.3.446-447.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaffner W, Standaert S M. Ehrlichiosis—in pursuit of an emerging infection. N Engl J Med. 1996;334:262–263. doi: 10.1056/NEJM199601253340410. [DOI] [PubMed] [Google Scholar]
- 36.Storey J R, Doros-Richert L A, Gingrich-Baker C, Munroe K, Mather T N, Coughlin R T, Beltz G A, Murphy C I. Molecular cloning and sequencing of three granulocytic Ehrlichiagenes encoding high-molecular-weight immunoreactive proteins. Infect Immun. 1998;66:1356–1363. doi: 10.1128/iai.66.4.1356-1363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima T, Zhi N, Lin Q, Rikihisa Y, Horowitz H W, Ralfalli J, Wormser G P, Hechemy K E. Comparison of two recombinant major outer membrane proteins of the human granulocytic ehrlichiosis agent for use in an enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 2000;7:652–657. doi: 10.1128/cdli.7.4.652-657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Telford S R, III, Dawson J E, Katavolos P, Warner C K, Kolbert C P, Persing D H. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc Natl Acad Sci USA. 1996;93:6209–6214. doi: 10.1073/pnas.93.12.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walls J J, Caturegli P, Bakken J S, Asanovich K M, Dumler J S. Improved sensitivity of PCR for diagnosis of human granulocytic ehrlichiosis using epank1 genes of Ehrlichia phagocytophila-group ehrlichiae. J Clin Microbiol. 2000;38:354–356. doi: 10.1128/jcm.38.1.354-356.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wormser G P, Horowitz H W, Dumler J S, Schwartz I, Aguero-Rosenfeld M. False-positive Lyme disease serology in human granulocytic ehrlichiosis. Lancet. 1996;347:981–982. doi: 10.1016/s0140-6736(96)91475-0. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J-R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 42.Zhi N, Ohashi N, Rikihisa Y. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J Biol Chem. 1999;274:17828–17836. doi: 10.1074/jbc.274.25.17828. [DOI] [PubMed] [Google Scholar]
- 43.Zhi N, Ohashi N, Rikihisa Y, Horowitz H W, Wormser G P, Hechemy K. Cloning and expression of the 44-kilodalton major outer membrane protein of the human granulocytic ehrlichiosis agent and application of the recombinant protein to serodiagnosis. J Clin Microbiol. 1998;36:1666–1673. doi: 10.1128/jcm.36.6.1666-1673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhi N, Rikihisa Y, Kim H Y, Wormser G P, Horowitz H W. Comparison of major antigenic proteins of six strains of the human granulocytic ehrlichiosis agent by Western immunoblot analysis. J Clin Microbiol. 1997;35:2606–2611. doi: 10.1128/jcm.35.10.2606-2611.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]