Abstract
For community-level monitoring, the European Commission under the EU Sewage Sentinel System recommends wastewater-based SARS-CoV-2 surveillance. Tracking SARS-CoV-2 variants in a community is pivotal for appropriate public health response. Genome sequencing of SARS-CoV-2 in wastewater samples for tracking variants is challenging, often resulting in low coverage genome sequences, thereby impeding the detection of the SARS-CoV-2 mutations. Therefore, we aimed at high-coverage SARS-CoV-2 genome sequences from sewage samples which we successfully accomplished. This first pan-European surveillance compared the mutation profiles associated with the variants of concerns: B.1.1.7, P.1, B.1.351 and B.1.617.2 across 20 European countries, including 54 municipalities. The results highlight that SARS-CoV-2 variants detected in the wastewater samples mirror the variants profiles reported in clinical data. This study demonstrated that >98% coverage of SARS-CoV-2 genomic sequences is possible and can be used to track SARS-CoV-2 mutations in wastewater to support identifying variants circulating in a city at the community level.
Keywords: High-coverage sequencing, SARS-CoV-2 RNA, Wastewater-based epidemiology, European sewage, Metagenomics
Graphical abstract
1. Introduction
Undeniably, the sudden emergence of SARS-CoV-2, which has caused a global pandemic, is a significant and in many regards unprecedented threat to public health. SARS-CoV-2 rapidly resulted in a high number of people requiring hospitalization, casualties and major socioeconomic disruptions, with consequences which we still do not fully oversee. Consequently, most countries have been forced to implement severe lockdown measures to ensure the physical distance between people and interrupt virus transmission (Martin et al., 2020). Overall, the high transmission rate and the rapidly evolving nature of the virus, leading to the emergence of new variants that may transmit more readily and evade the immune response, raise broad concerns about SARS-CoV-2 (Harvey et al., 2021; McCormick et al., 2021; Worobey et al., 2020).
The current phase of the COVID-19 pandemic is shaping into an era of genomic surveillance to track the genomic changes in the SARS-CoV-2 virus, which belongs to the family Coronaviridae, genus Betacoronavirus. According to PANGO lineages as of now, 1575 SARS-CoV-2 variants are known since the initial detection of SARS-CoV-2 by sequencing (Rambaut et al., 2020). In the last few months, genomic epidemiology, the analysis of genome sequences, has revealed some fast-spreading and highly virulent SARS-CoV-2 variants (Cyranoski, 2021; McCormick et al., 2021; Priesemann et al., 2021), making them variants of concern (VOC) (European Centre for Disease Prevention and Control, 2021a; World Health Organization 2021). This development underlines the importance of sequencing analyses. Although the European Commission recommended to sequence 5–10% of the SARS-CoV-2 positive patient samples, as of 22 January 2021, most Member States are below this recommended sequencing target (European Centre for Disease Prevention and Control, 2021b).
Wastewater-based epidemiology (WBE) is an emerging paradigm for monitoring the circulation of SARS-CoV-2 in a community. Several research groups across the globe have shown that WBE provides additional information about the dynamics of SARS-CoV-2 at community level (Agrawal et al., 2021a; Ahmed et al., 2020; Kitajima et al., 2020; Medema et al., 2020; La Rosa et al., 2020). WBE efforts have primarily focused on quantitative polymerase chain reaction (qPCR), determining the titers of SARS-CoV-2 in the sewage and its correlation to the reported number of SARS-CoV-2 positive cases. At present, very few studies are available that looked for the combined potential of genomic epidemiology and WBE to determine the SARS-CoV-2 genomic variants circulating in a specific region. These studies are based on the amplicon sequencing approach to perform targeted sequencing of the complete SARS-CoV-2 genome using multiple primers. Crits-Christoph et al. (2021) used the Illumina Respiratory Virus Oligo Panel generating 2 × 75 bp paired-end reads and achieved a SARS-CoV-2 genome coverage of >99% for 7 out of 22 samples. Some studies performed Illumina sequencing on amplicons generated using the ARCTIC Network Protocol, Hillary et al. (2021) recovered between 25 and 75% of the SARS-CoV-2 genome, whereas, Pérez-Cataluña et al. (2022) recovered a genome coverage higher than 90% for 11 out of 76 samples. Agrawal et al. (2021a) used the Ion AmpliSeq SARS-CoV-2 research panel together with the Ion Torrent platform and achieved >90% SARS-CoV-2 genome coverage. Other studies have used the Oxford Nanopore sequencing platform generating approximately 400 bp reads based on the ARCTIC Network Protocol. Nemudryi et al. (2020) generated a consensus genome with a coverage of 98.51% at 6,875X average sequencing depth. Swift et al. (2021) sequenced 30 samples with the SARS-CoV-2 genome coverage ranging between 17 and 99%, with seven samples having >90% SARS-CoV-2 genome coverage. Izquierdo-Lara et al. (2021) performed Illumina as well as Nanopore sequencing of 55 samples, and only for 12 samples they achieved a genome coverage >90% using nanopore sequencing. All of these studies reported a wide range of SARS-CoV-2 genome coverages (i.e. 0 - 98%) for the analyzed wastewater samples, which emphasizes the high impact of the variation in the complex composition of wastewater samples. Low genomic coverage could result in a lack of necessary genomic information to identify mutations and, consequently, SARS-CoV-2 variants. It is well known that analyzing SARS-CoV-2 in wastewater is challenging (Michael-Kordatou et al., 2020). In particular, these challenges arise from the inherent complexity of the wastewater matrix hindering sufficient recovery of the SARS-CoV-2 RNA, and consequently resulting in poor coverage of the SARS-CoV-2 genomes. Moreover, variation in the wastewater composition among different regions makes it difficult to devise a common approach for sequencing SARS-CoV-2 in wastewater.
This first pan-European study was conducted to address the following questions: (1) although the wastewater composition and the SARS-CoV-2 titer in wastewater differ across Europe, would it be possible to use one common approach for sequencing of SARS-CoV-2 in wastewater samples across all European countries to obtain high coverage SARS-CoV-2 genomes? (2) Can next-generation sequencing (NGS) of wastewater samples provide ad-hoc information about the diversity of the SARS-CoV-2 variants and associated mutations at the community level circulating in European countries? This study, which was conducted under the umbrella of the EU Sewage Sentinel System for SARS-CoV-2 (Gawlik et al., 2021), a direct result of what is called “The HERA Incubator” (European Commission 2021a, European Commission 2021b), shows a clear coherence between clinical and wastewater SARS-CoV-2 mutation profiles associated with the variants of concerns at the time: B.1.1.7, P.1, B.1.351 and B.1.617.2, circulating in 20 European countries. This comprehensive study presents the possibility of using a common protocol for different wastewater matrices across Europe and maybe even worldwide to deliver high coverage NGS data of SARS-CoV-2.
2. Material and methods
2.1. Sequencing analysis
For this study, 24 h composite wastewater samples were collected, from 54 wastewater treatment plants across 20 European countries, between weeks 10 – 13 of 2021 (i.e. 10th to 30th March 2021) and shipped TU Darmstadt (Darmstadt, Germany), packed with icepacks (approximately at 6 °C), for sequencing analysis. In Darmstadt, we used two different methods to concentrate and extract the SARS-CoV-2 RNA to overcome biases (if any) caused by the different wastewater matrices in combination with the chosen concentration and extraction methods. One liter of the untreated wastewater was filtered through a 0.45 μm electronegative membrane filter to concentrate the SARS-CoV-2 RNA, followed by extraction using the Fast RNA Blue Kit (MP Biomedicals) according to the manufacturer's protocol. Another 500 ml of the untreated wastewater was concentrated by ultrafiltration in 100 kDa Centricon® Plus-70 centrifugal ultrafilters (Merck) and RNA was extracted using the Ultra Microbiome kit (Thermofisher Scientific) according to the manufacturer's protocol. With the ultrafiltration method, it was only possible to concentrate 500 ml of wastewater sample at maximum, which explains the difference in sample volume used for each concentration method. Both RNA extracts were pooled together, to have the largest possible amount of the SARS-CoV-2 RNA available for downstream analysis. From the pooled RNA, cDNA was synthesized using SuperScript™ VILO™ Master Mix (Thermofisher Scientific), followed by library preparation using the Ion AmpliSeq SARS-CoV-2 Research Panel (Thermofisher Scientific) according to manufacturer's instructions. This panel consists of 237 primer pairs, resulting in an amplicon length range of 125–275 bp, which cover the near-full genome of SARS-CoV-2. We performed multiple sequencing runs to achieve a high number of reads per sample. For each sequencing run, eight libraries were multiplexed and sequenced using an Ion Torrent 530 chip on an Ion S5 sequencer (Thermofisher Scientific) according to manufacturer's instructions.
We used the SARS-CoV-2 Research Plug-in Package, which we installed in our Ion Torrent Suite software (v5.12.2) of Ion S5 sequence. We used the SARS_CoV_2_coverageAnalysis (v5.16) plugin, which maps the generated reads to a SARS-CoV-2 reference genome (Wuhan-Hu-1-NC_045512/MN908947.3), using TMAP software included in the Torrent Suite. The summary of mapping of each sample is provided in Table S1. For mutation calls, additional Ion Torrent plugins were used, similar to our previous study (Agrawal et al., 2021b). First, all single nucleotide variants (SNVs) were called using Variant Caller (v5.12.0.4) with “Generic - S5/S5XL (510/520/530) - Somatic - Low Stringency” default parameters. Then, for annotation and determination of the base substitution effect, COVID19AnnotateSnpEff (v1.3.0.2), a plugin developed explicitly for SARS-CoV-2, was used.
2.2. qPCR analysis
Samples were received at the KWR laboratory and processed as previously described (Medema et al., 2020). The N2 assay targeting a fragment of the nucleocapsid gene, as published by US CDC (US-CDC 2020), was used to quantify SARS-CoV-2 RNA in the sewage samples. All RT-PCR's were run as technical duplicates on 5 µl extracted nucleic acid. RT-qPCR reactions on serial dilutions containing RT-ddPCR calibrated EURM-019 single stranded RNA (provided by the Joint Research center) were used to construct calibration curves that subsequently were used to quantify N2 in RNA extracted from the sewage samples. Reactions were considered positive if the cycle threshold was below 40 cycles.
CrAssphage CPQ_064 specific PCR (Stachler et al., 2017) was used to quantify this DNA-virus that is ubiquitously present exclusively occurs in human intestinal tracts in high concentrations. Assays were performed in duplicate on 5 µl 1:10 diluted extracted nucleic acid. Quantification was performed using PCR assays on dilution series of a synthetic quantified gBlock (obtained from IDT, Leuven, Belgium) containing the CPQ_064 gene fragment.
2.3. Data analysis
We downloaded the variant surveillance data package from GISAID on 31th May 2021. This data package consists of information about the identified variants, the corresponding amino acid (AA) mutations and sample location. We filtered the dataset, limiting it to human samples with complete coverage. This dataset was used to determine associations between the amino acid (AA) mutations detected in wastewater samples and AA mutations, with their corresponding pangolin lineage, reported from clinical samples. From the GISAID data package, we also determined the fraction of clinical samples reporting the current variants of concern (VOC): (1) B.1.1.7, (2) P.1, (3) B.1.351, (4) and B.1.617.2 (European Centre for Disease Prevention and Control, 2021c, 2021a; World Health Organization 2021). Data analysis was performed in R (v3.6.2) using the ggplot (v3.3.3) package for data visualization, and pheatmap (v1.0.12) for hierarchy clustering and heatmap construction.
2.4. Data and materials availability
All data are available in the main text or the supplementary materials. Raw metagenomic sequence data are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under Submission ID SUB9829162, BioProject number PRJNA736964.
3. Results
3.1. SARS-CoV-2 Variants and mutations of concern in Europe
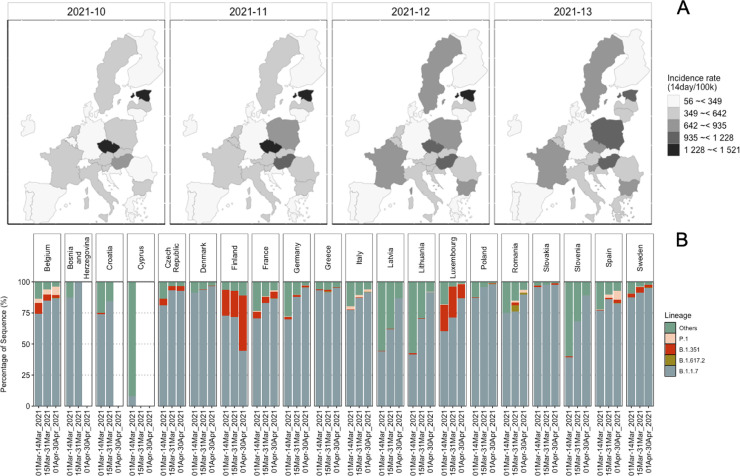
During March 2021, most countries in Europe reported an increase in the COVID-19 positive cases. To get an overview of the clinical situation, we determined the count of sequences associated with current VOCs (i.e., B.1.1.7, B.1.351, P.1, and B.1.617.2) (European Centre for Disease Prevention and Control, 2021d; World Health Organization 2021) in the 20 European countries that had provided wastewater samples (Fig. 1 ). The increased COVID-19 incidence rate corresponded with the increase in sequences of B.1.1.7 among the clinical samples sequenced in these countries. During this time, B.1.351 cases were reported in 18 out of 20 countries and P.1 cases in 10 countries. Also, very few B.1.617.2 sequences were reported (Fig. 1). While looking at the emergence of the mutations during March 2021, we found that among the most abundant mutations across all countries, the D614G (spike protein) and P323L (non-structural protein, Nsp12) mutations were detected in all the countries (Fig. S2).
Fig. 1.
COVID-19 situation in 20 European countries from clinical sequencing data. (A) Maps showing the incidence rate of the COVID-19 positive cases reported in the countries, during weeks 10 to 13, from when the wastewater samples originated. (B) Relative abundance of all sequences for the countries from where wastewater samples were analyzed, available in GISAID (https://www.gisaid.org) on 31–05–2021. All the sequences except B.1.1.7, B.1.351, P.1, and B.1.617 were categorized as “Others”.
Co-occurrences of D614G and P323L have often been reported (Vilar and Isom, 2021), however, some samples from Germany and China lacked P323L (Korber et al., 2020). The spike protein N501Y and H69del mutations were also highly abundant in most of the countries, earlier associated with B.1.1.7 and later with all VOCs except B.1.617.2 (Fig. S1, Fig. S2). In some countries the S106del, G107del, F108del mutations, which are ORF1b signature mutations of the VOCs P.1, B.1.1.7 and B.1.351 (Naveca et al., 2021), were also among the abundant mutations (Fig. S2). Across the twenty European countries twenty-six ORF1ab mutations, fourteen spike protein, eight nucleocapsid (N) protein, six ORF8, and three ORF3 mutations were among the dominant mutations, exhibiting spatial and temporal variation (Fig. S2).
3.2. Mutations associated with variants of concern in wastewater samples
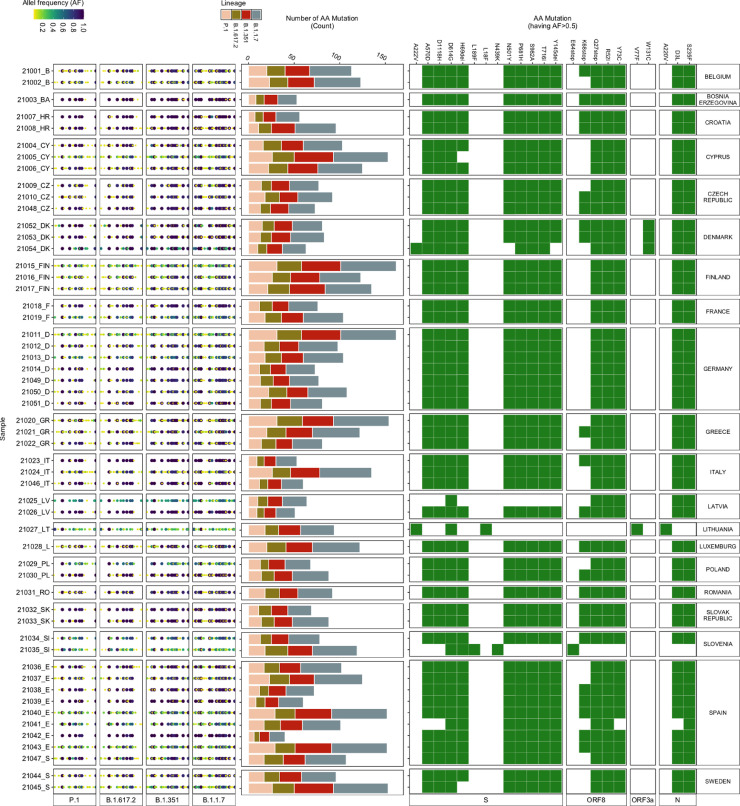
For this study, wastewater influent 24 h composite samples from 54 wastewater treatment plants (WWTPs) across 20 European countries were collected between weeks 10 – 13 of 2021 (i.e. 10th to 30th March 2021). Using protocols employed in this study, we were able to achieve more than 98% genome coverage for all the samples (Table S1, Data S1). We identified the mutations, which vary in their association with the VOCs, in each wastewater sample, by mapping the reads generated to the SARS-CoV-2 reference genome (Wuhan-Hu-1 [GenBank accession numbers NC_045512 and MN908947.3]). In total, 711 different mutations were identified across all the samples, out of which 633 mutations were associated with the VOCs. These 633 VOC associated mutations include mutations that are also associated with other SARS-CoV-2 lineages. Out of these mutations, 619 mutations were observed at >2.5%, 311 mutations at >5% and 23 mutations at >50% allele frequency (Fig. 2 ). Most of the 23 mutations were present in all but a few samples. For example, the W131C mutation was only detected in wastewater samples from Denmark (Fig. 2). W131C is one of the important mutations in ORF3a, which is found to assist the ion channel formation and thereby supports the virus in its infectivity (Hassan et al., 2021). The A220V mutation was only identified in samples from Lithuania (Fig. 2), which corresponds to the high count of A220V reported in clinical patient samples (Fig. S2).
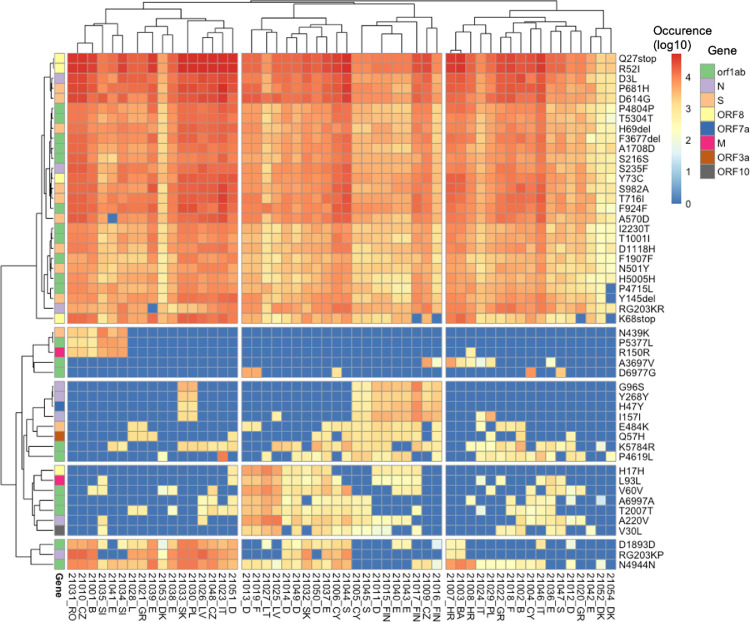
Fig. 2.
Amino Acid Mutations detected in wastewater samples. (Left) Allele frequency of the mutations (relative to the reference genome Wuhan-Hu-1 [GenBank accession numbers NC_045512 and MN908947.3]), associated with the variant of concern (VOCs), in each wastewater sample. (Middle) Number of mutations (count) detected in each sample corresponding to each VOC. (Right) Heatmap showing the presence (green) / absence (white) of mutations having more than 50% allele frequency. AA mutation: Amino acid mutations; AF: Allele Frequency; S: spike Protein; ORF8: open reading frame 8; ORF3a: open reading frame 3a; and N: nucleocapsid protein.
Although many low-frequency mutations were observed, the read abundance of low-frequency mutations was mostly similar to the abundance of the high-frequency mutations (Fig. S4). The highest count (ranging between 16 and 60) was observed for mutations associated with B.1.1.7 in all the samples (Fig. 3 ), followed by B.1.351. P.1 and B.1.617.2, which had a low count of associated mutations but included signature mutations. For example, signature spike protein mutations (i.e. L452R, T478K, P681R, D950N) (European Centre for Disease Prevention and Control, 2021d; Winger and Caspari, 2021) of B.1.617.2 were identified in some of the wastewater samples.
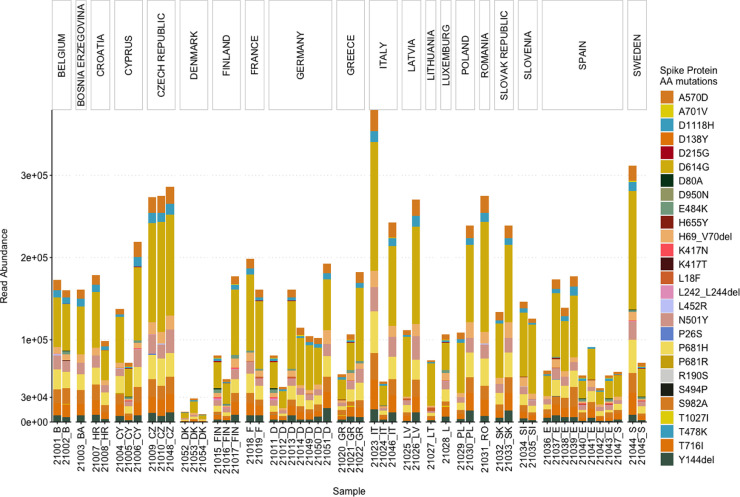
Fig. 3.
Abundance of the reads associated with Spike protein AA mutations used by the ECDC for the characterization of the VOCs.
3.3. Abundance of spike protein AA mutations
As the spike protein AA mutations have been associated with changes to characteristics of SARS-CoV-2, leading to an increase in transmissibility and reduced efficacy of treatments, a particular list of spike protein AA mutations is used by the European center for Disease Prevention and Control (ECDC) for characterizing the VOCs (European Centre for Disease Prevention and Control, 2021c, 2021a). Therefore, we also assessed the read abundance of these spike protein AA mutations in our wastewater samples (Fig. 3).
Overall, D614G was most abundant, followed by: P681H, T716I, A570D, S982A, H69del, Y144del, D1118H, N501Y, K417N, E484K and others, in decreasing order. Only six out of the 27 AA mutations (i.e. D1118H, D614G, H69del, N501Y, P681H, S982A, and T716I) were present in all the samples (Fig. 3). A570D and Y144del were identified in 53 samples. D1118H, S982A, T716I, H69del, P681H and A570D have been mainly found in the B.1.1.7 variant, whereas N501Y has been associated with B.1.1.7, B.1.351, and P.1 (Peacock et al., 2021). For 31 samples, the total read abundance of these AA mutations was above 1e+05 reads, the lowest values in all three sample were detected in the samples from Denmark ranging between 1e+04 and 3e+04 reads (Fig. 3).
3.4. Abundance and prevalence of dominant AA mutations
Most of the attention is given to spike protein AA mutations, especially since the emergence of B.1.1.7, because the spike protein mediates virus to host cell-surface attachment, and it is also the principal target of neutralizing antibodies (Harvey et al., 2021). However, mutations in other regions of the SARS-CoV-2 genome are also relevant (Nelson et al., 2020; Peacock et al., 2021). Therefore, we analysed the most abundant AA mutations found across all the samples for a comprehensive insight into the mutations detected in the wastewater samples (Fig. 4 ). Three mutations (Q27stop, R52I, and Y73C) in ORF8; D3L and S235F in the N protein; P681H, D614G, H69del, S982A, T716I, D1118H, and N501Y in the spike protein; P4804P, T5304T, F3677del, A1708D, S216S, F924F, I2230T, T1001I, F1907F, and H5005H in ORF1ab appeared to be most dominant and prevalent in the samples (Fig. 4). The presence of Q27stop and R52I mutations, along with the spike protein mutations, is reported to likely increase the transmissibility of SARS-CoV-2 (Pereira, 2021). Q27stop and R52I are known as characteristic mutations for B.1.1.7 (Emma, 2021) and are unique to B.1.1.7 (Singer et al., 2020). Also, F1907F, H5005H, A1708D and T1001I seem unique to B.1.1.7 (Singer et al., 2020). Another two ORF8 protein mutations, Y73C and K68stop, were present in most of the samples. The Y73C mutation is known to be unique in B.1.1.7 (Singh et al., 2021). Although no clear relevance of K68stop is known, it is reported to be present in SARS-CoV-2 genomes with the highest number of spike protein mutations (Pereira, 2021) and also seems unique to B.1.1.7 (Singer et al., 2020). The P681H spike protein mutation was the third most abundant across the samples. In total, 13 spike protein mutations were amongst the dominant mutations (Fig. 4).
Fig. 4.
Heatmap showing the read abundance of the top 50 AA mutations found across all the samples.
All spike protein mutations, along with the earlier mentioned ORF8 mutations, have been found in the B.1.1.7 variant in clinical samples (Pereira, 2021). E484K was observed in all wastewater samples from France and Sweden. This E484K mutation has been found in B.1.1.7, B.1.351, and P.1 (Tegally et al., 2020). Eight mutations of the nucleocapsid protein were also dominant in the samples, especially D3L and S235F. Both of these mutations are known to likely alter the stability and immunogenic properties of the N protein (Azad, 2021) and are signature mutations of B.1.1.7 (Galloway, 2021). In the ORF1ab region, 21 mutations were abundant in the samples. The F924F and P4715L ORF1ab mutations have been reported to have a strong allelic association with D614G in variants dominant in Europe (Koyama et al., 2020; Yang et al., 2020). The Q57H mutation of B.1.351 was also among the dominant mutation but was detected in only few samples (Fig. 4).
4. Discussion
Although previous studies have proven the usability of genome sequencing of SARS-CoV-2 in wastewater, there appears to be a common challenge, i.e., achieving high SARS-CoV-2 genome coverage across all the samples in each study (Crits-Christoph et al., 2021; Fontenele et al., 2021; Izquierdo-Lara et al., 2021; Nemudryi et al., 2020). High coverage is important to avoid loss of information about the mutations detected in wastewater because mutation profiles serve as the backbone to determine the variants present in wastewater samples. For example, some previous studies used detected mutations to determine variants (Fontenele et al., 2021; Pérez-Cataluña et al., 2022). The data presented in this study clearly shows that it is possible to obtain high coverage SARS-CoV-2 genomes, irrespective of variability in the composition of wastewater and the concentration of the SARS-CoV-2 RNA in the wastewater from different locations (Table S1). It is important to note that, there might be a need to optimize the primer pairs used for amplicon sequencing with emergence of new SARS-CoV-2 variants to achieve: (1) good coverage of the SARS-CoV-2 genome, and (2) better detection of emerging variants with new mutations, for example the Omicron variant with multiple new mutations. One of the approaches to overcome this challenge with emerging variants could be to include primers for more redundant amplicon coverage of all the regions of the SARS-CoV-2 genome.
According to the results shown in Fig. 1, showing the incidence rate of the COVID-19 positive cases reported in the sampled countries during weeks 10 to 13, variant B.1.1.7 was prevalent in all the countries except Cyprus. Cyprus shows an atypical picture, with relatively little B.1.1.7 and "other" variants prevalent. Similarly, based on wastewater sequencing data, B.1.1.7 seemed to be more prevalent in all the countries (Fig. S5). The difference between clinical and wastewater sequencing data of Cyprus most likely resulted from fewer clinical samples being sequenced during weeks 10 to 13. Variant B.1.351 seemed to be circulating more in Finland than in other countries based on clinical and wastewater sequencing data. B.1.351 was observed with relatively similar read abundance in all three wastewater samples, originating from three different WWTPs in Finland.
The pattern of genomic variants and the abundance of VOCs were consistent between the clinical and the wastewater sequencing data, especially for dominant mutations (Fig. S2, Fig. 4). For example, both data-sets show a high read abundance of the D614G mutation. Similarly, P681H was also dominant in clinical and wastewater samples (Fig. S2, Fig. 4). On the other hand, wastewater sequencing data can also reveal genomic variants which are not reported as dominant in clinical data. For example, Q27stop was one of the dominant mutations in wastewater samples but not in the clinical samples of all countries (Fig. S2, Fig. 4). The ORF8 mutations (such as Q27stop, R52I, K68stop) have been emphasized to be relevant and require closure attention (Jungreis et al., 2021; Pereira, 2021), especially because they occur recurrently (Peacock et al., 2021; Pereira, 2021). However, at the time of this study the count of the ORF8 mutations in genomes deposited in GISAID (https://www.gisaid.org) was low, which according to a previous study is due to bias in samples sequenced (Pereira, 2021). Across most of the wastewater samples, we detected a high occurrence of ORF8 mutations (i.e. Q27stop, R52I) (Fig. 4), which provides evidence for the circulation of SARS-COV-2 variants containing these mutations in the sampled regions. Also, wastewater sequencing data can reveal spatial prevalence of mutations. For example, the Q57H mutation was dominant in all three samples from Finland, which is consistent with clinical genomic data of Finland (Lam et al., 2020). This emphasizes that sequencing for SARS-CoV-2 in wastewater can provide additional information about the prevalence of mutations (such as Q27stop), which might not currently appear abundant based on clinical data. Also, relative abundance data grouping the mutations that are associated with a particular variant revealed a clear dominance of B.1.1.7 among the samples, followed by mutations associated with B.1.351, P.1, and B.1.617 (Fig. S5), which is similar to the clinical sequencing data. Wastewater sequencing can also provide information about those mutations that have not been detected in clinical samples of a region but detected in clinical samples of other regions. For example, Swift et al. (2021) reported detecting some mutations in the wastewater samples of South Carolina, which were not present in clinical samples of the same region but were present in the global sequence database of clinical samples.
5. Conclusion
The results obtained in this study clearly show that surveillance of SARS-CoV-2 mutation profiles associated with VOCs in wastewater samples is possible using NGS. The study also highlights that genomic surveillance of SARS-CoV-2 in wastewater could track clinically relevant mutations, which might be underrepresented in clinical sequencing data. The data generated also presents the possibility to attain above 98% coverage of the SARS-CoV-2 genome from the wastewater samples. In wastewater samples, a mixture of genomic material of multiple SARS-CoV-2 variants may be present. Nevertheless, it is still possible to obtain information about variants based on the prevalence of key mutations of the respective variant. However, it is essential to note that sequencing surveillance of wastewater samples should be considered complementary information to whole-genome sequencing of clinical samples.
CRediT authorship contribution statement
Shelesh Agrawal: Methodology, Investigation, Visualization, Supervision, Writing – original draft. Laura Orschler: Methodology, Investigation, Supervision, Writing – original draft. Selina Schubert: Investigation. Kira Zachmann: Investigation. Leo Heijnen: Methodology, Investigation. Simona Tavazzi: Conceptualization, Investigation, Project administration. Bernd Manfred Gawlik: Conceptualization, Investigation, Supervision, Project administration. Miranda de Graaf: Investigation, Supervision. Gertjan Medema: Conceptualization, Supervision, Supervision, Investigation. Susanne Lackner: Investigation, Project administration, Supervision, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We gratefully acknowledge the contribution from the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative (https://www.gisaid.org). We thank all WWTP operators for providing wastewater samples. We also thank Ray Izquierdo-Lara for critically reading the manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.watres.2022.118162.
Appendix. Supplementary materials
References
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021;11:5372. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S., Orschler L., Lackner S. Metatranscriptomic analysis reveals SARS-CoV-2 mutations in wastewater of the frankfurt metropolitan area in Southern Germany. Microbiol. Resour. Announc. 2021;10 doi: 10.1128/MRA.00280-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad G.K. Identification and molecular characterization of mutations in nucleocapsid phosphoprotein of SARS-CoV-2. PeerJ. 2021;9:e10666. doi: 10.7717/peerj.10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crits-Christoph A., Kantor R.S., Olm M.R., Whitney O.N., Al-Shayeb B., Lou Y.C., Flamholz A., Kennedy L.C., Greenwald H., Hinkle A., Hetzel J., Spitzer S., Koble J., Tan A., Hyde F., Schroth G., Kuersten S., Banfield J.F., Nelson K.L. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio. 2021;12 doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyranoski D. Alarming COVID variants show vital role of genomic surveillance. Nature. 2021;589:337–338. doi: 10.1038/d41586-021-00065-4. [DOI] [PubMed] [Google Scholar]
- Emma B. Hodcroft, 2021. CoVariants: SARS-CoV-2 Mutations and Variants of Interest [WWW Document]. URL https://covariants.org,(accessed 26.1.22).
- European Centre for Disease Prevention and Control, 2021a. SARS-CoV-2 variants of concern as of 3 June 2021 [WWW Document]. European Centre for Disease Prevention and Control. URL https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed 6.6.21).
- European Centre for Disease Prevention and Control, 2021b. Detection and characterisation capability and capacity for SARS-CoV-2 variants within the EU/EEA [WWW Document]. European Centre for Disease Prevention and Control. URL https://www.ecdc.europa.eu/en/publications-data/detection-and-characterisation-capability-and-capacity-sars-cov-2-variants (accessed 5.4.21).
- European Centre for Disease Prevention and Control, 2021c. Risk realted to the spread of new SARS-CoV-2 variants of concern in the EU/EEA - first update. Stockholm.
- European Centre for Disease Prevention and Control, 2021d SARS-CoV-2 variants of concern as of 3 June 2021 [WWW Document]. European Centre for Disease Prevention and Control. URL https://www.ecdc.europa.eu/en/covid-19/variants-concern (accessed 7.6.21).
- European Commission, 2021a. Commission Recommendation (EU) 2021/472 on a common approach to establish a systematic surveillance of SARS-CoV-2 and its variants in wastewaters in the EU (No. C/2021/1925).
- European Commission, 2021b. Communication from the commission to the European parliament, the European council and the council HERA Incubator: Anticipating together the threat of COVID-19 variants (No. COM/2021/78 final).
- Fontenele R.S., Kraberger S., Hadfield J., Driver E.M., Bowes D., Holland L.A., Faleye T.O.C., Adhikari S., Kumar R., Inchausti R., Holmes W.K., Deitrick S., Brown P., Duty D., Smith T., Bhatnagar A., Yeager R.A., Holm R.H., von Reitzenstein N.H., Wheeler E., Dixon K., Constantine T., Wilson M.A., Lim E.S., Jiang X., Halden R.U., Scotch M., Varsani A. High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway S.E. Emergence of SARS-CoV-2 B1.1.7 Lineage — United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70 doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawlik B., Tavazzi S., Mariani G., Skejo H., Sponar M., Higgins T., Medema G., Wintgens T. SARS-CoV-2 Surveillance employing Sewage - Towards a Sentinel System, EUR 30684 EN. Publications Office of the European Union; Luxembourg: 2021. ISBN 978-92-76-36887-8. [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021:1–16. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan Sk.S., Attrish D., Ghosh S., Choudhury P.P., Roy B. Pathogenic perspective of missense mutations of ORF3a protein of SARS-CoV-2. Virus Res. 2021;300 doi: 10.1016/j.virusres.2021.198441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary L.S., Farkas K., Maher K.H., Lucaci A., Thorpe J., Distaso M.A., Gaze W.H., Paterson S., Burke T., Connor T.R., McDonald J.E., Malham S.K., Jones D.L. Monitoring SARS-CoV-2 in municipal wastewater to evaluate the success of lockdown measures for controlling COVID-19 in the UK. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo-Lara, R., Elsinga, G., Heijnen, L., Munnink, B.B.O., Schapendonk, C.M.E., Nieuwenhuijse, D., Kon, M., Lu, L., Aarestrup, F.M., Lycett, S., Medema, G., Koopmans, M.P.G., de Graaf, M., 2021. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, The Netherlands and Belgium. Emerg Infect Dis 27, 1405–1415. 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed]
- Joshua Singer, Robert Gifford, Matthew Cotten, David Robertson, 2020. CoV-GLUE-Viz Mutations [WWW Document]. CoV-GLUE: a Web application for tracking SARS-CoV-2 genomic variation. http://cov-glue-viz.cvr.gla.ac.uk/mutations.php (accessed 1.26.22).
- Jungreis I., Sealfon R., Kellis M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 2021;12:2642. doi: 10.1038/s41467-021-22905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;139076 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Science of The Total Environment. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J.Y., Yuen C.K., Ip J.D., Wong W.M., To K.K.-W., Yuen K.Y., Kok K.H. Loss of orf3b in the circulating SARS-CoV-2 strains. Emerg. Microbes Infect. 2020;9:2685–2696. doi: 10.1080/22221751.2020.1852892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Klapsa D., Wilton T., Zambon M., Bentley E., Bujaki E., Fritzsche M., Mate R., Majumdar M. Tracking SARS-CoV-2 in sewage: evidence of changes in virus variant predominance during COVID-19 Pandemic. Viruses. 2020;12:1144. doi: 10.3390/v12101144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick K.D., Jacobs J.L., Mellors J.W. The emerging plasticity of SARS-CoV-2. Science. 2021;371:1306–1308. doi: 10.1126/science.abg4493. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8 doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca F.G., Nascimento V., de Souza V.C., Corado A.de L., Nascimento F., Silva G., Costa Á., Duarte D., Pessoa K., Mejía M., Brandão M.J., Jesus M., Gonçalves L., da Costa C.F., Sampaio V., Barros D., Silva M., Mattos T., Pontes G., Abdalla L., Santos J.H., Arantes I., Dezordi F.Z., Siqueira M.M., Wallau G.L., Resende P.C., Delatorre E., Gräf T., Bello G. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021:1–9. doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- Nelson C.W., Ardern Z., Goldberg T.L., Meng C., Kuo C.H., Ludwig C., Kolokotronis S.O., Wei X. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. eLife. 2020;9:e59633. doi: 10.7554/eLife.59633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;100098 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Penrice-Randal R., Hiscox J.A., Barclay W.S. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J. General Virol. 2021;102 doi: 10.1099/jgv.0.001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F. SARS-CoV-2 variants combining spike mutations and the absence of ORF8 may be more transmissible and require close monitoring. Biochem. Biophys. Res. Commun. 2021;550:8–14. doi: 10.1016/j.bbrc.2021.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Chiner-Oms Á., Cuevas-Ferrando E., Díaz-Reolid A., Falcó I., Randazzo W., Girón-Guzmán I., Allende A., Bracho M.A., Comas I., Sánchez G. Spatial and temporal distribution of SARS-CoV-2 diversity circulating in wastewater. Water Res. 2022;211 doi: 10.1016/j.watres.2021.118007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priesemann V., Balling R., Brinkmann M.M., Ciesek S., Czypionka T., Eckerle I., Giordano G., Hanson C., Hel Z., Hotulainen P., Klimek P., Nassehi A., Peichl A., Perc M., Petelos E., Prainsack B., Szczurek E. An action plan for pan-European defence against new SARS-CoV-2 variants. Lancet. 2021;397:469–470. doi: 10.1016/S0140-6736(21)00150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Samal J., Kumar V., Sharma J., Agrawal U., Ehtesham N.Z., Sundar D., Rahman S.A., Hira S., Hasnain S.E. Structure-function analyses of new SARS-CoV-2 Variants B.1.1.7, B.1.351 and B.1.1.28.1: clinical, diagnostic, therapeutic and public health implications. Viruses. 2021;13:439. doi: 10.3390/v13030439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51:9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift C.L., Isanovic M., Correa Velez K.E., Norman R.S. Community-level SARS-CoV-2 sequence diversity revealed by wastewater sampling. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally, H., Wilkinson, E., Giovanetti, M., Iranzadeh, A., Fonseca, V., Giandhari, J., Doolabh, D., Pillay, S., San, E.J., Msomi, N., Mlisana, K., von Gottberg, A., Walaza, S., Allam, M., Ismail, A., Mohale, T., Glass, A.J., Engelbrecht, S., Van Zyl, G., Preiser, W., Petruccione, F., Sigal, A., Hardie, D., Marais, G., Hsiao, M., Korsman, S., Davies, M.A., Tyers, L., Mudau, I., York, D., Maslo, C., Goedhals, D., Abrahams, S., Laguda-Akingba, O., Alisoltani-Dehkordi, A., Godzik, A., Wibmer, C.K., Sewell, B.T., Lourenço, J., Alcantara, L.C.J., Pond, S.L.K., Weaver, S., Martin, D., Lessells, R.J., Bhiman, J.N., Williamson, C., de Oliveira, T., 2020. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) Lineage with multiple spike mutations in South Africa (preprint). Epidemiology. 10.1101/2020.12.21.20248640.
- Vilar S., Isom D.G. One Year of SARS-CoV-2: how much has the virus changed? Biology (Basel) 2021;10:91. doi: 10.3390/biology10020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger, A., Caspari, T., 2021. The spike of concern—the novel variants of SARS-CoV-2. Viruses 13, 1002. 10.3390/v13061002. [DOI] [PMC free article] [PubMed]
- World Health Organization, 2021. Weekly epidemiological update on COVID-19 - 1 June 2021 [WWW Document]. https://www.who.int/publications/m/item/weekly, epidemiological-update-on-covid-19—1-june-2021 (accessed 6.6.21).
- Worobey M., Pekar J., Larsen B.B., Nelson M.I., Hill V., Joy J.B., Rambaut A., Suchard M.A., Wertheim J.O., Lemey P. The emergence of SARS-CoV-2 in Europe and North America. Science. 2020;370:564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.C., Chen C., Wang J.H., Liao H.C., Yang C.T., Chen C.W., Lin Y.C., Kao C.H., Lu M.Y.J., Liao J.C. Analysis of genomic distributions of SARS-CoV-2 reveals a dominant strain type with strong allelic associations. PNAS. 2020;117:30679–30686. doi: 10.1073/pnas.2007840117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.