Abstract
This study aims to elucidate the variations in spatiotemporal patterns and sociodemographic determinants of SARS-CoV-2 infections in Helsinki, Finland. Global and local spatial autocorrelation were inspected with Moran's I and LISA statistics, and Getis-Ord Gi* statistics was used to identify the hot spot areas. Space-time statistics were used to detect clusters of high relative risk and regression models were implemented to explain sociodemographic determinants for the clusters. The findings revealed the presence of spatial autocorrelation and clustering of COVID-19 cases. High–high clusters and high relative risk areas emerged primarily in Helsinki's eastern neighborhoods, which are socioeconomically vulnerable, with a few exceptions revealing local outbreaks in other areas. The variation in COVID-19 rates was largely explained by median income and the number of foreign citizens in the population. Furthermore, the use of multiple spatiotemporal analysis methods are recommended to gain deeper insights into the complex spatiotemporal clustering patterns and sociodemographic determinants of the COVID-19 cases.
Keywords: SARS-CoV-2, COVID-19, Space-time clusters, Spatial autocorrelation (Moran's I, LISA), Regression
1. Introduction
Coronavirus disease (COVID-19), caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), was first identified on 31 December 2019 in the Wuhan prefecture in the Hubei Province of China (WHO, 2020a), where the first cases were linked to the Huanan Seafood Wholesale Market (Hui et al., 2020). However, the origin of the pandemic has not yet been determined. On 11 March 2020, the World Health Organization (WHO) declared COVID-19 a pandemic (WHO, 2020b). In Finland, the first coronavirus case was diagnosed already on 29 January 2020 in Lapland (Haveri et al., 2020), while the first positive case in the City of Helsinki was diagnosed on February 25th (Jarva et al., 2021). Thereafter, the disease spread quickly after winter holiday travelers to Austria, Italy and Spain returned to Finland (Nguyen et al., 2021). In April and early May, the daily number of new cases remained relatively high (5.6% of tested individuals were positive) in the Greater Helsinki area (Jarva et al., 2021). In June and July, the COVID-19 epidemic eased temporarily in Finland and infections dropped steadily. In autumn 2020, a total number of new COVID-19 cases was on the rise and the second wave of coronavirus hit Finland, mainly seeded by new imported SARS-CoV-2 strains (Nguyen et al., 2021). The incidence of the second wave peaked in early December, and after a brief descent, started to rise again in mid-January. This heralded the beginning of a third wave driven by the more transmissible alpha (B.1.1.7) and beta (B.1.351) SARS-CoV-2 variants (Kant et al., 2021), which peaked in late March. This study focuses on the second and early third wave of the COVID-19 epidemic in the City of Helsinki from 28th October 2020 to 24th March 2021.
Since the outbreak of the COVID-19 pandemic in early 2020, there has been growing interest in spatial modeling of COVID-19 to understand spatial patterns and spatiotemporal dimensions of the disease spread (Fatima et al., 2021). Clustering, hot spot analysis, space-time scan statistics, and regression modeling have been the most commonly used spatial methods (Franch-Pardo et al., 2020). Global Moran's I and Local Indicators of Spatial Association (LISA) statistics have been used in previous COVID-19 studies to explore spatial epidemic dynamics of the virus (Dutta et al., 2021; Kang et al., 2020), and to examine spatial patterns of COVID-19 incidence cases and death rates (Cavalcante and Abreu, 2020, Kim et al., 2021). According to Fatima et al. (2021), there has been less attention to analyze COVID-19 hot spots with Getis-Ord Gi* statistics (see e.g. Das et al., 2021; Lakhani, 2020; Mollalo et al., 2020). Previous COVID-19 studies, to the best of our knowledge, did not include a comparison of LISA and Getis-Ord Gi* statistics.
Space-time scan statistic (Kulldorff, 1997) has been used to analyze space-time clusters and risk areas of COVID-19 in a number of studies (Andrade et al., 2020; Cordes and Castro, 2020; Desjardins et al., 2020), which mainly employed the prospective Poisson space–time scan statistic method. However, when using a large maximum scanning window in this method, a large cluster can hide several smaller distinct clusters (Han et al., 2016). To overcome this problem, Gini Optimization parameter was introduced in SaTScan software (Kulldorff, 2021) but it is not yet widely utilized.
Traditional OLS regression methods and spatial regression models have been used to understand the contribution of socioeconomic, demographic and environmental determinants to explain spatial variability of COVID-19 incidences and mortality in the epidemic (Mansour et al., 2021; Maiti et al., 2021; Mena et al., 2021; Middya and Roy 2021; Mollalo et al., 2020; Zhang and Schwartz, 2020; Snyder and Parks, 2020). However, in the majority of these studies, only one regression-based method was employed. Furthermore, most of the earlier geospatial COVID-19 studies have been conducted at global-continental or country scale (McKenzie and Adams, 2020; Melin et al., 2020; Moonsammy et al., 2021; Pourghasemi et al., 2020; Sannigrahi et al., 2020; Sobral et al., 2020) or at county/municipality level (Han et al., 2021, Liu et al., 2021; Imdad et al., 2021; Martines et al., 2021; Rahman et al., 2021; Sun et al., 2020; Sun et al., 2021). Less research has been carried out at postal/ZIP code level (Cordes and Castro, 2020; DiMaggio et al., 2020, Kim et al., 2021). Furthermore, the majority of previous studies have investigated COVID-19 with only one temporal time slot. This study represents a one-of-a-kind effort to contribute to the existing geospatial COVID-19 research by analysing second and early third wave COVID-19 cases with multitemporal postal code level data using GIS and spatial modeling methods.
1.1. Purpose of the study
This study aims to identify and map the significant clusters of COVID-19 cases and significantly elevated disease risk areas in Helsinki, Finland. As recommended in previous studies, we used the prospective Poisson space–time scan statistic. Besides that, there is a need to investigate the Poisson space-time scan approach using Gini Optimization further, so we tested Gini Optimized cluster detection with retrospective purely spatial Poisson scan statistics. Furthermore, the goal of this research is to determine sociodemographic factors that influence the spread of COVID-19. We employed three different regression methods: Ordinary Least Squares (OLS) (Ward and Gleditsch, 2018), Geographically Weighted Regression (GWR) (Brunsdon et al., 1996), and Multiscale Geographically Weighted Regression (MGWR) (Fotheringham et al., 2017) with sociodemographic determinants to explain spatial variability of COVID-19 infection rates.
This is the first comprehensive spatial analysis study of COVID-19 (SARS-CoV-2) infections in Finland, and it provides new insights into the disease's spatial spread, temporal trends and sociodemographic correlates. The findings could help public health services better target intervention locations and control disease spread. The spatiotemporal analysis methods presented here could be used to investigate and provide information to improve management and control of the ongoing COVID-19 crisis and future pandemics in other parts of the world.
2. Materials
2.1. Study area and COVID-19 infection data
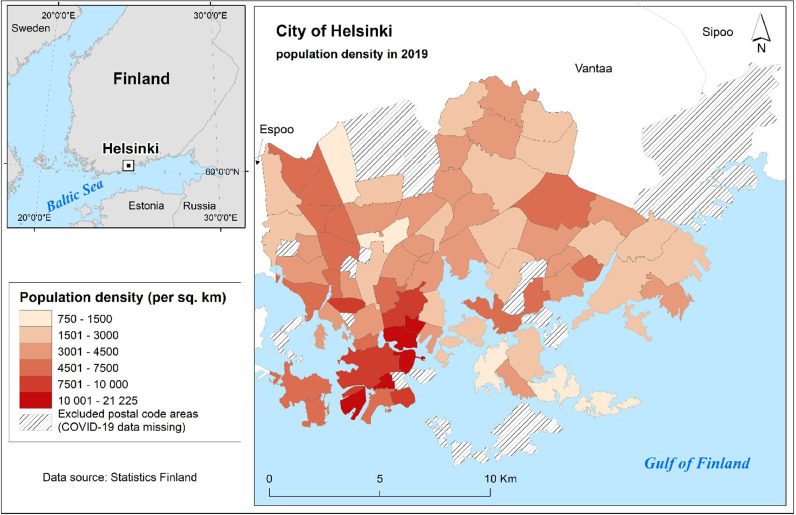
Finland is situated in northern Europe, bordering Sweden, Norway and Russia. Helsinki, the capital of Finland, is located in the southern part of the country on the shore of the Gulf of Finland (Fig. 1 ). Helsinki is the most densely populated city in Finland (2934 people/km2), with 653 835 inhabitants (31 December 2019) of whom 9.6% are of foreign origin (City of Helsinki, 2020a). The City of Helsinki is officially divided into 60 neighborhoods, and these neighborhoods have altogether 84 postal code areas (Fig. 1).
Fig. 1.
Study area showing the population density (per sq. km) within 69 postal code areas in the City of Helsinki.
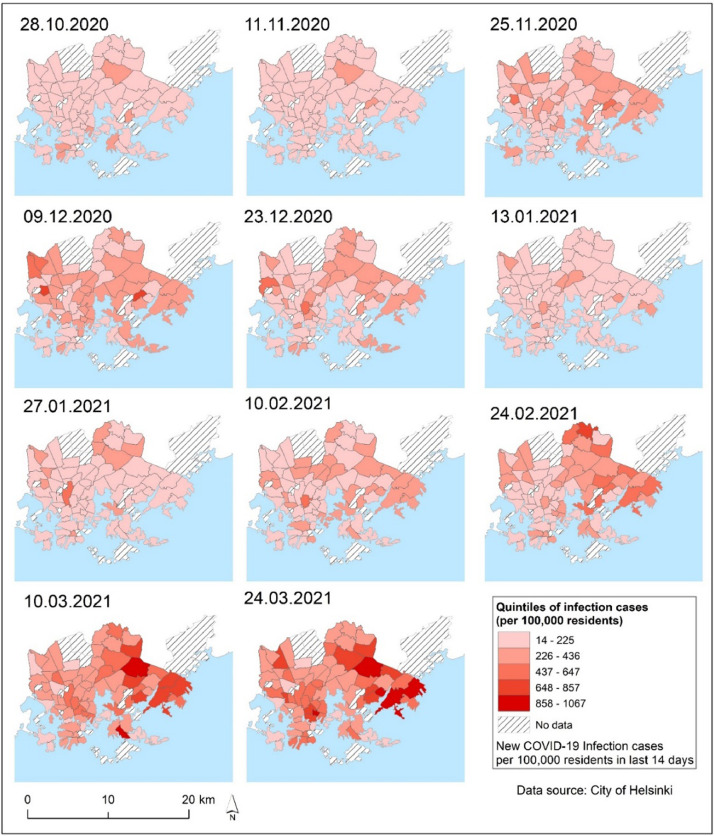
The City of Helsinki provided the dataset of COVID-19 infections at the postal code level. The data are publicly available online and include information on new COVID-19 infections and new cases per 100,000 residents at approximately 14-day intervals (14-day notification rate) (City of Helsinki, 2020b). This study used data from 28th October 2020 to 24th March 2021. During this period, a total of 21,668 COVID-19 infections were diagnosed in Helsinki comprising 29.5% of all cases in Finland during the period ((City of Helsinki, 2020b). There were 69 postal code areas in this study for which COVID-19 data were available for 11 time periods (Fig. 2 ). Due to sensitivity issues, data for 15 postal code areas were missing (less than five COVID-19 infection cases in postal code areas cannot be published) (City of Helsinki, 2020b).
Fig. 2.
Spatial distribution patterns of the new COVID-19 infection cases per 100,000 residents in postal code areas for each previous 14-day time period (14-day notification rate).
2.2. Sociodemographic data for regression analysis
Statistics Finland (2020) provided sociodemographic data, and the Helsinki Region Environmental Services SeutuData'19 database provided data on foreign citizens (HSY, 2019). Based on previous COVID-19 studies (Das et al., 2021; DiMaggio et al., 2020; Li et al., 2021, Liu et al., 2021; Mansour et al., 2021; Mollalo et al., 2020), seven potential predictor variables for regression analyses were chosen as outlined in Table 1 .
Table 1.
Spatiotemporal dataset including dependent COVID-19 infections per 100,000 residents variables and seven sociodemographic predictors.
| Variable | Description | Source |
| Dependent | ||
| Case028 to Case0324 | Number of new COVID-19 infections per 100,000 residents in postal code areas during the previous 14 days (28.10.2020–24.3.2021) | City of Helsinki |
| Predictors | ||
| ulkoka19 | Number of foreign citizens in residential building aggregated to postal code areas | SeutuData'19 - HSY |
| ko_perus | Basic level studies, 2018 – no qualification after basic level or qualification unknown | Statistics Finland |
| hr_mtu | Median income of inhabitants, 2017 | Statistics Finland |
| hr_pi_tul | Inhabitants belonging to the lowest income category, 2017 | Statistics Finland |
| tr_pi_tul | Households belonging to the lowest income category, 2017 | Statistics Finland |
| pt_tyott | Unemployed, 2017 – (people aged 15–64 years) | Statistics Finland |
| pt_elakel | Pensioners, 2017 | Statistics Finland |
3. Methods
3.1. Spatial association of COVID-19 infections in the City of Helsinki
First, we used global Moran's I to test the spatial independence of COVID-19 infection cases per 100,000 residents in Helsinki (Moran, 1950). Global Moran's I computes the degree of spatial autocorrelation across the entire study area. Moran's I values range from −1 to +1, with a positive Moran's I value indicating similar value clustering, a negative Moran's I value indicating dissimilar value clustering, and a value of 0 indicating random distribution.
Second, two analyses of local spatial autocorrelation were performed: the Local Indicators of Spatial Association (LISA) (Anselin, 1995) and the Getis-Ord Gi* statistic (Getis and Ord, 1992). LISA analysis was used to detect statistically significant local spatial clusters with high values high–high (H–H) or low values low–low (L–L), as well as spatial outliers with high–low (H–L) or low–high (L–H). The cluster type distinguishes a statistically significant cluster of high values (H–H), a statistically significant cluster of low values (L–L), an outlier in which a high value is surrounded primarily by low values (H–L), and an outlier in which a low value is surrounded primarily by high values (L–H). The Getis-Ord Gi* statistic was used to determine the geographic distribution of potential COVID-19 infection hot spots (high values) and cold spots (low values) per 100,000 residents.
GeoDa (version 1.18.0) was used to compute global Moran's I and LISA statistics (Anselin et al., 2010). A Monte Carlo simulation of 999 random iterations yielded a pseudo p-value and z-score of the global Moran's I for each variable. ArcGIS Desktop (version 10.8) was used to compute Getis-Ord Gi* hot spot statistics (ESRI, 2020). Supplementary Materials 1 contains the statistics equations for Moran's I, LISA, and Getis-Ord Gi* hot spots.
3.2. Space-time clustering patterns and epidemic curve
The space-time patterns of COVID-19 infections in Helsinki were examined in SaTScan software (version 10.0) to detect significant (p-value<0.05) space-time clusters using the prospective Poisson space-time scan statistic and the retrospective purely spatial Poisson scan statistic (Kulldorff, 2021). First, we ran purely spatial scan statistics for three time periods: 28.10.2020–9.12.2020, 23.12.2020–10.2.2021, and 24.2.2021–24.3.2021. We computed pure spatial scan statistics with and without Gini optimization to see if there were any 'Gini clusters' in the data. Then, for the same time periods, we used prospective Poisson space-time scan statistics to detect active and emerging clusters. We restricted the spatial and temporal scanning windows to include ≤20% of the population at-risk and ≤50% of the study period. In addition, each candidate must contain at least 5 infection cases at a minimum duration of 2 days (Desjardins et al., 2020; Hohl et al., 2020). To avoid detecting extremely large clusters, larger spatial and temporal windows were not chosen (Desjardins et al., 2020). To determine the statistical significance of space-time clusters, we used Monte Carlo testing with 999 simulations. The relative risk maps for COVID-19 infections were then reported and visualized for three different time periods, presenting spatiotemporal variation of significant (p-value<0.05) relative risk (RR) clusters with significantly higher observed cases compared to expected, and relative risk values for each postal code area. In addition, an epidemic curve for COVID-19 infections in seven major districts of Helsinki was created.
3.3. Regression modeling
For each of the 14-day datasets, the ArcGIS Exploratory Regression data-mining tool was used prior to regression modeling. All possible combinations of the seven sociodemographic input candidate explanatory variables for regression models were evaluated by the tool. The tool looks for the best ordinary least squares (OLS) models that meet certain criteria. Refer to Supplementary Materials 1 for more information. Variable combinations with the lowest corrected Akaike Information Criterion (AICc) (Akaike, 1974) value were chosen for regression analyses for each model. To identify the significant sociodemographic determinants of COVID-19 infection rates, we used three regression methods: OLS, GWR, and MGWR. Their detailed discussions can be found elsewhere, for example (Brunsdon et al., 1996; Fotheringham and Oshan, 2016; Ward and Gleditsch, 2018; Oshan et al., 2019) and their equations are presented in Supplementary Materials 1.
We may assume that COVID-19 infections are spatially autocorrelated, which violates the implicit assumptions of OLS. In order to allow parameters varying spatially, we used GWR, which calculates regression coefficients for each individual data entity (in this case, postal code areas) separately rather of estimating global values for regression parameters (Fotheringham and Oshan, 2016). However, GWR assumes that the scale of relationships remains constant across space, which may not be the case in spatial models. To overcome this implicit assumption, we applied MGWR (Fotheringham et al., 2017; Yu et al., 2020). For comparison, all regression models (OLS, GWR, and MGWR) were implemented with the same variables in the MGWR 2.2 software (Oshan et al., 2019). To compare the model performances, the adjusted R2 and AICc were used. A higher adjusted R2 value and a lower AICc value indicated the best model fit. Moran's I was used to see if there was any significant spatial autocorrelation in the regression model residuals.
4. Results
4.1. Global spatial autocorrelation
This study examined whether a spatial association occurred in the new infection cases of COVID-19 per 100,000 residents in the City of Helsinki. Results, presented in Table 2 , indicate that positive spatial autocorrelation was detected during the whole study period from 28.10.2020 to 24.3.2021 (Moran's I = 0.1393, p<0.029). Positive spatial autocorrelation, on the other hand, was relatively low for the majority of the time periods. Global Moran's I statistic reveals that the distribution of COVID-19 infections is not random for most of the studied periods, and spatial clustering was detected. Moran's I statistics with the highest values were 0.3243 for 11.11.2020, 0.2992 for 24.02.2021, 0.2645 for 24.03.201, and 0.1866 for 10.03.2021. Moran's I produced lower values for other dates. Global Moran's I statistics in the City of Helsinki were only moderate at best, indicating spatial heterogeneity of COVID-19 case rates between postal code areas. As a result, it was critical to investigate local spatial autocorrelation in greater depth.
Table 2.
Global Moran's I values with z-values and pseudo p-values of the distribution of COVID-19 infection rates at different time period.
| Time period | Moran's I value | Z-score | pseudo p-value |
| 28.10.2020–24.03.2021 | 0.1393 | 2.0099 | 0.029 |
| 28.10.2020 | 0.0536 | 0.9241 | 0.054 |
| 11.11.2020 | 0.3243 | 4.6738 | 0.001 |
| 25.11.2020 | 0.0206 | 0.4786 | 0.305 |
| 09.12.2020 | 0.0893 | 1.4087 | 0.082 |
| 23.12.2020 | 0.0662 | 1.0172 | 0.148 |
| 13.01.2021 | 0.0377 | 0.7181 | 0.228 |
| 27.01.2021 | 0.0939 | 1.4653 | 0.084 |
| 10.02.2021 | −0.0147 | −0.3024 | 0.395 |
| 24.02.2021 | 0.2992 | 4.0603 | 0.001 |
| 10.03.2021 | 0.1866 | 2.6119 | 0.008 |
| 24.03.2021 | 0.2645 | 3.7255 | 0.001 |
4.2. Local spatial autocorrelation
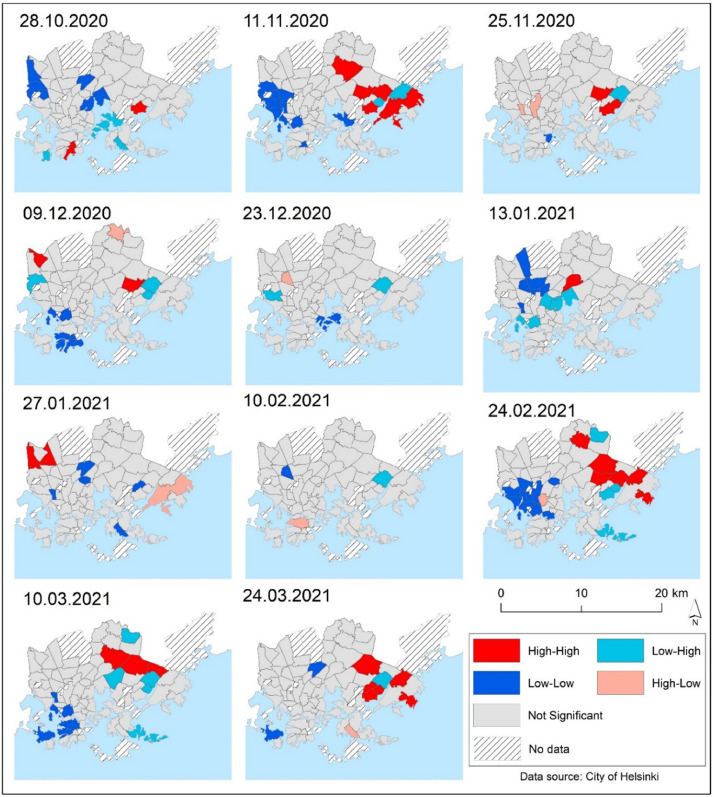
LISA cluster maps were used to indicate the locations of significant spatial clusters and outliers of COVID-19 infection rates. Fig. 3 depicts the postal code areas associated with high–high (H–H), high–low (H–L), low–high (L–H), and low–low (L–L) values of COVID-19 infection rates in the LISA cluster map. Throughout the study period of 28th October 2020 to 24th March 2021 high–high (H–H) clustering areas were mostly found in the eastern parts of the city, while low–low (L–L) clusters were mostly found in the city's western postal code areas (Fig. 3). When the COVID-19 epidemic eased temporarily in Helsinki in December 2020 and January 2021, there were few exceptions to the general pattern. Only few high–high (H–H) clusters emerged as local outbreaks. Many high–high (H–H) clusters were identified in the eastern parts of the city where SARS-CoV-2 infections spread rapidly in late February and early March 2021. Low–low (L–L) clusters were mostly observed in the city's western and southern outskirts. Low–high (L–H) outliers were mostly found near high–high (H–H) clusters, whereas high–low (H–L) outliers were evenly distributed across Helsinki (Fig. 3).
Fig. 3.
Local indicators of spatial autocorrelation of the COVID-19 infection rates at the studied time periods (14-day notification rate). The cluster type distinguishes a statistically significant cluster of high values (H–H), a statistically significant cluster of low values (L–L), an outlier in which a high value is surrounded primarily by low values (H–L), and an outlier in which a low value is surrounded primarily by high values (L–H).
4.3. Hot spot analysis of COVID-19 infections
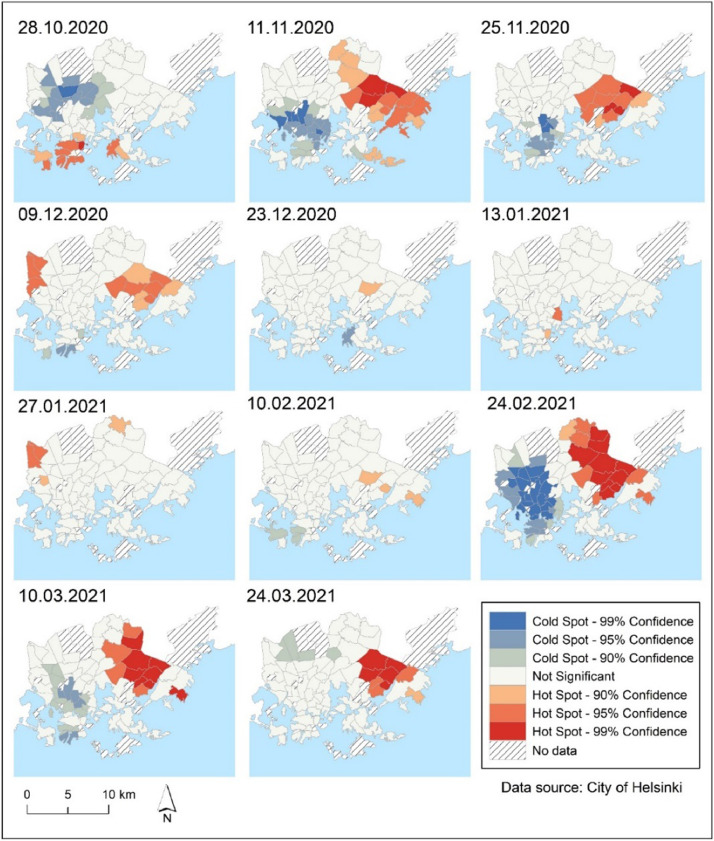
To detect hot spot and cold spot clusters of new COVID-19 cases in Helsinki, Getis-Ord Gi* hot spot analysis was used as an alternative method to LISA. Overall, the patterns were similar to those found with LISA analysis, indicating that the eastern suburbs were hot spots during the majority of the studied periods, while the western parts of Helsinki were cold spots (Fig. 4 ). However, a comparison of the LISA map (Fig 3) and the hot spots map (Fig 4) reveals certain differences in recognition of COVID-19 hot and cold spots that must be considered when interpreting the results.
Fig. 4.
Locations of hot spots and cold spots of COVID-19 infection rates per 100,000 residents in the City of Helsinki during 11 time periods (14-day notification rate) identified by Getis-Ord Gi* statistic analysis.
4.4. Spatial distribution of pure spatial and space-time clusters of COVID-19 and epidemic curve
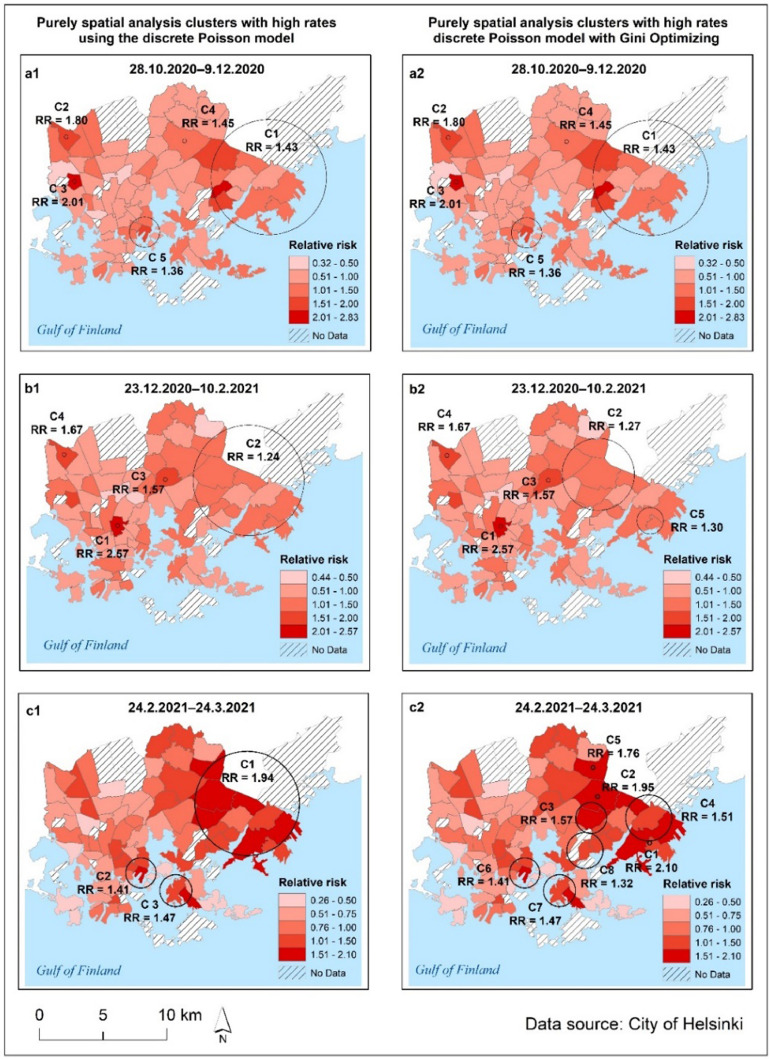
The results of the retrospective pure spatial Poisson scan statistic and the prospective Poisson space–time scan statistic show that the detected COVID-19 clusters varied over time and space in Helsinki (Fig. 5, Fig. 6 ). Tables S1, S2 and S3 in Supplementary Materials 2, provide the characteristics of the statistically significant (p<0.05) pure spatial scan statistic and space-time scan results of the COVID-19 infection rates at the postal code level at three different aggregated time periods, and from October 28th 2020 to March 24th 2021.
Fig. 5.
Patterns of significant pure spatial scan statistic clusters (indicated by black circles with the text C on the maps) of COVID-19 at the postal code level and the relative risk in the City of Helsinki for three aggregated time periods: 28.10.2020–9.12.2020, 23.12.2020–10.2.2021 and 24.2.2021–24.3.2021. Maps on the left present pure spatial scan statistics (a1, b1 and c1) and maps on the right (a2, b2 and c2) present pure spatial scan statistic with “Gini optimized” clusters.
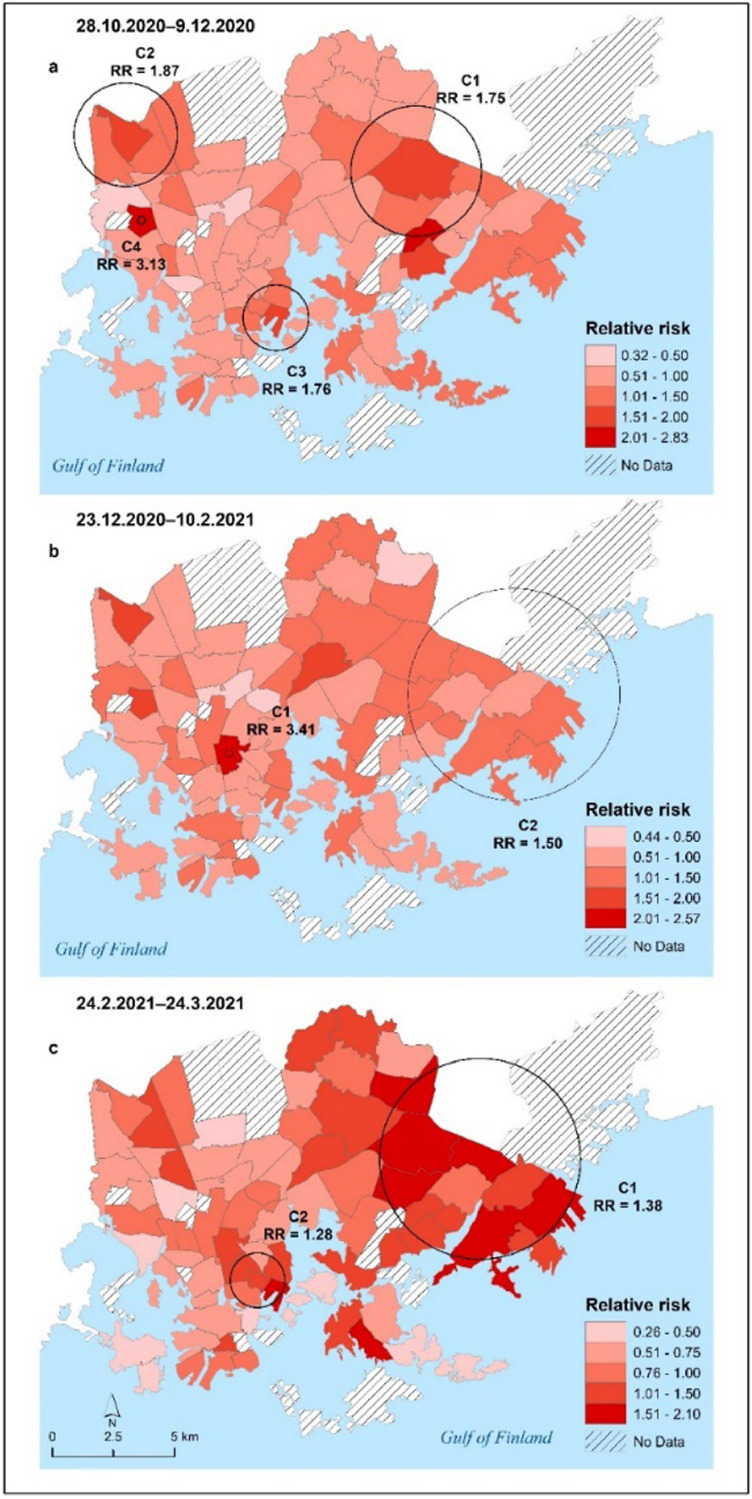
Fig. 6.
Patterns of significant space-time clusters (indicated by black circles with the text C on the maps) of COVID-19 at the postal code level and propagation of COVID-19 relative risk in the City of Helsinki for three aggregated time periods: 28.10.2020–9.12.2020 (map a), 23.12.2020–10.2.2021 (map b) and 24.2.2021–24.3.2021 (map c).
4.4.1. Pure spatial scan statistic results
From late October to mid-December 2020 (28.10.2020–9.12.2020), two clusters (C1, C4) were detected in the eastern parts of Helsinki, two clusters (C2, C3) were identified in northwestern areas and one cluster (C5) was found closer to the city center (Fig. 5 a1–2). A similar pattern was observed in the 23.12.2020–10.2.2021 data (Fig. 5 b1–2), with the main cluster detected in the eastern parts of Helsinki and smaller clusters detected in northwestern areas and in the central parts of Helsinki. The smaller clusters had a high relative risk of COVID-19 infection. It is worth noting that one 'Gini cluster' (C5) was discovered in the eastern suburbs (Fig. 5 b2). The COVID-19 situation deteriorated in February and March 2021, as the number of infections increased rapidly, particularly in the eastern suburbs, where the main cluster with a high relative risk (RR 1.94) was discovered (Fig. 5 c1). Two more clusters (C2 and C3) were detected in the central and southeastern regions (Fig. 5 c1). Furthermore, the eastern suburbs had a high relative risk (RR) and in the eastern parts of Helsinki, many non-overlapping 'Gini clusters' were detected (Fig. 5 c2).
4.4.2. Space-time scan statistic results
According to the results of prospective space-time scan statistics of COVID-19 clustering, the main clusters were active or emerged in the eastern parts of Helsinki, and smaller clusters were detected in other areas, possibly as a result of local outbreaks (Fig. 6). Space-time scan statistics, on the other hand, detected less significant clusters than pure spatial scan statistics. Table S3 in Supplementary Materials 2 describes the characteristics of the statistically significant active and emerging space-time clusters of COVID-19 infection rates at the postal code level over three time periods.
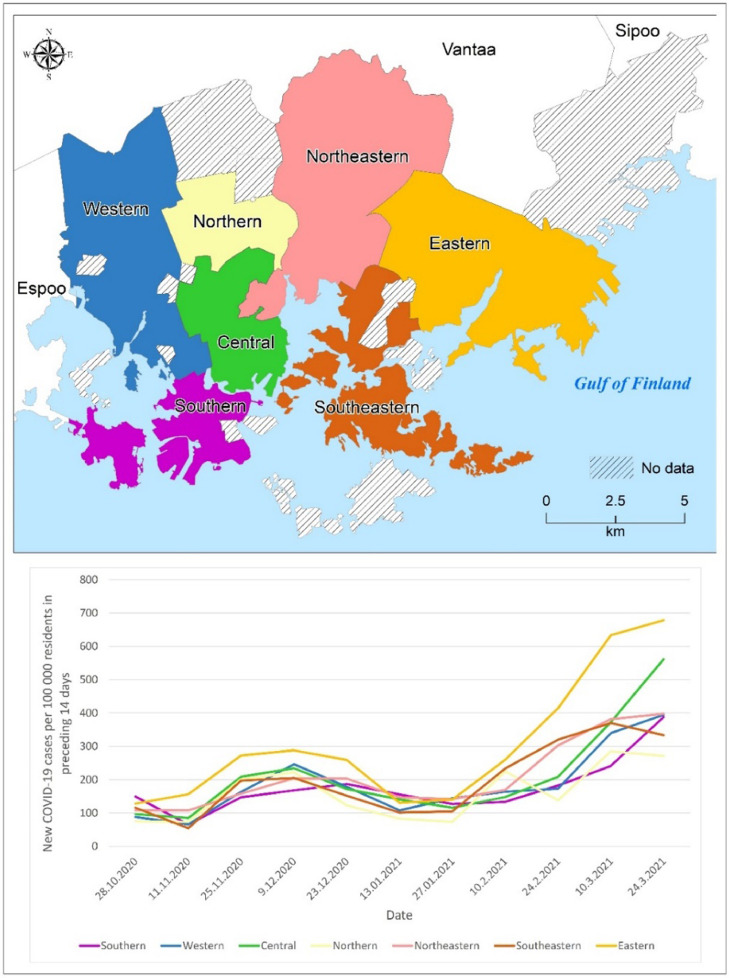
Fig. 7 depicts the epidemic curve of new COVID-19 infection cases per 100,000 Helsinki residents based on major districts. The eastern district had the highest infection rates throughout the study period, while the northern and southeastern districts had the lowest. Infection rates were relatively low in all districts in October 2020, but began to rise in November 2020 and continued to rise in December 2020. The spread of COVID-19 slowed in January 2021, but infection rates began to rise again in February 2021, particularly in the eastern district. Infections peaked in all districts in March 2021.
Fig. 7.
The epidemic curve indicating the new COVID-19 infection cases per 100,000 residents per two weeks period in different major districts of Helsinki.
4.5. Performance of global and local regression models
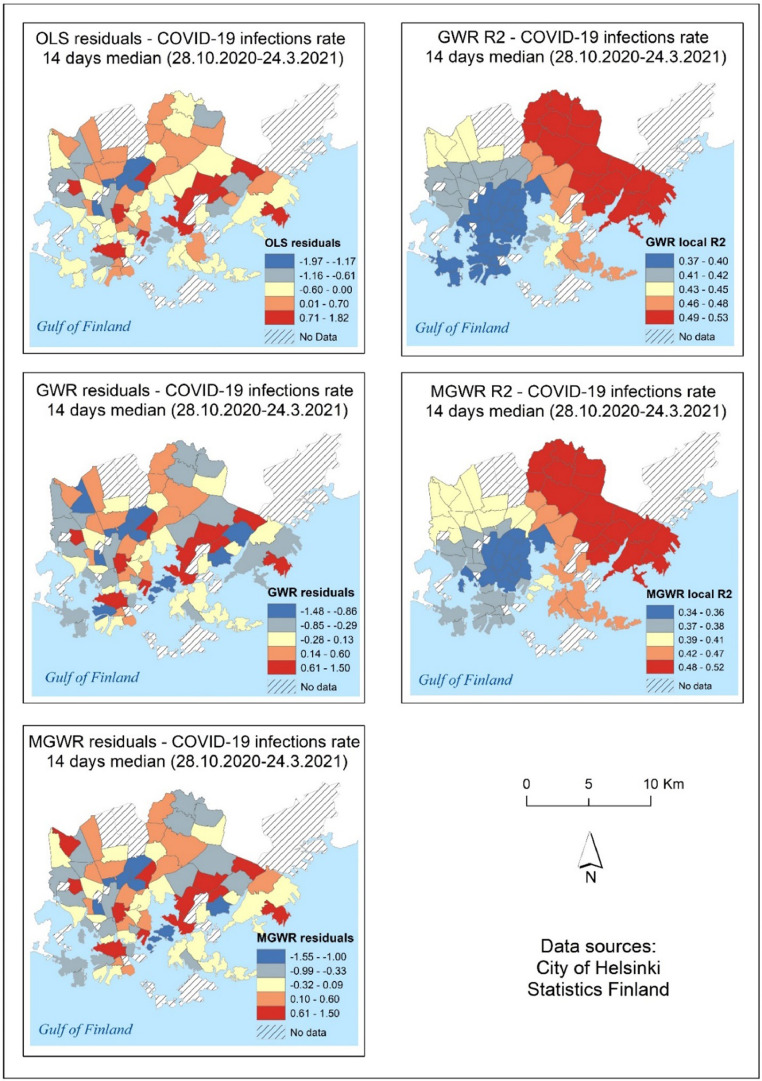
Different regression model methods produced slightly different model-fit results. Models were unable to explain variation in COVID-19 data over many time periods, resulting in very low adjusted R2 values (Table 3 ). Overall, MGWR models outperformed all other models, with the highest adjusted R2 and lowest AICc values. Unexpectedly, at three time periods: 9.12.2020, 23.12.2020, and 13.01.2021, OLS models outperformed MGRW and GWR models. GWR models performed best on two occasions: 10.02.2021 and 24.02.2021. Supplementary Materials 2 shows the regression variables and coefficients of the final multivariate OLS models for COVID-19 (Tables S4 and S5). Supplementary Materials 3 contains the results of all regression modeling (OLS, GWR, and MGWR). According to Supplementary Materials 3, for the entire study period (28.10.2020–24.03.2021), the OLS model with the variables: median income of inhabitants, number of foreign citizens, and pensioners could explain approximately 40% of the variation in COVID-19 infection rates data (adjusted R2=0.401), for GWR adjusted R2 was 0.453, and for MGWR (adjusted R2=0.436). Moran's I statistics were computed in all OLS, GWR, and MGWR regression model residuals, and Moran's I test revealed significant spatial autocorrelation in some of the OLS models. Spatial autocorrelation was absent in all GWR and MGWR regression models. Fig. 8 show the residuals and local R2 values of the OLS, GWR, and MGWR models for COVID-19 infection rate data from 28.10.2020 to 24.3.2021. Generally, local R² values for GWR and MGWR models are high in the eastern part of the study area, decreasing gradually towards the western part of the city.
Table 3.
Measures of goodness-of-fit for OLS, GWR, and MGWR in modeling COVID-19 infection rates in the City of Helsinki. The best-fitting model (highest adjusted R2 value) for each 14 days time period and for 28.10.2020–24.03.2021is bolded.
| OLS | GWR | MGWR | ||||
| Time period | Adjusted R2 | AICc | Adjusted R2 | AICc | Adjusted R2 | AICc |
| 28.10.2020–24.03.2021 | 0.401 | 168.321 | 0.453 | 166.087 | 0.436 | 168.363 |
| 28.10.2020 | 0.162 | 191.454 | 0.211 | 191.738 | 0.238 | 191.348 |
| 11.11.2020 | 0.163 | 190.139 | 0.18 | 190.159 | 0.191 | 189.639 |
| 25.11.2020 | 0.079 | 195.501 | 0.085 | 197.374 | 0.115 | 197.197 |
| 09.12.2020 | 0.183 | 187.184 | 0.17 | 189.864 | 0.171 | 190.019 |
| 23.12.2020 | 0.137 | 192.231 | 0.112 | 196.517 | 0.107 | 197.236 |
| 13.01.2021 | 0.061 | 198.071 | 0.041 | 201.659 | 0.047 | 142.504 |
| 27.01.2021 | 0.047 | 197.854 | 0.067 | 197.935 | 0.087 | 198.254 |
| 10.02.2021 | 0.132 | 193.907 | 0.163 | 196.711 | 0.131 | 197.266 |
| 24.02.2021 | 0.289 | 180.074 | 0.456 | 171.307 | 0.42 | 170.887 |
| 10.03.2021 | 0.548 | 150.263 | 0.592 | 148.309 | 0.615 | 146.05 |
| 24.03.2021 | 0.489 | 157.289 | 0.546 | 156.83 | 0.571 | 151.517 |
Fig. 8.
Spatial distribution of OLS residuals and local R2 of GWR and MGWR models for COVID-19 infection rate data from 28.10.2020 to 24.3.2021.
4.6. Sociodemographic variables explaining variation in COVID-19 infection data
In this study, sociodemographic variables were investigated to determine the best predictors of COVID-19 infections in postal code areas in the City of Helsinki. To identify the best predictors, we used linear regression with the dependent variable being the COVID-19 case median rate for the entire study period of 28.10.2020–24.03.2021 (Table 4 ). Each independent sociodemographic variable was tested separately. Table 4 also shows how frequently each sociodemographic variable was chosen for the OLS regression models out of a total of 11 regression models run for different 14-day time periods to explain the variation in SARS-CoV-2 infection rates data.
Table 4.
Linear regression performance of sociodemographic variables explaining independently the variation in median COVID-19 infection rates data for the time period 28.10.2020–24.03.2021, and the total number of times each variable was selected to the OLS models out of a total of 11 regression models run for different 14-day time periods.
| Predictor | Description | Adjusted R Square | Standard Error | Number of times selected to OLS models |
| hr_mtu | Median income of inhabitants, 2017 | 0.30 | 49.954 | 8 |
| ulkoka19 | Number of foreign citizens in residential building summed to postal code area | 0.23 | 52.255 | 5 |
| ko_perus | Basic level studies, 2018 – no qualification after basic level or qualification unknown | 0.22 | 52.581 | 4 |
| pt_tyott | Unemployed, 2017 – (people aged 15–64 years who were unemployed) | 0.18 | 53.947 | 3 |
| hr_pi_tul | Inhabitants belonging to the lowest income category, 2017 | 0.09 | 56.872 | 2 |
| pt_elakel | Pensioners, 2017 | 0.04 | 58.477 | 2 |
| tr_pi_tul | Households belonging to the lowest income category, 2017 | 0.11 | 56.097 | 1 |
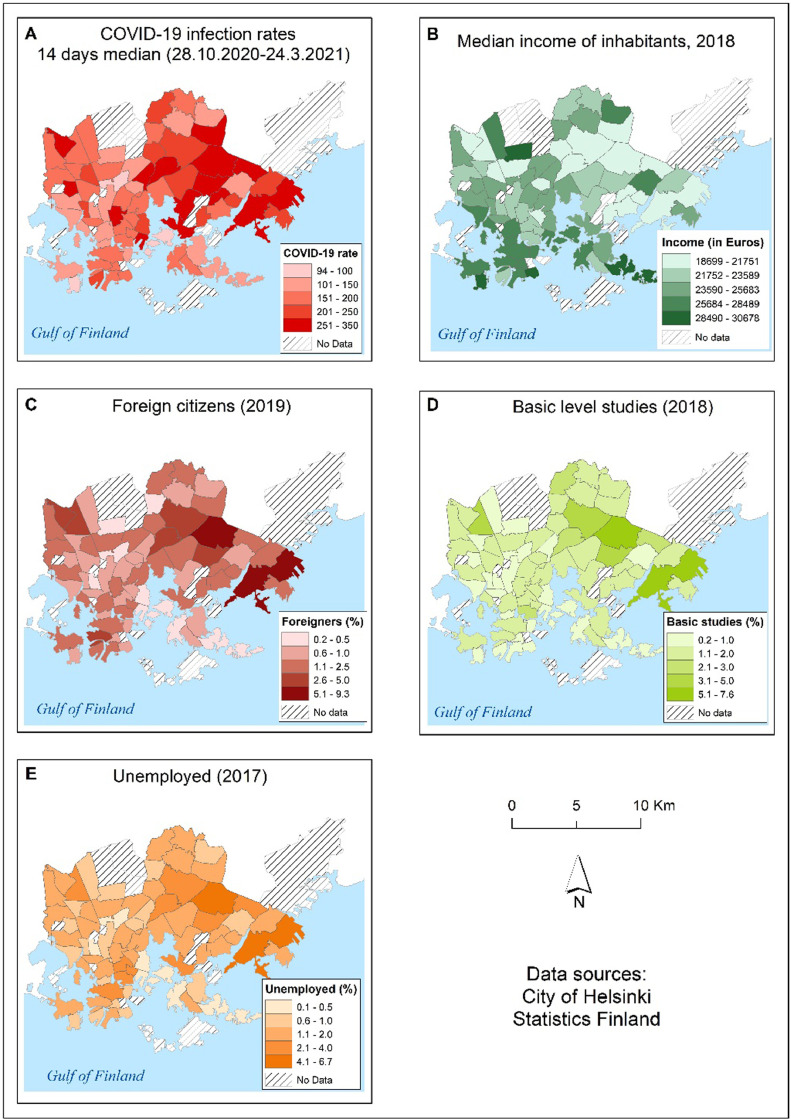
Fig. 9 presents the spatial distribution of median COVID-19 infection rates and sociodemographic variables that best explain the variation in median COVID-19 infection rates in the City of Helsinki from October 28th, 2020 to March 24th, 2021. The maps show that COVID-19 infections are concentrated in areas with a lower median income, a relatively high number of foreign citizens, a low level of education, and a high number of unemployed citizens (Fig. 9).
Fig. 9.
Map presents the distribution of COVID-19 infection rates (14 days median) for time period 28.10.2020–24.03.2021 (A), and the distribution of demographic and socioeconomic variables in the City of Helsinki by postal code area; median income of inhabitants in Euros (B), percentage of foreign citizens (C), percentage of people with only basic education (D) and percentage of unemployed citizens (E).
5. Discussion
Throughout the epidemic, Helsinki has been the COVID-19 disease epicenter in Finland. Approximately 30% of all COVID-19 infections in Finland were diagnosed in Helsinki between the 28th of October 2020 and the 24th of March 2021. This study sought to understand the spatiotemporal clustering patterns and sociodemographic determinants of the second and early third wave of COVID-19 (SARS-CoV-2) infections. We demonstrate a holistic approach to analyze COVID-19 epidemic at local level using four spatial and spatiotemporal techniques; global and local spatial autocorrelation (Moran's I and LISA), Getis-Ord Gi* hot spot analysis, space-time scan statistics, and three regression modeling methods (OLS, GWR and MGWR).
5.1. Global and local spatial autocorrelation of COVID-19 in the City of Helsinki
Global Moran's I analysis revealed that there was a moderate positive spatial autocorrelation (Moran's I = 0.1393, pseudo p-value=0.029) between 28th October 2020 and 24th March 2021, indicating that COVID-19 infection rates were not randomly distributed in Helsinki with clear variations in different time periods (Table 2). The LISA map (Fig. 3) shows high–high COVID-19 clusters in the eastern parts of Helsinki. With a few exceptions, the Getis-Ord Gi* hot spot map yields essentially similar results (Fig 4). Moran's I value was relatively low from late November 2020 to February 2021, with the absence of statistically significant clusters from the majority of Helsinki (Figs. 3 and 4). There were only few significant COVID-19 clusters that could be identified as local outbreaks during that time period. COVID-19 infections began to rise with the third wave of coronavirus in mid-February 2021. Moran's I value was statistically significant from data on February 24, 2021 to the end of the study period. LISA and Getis-Ord Gi* hotspot maps (Figs 3 and 4) shows that COVID-19 infections have strong clusters in eastern part of Helsinki. Previous research has shown that Moran's I is a useful method for understanding the overall spatial dependency of COVID-19 and for presenting reliable information about the disease's spatiotemporal patterns to local health authorities and policymakers (Dutta et al., 2021; Kang et al., 2020, Kim et al., 2021). However, global Moran's I cannot reveal spatial heterogeneity of the studied phenomenon, and local spatial autocorrelation analysis i.e. LISA statistics analysis is recommended to map the local variation of COVID-19-related phenomena. (Dutta et al., 2021, Liu et al., 2021, Sun et al., 2021). We utilized Getis-Ord Gi* hot spot analysis to compare clustering patterns from the LISA statistic. Interestingly, LISA clusters and Getis-Ord Gi* hot spot analysis maps yielded partly different clustering results (Figs. 3 and 4). This may imply that both cluster analysis methods, as observed in the previous study (Sánchez-Martn et al., 2019), are required to gain a deeper understanding of the spatiotemporal clustering of the studied phenomena. Overall, our findings are consistent with previous studies in which LISA analysis of COVID-19 infection or mortality cases revealed local clustering patterns (Dutta et al., 2021, Liu et al., 2021, Sun et al., 2021). Unlike previous studies, we were able to examine short-term variations in COVID-19 cases at 14-day intervals (14-day notification rate). We discovered spatiotemporal variations in which COVID-19 clusters emerged at short intervals, such as high–high (H–H) and low–low (L–L) clusters most likely related to local outbreaks (Figs. 3 and 4).
5.2. Space-time scan statistics, spatiotemporal trends and epidemic curve
Space-time scan analysis (Kulldorff, 1997) was performed over three aggregated time periods, yielding statistically significant COVID-19 clusters. Throughout the study period, clusters were found in the eastern areas of the City of Helsinki, though local clusters emerged in other parts of the city as well. A similar pattern was discovered in the relative risk of COVID-19 in postal code areas (Figs. 5 and 6). The prospective Poisson Space-time scan analysis method has been found to be a valuable method for detecting COVID-19-related clusters and relative risk areas (Desjardins et al., 2020; Masrur et al., 2020; Xu et al., 2021). In order to detect smaller secondary clusters, we tested Gini Optimized cluster detection with the retrospective purely spatial Poisson scan statistic method, rarely used in previous COVID-19 studies. We discovered a number of secondary clusters in eastern Helsinki, particularly between 24.2.2021 and 24.3.2021 (Fig 5, c2), coinciding with the third wave of the COVID-19 epidemic, when the disease caused by alpha and beta variants spread rapidly in the eastern suburbs, as shown in Fig. 7. In addition to space-time scan analysis, the multitemporal quintiles map visualizes spatial and temporal trends for COVID-19 incidence diffusion, allowing comparison of the epidemic over time (Fig. 2). In most postal code areas, the number of infection cases per 100 000 residents remained low in autumn 2020. The virus began to spread throughout Helsinki in November and December 2020, but the number of infections remained relatively low, with only a few postal code areas having a high number of infection cases. The epidemic subsided temporarily in December 2020 and January 2021, but with the third wave of coronavirus, the number of COVID-19 infection cases began to rapidly increase in mid-February 2021. The third coronavirus wave exacerbated the situation, particularly in the eastern and northeastern suburbs (Fig 2). Interestingly, high population density in postal code areas did not appear to be the primary cause of high clusters, as shown in Figs. 1 and 2. This suggests that other sociodemographic factors could account for COVID-19 clustering. The sociodemographic factors associated with the previously mentioned patterns are discussed in greater detail in the forthcoming section. The epidemic curve, along with an accompanying map, may reveal valuable information about the epidemic's spatial progression (Fig. 7). However, due to reporting delays, it may be difficult in some cases to determine the progression of epidemics.
5.3. Performance of OLS, GWR, and MGWR regression models
In this study, we discovered spatiotemporal clustering patterns of COVID-19 infections in Helsinki, but spatial analysis only revealed "half of the story." Therefore, we attempted to determine the factors underlying the spatial patterns of COVID-19 infection rates. To explain the variation in COVID-19 data, we used the most important sociodemographic determinants identified in previous studies. Finally, we used only seven sociodemographic predictors to reduce predictor multicollinearity, also observed in previous studies (Mollalo et al., 2020; Snyder and Parks, 2020). However, Souza et al. (2020) discovered that reducing multicollinearity may compromise the study's quality. Similarly, in this study, reducing multicollinearity and then using only seven predictors could explain why some regression models had low explanatory power at certain time periods. Low model performance may also result from more efficient virus transmission throughout Helsinki, such as during the period 13.01.2021–27.01.2021, reducing the ability of sociodemographic factors to explain variation in COVID-19 infections. In addition to sociodemographic predictors, other variables not included in our study, such as environmental, distance-based, and behavioral factors, could improve the models' explanatory power. The best performance of regression models appears to have been achieved during time periods with positive local and global spatial autocorrelation and scan statistics detecting significant clusters in the COVID-19 data, such as on 10.03.2021 and 24.03.2021.
5.4. Sociodemographic predictors explaining variation in COVID-19 infections in Helsinki
In general, median income of inhabitants and a high number of foreign citizens were the best predictors of variation in COVID-19 infection data, followed by a low level of education and a relatively high number of unemployed in postal code areas (Table 4). Our findings are in line with previous socioeconomic studies in Finland, which found that COVID-19 infections were most common in adults with low incomes and a low level of education (Helsinki GSE, 2021), and one out of every four coronavirus infections in Finland has been diagnosed among foreign citizens (Holmberg et al., 2022, THL, 2020). Furthermore, health authorities in Helsinki region announced in 2020 that high number of COVID-19 infections had been detected among the foreign population in the metropolitan area (Rantavaara, 2020). According to THL's MigCOVID survey (Skogberg et al., 2021), there is an increased risk of imported SARS-CoV-2 virus infection among people of migrant origin due to the following factors; working conditions, lower education and income, and crowded housing. However, one factor stood out above the rest: working conditions. According to Skogberg et al. (2021), foreigners do a lot of work that cannot be done remotely. Many professions, for example, are in the service sector, and only one-third of foreign citizens had the opportunity to work remotely. In addition, half of the foreign-speaking respondents said it was impossible to observe safety intervals at work. Skogberg et al. (2021) also discovered that during the pandemic, foreign citizens traveled more than Finnish native citizens, primarily to visit relatives living in other countries. This could be another reason for the virus's rapid spread among foreign communities. These postal code areas in Helsinki with lower median incomes and higher proportions of foreign citizens are mostly found in the eastern and northeastern suburbs (Fig 9). In these areas, there are pockets of poverty neighborhoods with low median income, low levels of employment, and low levels of education (Kortteinen and Vaattovaara, 2015). Furthermore, in the eastern and northeastern suburbs, there may be concentrations of cramped condominiums with circular migration workers, such as construction workers from the Baltic and Eastern European countries. Many of the COVID-19 infections in Helsinki and the surrounding area were linked to these migrant worker housing conditions, and a large number of infections were observed on construction sites (Ervasti, 2020).
5.5. Targeting future interventions and control of the disease spread in the City of Helsinki
Based on the study's findings that COVID-19 infections are concentrated in areas with lower income, relatively high number of foreign citizens, a low level of education and a high number of unemployed citizens; future disease-control interventions should be geographically targeted particularly to the eastern and norther eastern suburbs, which have served as hotspots during most epidemics. Furthermore, as the virus was discovered to be spreading strongly among foreign citizens, public health authorities should be ready to co-operate with foreign communities and organizations. Active efforts should be emphasized to disseminate COVID-19-related information in the City of Helsinki with multilingual and multi-channel communication and counseling. Moreover, COVID-19 infections were detected in foreign citizen communities, particularly among the younger generations; thus, low-threshold tracking and testing, as well as mobile vaccination points, are required particularly in these target groups and intervention areas.
5.6. Holistic approach to analyze spatiotemporal patterns of COVID-19
This study takes a holistic approach to analysing spatiotemporal clustering patterns and sociodemographic determinants of COVID-19 infections in Helsinki. There are only a few studies that we are aware of that take a holistic approach to study the spatiotemporal aspects of COVID-19 transmission (Liu et al., 2021, Wang et al., 2021). However, these studies were conducted on a small scale and focused on factors such as population movement, meteorological parameters, and air pollutants. Whereas, for the first time, our study was able to provide holistic insights into the spatial patterns and sociodemographic determinants of COVID-19 infections in Helsinki at the postal scale level. Based on our study results, spatial analysis techniques can identify neighborhoods and communities where public health interventions should be targeted to reduce local COVID-19 outbreaks. Furthermore, these techniques could be utilized in contacts tracing in healthcare units to more efficient action. However, these techniques were not used during the first, second, and third waves of the COVID-19 epidemic in Finland as the governmental and local authorities in Finland were not fully aware of the benefits of geospatial analysis in fighting the pandemic. In future, in order to study quickly spatiotemporal phenomena of emerging diseases and epidemics, spatial and molecular epidemiology data should be available for researchers and aggregated more efficiently.
5.7. Limitations of spatiotemporal analysis in COVID-19 studies
Although our findings indicated that the overall spatial patterns of second-wave COVID-19 infections in the City of Helsinki could be analysed, as well as space-time clusters and risk areas identified and mapped, this study has limitations. In 15 postal code areas, low numbers of COVID-19 infection cases prohibited the City of Helsinki from publishing data due to privacy reasons. These postal code areas could not be analysed, which may have influenced the results. Furthermore, spatial analysis at postal code level aggregates information, hindering understanding of the local variations inside these areal units. Thus, interpretation of the results is limited to postal code scale due to ecological fallacy and modifiable areal unit problem (MAUP) (Wang and Di, 2020). In addition, misinterpretations of the resulted maps of COVID-19 infections may arise from the way in which COVID-19 infection data were collected. Postal code information is based on the infected person's home address, not the location of the initial infection, which is often difficult to determine and may differ. As a result, study findings from spatial analysis of COVID-19 infection data collected at the postal code level must be interpreted with caution.
Previous spatiotemporal studies on COVID-19 infections have encountered similar difficulties in other parts of the world. Kim and Castro (2020) mentioned that more detailed information about COVID-19 infection cases would have allowed them to fine-tune their analysis to a finer-scale than district level in South Korea. According to Fatima et al. (2021), COVID-19 data quality and access to fine-scale data are crucial and are the main limitations in spatial analysis of COVID-19 epidemics. There are also limitations in sociodemographic data, as we were unable to obtain the most recent data for postal code areas because it was not freely available. Furthermore, we used sociodemographic predictors that were available, but other variables may explain the variation in COVID-19 infections better. It is also worth noting that because vaccinations were only started in January 2021 in Helsinki, a sufficiently rapid accumulation of population immunity by disease, or vaccine immunity in a given area, could not be counted in any way in this study.
6. Conclusion
Our findings show that open datasets, such as the City of Helsinki's postal code level COVID-19 infection data and Statistics Finland's open sociodemographic dataset, can be used in spatial analysis to gain a better understanding of spatiotemporal patterns and sociodemographic determinants even without access to individual-level data. The holistic approach used in this study, including global- and local spatial autocorrelation (Moran's I and LISA); Getis-Ord Gi* hot spot analysis, and regression models (OLS, GWR, MGWR), can be applied to any other global location with similar datasets to contribute to the existing geospatial knowledge of the COVID-19 pandemic at the local scale.
However, acquisition of real-time, fine-scale COVID-19 infection and sociodemographic data are often challenging, making spatial analyses difficult to conduct. To be better prepared for future pandemic waves and to guide policymakers and local health authorities in implementing mitigation strategies, it is critical to understand the benefits of the holistic approach in spatial epidemic analyses in Finland and elsewhere in the globe. Future research should be conducted using fine-scaled COVID-19 surveillance data in collaboration with health authorities, who should be encouraged to elucidate these complex spatiotemporal patterns to inform mitigation and control efforts of the ongoing as well as future pandemics.
Acknowledgements
The authors would like to acknowledge the funding from the Academy of Finland for the VECLIMIT project (decision No #329323) and SARS-CoV-2 in space and time: Molecular epidemiology and phylogeography for disease prevention and mitigation sub-project.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.sste.2022.100493.
Appendix. Supplementary materials
References
- Akaike H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Andrade L.A., Gomes D.S., Góes MA de O., de Souza M.S.F., Teixeira D.C.P., Ribeiro C.J.N., et al. Surveillance of the first cases of COVID-19 in sergipe using a prospective spatiotemporal analysis: The spatial dispersion and its public health implications. Rev. Soc. Bras. Med. Trop. 2020;53:1–5. doi: 10.1590/0037-8682-0287-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselin L. Local Indicators of Spatial Association—LISA. Geogr. Anal. 1995;27:93–115. doi: 10.1111/j.1538-4632.1995.tb00338.x. [DOI] [Google Scholar]
- Anselin L., Syabri I., Kho Y. In: Handb. Appl. Spat. Anal. Softw. Tools, Methods Appl. Fischer MM, Getis A, editors. Springer; 2010. GeoDa: an introduction to spatial data analysis; pp. 73–89. [Google Scholar]
- Brunsdon C., Fotheringham A.S., Charlton M.E. Geographically weighted regression. Geogr. Anal. 1996;28 doi: 10.4135/9781412939591.n478. [DOI] [Google Scholar]
- Cavalcante J.R., Abreu A.J.L. COVID-19 in the city of Rio de Janeiro: spatial analysis of first confirmed cases and deaths. Epidemiol. Serv. Saude. 2020;29 doi: 10.5123/S1679-49742020000300007. Brasília2020. [DOI] [PubMed] [Google Scholar]
- City of Helsinki. Coronavirus updates from Helsinki 2020b. https://www.hel.fi/helsinki/coronavirus-en/social-and-health/corona-situation. (accessed 24 March 2021).
- Cordes J., Castro M.C. Spatial analysis of COVID-19 clusters and contextual factors in New York City. Spat. Spatiotemporal. Epidemiol. 2020;34 doi: 10.1016/j.sste.2020.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Ghosh S., Das K., Basu T., Dutta I., Das M. Living environment matters: Unravelling the spatial clustering of COVID-19 hotspots in Kolkata megacity, India. Sustain. Cities Soc. 2021;65 doi: 10.1016/j.scs.2020.102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins M.R., Hohl A., Delmelle E.M. Rapid surveillance of COVID-19 in the United States using a prospective space-time scan statistic: Detecting and evaluating emerging clusters. Appl. Geogr. 2020;118 doi: 10.1016/j.apgeog.2020.102202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaggio C., Klein M., Berry C., Frangos S. Black/African American Communities are at highest risk of COVID-19: spatial modeling of New York City ZIP Code–level testing results. Ann. Epidemiol. 2020;51:7–13. doi: 10.1016/j.annepidem.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta I., Basu T., Das A. Spatial analysis of COVID-19 incidence and its determinants using spatial modeling: A study on India. Environ. Challeng. 2021 doi: 10.1016/j.envc.2021.100096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ervasti A.-.E. Ei täällä voi mitään etäisyyttä pitää” – HS tutustui rakennustyöläisten yhteisasuntoihin. joita on syytetty koronapesäkkeiksi. 2020 https://www.hs.fi/kotimaa/art-2000006562993.html accessed 3 February 2021. [Google Scholar]
- ESRI, 2020https://support.esri.com/en/Products/Desktop/arcgis-desktop/arcmap/10-8.
- Fatima M., KJ O ’keefe, Wei W., Arshad S., Gruebner O. Geospatial analysis of covid-19: A scoping review. Int. J. Environ. Res. Public Health. 2021;18:1–14. doi: 10.3390/ijerph18052336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotheringham A.S., Oshan T.M. Geographically weighted regression and multicollinearity: dispelling the myth. J. Geogr. Syst. 2016;18:303–329. doi: 10.1007/s10109-016-0239-5. [DOI] [Google Scholar]
- Fotheringham A.S., Yang W., Kang W. Multiscale Geographically Weighted Regression (MGWR) Ann. Am. Assoc. Geogr. 2017;107:1247–1265. doi: 10.1080/24694452.2017.1352480. [DOI] [Google Scholar]
- Franch-Pardo I., Napoletano B.M., Rosete-Verges F., Billa L. Spatial analysis and GIS in the study of COVID-19. A review. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.140033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getis A., Ord J.K. The analysis of spatial association by use of distance statistics. Geogr. Anal. 1992;24:189–206. doi: 10.1111/j.1538-4632.1992.tb00261.x. [DOI] [Google Scholar]
- Han J., Zhu L., Kulldorff M., Hostovich S., Stinchcomb D.G., Tatalovich Z., et al. Using Gini coefficient to determining optimal cluster reporting sizes for spatial scan statistics. Int. J. Health Geogr. 2016;15 doi: 10.1186/s12942-016-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Yang L., Jia K., Li J., Feng S., Chen W., et al. Spatial distribution characteristics of the COVID-19 pandemic in Beijing and its relationship with environmental factors. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveri A., Smura T., Kuivanen S., Österlund P., Hepojoki J., Ikonen N., et al. Serological and molecular findings during SARS-CoV-2 infection: The first case study in Finland, January to February 2020. Eurosurveillance. 2020;25:1–6. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsinki GSE . Helsinki Graduate School of Economics; 2021. (Helsinki Graduate School of Economics). Situation Room Report: The Corona Virus and Health Differences – In Which Socioeconomic Groups Have the Most Infections Been Observed in Finland?https://www.helsinkigse.fi/covid19-data-en/situation-room-report-the-corona-virus-and-health-differences-in-which-socioeconomic-groups-have-the-most-infections-been-observed-in-finland/ (accessed 24 March 2021) [Google Scholar]
- Hohl A., Delmelle E.M., Desjardins M.R., Lan Y. Daily surveillance of COVID-19 using the prospective space-time scan statistic in the United States. Spat. Spatiotemporal. Epidemiol. 2020;34 doi: 10.1016/j.sste.2020.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg V., Salmi H., Kattainen S., Ollgren J., Kantele A., Pynnönen J., et al. Association between first language and SARS-CoV-2 infection rates, hospitalization, intensive care admissions and death in Finland: a population-based observational cohort study. Clin. Microbiol. Infect. 2022;28:107–113. doi: 10.1016/j.cmi.2021.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HSY. Helsingin kaupunkimittauspalvelut, Helsingin seudun kunnat ja HSY, 2019.
- Hui DS I., Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imdad K., Sahana M., Rana M.J., Haque I., Patel P.P., Pramanik M. A district-level susceptibility and vulnerability assessment of the COVID-19 pandemic’s footprint in India. Spat. Spatiotemporal. Epidemiol. 2021;36 doi: 10.1016/j.sste.2020.100390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarva H., Lappalainen M., Luomala O., Jokela P., Jääskeläinen A.E., Jääskeläinen A.J., et al. Laboratory-based surveillance of COVID-19 in the Greater Helsinki area, Finland, February–June 2020. Int. J. Infect. Dis. 2021;104:111–116. doi: 10.1016/j.ijid.2020.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Choi H., Kim J.H., Choi J. Spatial epidemic dynamics of the COVID-19 outbreak in China. Int. J. Infect. Dis. 2020;94:96–102. doi: 10.1016/j.ijid.2020.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant R., Nguyen P.T., Blomqvist S., Erdin M., Alburkat H., et al. Incidence Trends for SARS-CoV2 Alpha and Beta Variants, Finland, Spring 2021. Emerg. Infect. Dis. 2021;(12):3137–3141. doi: 10.3201/eid2712.211631. Dec; 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Castro M.C. Spatiotemporal pattern of COVID-19 and government response in South Korea (as of May 31, 2020) Int. J. Infect. Dis. 2020;98:328–333. doi: 10.1016/j.ijid.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Rundle A.G., Goodwin A.T.S., Morrison C.N., Branas C.C., El-Sadr W., et al. COVID-19 testing, case, and death rates and spatial socio-demographics in New York City: An ecological analysis as of June 2020. Health & Place. 2021;68 doi: 10.1016/j.healthplace.2021.102539. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortteinen M., Vaattovaara M. Segregaation aika. Yhteiskuntapolitiikka. 2015;80:562–574. [Google Scholar]
- Kulldorff M. A spatial scan statistic. Commun. Stat. Theo. Methods. 1997;26:1481–1496. doi: 10.1080/03610929708831995. [DOI] [Google Scholar]
- Kulldorff M., SaTScan User Guide v10.0 2021:1–121. https://www.satscan.org/.
- Lakhani A. Which Melbourne metropolitan areas are vulnerable to COVID-19 based on age, disability, and access to health services? J. Pain Symptom Manage. 2020;60:e41–e44. doi: 10.1016/j.jpainsymman.2020.03.041. Using Spatial Analysis to Identify Service Gaps and Inform Delivery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhang Z., Cao W., Liu Y., Du B., Canping Chen, et al. Identifying novel factors associated with COVID-19 transmission and fatality using the machine learning approach. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Liu M., Li Z., Zhu Y., Liu Y., Wang X., et al. The spatial clustering analysis of COVID-19 and its associated factors in mainland China at the prefecture level. Sci. Total Environ. 2021;777 doi: 10.1016/j.scitotenv.2021.145992. [DOI] [Google Scholar]
- Maiti A., Zhang Q., Sannigrahi S., Pramanik S., Chakraborti S., Cerda A., et al. Exploring spatiotemporal effects of the driving factors on COVID-19 incidences in the contiguous United States. Sustain. Cities Soc. 2021;68 doi: 10.1016/j.scs.2021.102784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S., A Al Kindi, Alkhattab Al-Said, Adham Al-Said, Atkinson P. Sociodemographic determinants of COVID-19 incidence rates in Oman: Geospatial modelling using multiscale geographically weighted regression (MGWR) Sustain. Cities Soc. 2021;65 doi: 10.1016/j.scs.2020.102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines M.R., Ferreira R.V., Toppa R.H., Assunção L.M., Desjardins M.R., Delmelle E.M. Detecting space–time clusters of COVID-19 in Brazil: mortality, inequality, socioeconomic vulnerability, and the relative risk of the disease in Brazilian municipalities. J. Geogr. Syst. 2021;23:7–36. doi: 10.1007/s10109-020-00344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masrur A., Yu M., Luo W., Dewan A. Space-time patterns, change, and propagation of COVID-19 risk relative to the intervention scenarios in Bangladesh. MedRxiv. 2020 doi: 10.1101/2020.07.15.20154757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie G., Adams B. A country comparison of place-based activity response to COVID-19 policies. Appl. Geogr. 2020;125 doi: 10.1016/j.apgeog.2020.102363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin P., Monica J.C., Sanchez D., Castillo O. Analysis of Spatial Spread Relationships of Coronavirus (COVID-19) Pandemic in the World using Self Organizing Maps. Chaos, Solitons and Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena G.E., Martinez P.P., Mahmud A.S., Marquet P.A., Buckee C.O., Santillana M. Socioeconomic status determines COVID-19 incidence and related mortality in Santiago, Chile. Science (80-) 2021;372:eabg5298. doi: 10.1126/science.abg5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middya A.I., Roy S. Geographically varying relationships of COVID-19 mortality with different factors in India. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-86987-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollalo A., Vahedi B., Rivera K.M. GIS-based spatial modeling of COVID-19 incidence rate in the continental United States. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonsammy S., Oyedotun T.D.T., Renn-Moonsammy D.M., Oyedotun T.D. COVID-19 modelling in the Caribbean: Spatial and statistical assessments. Spat. Spatiotemporal. Epidemiol. 2021;37 doi: 10.1016/j.sste.2021.100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P.A. Notes on continuous stochastic phenomena. Biometrika. 1950;37:17–23. doi: 10.1093/biomet/37.1-2.17. [DOI] [PubMed] [Google Scholar]
- Nguyen P.T., Kant, R., Van den Broeck, F. et al. The phylodynamics of SARS-CoV-2 during 2020 in Finland — Disappearance and re-emergence of introduced strains. 2021 https://www.researchsquare.com/article/rs-753457/v1. [DOI] [PMC free article] [PubMed]
- Oshan T.M., Li Z., Kang W., Wolf L.J., Fotheringham Stewart, MGWR A. A python implementation of multiscale geographically weighted regression for investigating process spatial heterogeneity and scale. ISPRS Int. J. Geo-Inform. 2019;8 doi: 10.3390/ijgi8060269. [DOI] [Google Scholar]
- Pourghasemi H.R., Pouyan S., Heidari B., Farajzadeh Z., Fallah Shamsi S.R., Babaei S., et al. Spatial modeling, risk mapping, change detection, and outbreak trend analysis of coronavirus (COVID-19) in Iran (days between February 19 and June 14, 2020) Int. J. Infect. Dis. 2020;98:90–108. doi: 10.1016/j.ijid.2020.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.H., Zafri N.M., Ashik F.R., Waliullah M., Khan A. Identification of risk factors contributing to COVID-19 incidence rates in Bangladesh: A GIS-based spatial modeling approach. Heliyon. 2021;7:e06260. doi: 10.1016/j.heliyon.2021.e06260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantavaara M. 2020. Tartunnat Ovat Lisääntyneet Viime Aikoina Itä-Helsingin lähiöissä, Syinä Muun Muassa Suuret Perhekoot Ja Ahtaammat Kotiolot.https://www.hs.fi/kaupunki/art-2000007629544.html (accessed 12 December 2020) [Google Scholar]
- Sánchez-Martín J.M., Rengifo-Gallego J.I., Blas-Morato R. Hot Spot Analysis versus Cluster and Outlier Analysis: An enquiry into the grouping of rural accommodation in Extremadura (Spain) ISPRS Int. J. Geo-Inform. 2019;8 doi: 10.3390/ijgi8040176. [DOI] [Google Scholar]
- Sannigrahi S., Pilla F., Basu B., Basu A.S. The overall mortality caused by COVID-19 in the European region is highly associated with demographic composition: A spatial regression-based approach. ArXiv. 2020;62 doi: 10.1016/j.scs.2020.102418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogberg N., Koponen P., Lilja E., Austero S., Achame S., Castaneda A.E. 2021. Discussion Paper 8/2021; p. 38. [Google Scholar]
- Snyder B.F., Parks V. Spatial variation in socio-ecological vulnerability to Covid-19 in the contiguous United States. Heal Place. 2020;66 doi: 10.1016/j.healthplace.2020.102471. [DOI] [PubMed] [Google Scholar]
- Sobral M.F.F., Duarte G.B., da Penha Sobral A.I.G., Marinho M.L.M., de Souza Melo A. Association between climate variables and global transmission oF SARS-CoV-2. Sci. Total Environ. 2020;729 doi: 10.1016/j.scitotenv.2020.138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza C.D.F., Machado M.F., Do Carmo R.F. Human development, social vulnerability and COVID-19 in Brazil: A study of the social determinants of health. Infect. Dis. Poverty. 2020;9:4–13. doi: 10.1186/s40249-020-00743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Matthews S.A., Yang T.C. Hu MH. A spatial analysis of the COVID-19 period prevalence in U.S. counties through June 28, 2020: where geography matters? Ann. Epidemiol. 2020;52:54–59. doi: 10.1016/j.annepidem.2020.07.014. e1https://doi.org/10.1016/j.annepidem.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Hu X., Xie J. Spatial inequalities of COVID-19 mortality rate in relation to socioeconomic and environmental factors across England. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- City of Helsinki, 2020a. Helsinki facts and figures 2020. City of Helsinki, Executive Office, Urban Research and Statistics 2020. https://www.hel.fi/hel2/tietokeskus/julkaisut/pdf/20_06_18_HKI_taskutilasto2020_eng.pdf (accessed 3 February 2021).
- THL, 2020. (The Finnish Institute for Health and Welfare). Diverse communications and collaboration with key community representatives as strategies to prevent covid-19 among migrant origin persons. https://thl.fi/en/web/thlfi-en/-/diverse-communications-and-collaboration-with-key-community-representatives-as-strategies-to-prevent-covid-19-among-migrant-origin-persons 2020. (accessed 4 December 2020).
- Statistics Finland, 2020. Paavo postal code area statistics. https://www.stat.fi/tup/paavo/index_en.html (accessed 20 November 2020).
- Wang Q., Dong W., Yang K., Ren Z., Huang D., Zhang P., et al. Temporal and spatial analysis of COVID-19 transmission in China and its influencing factors. Int. J. Infect. Dis. 2021;105:675–685. doi: 10.1016/j.ijid.2021.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Di Q. Modifiable areal unit problem and environmental factors of COVID-19 outbreak. Sci. Total Environ. 2020;740 doi: 10.1016/j.scitotenv.2020.139984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M.D., Gleditsch K.S. 2018. Spatial Regression Models. second edSage, London. [Google Scholar]
- World Health Organization (WHO) 2020. Novel Coronavirus (2019-nCoV) Situation report - 1. Geneva: WHO, 21 Jan 2020.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf accessed 15 January 2021. [Google Scholar]
- World Health Organization (WHO) 2020. Virtual Press Conference On COVID-19 –11 March 2020.https://www.who.int/docs/default-source/coronaviruse/transcripts/who-audio-emergencies-coronavirus-press-conference-full-and-final-11mar2020.pdf?sfvrsn=cb432bb32 accessed 15 January 2021. [Google Scholar]
- Xu M., Cao C., Zhang X., Shea D.R., Lin H., Yao Z., et al. Fine-scale space-time cluster detection of covid-19 in mainland china using retrospective analysis. Int. J. Environ. Res. Public Health. 2021;18 doi: 10.3390/ijerph18073583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Fotheringham A.S., Li Z., Oshan T., Kang W., Wolf L.J. Inference in multiscale geographically weighted regression. Geogr. Anal. 2020;52:87–106. doi: 10.1111/gean.12189. [DOI] [Google Scholar]
- Zhang C.H., Schwartz G.G. Spatial disparities in coronavirus incidence and mortality in the United States: an ecological analysis as of May 2020. J. Rural. Heal. 2020;36:433–445. doi: 10.1111/jrh.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.