Abstract
Detection and epidemiologic characterization of infectious disease outbreaks are key for early identification and response to potential pandemic threats. The rapid global spread of severe SARS-CoV-2 in 2020 highlighted the critical role of diagnostics in understanding the epidemiology of the virus early in the pandemic. As a natural extension of Abbott's work in diagnostics, virus discovery, and virus surveillance, the Abbott Pandemic Defense Coalition (APDC) was launched in early 2021. The APDC is a global multisector scientific and public health partnership whose primary objective is the early detection and mitigation of infectious disease threats of pandemic potential. As of January 2022, the APDC network has partners on 5 continents including academic institutions, governmental, and nongovernmental organizations. A novel element of the APDC is the capacity for early development and rapid deployment of scalable, quality diagnostics targeting newly identified pathogens of pandemic potential.
Keywords: Pandemic preparedness; emerging pathogens, virus discovery, surveillance, public health
Introduction
In recent decades, the world has experienced the emergence and reemergence of numerous infectious disease threats. Viruses originating from animal reservoirs, such as HIV, influenza A (H1N1), Ebola virus, yellow fever virus, Zika virus, chikungunya virus (CHIKV), SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV), have taken enormous tolls on health and economies (Morens et al. 2020). Globalization, international travel, displacement of peoples, changes in the environment, and encroachment of humans into natural habitats are all factors contributing to outbreaks and facilitating their global spread (Morens et al. 2020). International capacity to respond to emerging disease threats was tested most recently with the emergence of SARS-CoV-2. One success in the response to SARS-CoV-2 was the unprecedented development of several safe and effective vaccines less than 1 year after the SARS-CoV-2 genetic sequence became available (United States Government Accountability Office, 2021). This success can be attributed in large part to the US government funding made available through Operation Warp Speed, mitigating the financial risks to the pharmaceutical industry in vaccine development. In contrast, diagnostics, the “first line of defence” in the response to SARS-CoV-2, were developed by industry with limited government support. Although the development of diagnostics for SARS-CoV-2 has been a success story as well, a public-private global disease surveillance network could have potentially helped identify SARS-CoV-2 earlier, understand the epidemiology of the virus (eg, person-to-person spread, asymptomatic infections) earlier, and developed and distributed authorized sensitive and specific diagnosics earlier, perhaps resulting in hundreds of thousands of lives saved.
In 2005, amid growing concerns about global health security and preparedness and to engage governments and ministries of health in pandemic preparedness and control, the World Health Organization (WHO) and member states formulated and adopted the revised International Health Regulations (IHR) (Gostin and Katz 2016). The IHR mandate that countries develop and maintain defined core capacities for national surveillance and response to diseases of epidemic potential. The IHR highlights the global importance placed on early recognition of disease events so that measures can be taken promptly to control the threat at its source before the disease can spread to other countries. SARS-CoV-2, with its rapid global spread, clearly exposed this preparedness gap. The Abbott Pandemic Defense Coalition (APDC) aims to support the IHR and narrow this gap in early discovery and detection of emerging pathogens and diagnostics availability in preparation for the next pandemic. In addition to pathogen discovery, a novel and core tenet of the coalition includes the early and scalable development and rapid deployment of quality diagnostics for emerging infectious disease threats. Early distribution of diagnostics may provide the world with the ability for more timely understanding of the epidemiology and spread of the next pandemic- key to mitigation and control efforts, including preventive (including nonpharmaceutical) interventions, therapeutics, and vaccine development.
A Legacy of Infectious Diseases Research at Abbott
More than 60% of the world's blood supply is screened through Abbott instruments, and Abbott has been producing blood screening technologies for more than 40 years (Abbott Diagnostics 2021a). To ensure the safety of the blood supply and accuracy of diagnostic tests for infectious diseases, Abbott has been conducting global surveillance of known infectious diseases since 1994, with a focus on highly diverse pathogens, including HIV, hepatitis B virus (HBV), and hepatitis C virus (HCV) (Abbott Diagnostics 2021b). The aim of this program is to ensure that variant and recombinant strains do not escape detection. This research program has not only helped ensure a safe blood supply, but has also led to the discovery of new, rare strains of HIV-1 (Rodgers et al. 2017 Yamaguchi et al. 2020) and identified a group of HIV elite controllers in the Democratic Republic of Congo (Berg et al. 2021). In addition, the Abbott Global Surveillance Program has also supported important public health efforts including viral hepatitis research in Cameroon, where a previously unrecognized, large burden of hepatitis D virus among persons with HBV infection was discovered (Butler et al. 2018), and HCV elimination efforts in Georgia and in Pakistan (Abbott Laboratories 2020 Abid et al. 2021).
A natural extension of this work is the Virus Discovery Program, wherein specimens from individuals with illnesses of unknown etiology are characterized by metagenomic next-generation sequencing for known pathogens and potentially novel viruses. The discovery of human pegivirus-2 (HPgV-2) (Berg et al. 2015) led to the development of molecular and serologic diagnostic tests and the launch of a global effort to describe the epidemiology of the virus (Coller et al. 2016 Frankel et al. 2017). These studies found HPgV-2 to be a blood-borne virus associated with HCV and may result in chronic infection, although no discernible threat to human health has been identified (Berg et al. 2015 Kandathil et al. 2017). This successful industry-academia collaboration illustrated the cascade from discovery to test development to translational research needed to understand the nature of the discovery, a model that serves as the foundation for the APDC.
In addition, since 2020, Abbott has expended considerable resources in developing and launching SARS-CoV-2 diagnostics, launching 12 tests worldwide, including molecular, antigen, and antibody detection tests (Abbott Laboratories 2021). Surveillance for SARS-CoV-2 has been woven into Abbott's global surveillance efforts, including monitoring for new variants (Mary A Rodgers 2021). These efforts have allowed Abbott to respond quickly to the evolving SAR-CoV-2 pandemic, ensuring the development of critical diagnostics and contributing to the understanding and detection of variant strains.
Launch of the APDC
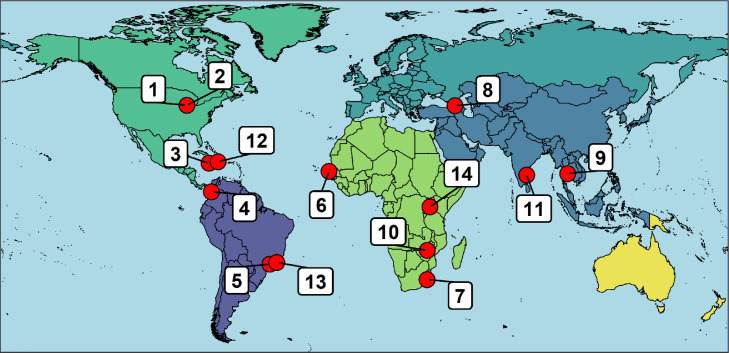
The Abbott Pandemic Defense Coalition (APDC) is a first-of-its-kind global scientific and public health partnership with the goals of early detection and rapid response to future pandemic threats. By connecting global centers of excellence with expertise in infectious diseases, laboratory testing, genetic sequencing and epidemiology with industry, the program aims to identify new pathogens, determine their potential for pandemic spread, rapidly develop and deploy new diagnostic testing, if indicated, and assess public health impact in near real-time. Including Abbott, there are currently 14 APDC partners on 5 continents (Figure 1 ). The APDC network shares expertise in epidemiology, diagnostics, next-generation sequencing (NGS), and bioinformatics for the investigation of emerging pathogens. This capacity building will facilitate pathogen discovery and surveillance in the country and region where institutions are located. Collaboration with sister institutions within the network will allow for timely investigation and communications with the WHO, Centers for Disease Control and Prevention (CDC), and other public health institutions, enabling early response to potential threats. Furthermore, the APDC partners will benefit from shared experiences and opportunities to collaborate on infectious disease research across the network. Beyond pathogen discovery, priority activities will include SARS-CoV-2 variant surveillance, blood-borne pathogen surveillance, scientific and epidemiologic capacity building, and support for assay development and quality assurance; Table 1 summarizes the goals and activities of the APDC.
Figure 1.
Abbott Pandemic Defense Coalition member institutions:
1. Abbott Diagnostics, United States
2. Rush University Medical Center, United States
3. University of the West Indies
4. Colombia Wisconsin One Health, National University of Colombia, Colombia
5. University of Sao Paulo, Brazil
6. Institute for Health Research, Epidemiological Surveillance, and Training (IRESSEF), Senegal
7. Centre for Epidemic Response and Innovation, Stellenbosch University, South Africa
8. National Center for Disease Control, Georgia
9. Mahidol University, Thailand
10. University of Zimbabwe, Zimbabwe
11. YRG Cares, India
12. Quisqueya University, Haiti
13. Foundation Oswaldo Cruz (Fiocruz), Brazil
14. Uganda Virus Research Institute, Uganda
Table 1.
Goals and priority activities of the Abbott Pandemic Defense Coalition (APDC)
| Goals |
| Early identification of emerging pathogens: define epidemiology and notify key public health institutions (WHO, CDC, etc) |
| Build capacity in diagnostics, next-generation sequencing, bioinformatics and epidemiology in low- and middle-income countries (LMICs) |
| Molecular surveillance of known pathogens of public health significance (eg, SARS-CoV-2, HIV, HBV) and increased understanding of endemic infectious diseases |
| Timely development, validation, and dissemination of diagnostic assays targeting emerging pathogens |
| Activities |
| Recruit academic, government, and non-government institutions in key LMICs to join the APDC |
| Provide resources to APDC members to conduct emerging pathogen and public health surveillance in country |
| Establish facility-based surveillance to identify and recruit case patients and ensure quality specimen collection and testing, epidemiologic data collection, data management, and analysis |
| Provide and participate in virtual and in-person training and capacity building sessions |
| Standardize laboratory protocols for virus discovery and diagnostic assay development |
| Provide access to bioinformatics tools and expertise |
| Promote collaborative research among APDC institutions and partners and dissemination of findings including presentations at scientific meeting and publications in the peer-reviewed literature |
| Develop partnerships with key global institutions and networks in infectious disease and emerging pathogen prevention, surveillance, and research |
| Ensure sustainability of the APDC with resources from Abbott and external sources by demonstrating value |
Abbreviations: APDC, Abbott Pandemic Defense Coalition; CDC, Centers for Disease Control and Prevention; LMICs, low- and middle-income countries; WHO, World Health Organization.
For emerging pathogen discovery, data collection will be harmonized across the APDC, facilitating collaborative research and analysis of findings. Case definitions for acute febrile illness, influenza-like illness, severe acute respiratory infection and other syndromes of interest will be incorporated. Clinical assessment will be conducted and include diagnostic testing for a range of bacterial, parasitic, mycotic, and viral pathogens as well as noninfectious etiologies. Case patients without a recognized etiology will be enrolled into the discovery program where demographic, clinical, and epidemiologic data will be collected, anonymized, and entered into a centralized, cloud-based database. Enrolled patients will be followed up through hospitalization to gather key laboratory and clinical outcomes (eg, diagnostic test results, specimen collection, imaging results, admission to the intensive care unit, use of mechanical ventilation, in-hospital death). Appropriate specimens will be collected and stored at −70°C for NGS or other analyses. Institutions will be supported by advanced bioinformatics technology and specimen repositories will be created to share and cross-reference between partners. The network will therefore provide access to geographically diverse specimens as member sites leverage relationships with local hospitals and clinics to enroll appropriate case patients beyond their institutions.
The program has entered into a first-of-its-kind industry partnership with the Task Force for Global Health's Training Programs in Epidemiology and Public Health Interventions Network (TEPHINET) to support epidemiologic capacity in pandemic preparedness in low- and middle-income countries (LMICs). TEPHINET is a global network of 75 Field Epidemiology Training Programs (FETPs) typically associated with National Ministries of Health and the US CDC Country Offices dedicated to strengthening public health capacity (Martin and Fall 2021). Abbott and TEPHINET will collaborate by providing funding and mentoring to FETP Fellows to support projects in pathogen discovery and other priority public health research in collaboration with the APDC partners in their home country.
Discussion
Although it cannot be predicted when or where the next pandemic will arise, or how the next pandemic pathogen will be transmitted, the SARS-CoV-2 pandemic has made it evident that complacency and inaction will result in great loss of human lives and economic disruptions globally. Human-to-human respiratory transmission poses the greatest risk for rapid dissemination of novel pathogens; however, the toll of HIV, viral hepatitis, malaria, and diarrheal illnesses serve as a reminder of the need to not simply focus on respiratory pathogens. We cannot underestimate the ability of emerging pathogens to surprise us and spread through blood, fecal-oral, sexual contact, or other modes, including vector-borne. To maintain readiness for the next pandemic, the APDC will track and monitor SARS-CoV-2 variants while continuing surveillance for new and variant blood, respiratory vector-borne, and water-borne pathogens, to identify novel pathogens and collaboratively respond to these threats promptly to prevent and /or mitigate the impact of the next pandemic.
The APDC offers some unique attributes in global pandemic preparedness. The strengths of the APDC include the industry-academic-government collaborations established, the emphasis on building laboratory, and epidemiologic capacity in LMICs, and the ability to rapidly develop and deploy scalable diagnostic tests following the discovery of a new pandemic threat; all will be key to a timely response to the next pandemic. For the APDC to have the greatest impact in pandemic mitigation and control, its efforts should align and be coordianted with other pandemic preparedness networks, forming a “network of networks.”
Preparedness funding tends to follow a cyclical pattern flowing after the emergence of a new outbreak, and then drying up after a period of time, usually not more than a few years, until a new threat triggers another wave of funding (United Kingdom G7 2021). Over the last century, only 2 pandemic viruses have killed more people than SARS-CoV-2 (estimated 5.9 million deaths as of February 2022)—the 1918 Influenza Pandemic with an estimated 50 million deaths and HIV with an estimated 32 million deaths (Morens et al. 2020). The enormous economic and human costs of SARS-CoV-2 highlight the need to rethink the paradigm of “feast or famine” funding, and that pandemic preparedness is a shared responsibility where governments work together with civil society, academia, and the private sector. The APDC, as a multisector partnership, is modeling this approach, and with capacity in diagnostics, epidemiology, and diagnostics development brings some novel strengths to pandemic preparedness efforts.
Acknowledgments
Conflicts of Interest
FA, MB, MR SO, XL, MA, TM, and GC receive salaries from and/or own stock in Abbott. AL, EK, AG, JO, JL, JA. JM, MA, MS, SS, TdO, KC, JP, JHH, JD, PK, RCA, and YS and/or their institutions receive funding from Abbott Diagnostics.
Funding Sources
This work is supported by Abbott Diagnostics.
Ethical Approvals
Ethical approval was not required for this submission
Role of the funder/sponsor
Abbott Diagnostics reviewed and approved this manuscript before submission.
Patient Consent Statement
N/A
Author contributions
FA, GC, and MB conceived the manuscript. FA and MB wrote the manuscript. All authors contributed and reviewed the manuscript.
Acknowledgments
The authors wish to thank Gregory Orf for working on developing the figure included in the manuscript. This work is supported by Abbott Diagnostics.
References
- Abbott Diagnostics, ADD. 2021a. ‘https://www.abbott.com/corpnewsroom/strategy-and-strength/delivering-accurate-results-faster-than-ever.html’.
- Abbott Diagnostics, ADD. 2021b. ‘https://www.corelaboratory.abbott/us/en/about-us/history-heritage’.
- Abbott Laboratories, Abbott. 2020. ‘https://www.abbott.com/careers/working-with-us/changing-lives/help-eliminate-hepatitis-c.html’.
- Abbott Laboratories, Abbott. 2021. ‘https://www.abbott.com/coronavirus.html’.
- Abid A., Uddin M., Muhammad T., Awan S., Applegate T., Dore G.J., Cloherty G., Hamid S. Evaluation of Hepatitis C Virus Core Antigen Assay in a Resource-Limited Setting in Pakistan. Diagnostics (Basel) 2021:11. doi: 10.3390/diagnostics11081354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M.G., Lee D., Coller K., Frankel M., Aronsohn A., Cheng K., Forberg K., Marcinkus M., Naccache S.N., Dawson G., Brennan C., Jensen D.M., Hackett J., Jr., Chiu C.Y. Discovery of a Novel Human Pegivirus in Blood Associated with Hepatitis C Virus Co-Infection’. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M.G., Olivo A., Harris B.J., Rodgers M.A., James L., Mampunza S., Niles J., Baer F., Yamaguchi J., Kaptue L., Laeyendecker O., Quinn T.C., McArthur C., Cloherty G.A. A high prevalence of potential HIV elite controllers identified over 30 years in Democratic Republic of Congo. EBioMedicine. 2021;65 doi: 10.1016/j.ebiom.2021.103258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler E.K., Rodgers M.A., Coller K.E., Barnaby D., Krilich E., Olivo A., Cassidy M., Mbanya D., Kaptue L., Ndembi N., Cloherty G. High prevalence of hepatitis delta virus in Cameroon. Sci Rep. 2018;8:11617. doi: 10.1038/s41598-018-30078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller K.E., Berg M.G., Frankel M., Forberg K., Surani R., Chiu C.Y., Hackett J., Jr., Dawson G.J. Antibodies to the Novel Human Pegivirus 2 Are Associated with Active and Resolved Infections. J Clin Microbiol. 2016;54:2023–2030. doi: 10.1128/JCM.00515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel M., Forberg K., Coller K.E., Berg M.G., Hackett J., Jr., Cloherty G., Dawson G.J. Development of a high-throughput multiplexed real time RT-PCR assay for detection of human pegivirus 1 and 2. J Virol Methods. 2017;241:34–40. doi: 10.1016/j.jviromet.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Gostin L.O., Katz R. The International Health Regulations: The Governing Framework for Global Health Security. Milbank Q. 2016;94:264–313. doi: 10.1111/1468-0009.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandathil A.J., Breitwieser F.P., Sachithanandham J., Robinson M., Mehta S.H., Timp W., Salzberg S.L., Thomas D.L., Balagopal A. Presence of Human Hepegivirus-1 in a Cohort of People Who Inject Drugs. Ann Intern Med. 2017;167:1–7. doi: 10.7326/M17-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Fall I.S. Field Epidemiology Training Programs to accelerate public health workforce development and global health security. Int J Infect Dis. 2021 doi: 10.1016/j.ijid.2021.08.021. [DOI] [PubMed] [Google Scholar]
- Rodgers Mary A, Olivo Ana, Vallari Ana, Harris Barbara, Lark Chris, Luo Xinxin, Berg Michael G, Meyer Todd V, Mohaimani Aurash, Orf Gregory S, Goldstein Yitz, Fox Amy S, Hirschhorn Julie, Glen William B, Nolte Frederick, Landay Alan, Jennings Cheryl, Moy James, Servellita Venice, Chiu Charles, Batra Rahul, Snell Luke B, Nebbia Gaia, Douthwaite Sam, Tanuri Amilcar, Singh Lavanya, Oliveira Tulio de, Ahouidi Ambroise, Mboup Souleymane, Daghfal David, Roth Richard, Huang Shihai, Kovacs Stephen, Cloherty Gavin A. Detection of SARS-CoV-2 variants by Abbott molecular, antigen, and serological tests. Journal of Clinical Virology (in review) 2021 doi: 10.1016/j.jcv.2022.105080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M., Daszak P., Markel H., Taubenberger J.K. Pandemic COVID-19 Joins History’s Pandemic Legion. mBio. 2020:11. doi: 10.1128/mBio.00812-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers M.A., Vallari A.S., Harris B., Yamaguchi J., Holzmayer V., Forberg K., Berg M.G., Kenmenge J., Ngansop C., Awazi B., Mbanya D., Kaptue L., Brennan C., Cloherty G., Ndembi N. Identification of rare HIV-1 Group N, HBV AE, and HTLV-3 strains in rural South Cameroon. Virology. 2017;504:141–151. doi: 10.1016/j.virol.2017.01.008. [DOI] [PubMed] [Google Scholar]
- United Kingdom G7, UKG7. 2021. ‘100 Days Mission to respond to future pandemics: a report to the G7’, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/992762/100_Days_Mission_to_respond_to_future_pandemic_threats__3_.pdf.
- US Government Accountability Office, GAO. 2021. ‘Operation Warp Speed: Accelerated COVID-19 Vaccine Development Status and Efforts to Address Manufacturing Challenges’, GAO-21-319.
- Yamaguchi J., Vallari A., McArthur C., Sthreshley L., Cloherty G.A., Berg M.G., Rodgers M.A. Brief Report: Complete Genome Sequence of CG-0018a-01 Establishes HIV-1 Subtype L’. J Acquir Immune Defic Syndr. 2020;83:319–322. doi: 10.1097/QAI.0000000000002246. [DOI] [PMC free article] [PubMed] [Google Scholar]