Abstract
A total of 150 fecal and water samples from four swine farms were tested for the presence of Salmonella enterica using different enrichment techniques as follows: (i) 92 fecal samples from nursery and farrowing barns at three swine farms were preenriched overnight in tryptic soy broth (TSB) at 37°C followed by overnight enrichment in Rappaport-Vassiliadis 10 broth (RV10) at 42°C; (ii) 24 water samples from the third farm were preenriched overnight in 3MC broth at 37°C followed by overnight enrichment in RV10 at 42°C; and (iii) 34 fecal samples from a fourth farm, a finishing farm, were enriched overnight in RV10 at 42°C with no additional enrichment. Following each of the enrichment techniques, samples were subcultured onto modified semisolid Rappaport-Vassiliadis (MSRV) agar prior to transfer to Hektoen Enteric agar plates for the recovery of viable Salmonella bacteria. Presumptive Salmonella isolates were biochemically and serologically confirmed. For the PCR detection of Salmonella, a 1-ml portion was removed from each sample after the first overnight enrichment and the DNA was extracted using a Sepharose CL-6B spin column. Amplicons (457 bp) derived from primers to the invA and invE genes were confirmed as Salmonella specific on ethidium bromide-stained agarose gels by Southern hybridization with a 20-mer oligonucleotide probe specific for the Salmonella invA gene. Neither the standard microbiological method nor the molecular method detected all of the 65 samples that tested positive by both methods or either method alone. Salmonella bacteria were detected by both cultivation and PCR-hybridization in 68% (17 of 25) of the positive samples that were preenriched in TSB, in 73% (11 of 15) of the positive samples preenriched in 3MC broth, and in 24% (6 of 25) of the positive samples enriched in RV10. Agreement between Salmonella detection using cultivation with preenrichment and detection by PCR was 76% using the kappa statistic. However, agreement between Salmonella detection using cultivation without preenrichment and detection by PCR was about 6%; the PCR assay detected 80% (20 of 25) of the 25 positive samples, while Salmonella bacteria were recovered from only 44% (11 of 25) by cultivation. Our results indicate that the PCR-hybridization approach is equivalent to or better than cultivation for detecting Salmonella in swine feces or water samples from swine farms when using the medium combinations evaluated in this study.
Samonella bacteria shed in the feces of asymptomatic swine are important sources of environmental and carcass contamination (3, 19, 29, 30). They can be shed from swine at levels of less than 10 CFU/g of feces (19) and remain viable in soil for more than a year (28). Detection of Salmonella enterica in fecal or water samples can be limited by low numbers of the bacterium (e.g., less than 10 CFU/g), thus necessitating the use of an enrichment step. Enrichment and preenrichment broths can dilute inhibitory compounds produced by competing bacteria in the sample, as well as aid in recovery of injured, stressed, or lag-phase bacterial cells (6, 21). Generally, isolation of Salmonella from samples, such as feces, that contain approximately >107 aerobic bacterial cells/g requires a selective enrichment medium that permits the growth of Salmonella while inhibiting the growth of other aerobic bacteria (21, 24). However, some selective enrichment media also inhibit the growth of Salmonella bacteria that are stressed or damaged (4, 6).
Although bacteriological assays have historically been the method of choice for the recovery of Salmonella from feces and environmental samples, PCR has become an important technique for more-rapid detection of pathogens in feces and environmental samples when an isolate is not required (5, 7, 37, 38). Like bacteriological assays, PCR often requires enrichment of fecal samples to increase Salmonella numbers and to aid in the dilution of compounds that may interfere with the PCR (5, 7, 22, 23, 37, 38, 40, 42).
Discrepancies between detection of Salmonella nucleic acid sequences in feces by the PCR and recovery of the bacterium on synthetic media have been noted in favor of the PCR; however, one comparative study did not use the same enrichment broth for comparing the two approaches, bacteriological cultivation and PCR, and some studies did not include a preenrichment step in the cultivation procedure (5, 7, 38). Also, the PCR primers selected can lead to inaccurate results. False-positive results arise from mispriming of nucleic acid sequences that are similar to target DNA, particularly when samples contain DNA from ingested material, fecal flora, and/or mammalian cells (31). For instance, there is over 90% homology between the genomes of Salmonella and Escherichia coli (34), and this can cause considerable mispriming and lead to false-positive amplicons. Misprimed amplicons similar in length to the amplicon of interest are also difficult to distinguish on agarose gels but can be identified by Southern hybridization with internal nucleotide probes (31, 38, 43).
An improved understanding of the agreement between the bacteriological recovery of Salmonella, the current “gold standard” to which all other Salmonella detection tests are compared, and the PCR-based detection of Salmonella is warranted to determine if the latter could be used as a definitive or a presumptive test. As a presumptive test, the PCR could be used to determine samples from which a representative isolate should be obtained for further characterization. For this use, results from the PCR must be known within 18 to 24 h so that an isolate can be obtained before appreciable cell death occurs in the retained sample or in the primary enrichment broth (32). As a definitive test, the PCR, followed by Southern hybridization for the confirmation of amplicons, could be used to determine which samples contain Salmonella DNA.
The purpose of this research was to compare cultivation- and PCR-based methods for the detection of Salmonella in porcine fecal and water samples from swine farms. The null hypothesis was that no difference would be measured between these methods for detecting Salmonella.
MATERIALS AND METHODS
Sample collection and bacterial isolation.
Samples were enriched by one of three methods: (i) overnight preenrichment in tryptic soy broth (TSB; Difco Laboratories Inc., Detroit, Mich.) at 37°C and overnight enrichment in Rappaport-Vassiliadis 10 broth (RV10; Difco) at 42°C (TSB-RV10 enrichment method), (ii) overnight preenrichment in a laboratory-prepared medium previously designated 3MC broth (2, 20) at 37°C and overnight enrichment in RV10 at 42°C (3MC-RV10 enrichment method), and (iii) direct overnight enrichment in RV10 at 42°C (with no secondary enrichment) (RV10 enrichment method). All samples were collected within 100 mi of the laboratory. The samples were placed directly into 10 ml of TSB, 3MC broth, or RV10 and stored on ice in a cooler until processed later the same day.
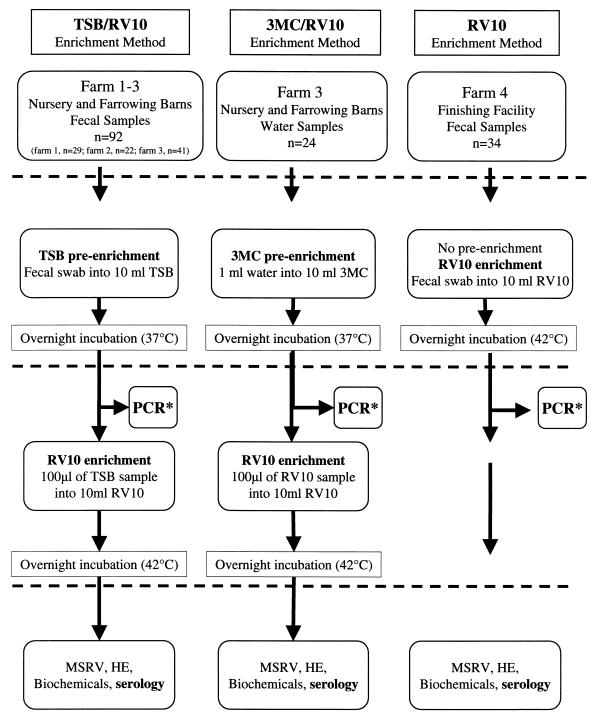
In the first trial, 92 swabs of fresh swine feces, consisting of rectally inserted, cotton-tipped applicators or cotton-tipped applicators that had been inserted into fresh swine feces on the floor, collected from the nursery and farrowing barns at three swine farms (farm 1, 4 March 1998, n = 29; farm 2, 10 March 1998, n = 22; and farm 3, 6 April 1998, n = 41) were placed into 10 ml of TSB and preenriched overnight at 37°C (Fig. 1). One hundred microliters of each sample was then transferred into 10 ml of RV10 and enriched overnight at 42°C (Fig. 1). In the second trial, 24 water samples (1 ml), collected from bowl waterers or mud holes from farm 3 on the same day that the fecal samples were collected, were placed into 10 ml of 3MC broth and preenriched overnight at 37°C (Fig. 1). One hundred microliters of each sample was then transferred into 10 ml of RV10 and enriched overnight at 42°C (Fig. 1). In the third trial, 34 swabs of fresh swine feces collected from the pen floors of a swine finishing facility at a fourth farm (13 April 1998) were placed directly into 10 ml of RV10 (Fig. 1) and incubated overnight at 42°C. After enrichment in RV10, all 150 samples were subcultured onto modified semisolid Rappaport-Vassiliadis (Difco) agar plates (Fig. 1) by drop (50-μl) inoculation (11) and incubated for 24 h at 42°C. Samples displaying motile growth around the initial drop of the inoculum were subcultured onto Hektoen Enteric (Difco) agar plates for the isolation of single colonies and incubated at 37°C for 24 h (Fig. 2). Bacterial colonies with typical Salmonella morphology (i.e., clear with black centers for most Salmonella serovars or clear without black centers for S. enterica serovar Choleraesuis) were tested by slide agglutination with Salmonella antigen group A-I, Vi polyvalent antiserum (Difco). In addition, Salmonella isolates that tested positive using the polyvalent antiserum were forwarded to the National Veterinary Service Laboratory (Ames, Iowa) for further authentication and identification to species level using antisera specific for O type and H type (i.e., univalent antiserum).
FIG. 1.
Flow diagram of the methods used to isolate or detect Salmonella in fecal or water samples from swine feces. ∗, after DNA extraction, 10 μl of sample DNA was used per PCR. MSRV, modified semisolid Rappaport-Vassiliadis agar; HE, Hektoen Enteric agar.
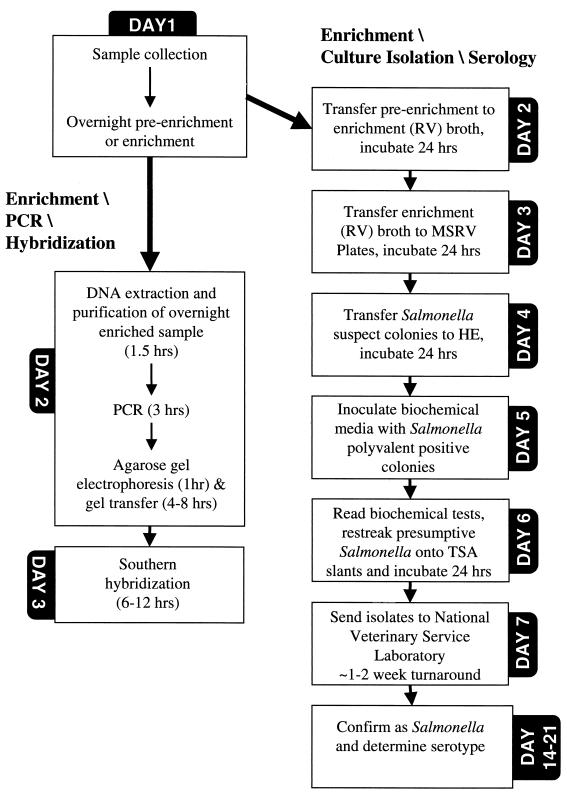
FIG. 2.
Comparison of the time to Salmonella detection via Southern hybridization with that by serotyping of a single colony isolate. MSRV, modified semisolid Rappaport-Vassiliadis agar; HE, Hektoen Enteric agar; TSA, tryptic soy agar.
DNA extraction and purification.
Samples for DNA extraction and subsequent PCR amplification were retrieved directly from the broths after the initial enrichment. The DNA was extracted as described previously by Lou et al. (23) with some modifications. The 10-ml overnight samples were vortexed for 2 min, and particulate matter was allowed to settle for 10 min at room temperature. A 1.5-ml portion of the resultant supernatant was transferred to a microcentrifuge tube and centrifuged at room temperature for 2 min at 2,800 × g. One milliliter of this supernatant was removed to a second microcentrifuge tube and centrifuged at room temperature for 15 to 20 min at 12,000 × g. The supernatant was discarded, and the pellet was resuspended in 1 ml of sterile high-pressure liquid chromatography-grade H2O (Fisher Scientific, St. Louis, Mo.) and centrifuged at room temperature for 5 min at 12,000 × g. This wash step was repeated, and the pellet was resuspended in 500 μl of sterile H2O, boiled for 5 min, and then centrifuged at room temperature for 5 min at 12,000 × g. Eighty microliters of sample supernatant and 20 μl of a 5× stop solution (2% sodium dodecyl sulfate, 25% glycerol, 0.05% bromophenol blue, and 0.05% xylene cyanol) were added to 60 μl of sterile H2O and incubated for 15 to 20 min at 70°C. Ninety microliters of this supernatant-stop mixture was added to a Sepharose CL-6B gel filtration spin column and centrifuged at 700 × g for 2 min at room temperature. If blue dye was noted in the resultant filtrate, centrifugation was repeated using a fresh spin dialysis column. The resultant filtrate was used for the PCR.
Sepharose CL-6B spin columns were prepared essentially as described by Maniatis et al. (25). Sepharose CL-6B (Sigma, St. Louis, Mo.) was washed three times with equal amounts of 10 mM Tris-Cl–1 mM EDTA (TE) buffer (pH 8.0). Buffer was removed for a final concentration of 60% Sepharose CL-6B. Autoclaved 60% Sepharose CL-6B in TE buffer (800 μl) was added to a 1-ml tuberculin syringe packed with glass wool to about 1 cm. The spin column was centrifuged at 700 × g for 2 min to remove excess TE buffer. Additional 60% Sepharose CL-6B was added, and centrifugation was repeated until the Sepharose CL-6B reached a final height of the 1-ml mark on the tuberculin syringe.
PCR.
To optimize the efficacy of target DNA amplification, thawed reagents were held on ice during setup for the PCR. All reactions included a hot start technique (31) in HOTSTART reaction tubes (HOTSTART Micro 50; Molecular Bio-Products, Inc., San Diego, Calif.). All PCR assays were performed using 10 μl of template and 90 μl of the PCR mixture. Templates were heated to 94°C for 5 min before addition of PCR core reagents. Final concentrations of PCR reagents were 1.8 mM MgCl2; 1× PCR buffer II at 10% of the total volume; 200 μM (each) deoxynucleoside triphosphate; 2.5 U of Taq DNA polymerase (Gene Amp core reagents; Perkin-Elmer Corporation, Foster City, Calif.) per 100 μl; and 0.3 μM Salmonella oligonucleotide primers (37, 38) (Gibco BRL, Life Technologies, Rockville, Md.). The DNA templates were amplified by use of one denaturation cycle at 94°C for 2 min; 5 amplification cycles at 94°C for 60 s, 52°C for 30 s, and 72°C for 30 s; 40 cycles at 94°C for 30 s, 52°C for 15 s, and 72°C for 30 s, followed by an extension cycle at 72°C for 15 min; and a hold cycle at 4°C.
S. enterica serovar Choleraesuis DNA extracted by the Sepharose CL-6B method from RV10, 3MC broth, and TSB was used to determine optimal MgCl2 concentrations for PCR amplification. Negative controls, uninoculated TSB, 3MC broth, and RV10 samples and E. coli grown in TSB, were subjected to Sepharose purification. Positive controls consisted of Sepharose purifications from a pure culture of S. enterica serovar Choleraesuis grown overnight separately in TSB, RV10, and 3MC broth seeded with a swab of swine feces.
Detection of amplified products.
Twelve-microliter aliquots of a PCR sample and 3 μl of sample application buffer (0.25% bromophenol blue, 40% [wt/vol] sucrose in water) were analyzed using ethidium bromide (EtBr)-stained agarose gels. To confirm amplification of PCR products detected on agarose gels as Salmonella, Southern hybridization (36) was conducted using a previously described digoxigenin-labeled, internal, oligonucleotide probe corresponding to nucleotides 106 to 125 of the Salmonella invA gene (37, 38). Nylon membranes (MSI, Westboro, Mass.) were prehybridized followed by hybridization with the probe as specified by Boehringer Mannheim (Indianapolis, Ind.). The DIG Easy Hyb buffer (Boehringer Mannheim) was used for the prehybridization and hybridization steps. Using the DIG Wash and Block buffer set (Boehringer Mannheim), probe that hybridized to the amplified Salmonella nucleotide segment was detected using the antidigoxigenin-alkaline phosphatase antibody (α-DIG-Alkaline Phosphatase-Fab fragments; Boehringer Mannheim), a chemiluminescent substrate for alkaline phosphatase (CSPPD; Boehringer Mannheim), and radiographic film (medical X-ray film; Fuji, Tokyo, Japan). The nylon membrane was exposed to radiographic film for 2 h. The PCR amplicons observed as bands on EtBr-stained agarose gels were compared to the corresponding bands on the radiographic film.
Data analysis.
For the purposes of this study, samples that were positive by either cultivation (i.e., bacterial isolation with serological confirmation) or PCR (i.e., the PCR with confirmation using Southern hybridization) were considered positive, even if the samples were negative using either one of these detection methods alone. This assumption, using the combinatorial results of the PCR and cultivation to identify Salmonella present in a sample, enabled us to estimate the false negatives for each test. For example, a false negative would be defined as a sample that tested negative by cultivation and positive by the PCR and vice versa. Since the true-negative status of the samples was not known, specificity could not be reported. For this reason, we used the term “relative specificity” to approximate specificity. The relative specificity and sensitivity (26) of the PCR were determined by comparison to bacteriological results. For statistical analysis, a 2 × 2 comparison table was constructed to show the association of Salmonella-positive results between the two detection strategies. The three different enrichment methods used were not compared because different samples were used for each enrichment scheme. The relative specificity and the sensitivity of PCR-hybridization and EtBr-stained agarose gels were calculated with the total number of positive samples as the standard by using a 2 × 2 table as described previously (26), and the differences between the two methods were compared.
Agreement between the cultivation- and PCR-based methods for detection of Salmonella was evaluated by use of the kappa statistic (17, 27, 35). The kappa statistic measures agreement between two tests that is beyond chance, with chance being a value of zero and with 1.00 being complete agreement, not by chance (8). Agreement between tests on positive samples is given as much weight by the kappa statistic as agreement between tests on negative samples. Kappa values were summarized as unacceptable (<0.3), acceptable (0.3 to ≤0.5), good (0.5 to 0.7), and excellent (>0.7) (27).
RESULTS
Detection of Salmonella by either cultivation- or PCR-based methods.
Of the 150 samples tested in this study, 65 (43%) were positive for Salmonella by either cultivation or PCR-hybridization (data not shown). No Salmonella bacteria were detected on farm 1 or farm 2 by either approach alone (Table 1). Salmonella bacteria were detected in 61% (25 of 41) of the fecal samples from farm 3 (Table 1). On this farm, Salmonella bacteria were also detected in 63% (15 of 24) of the water samples (Table 1). Detection of Salmonella in fecal samples from the finishing farm (i.e., farm 4) was 74% (25 of 34) (Table 1). Of the 65 Salmonella-positive samples from all of the farms tested, 47 (72%) were positive by cultivation, 52 (80%) were positive by PCR-hybridization, and 51 (79%) were positive by visualization of the PCR amplicons on EtBr-stained agarose gels (data not shown). Therefore, there were 18 false negatives with respect to cultivation, 13 false negatives with respect to the PCR-hybridization detection, and 14 false negatives with respect to EtBr-stained agarose gels. Only 34 (52%) of the 65 positive samples were detected by both cultivation and PCR-hybridization (data not shown).
TABLE 1.
Detection of Salmonella using cultivation, PCR-hybridization, and a combination of cultivation and PCR-hybridization following enrichment using three different media
| Farm no. and sample type (no. of samples) | Enrichment medium (-a) | Detection (%) of Salmonella (no. positive/total no. of samples) by method:
|
||
|---|---|---|---|---|
| Cultivation | PCR-based detection | Cultivation–PCR-based detection | ||
| Farm 1, feces (29) | TSB-RV10 | 0.0 | 0.0 | 0.0 |
| Farm 2, feces (22) | TSB-RV10 | 0.0 | 0.0 | 0.0 |
| Farm 3, feces (41) | TSB-RV10 | 53.7 (22/41) | 48.8 (20/41) | 60.9 (25/41) |
| Total | 23.9 (22/92) | 21.7 (20/92) | 27.2 (25/92) | |
| Farm 3, water (24) | 3MC-RV10 | 58.3 (14/24) | 50.0 (12/24) | 62.5 (15/24) |
| Farm 4, feces (34) | RV10 | 32.4 (11/34) | 58.8 (20/34) | 73.5 (25/34) |
In the present study, five Salmonella serovars were detected from two of the four clinically normal swine herds tested (data not shown). At a nursery-farrowing farm, S. enterica serovar Choleraesuis biotype kunzendorf, S. enterica serovar Anatum, S. enterica serovar Derby, and S. enterica serovar Enteritidis were isolated from feces, whereas serovar Anatum and serovar Derby were isolated from water samples. At the finishing farm, S. enterica serovar Choleraesuis, S. enterica serovar Enteritidis, and S. enterica serovar Bredeney were isolated from fecal samples.
Comparison of detection methods.
Of the 65 positive Salmonella samples, only 52% (34 of 65) were detected by both cultivation and PCR-hybridization; overall agreement (kappa) between both methods was 53% (data not shown). For the TSB-RV10 enrichment method, Salmonella bacteria were detected by both cultivation and PCR-hybridization in 68% (17 of 25) of the positive samples. Likewise, for the 3MC-RV10 enrichment method, Salmonella bacteria were detected by both methods in 73% (11 of 25) of the positive samples. However, for the RV10 enrichment method, Salmonella bacteria were detected by both methods in only 24% (6 of 25) of the positive samples. Therefore, when a TSB preenrichment followed by enrichment in RV10 was used, cultivation failed to detect 3% (3 of 92) of the positive samples while PCR-hybridization failed to detect Salmonella in 5% (5 of 92) of the positive samples (Table 2). In contrast, when a preenrichment step was not performed and samples were directly enriched into RV10, there was a higher rate of false-negative reactions as cultivation failed to detect 41% (14 of 34) of the positive samples (Table 2). The agreement (kappa) between cultivation and PCR-hybridization was 76% for samples that were preenriched (data not shown). Agreement (kappa) between cultivation and PCR-hybridization was higher for the TSB-RV10 (71%) and 3MC-RV10 (63%) methods than for the RV10 enrichment method (6%) (data not shown). Compared to cultivation, PCR-hybridization displayed 77% sensitivity and 96% relative specificity with respect to the TSB-RV10 enrichment method, 79% sensitivity and 90% relative specificity with respect to the 3MC-RV10 enrichment method, and 55% sensitivity and 39% relative specificity with respect to the RV10 enrichment method. Also, in comparison to bacterial isolation and biochemical identification of presumptive Salmonella isolates, PCR-hybridization detected Salmonella 3 days earlier than if a preenrichment was used and 2 days earlier if preenrichment was not performed (Fig. 2).
TABLE 2.
A 2 × 2 comparison of Salmonella detection by cultivation and PCR-hybridization versus the combined results of cultivation and PCR-hybridization
| Enrichment method | No. of samples | Detection technique | Result type | % Detection by combination of cultivation and PCR-hybridization (no. with result/total no. of samples)
|
|
|---|---|---|---|---|---|
| + | − | ||||
| TSB-RV10 | 92 | Bacteriological | + | 23.9 (22/92) | 0 |
| − | 3.3 (3/92) | 72.8 (67/92) | |||
| PCR based | + | 21.7 (20/92) | 0 | ||
| − | 5.4 (5/92) | 72.8 (67/92) | |||
| 3MC-RV10 | 24 | Bacteriological | + | 58.3 (14/24) | 0 |
| − | 4.2 (1/24) | 37.5 (9/24) | |||
| PCR based | + | 50.0 (12/24) | 0 | ||
| − | 12.5 (3/24) | 37.5 (9/24) | |||
| RV10 | 34 | Bacteriological | + | 32.4 (11/34) | 0 |
| − | 41.2 (14/34) | 26.5 (9/34) | |||
| PCR based | + | 58.8 (20/34) | 0 | ||
| − | 14.7 (5/34) | 26.5 (9/34) | |||
Comparison between agarose gel electrophoresis and Southern hybridization for the detection of PCR amplicons.
Nine amplicons, three from each of the three enrichment methods, were visualized on EtBr-stained agarose gels but did not hybridize to the Salmonella-specific oligonucleotide probe (Table 3). Therefore, the corresponding false-positive rates for detection of Salmonella amplicons on agarose gels for each type of enrichment were 3, 13, and 9% for the TSB-RV10, 3MC-RV10, and RV10 enrichment methods, respectively (Table 3). Southern hybridization detected only one false-negative sample compared to EtBr-stained agarose gel electrophoresis detection of the PCR amplicons, as one band was seen on a radiograph of a Southern hybridization that was not seen on the corresponding EtBr-stained agarose gel when the RV10 enrichment method was used (Table 3).
TABLE 3.
A 2 × 2 comparison of Salmonella detection by PCR and EtBr-stained agarose gels with that by PCR-hybridization
| Enrichment method | No. of samples | Result type for PCR-EtBr | % Detection by PCR-hybridization (no. with result/total no. of samples)
|
|
|---|---|---|---|---|
| + | − | |||
| TSB | 92 | + | 21.7 (20/92) | 3.3 (3/92) |
| − | 0 | 75.0 (69/92) | ||
| 3MC broth | 24 | + | 50.0 (12/24) | 12.5 (3/24) |
| − | 0 | 37.5 (9/24) | ||
| RV10 | 34 | + | 55.9 (19/34) | 8.8 (3/34) |
| − | 2.9 (1/34) | 32.4 (11/34) | ||
DISCUSSION
Neither cultivation nor PCR-hybridization alone detected all of the 65 Salmonella-positive samples from swine farms. However, the results from this study indicate that the PCR is equivalent to cultivation for delineating positive samples, particularly when a preenrichment step is used. Additionally, our results and those of others (38, 43) establish the importance of nucleic acid probes to evaluate the PCR amplicons visualized on EtBr-stained gels. With respect to gel electrophoresis and the visualization of EtBr-stained gels, Southern hybridization improved both the sensitivity and relative specificity of the PCR. While we had only one false positive (0.67%; 1 of 150), two other studies, one using feces and one using blood as the biological samples, have shown higher false-positive rates (38, 43). More specifically, 13 of 21 (54%) fecal samples tested from a single colony of beagles had PCR amplicons that could be detected on an EtBr-stained agarose gel but not on the corresponding radiographic film after Southern hybridization. The higher false-positive rate with respect to feces was most likely due to the cross-reaction of the PCR primers with a commensal bacterium or other DNA within the fecal specimens of the beagle colony tested. As there is no way of determining background flora, or other background DNA, within a sample containing DNA that may cross-react, it is imperative to perform hybridization. For this reason, Southern hybridization was used to confirm presumptive Salmonella positives from PCR-based assays just as serotyping was used to confirm presumptive Salmonella from cultivation-based strategies. While the PCR-hybridization approach was equivalent to the bacteriology-based technique, the former only confirms the isolate at the genus level and does not delineate Salmonella species or serovar. For epidemiological purposes, the species or serovar of Salmonella may be of interest, particularly on swine farms where Salmonella prevalence is high and where more than one Salmonella species or serovar can be isolated (1, 9, 29, 33). In this regard, univalent O typing can be done within 1 day, whereas confirmation of Salmonella isolates by serotyping could take an additional 1 to 2 weeks depending on the difficulty of flagellar typing (i.e., H typing).
S. enterica serovar Choleraesuis biotype kunzendorf, the most common cause of salmonellosis in swine (16, 41), and serovar Choleraesuis, a rare serovar in swine in the United States (16), generally have been isolated only from clinically ill swine (14, 37, 41). Our ability to detect serovar Choleraesuis, an H2S-negative biotype, was aided by using modified semisolid Rappaport-Vassiliadis agar, a medium previously described for the enhanced recovery of Salmonella from human feces (13, 18). Detection by this medium is based on motility and not on H2S production as determined on Hektoen Enteric or XLT4 agar (11, 12, 13, 18). In contrast to our study, the predominant Salmonella serotypes recovered from swine feces in other studies included S. enterica serovar Heidelberg, S. enterica serovar Mbandaka, S. enterica serovar Typhimurium, S. enterica serovar Worthington, and S. enterica serovar Tennessee (1, 9, 10, 33).
At the farms where Salmonella was detected by either cultivation or the PCR technique, the detection of Salmonella ranged between 61% (25 of 41) and 74% (25 of 34). Previously, field trials using PCR-based and culture-based methods for the detection of Salmonella from nonsymptomatic pigs have not been reported. Detection of Salmonella from swine intestinal swabs was 40% (two of five) using either bacteriology-or PCR-based methods when intestinal specimens from five dead pigs suspected of having salmonellosis were tested (37). As another example, detection of Salmonella was about 8% (6 of 79) and 9% (7 of 79) using bacteriology-and PCR-based methods, respectively, when intestinal specimens from 79 pigs with diarrhea were tested (14). In a study of clinically normal swine, the detection of Salmonella in feces averaged 25% from the feces from 24 of 29 farms tested (10). In additional studies of clinically normal swine, the detection of Salmonella in feces was 37% (186 of 504) in feces from one farm (33) and ranged between 3 and 22% for seven other farms tested (9). Frequencies this high could challenge the Salmonella reduction practices at slaughter and could present an appreciable risk to public health.
RV10 was used as an enrichment medium for all samples in this study as Rappaport-Vassiliadis broths are optimal for isolating Salmonella from swine feces (1, 15, 39). The RV10 enrichment method (i.e., no preenrichment) did not perform as well as the other two methods that used a preenrichment step and subsequent RV10 enrichment. While the RV10 enrichment method may not be an ideal method for the isolation of some Salmonella species from feces, Salmonella could be detected by the PCR method after enrichment in RV10. As reported by Stone et al. (37), a Rappaport-Vassiliadis enrichment broth inhibited the detection of Salmonella by the PCR, whereas in the present study RV10 was not inhibitory to PCR amplification of DNA. Within the RV10 enrichment group, cultivation detected only 44% (11 of 25) of the positive samples even though detection of Salmonella in fecal samples at this finishing farm was 74% (25 of 34). Our ability to detect Salmonella using the PCR was independent of the enrichment medium and was most likely due to the DNA extraction technique used.
Unfortunately, the experimental design of this study did not permit comparisons among the different enrichment techniques, so we do not know if the addition of a preenrichment step would have improved the isolation of Salmonella in the RV10 enrichment group. Since the three enrichment methods used in this study were not directly compared, we also do not know if the enrichment protocol used for a sample affected the recovery of specific Salmonella serotypes. However, some media may be better than others for the recovery of Salmonella. Further studies to compare the efficacies of selective preenrichment, nonselective preenrichment, and no preenrichment using the same sample are warranted.
The kappa statistic was used to evaluate the agreement of results between bacteriology-and PCR-based methods. When a preenrichment was used (i.e., TSB or 3MC broth), agreement between bacteriology-and PCR-based methods was higher than the agreement between bacteriology-and PCR-based methods from the RV10 enrichment method. According to the kappa statistic, the PCR-hybridization approach was as effective as cultivation for detecting Salmonella for the TSB-RV10 and 3MC-RV10 enrichment groups. These results indicate that enrichments in TSB-RV10 and 3MC-RV10 were equivalent to the PCR-hybridization procedure, while enrichment in RV10 was not optimal. In addition, the PCR-based detection may have outperformed cultivation with respect to the RV10 enrichment group due to a high number of nonviable Salmonella bacteria. It can be argued that bacterial cells in field samples need a resuscitating step, as they can be damaged. These damaged cells may not grow or may die in selective media. Likewise, samples containing only dead cells that are diluted into a synthetic medium must have a final concentration of 4 × 102 cells/ml to have one cell/2.5 μl, the volume used in the PCR. Therefore, it is highly unlikely that the PCR-hybridization method detected dead cells in this study.
While used in this study to compare PCR- and bacteriology-based assays, the kappa statistic has some disadvantages. The kappa statistic measures agreement of negative results as well as positive results. As such, a high kappa value, such as 90%, could indicate that the compared tests are equivalent in missing a positive sample. Since the samples used in this study were from farms, we do not know if both bacteriology-and PCR-based assays missed any Salmonella-positive samples. Another weakness of the kappa statistic pertains to the cutoff value that should be used to distinguish between poor and good agreement. The kappa statistic cutoff values are strictly arbitrary and vary by source (8, 17, 27, 35). In spite of these weaknesses, the kappa statistic was used because the presence or absence of Salmonella in the samples was unknown, so a measure of agreement, not accuracy, between the two tests was required. When using the kappa statistic to evaluate the agreement between the two methods, the researcher must determine what is acceptable, particularly in the range between 0.3 and 0.7, as the scientist may reasonably find 0.3, as well as a higher value such as 0.60, to be unacceptable.
Salmonellosis in swine is an economic concern to the swine producer and a human health risk, and a reliable and rapid technique for Salmonella detection would be useful. Historically, the PCR has been used as a confirmatory test following bacterial isolation. We provide evidence here that the PCR could be used in combination with cultivation to improve Salmonella detection. For the synthetic medium combinations used in this study, the PCR worked as well or better at delineating positive samples. When an isolate is required, the PCR could be used as a presumptive or screening test. The organism could then be cultivated from the enrichment broth used for the PCR or from the original sample. However, the observation that the PCR detected only 72% (34 of 47) of those samples which cultivation identified as positive indicates that additional improvements are warranted before PCR replaces cultivation as the gold standard for detection of Salmonella from swine.
ACKNOWLEDGMENTS
This research was supported by the Kansas Agricultural Experiment Station through its funding of the NC-62 Committee “Enteric Diseases of Swine and Cattle: Prevention, Control, and Food Safety,” the Department of Diagnostic Medicine and Pathobiology, and the Recombinant DNA Laboratory of the Food Animal Health and Management Center, Kansas State University (KSU), Manhattan.
We thank Linda Cox (KSU) for technical support, George Stewart (KSU) and Doreene Hyatt (KSU) for reviewing early drafts of the manuscript, and Eileen Schofield (KSU) and James Smith (USDA, Agricultural Research Center, Eastern Regional Research Center) for editing the manuscript. We also thank the National Veterinary Diagnostic Laboratory for the confirmation and identification to species and serovar level of the Salmonella isolates.
Footnotes
Contribution 00-238-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Bager F, Peterson J. Sensitivity and specificity of different methods for the isolation of Salmonella from pigs. Acta Vet Scand. 1991;32:473–481. doi: 10.1186/BF03546947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blessman B H, Morse E V, Midla D A, Swaminathan B. The 24th Annual Proceedings of the American Association of Veterinary Laboratory Diagnosticians, St. Louis, Mo. 1981. Culture and identification of Salmonella typhisuis and related serotypes; pp. 1–10. [Google Scholar]
- 3.Borch E, Nesbakken T, Christensen H. Hazard identification in swine slaughter with respect to foodborne bacteria. Int J Food Microbiol. 1996;30:9–25. doi: 10.1016/0168-1605(96)00988-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Fraser A D, Yamazaki H. Evaluation of the toxicity of Salmonella selective media for shortening the enrichment period. Int J Food Microbiol. 1993;18:151–159. doi: 10.1016/0168-1605(93)90219-7. [DOI] [PubMed] [Google Scholar]
- 5.Chiu C-H, Ou J T. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture-multiplex PCR combination assay. J Clin Microbiol. 1996;34:2619–2622. doi: 10.1128/jcm.34.10.2619-2622.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark C W, Ordal Z J. Thermal injury and recovery of Salmonella typhimurium and its effect on enumeration procedures. Appl Microbiol. 1969;18:332–336. doi: 10.1128/am.18.3.332-336.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen N D, Martin L J, Simpson R B, Wallis D E, Neibergs H L. Comparison of polymerase chain reaction and microbiological culture for detection of salmonellae in equine feces and environmental samples. Am J Vet Res. 1996;57:780–786. [PubMed] [Google Scholar]
- 8.Cyr L, Francis K. Measures of clinical agreement for nominal and categorical data: the kappa coefficient. Comput Biol Med. 1992;22:239–246. doi: 10.1016/0010-4825(92)90063-s. [DOI] [PubMed] [Google Scholar]
- 9.Davies P R, Bovee F G, Funk J A, Morrow W E, Jones F T, Deen J. Isolation of Salmonella serotypes from feces of pigs raised in a multiple-site system. J Am Vet Med Assoc. 1998;212:1925–1929. [PubMed] [Google Scholar]
- 10.Davies P R, Morrow W E, Jones F T, Deen J, Fedorka-Cray P J, Harris I T. Prevalence of Salmonella in finishing swine raised in different production systems in North Carolina, USA. Epidemiol Infect. 1997;119:237–244. doi: 10.1017/s095026889700784x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Smedt J M, Bolderdijk R F. Dynamics of Salmonella isolation with modified semi-solid Rappaport-Vassiliadis medium. J Food Prot. 1987;50:658–661. doi: 10.4315/0362-028X-50.8.658. [DOI] [PubMed] [Google Scholar]
- 12.De Smedt J M, Bolderdijk R F, Rappold H, Lautenschlaeger D. Rapid Salmonella detection in foods by motility enrichment on a modified semi-solid Rappaport-Vassiliadis medium. J Food Prot. 1986;49:510–514. doi: 10.4315/0362-028X-49.7.510. [DOI] [PubMed] [Google Scholar]
- 13.Dusch H, Altwegg M. Evaluation of five new plating media for isolation of Salmonella species. J Clin Microbiol. 1995;33:802–804. doi: 10.1128/jcm.33.4.802-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder R O, Duhamel G E, Mathiesen M R, Erickson E D, Gebhart C J, Oberst R D. Multiplex polymerase chain reaction for simultaneous detection of Lawsonia intracellularis, Serpulina hyodysenteriae, and salmonellae in porcine intestinal specimens. J Vet Diagn Investig. 1997;9:281–286. doi: 10.1177/104063879700900309. [DOI] [PubMed] [Google Scholar]
- 15.Feder I, Nietfeld J C, Kelly B, Butine M D, McNamara P, Chengappa M M. Evaluation of enrichment techniques for the isolation of Salmonella choleraesuis from swine feces. J Microbiol Methods. 1998;33:143–151. [Google Scholar]
- 16.Ferris K E, Miller D A. The Proceedings of the 101st Annual Meeting of the United States Animal Health Association, Louisville, Ky. 1997. Salmonella serotypes from animals and related sources reported during July 1996-June 1997; pp. 419–443. [Google Scholar]
- 17.Fleis J L. Statistical methods for rates and proportions. 2nd ed. New York, N.Y: John Wiley and Sons; 1981. p. 218. [Google Scholar]
- 18.Goossens H, Wauters G, DeBoeck M, Janssens M, Butzler J-P. Semisolid selective-motility enrichment medium for isolation of salmonellae from fecal specimens. J Clin Microbiol. 1984;19:940–941. doi: 10.1128/jcm.19.6.940-941.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray J T, Fedorka-Cray P J, Stabel T J, Kramer T T. Natural transmission of Salmonella choleraesuis in swine. Appl Environ Microbiol. 1996;62:141–146. doi: 10.1128/aem.62.1.141-146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenwood D E, Swaminathan B, Morse E V. Two selective enrichment media for the isolation of Salmonella from mechanically deboned poultry meat. J Food Sci. 1980;45:1131–1135. [Google Scholar]
- 21.Jameson J E. A discussion of the dynamics of Salmonella enrichment. J Hyg. 1962;60:193–205. doi: 10.1017/s0022172400039462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lantz P-G, Matsson M, Wadström T, Rådström P. Removal of PCR inhibitors from human faecal samples through the use of an aqueous two-phase system for sample preparation prior to PCR. J Microbiol Methods. 1997;28:159–167. [Google Scholar]
- 23.Lou Q, Chong S K F, Fitzgerald J F, Siders J A, Allen S D, Lee C-H. Rapid and effective method for preparation of fecal specimens for PCR assays. J Clin Microbiol. 1997;35:281–283. doi: 10.1128/jcm.35.1.281-283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madigan M T, Martinko J M, Parker J, editors. Brock biology of microorganisms. 8th ed. Upper Saddle River, N.J: Prentice-Hall, Inc.; 1997. Host-parasite relationships; pp. 789–817. [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 26.Martin S W. The evaluation of tests. Can J Comp Med. 1977;41:19–25. [PMC free article] [PubMed] [Google Scholar]
- 27.Martin S W, Bonnett B. Clinical epidemiology. Can Vet J. 1987;28:318–325. [PMC free article] [PubMed] [Google Scholar]
- 28.Morse E V, Midla D A, Blessman B H. The 25th Annual Proceedings of the American Association of Veterinary Laboratory Diagnosticians, Nashville, Tenn. 1982. Survival of Salmonella spp. under natural simulated conditions in the swine environment; pp. 99–114. [Google Scholar]
- 29.Nietfeld J C, Feder I, Kramer T T, Schoneweis D, Chengappa M M. Preventing Salmonella infection in pigs with offsite weaning. Swine Health Prod. 1998;6:27–32. [Google Scholar]
- 30.Nietfeld J C, Kelly B, Dritz S S, Feder I, Galland J C. Comparison of conventional and delayed secondary enrichment for isolation of Salmonella spp. from swine samples. J Vet Diagn Investig. 1998;10:285–287. doi: 10.1177/104063879801000312. [DOI] [PubMed] [Google Scholar]
- 31.Nuovo G J. PCR in situ hybridization, protocols and applications. 2nd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 6–99. [Google Scholar]
- 32.Oberst R D, Hays M P, Bohra L K, Phebus R K, Yamashiro C T, Paszko-Kolva C, Flood S J, Sargeant J M, Gillespie J R. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Carroll J M, Davies P R, Correa M T, Slenning B D. Effects of sample storage and delayed secondary enrichment on detection of Salmonella spp. in swine feces. Am J Vet Res. 1999;60:359–362. [PubMed] [Google Scholar]
- 34.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington, D.C.: American Society for Microbiology; 1994. pp. 229–243. [Google Scholar]
- 35.Seigel D G, Podgor M J, Remaley N A. Acceptable values of kappa for comparison of two groups. Am J Epidemiol. 1992;135:571–578. doi: 10.1093/oxfordjournals.aje.a116324. [DOI] [PubMed] [Google Scholar]
- 36.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 37.Stone G G, Oberst R D, Hays M P, McVey S, Chengappa M M. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. J Clin Microbiol. 1994;32:1742–1749. doi: 10.1128/jcm.32.7.1742-1749.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stone G G, Oberst R D, Hays M P, McVey S, Galland J C, Curtiss III R, Kelly S M, Chengappa M M. Detection of Salmonella typhimurium from rectal swabs of experimentally infected beagles by short cultivation and PCR-hybridization. J Clin Microbiol. 1995;33:1292–1295. doi: 10.1128/jcm.33.5.1292-1295.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vassiliadis P. The Rappaport-Vassiliadis (RV) enrichment medium for the isolation of salmonellas: an overview. J Appl Bacteriol. 1983;54:69–76. doi: 10.1111/j.1365-2672.1983.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 40.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilcock B P, Schwartz K J. Salmonellosis. In: Leman A D, Straw B E, Mengeling W E, D'Allaire S, Taylor D J, editors. Diseases of swine. 7th ed. Ames: Iowa State University Press; 1992. pp. 570–583. [Google Scholar]
- 42.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J Clin Microbiol. 1990;28:1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zingg B C, LeFebvre R B. Polymerase chain reaction for detection of Borrelia coriaceae, putative agent of epizootic bovine abortion. Am J Vet Res. 1994;55:1509–1515. [PubMed] [Google Scholar]