Abstract
Introduction: The mesencephalic reticular formation, isthmic reticular formation, microcellular tegmental nucleus, ventral tegmental area-parabrachial pigmented nucleus complex, and caudal-rostral linear nucleus of the raphe are small brainstem regions crucially involved in arousal, sleep, and reward. Yet, these nuclei are difficult to identify with magnetic resonance imaging (MRI) of living humans. In the current work, we developed a probabilistic atlas of these brainstem nuclei in living humans, using noninvasive ultra-high-field MRI.
Methods: We acquired single-subject, multicontrast (diffusion and T2-weighted), 1.1-mm isotropic resolution, 7 Tesla MRI images of 12 healthy subjects. After preprocessing and alignment to the stereotactic space, these images were used to delineate (in each subject) the nuclei of interest based on the image contrast as well as on neighboring nuclei and landmarks. Nucleus labels were averaged across subjects to yield probabilistic labels. The latter were further validated by assessment of the label inter-rater agreement, internal consistency, and volume.
Results: Labels were delineated for each nucleus with good overlap across subjects. The inter-rater agreement and internal consistency were below (p < 10−8) the linear spatial imaging resolution (1.1 mm), thus validating the generated probabilistic atlas labels. The volumes of our labels did not differ from literature volumes (p < 0.05), further validating our atlas.
Discussion and Conclusion: The probabilistic atlas of these five mesopontine nuclei expands current in vivo brainstem nuclei atlases and can be used as a tool to identify the location of these areas in conventional (e.g., 3 Tesla) images. This might serve to unravel the brainstem structure-to-function link and thus improve clinical outcomes.
Impact statement
The mesencephalic reticular formation, isthmic reticular formation, microcellular tegmental nucleus, ventral tegmental area-parabrachial pigmented nucleus complex, and caudal-rostral linear nucleus of the raphe are small brainstem regions crucially involved in arousal, sleep, and reward. In the current work, we developed a probabilistic atlas of these brainstem nuclei in living humans, using noninvasive, ultra-high-field magnetic resonance imaging. The probabilistic atlas of these five mesopontine nuclei expands current in vivo brainstem nuclei atlases and can be used as a tool to identify the location of these areas in conventional (e.g., 3 Tesla) images. This might serve to unravel the brainstem structure-to-function link and thus improve clinical outcomes.
Keywords: brainstem nuclei atlas, caudal-rostral linear nucleus of the raphe, isthmic reticular formation, mesencephalic reticular formation, microcellular tegmental nucleus, ventral tegmental area-parabrachial pigmented nucleus complex
Introduction
To understand the basic organizational features of the entire brain, constant efforts are being made to generate brainstem atlases, for instance, to define gray matter nuclei of importance for arousal, sleep, and reward. Histology-based atlases of brainstem nuclei (Naidich et al., 2009; Olszewski and Baxter, 1954; Paxinos and Huang, 1995; Paxinos et al., 2012) are theoretically considered the gold standard because of their high spatial resolution and direct cytoarchitectural mapping; however, they are two dimensional (2D), based on only one or few specimens, and not easy to extrapolate to images of living humans. This leads to imprecision in the localization of these nuclei in in vivo studies.
To this end, probabilistic atlases based on living humans (Bianciardi et al., 2015, 2018; García-Gomar et al., 2019; Singh et al., 2019; Trutti et al., 2019; Xiao et al., 2014), using ultra-high-field (e.g., 7 Tesla) magnetic resonance imaging (MRI), provide information on the location of brainstem nuclei at a lower resolution than postmortem atlases, yet they are three dimensional (3D) and can be automatically mapped to images of living humans.
Continuing our current project of generating a probabilistic atlas of nuclei in the living human brainstem by the use of 7 Tesla multicontrast MRI, in the current study, we generated an original probabilistic atlas of the mesencephalic (i.e., midbrain) reticular formation nuclei (mRt, viz mRt dorsal, mRt lateral, and mRt anterior parts), isthmic reticular formation (isRt), microcellular tegmental nucleus–prabigeminal nucleus (MiTg-PBG), ventral tegmental area–parabrachial pigmented nucleus complex (VTA-PBP), and caudal-rostral linear nucleus of the raphe complex (CLi-RLi).
The mRt has been shown to be involved in vertical and horizontal components of gaze changes (Moruzzi and Magoun, 1949) and in arousal. In the current study, this region was subdivided into three nuclei based on the imaging contrast, location, and existing atlas of neighboring regions (the three subregions are described in the Methods section). Nuclei such as isRt and MiTg-PBG have been modified in location (Paxinos and Huang, 1995; Paxinos et al., 2012) over time due to their functional and structural overlap with neighboring cuneiform and pedunculotegmental nuclei (Paxinos and Huang, 1995; Paxinos et al., 2012).
In this study, we followed the definition given by Paxinos et al. (2012) to develop an in vivo atlas of these nuclei, which may help further define their function in future studies of living humans. VTA-PBP, a widely studied region (Pauli et al., 2018) connected with reward-related behavior, motivation, addiction, and motor functioning, has been added in the current study along with CLi-RLi to, respectively, provide a more comprehensive view of midbrain regions (e.g., VTA-PBP) neighboring the previously delineated red nucleus and substantia nigra (Bianciardi et al., 2015) and to update previous reports of CLi (Bianciardi et al., 2018).
This atlas might augment studies of the structural and functional networks of these nuclei. Moreover, the current atlas might also provide a reliable scaffold to analyze imaging results, to reveal the general network structure of the brainstem with the rest of the brain, and to predict the brainstem–brain information flow.
Methods
Data acquisition
To delineate nucleus labels of the mRt (mesencephalic reticular formation dorsal part [mRtd], mesencephalic reticular formation anterior part [mRta], mesencephalic reticular formation lateral part [mRtl]), isRt, MiTg-PBG, VTA-PBP, and CLi-RLi, we processed data obtained in our previous study (Bianciardi et al., 2015), as explained here. Briefly, we scanned 12 healthy subjects (6 m/6 f, mean ± standard error [SE] age: 28 ± 1 years) with 7 Tesla MRI in accordance with the Declaration of Helsinki after obtaining written informed consent. The study protocol was reviewed and approved by the Massachusetts General Hospital Institutional Review Board.
We used a custom-built 32-channel receive coil and volume transmit coil (Keil et al., 2010), providing enhanced sensitivity in the brainstem compared with commercial coils. Diffusion tensor images (DTIs) were acquired with 1.1-mm isotropic resolution in the sagittal plane, using common, single-shot, 2D echo-planar images (EPIs): matrix size = 180 × 240, GeneRalized Autocalibrating Partial Parallel Acquisition factor = 3, and nominal echo spacing = 0.82 msec. Nondiffusion-weighted EPIs of the DTI protocol were used as T2-weighted images with matched geometric distortions and spatial resolution to the DTIs. The use of T2-weighted EPIs also allowed overcoming specific absorption rate limits usually encountered with spin-warp T2-weighted MRI at ultra-high-magnetic field strength.
DTI and T2-weighted images were acquired using the spin-echo EPI sequence with the following parameters: echo time = 60.8 msec, repetition time = 5.6 sec, 61 slices, partial Fourier: 6/8, unipolar diffusion-weighting gradients (for DTI), 60 diffusion directions with b-value ∼1000 sec/mm (for DTI), seven interspersed nondiffusion-weighted “b0” images with b-value ∼0 sec/mm2 (also used as T2-weighted MRI), and four repetitions, with 6′43′′ acquisition time for each repetition. The acquisition time for the entire T2-weighted and DTI protocol was ∼27′. We used unipolar (Stejskal and Tanner, 1965) in lieu of bipolar diffusion gradients (Reese et al., 2003) for DTI acquisition, leading to reduced echo time and significantly improved sensitivity for tissue having a shorter T2 value at 7 Tesla.
Data processing and alignment to standard space
On a single-subject basis, data of four DTI repetitions were concatenated and distortion and motion corrected using the Diffusion Toolbox of FMRIB Software Library (FSL, Oxford, United Kingdom). Using this toolbox, we estimated the diffusion tensor at each voxel to calculate the diffusion fractional anisotropy (FA) from tensor eigenvalues. The 28 “b0” T2-weighted images (7 “b0” images per repetitions, 4 repetitions) were averaged after motion correction.
Next, the average T2-weighted image and FA map were precisely coregistered to the MNI space on a single-subject basis, as described in the study by Bianciardi et al. (2015). Briefly, the Advanced Normalization Tools (ANTs, Philadelphia, PA) (Avants et al., 2011) were used to precisely align the brainstem of each subject to an FA template in an MNI-based space (termed as IIT space; Illinois Institute of Technology Human Brain Atlas, v.3, Chicago, IL) (Varentsova et al., 2014). This template was used due to optimal coverage of the brainstem (including medulla), high contrast, and compatibility with diffusion-based tractography. For precise coregistration to standard space, generic affine transformation and high-dimensional, nonlinear warp transformation of images with the same modality (FA maps) were computed and concatenated (Bianciardi et al., 2015).
The transformations were merged using a linear single-interpolation step method, which was then applied to the single-subject FA map and average T2-weighted image. Additionally, for each subject, FA and T2-weighted images were also aligned to the MNI152 space (nonlinear sixth-generation MNI152_T1_1mm available in FSL; known as MNI152_1mm space), by using the linear single-interpolation step method and applying two concatenated transformations: (1) single subject to IIT space transformation and (2) IIT to MNI152_1mm nonlinear transformation. The latter transformation was computed (Bianciardi et al., 2015) to compensate for a slight misalignment between the MNI152_1mm space and the IIT space in lower brainstem regions.
Nucleus delineation and probabilistic atlas generation
Two independent raters (K.S. and M.B.) manually delineated nuclei of interest (mRtd, mRta, mRtl, isRt, and MiTg-PBG, VTA-PBP, and CLi-RLi) using multicontrast T2-weighted images and FA maps in the IIT space (using fslview; FSL) to yield single-subject labels. The intersection of the labels delineated by the two independent raters was considered as the final label. To precisely delineate each nucleus, raters used the image modality with the best contrast for that nucleus (FA map for mRtd, mRta, mRtl, isRt, and MiTg-PBG) and the other modality (T2-weighted MRI) to identify some neighboring nuclei as additional landmarks for delineation (e.g., red nucleus and substantia nigra for VTA-PBP). The postmortem brainstem atlas (Paxinos et al., 2012) assisted in manual delineation of these nuclei by providing anatomical landmarks and describing neighborhood rules.
A probabilistic atlas of each nucleus in the IIT space was prepared as an average probability map of that nucleus label across all subjects (i.e., a 100% overlap of nucleus labels throughout all 12 subjects was considered as highest probability). A similar atlas in MNI152_1mm was obtained by applying the IIT to MNI152 transformations described above to single-subject labels and then by averaging the labels across subjects. The current proposed atlas has been developed in both IIT and MNI152 standard spaces to facilitate its extrapolation to a broader range of modalities such as diffusion and functional MRI.
We calculated the nucleus label volume in native space for each label and subject (coregistered from the IIT space to single-subject native space through inverse registration, as described in the Data processing and alignment to standard space section). We computed the mean (±SE) volumes for all subjects and labels, which were later compared with values in literature (Paxinos et al., 2012), as described in the Atlas validation section.
Atlas validation
The resulting probabilistic nucleus atlas labels were validated by measuring the label (1) inter-rater agreement and (2) internal consistency. The inter-rater agreement was computed as the modified Hausdorff distance between nucleus labels delineated by the two independent raters. The modified Hausdorff distance (Dubuisson and Jain, 1994) is a measure of spatial overlap of two images, which has been used in recent studies (Bianciardi et al., 2015, 2018; Fischl et al., 2008; García-Gomar et al., 2019; Singh et al., 2019). Specifically, for each nucleus label, the least distance from every point on one nucleus label to the other nucleus label was calculated and averaged across all points, resulting in two distance values. The maximum of these two values was computed and termed as the modified Hausdorff distance.
The internal consistency was computed as the modified Hausdorff distance between each final label and the probabilistic atlas label (thresholded at 35%) generated by averaging labels across the other 11 subjects (leave-one-out cross-validation). Both the inter-rater agreement and internal consistency were then averaged across subjects.
For further probabilistic atlas validation, we computed (3) the volumes of the delineated final nuclei (i.e., intersection of the labels generated by two independent raters) in native space and compared them with manually computed, Paxinos atlas literature nucleus volumes (Paxinos et al., 2012). For Paxinos atlas nucleus volumes, we obtained images of brainstem plates ranging from 33 to 47 mm from the obex (figures 8.50–8.64 of the study by Paxinos et al., 2012) using Adobe Acrobat Reader. Subsequently, each plate image was translated to single-slice nifti images with proper spatial resolution and 1-mm slice thickness using MATLAB 2019b. Based on the coordinate system provided for each plate (Paxinos et al., 2012), we determined the in-plane isotropic spatial resolution of each plate by manually computing the number of pixels between adjacent coordinates. This varied between 0.037037 and 0.06667 mm in the examined plates.
Using ITK-SNAP (Yushkevich et al., 2006), we manually delineated subregions of nuclei of interest based on the Paxinos nomenclature (Paxinos et al., 2012) and combined their volumes to obtain the final nucleus volume (Table 1). To compute Paxinos volumes for these nuclei/subnuclei, we multiplied the number of voxels of each nucleus/subnucleus delineated in Paxinos images by the voxel volume of each slice and added these numbers across all the slices for each individual nucleus/subnucleus.
Table 1.
Detailed Computation of Nucleus Volumes from a Previous Histology Atlas (Paxinos et al., 2012) and from the Current Study
| Nucleus name (acronym) | Prior study (Paxinos et al., 2012) |
Current study |
|||
|---|---|---|---|---|---|
| Subnucleus name (acronym) | Volume (mm3) | Total volume (mm3) | Right nucleus volume (mm3) mean ± SE | Left nucleus volume (mm3) mean ± SE | |
| Mesencephalic reticular formation (mRt) | Mesencephalic reticular formation (mRt) | 237.28 | 389.13 | 373.46 ± 16.55 | 369.46 ± 16.83 |
| P1 reticular formation (p1RT) | 141.66 | ||||
| Central mesencephalic nucleus (CeMe) | 5.49 | ||||
| Retroparafascicular nucleus (RPF) | 4.70 | ||||
| Isthmic reticular formation (isRt) | Isthmic reticular formation (isRT) | 48.74 | 58.48 | 57.79 ± 2.42 | 53.91 ± 3.30 |
| Nucleus U (U) | 9.75 | ||||
| Microcellular tegmental nucleus–prabigeminal nucleus (MiTg-PBG) | Microcellular tegmental nucleus–spinothalamic tract (MiTg-spth) | 22.11 | 27.86 | 28.06 ± 1.22 | 26.18 ± 1.01 |
| Prabigeminal nucleus (PBG) | 5.75 | ||||
| Ventral tegmental area–parabrachial pigmented nucleus complex (VTA-PBP) | Parabrachial pigmented nucleus of the ventral tegmental area (PBP) | 117.60 | 267.88 | 274.63 ± 6.73 | 276.07 ± 5.87 |
| Ventral tegmental area (VTA) | 99.09 | ||||
| Interpeduncular nucleus, caudal subnucleus (IPC) | 20.95 | ||||
| Interpeduncular fossa (PIF) | 14.21 | ||||
| Parapeduncular nucleus (PaP) | 7.55 | ||||
| Paranigral nucleus of the ventral tegmental | 6.63 | ||||
| area (PN) | |||||
| Interpeduncular nucleus, dorsomedial | 1.22 | ||||
| subnucleus (IPDM) | |||||
| Interfascicular nucleus (IF) | 0.63 | ||||
| Caudal–rostral linear nucleus of the raphe (CLi-RLi) | Rostral linear nucleus (RLi) | 62.04 | 107.75 | 110.69 ± 3.59 | |
| Caudal linear nucleus of the raphe, zygoz | 31.90 | ||||
| part (CLiZ) | |||||
| Caudal linear nucleus of the raphe, azygoz | 13.81 | ||||
| part (CLiAz) | |||||
Statistical analysis: volumes of left nuclei did not significantly differ from volumes of right nuclei (paired one-sample t-test, p < 0.05, computed on mRt, isRt, MiTg-PBG, and VTA-PBP); the volumes of our labels did not differ from Paxinos' volumes (p < 0.05) (Paxinos et al., 2012).
isRt, isthmic reticular formation; MiTg-PBG, microcellular tegmental nucleus–prabigeminal nucleus; mRt, mesencephalic reticular formation; SE, standard error; VTA-PBP, ventral tegmental area–parabrachial pigmented nucleus complex.
Results
The probabilistic neuroimaging structural labels in the MNI space of mRt (including mRtd, mRta, and mRtl), isRt, MiTg-PBG, VTA-PBP, and CLi-RLi based on MRI contrast and neighborhood relationships are shown in Figures 1–5, together with labels of neighboring nuclei.
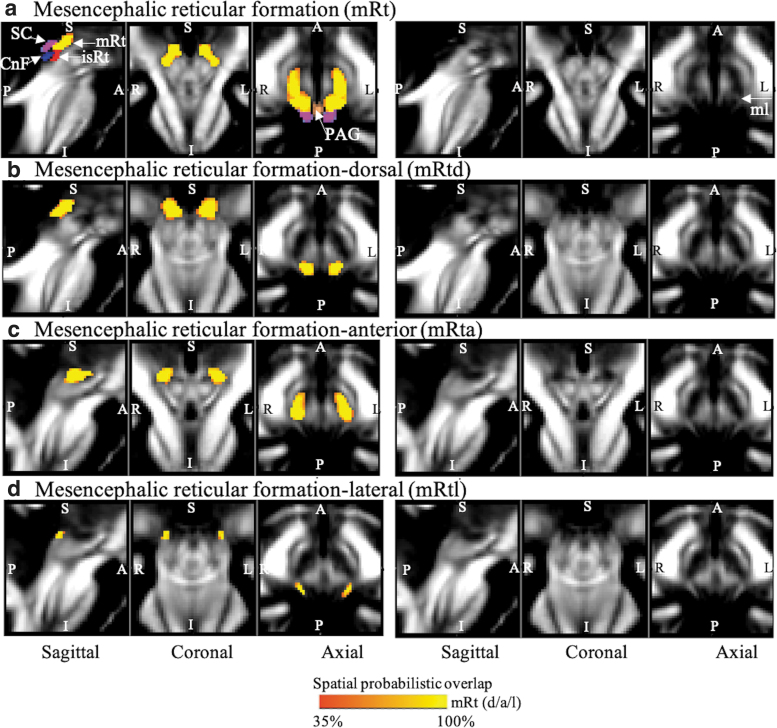
FIG. 1.
Probabilistic (n = 12) atlas label in the MNI space of the (a) entire mRt (both left and right nuclei: red-to-yellow). White arrows indicate neighboring nuclei such as SC (cool), CnF (blue), isRt (red), PAG (copper), and white matter tract of medial lemniscus (mL). Probabilistic (n = 12) atlas label in the MNI space of the (b) mRtd, (c) mRta, and (d) mRtl (both left and right nuclei: red-to-yellow). As a background image, we used the FA map in the MNI space. Very good (i.e., up to 100%) spatial agreement of labels across subjects was observed, indicating the feasibility of delineating the probabilistic labels of these nuclei. CnF, cuneiform nucleus; FA, fractional anisotropy; isRt, isthmic reticular formation; mRt, mesencephalic reticular formation; mRta, mesencephalic reticular formation anterior part; mRtd, mesencephalic reticular formation dorsal part; mRtl, mesencephalic reticular formation lateral part; PAG, periaqueductal gray; SC, superior colliculus.
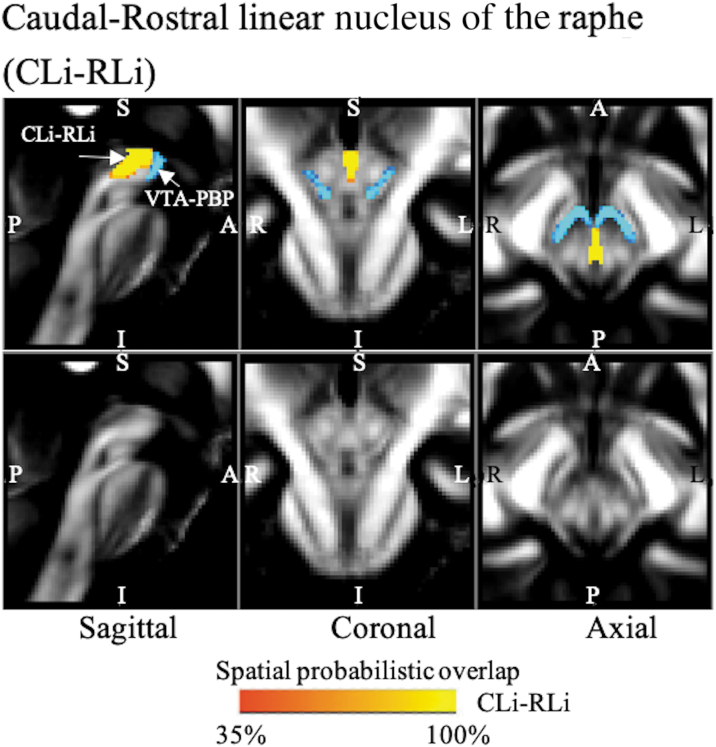
FIG. 5.
The probabilistic (n = 12) atlas label in the MNI space of the CLi-RLi nucleus complex is shown (red-to-yellow) with neighboring nuclei, such as VTA-PBP (blue-to-light blue). As a background image, we used the FA map in the MNI space. Very good (i.e., up to 100%) spatial agreement of labels across subjects was observed, indicating the feasibility of delineating the probabilistic label of this nucleus. CLi-RLi, caudal–rostral linear nucleus of the raphe.
Figure 1 shows the whole mRt and its three subdivisions, viz mRtd, mRta, and mRtl, based on the contrast seen on the MRI images and landmark neighboring regions. Overall, in our images, this set of three subnuclei was located in the dorsolateral part of the mesencephalic tegmentum, anterior to the superior quadrigeminal bodies (i.e., colliculi). It extended from the superior part of the inferior colliculus to the oral border of the superior colliculus (shown in our previous article [García-Gomar et al., 2019] as a T2-weighted hypointense region), progressively increasing in size. It was triangular or quadrangular in shape and medially surrounded by periaqueductal gray matter, dorsally by the superior colliculus, and laterally by the medial lemniscus—visible as a FA hyperintense stripe in our MRI images.
In our MRIs, mRtd was dorsally surrounded by the superior colliculus and dorsally–medially by the periaqueductal gray matter (shown in our previous article [Bianciardi et al., 2015; García-Gomar et al., 2019] as a FA hypointense region). mRta was seen more rostral and anterior to mRtd. It was bound inferiorly by the cuneiform/pedunculotegmental nuclei seen as hypointense regions on FA maps. mRtl appeared as a gray region in FA maps, as opposed to the darker mRta and mRtd regions, due to amalgamation of the white matter tract of the spinothalamic tract. mRtl was seen prominently as a subdivision of mRt, hyperintense on FA maps, and bounded dorsally by the inferior colliculus and medially by the cuneiform nucleus and isRt (described below).
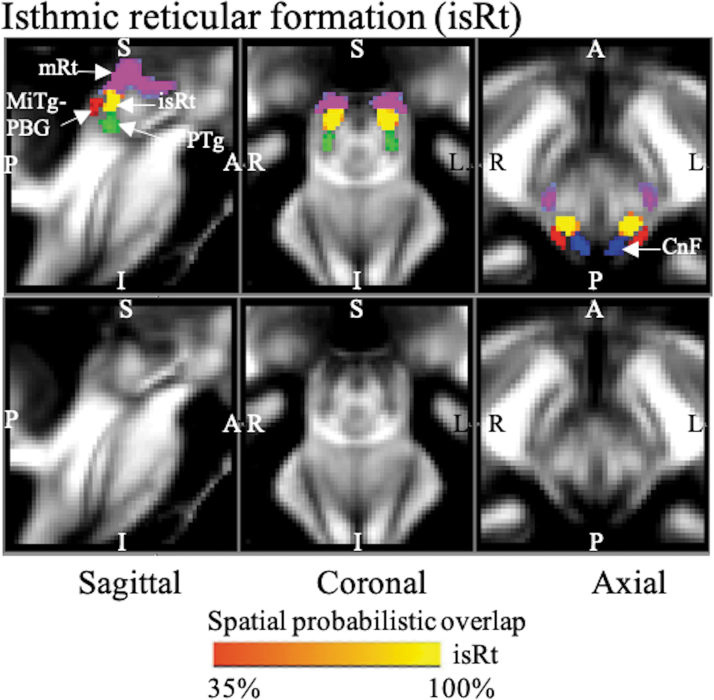
isRt was visible as an FA hypointense region (Fig. 2), a sandwiched region between the gray matter cuneiform nucleus dorsally and pedunculotegmental nucleus ventromedially. It was lined posterior/laterally by our new delineation of MiTg-PBG and superiorly by mRt.
FIG. 2.
The probabilistic (n = 12) atlas label in MNI space of the isRt is shown (red-to-yellow) with neighboring nuclei (white arrows), such as PTg (green), CnF (blue), entire mRt (cool), and MiTg-PBG (red). As a background image, we used the FA map in the MNI space. Very good (i.e., up to 100%) spatial agreement of labels across subjects was observed, indicating the feasibility of delineating the probabilistic labels of these nuclei. CnF, cuneiform nucleus; MiTg-PBG, microcellular tegmental nucleus–prabigeminal nucleus; PTg, pedunculotegmental nucleus.
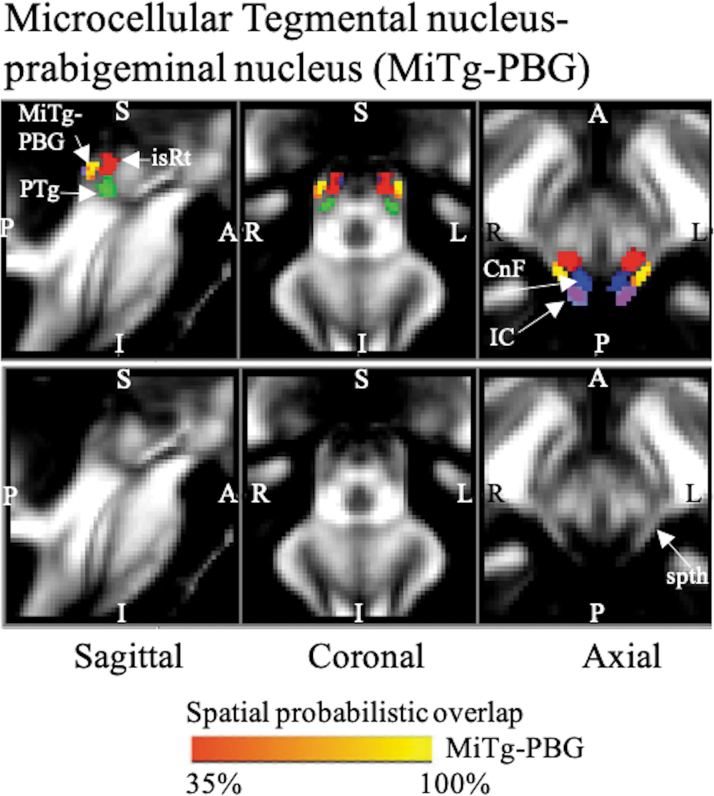
MiTg-PBG (Fig. 3) appeared as a thin stripe of FA hyperintense region as it contains the white matter region of the spinothalamic tract. It was seen lateral to the FA hypointense regions of cuneiform nucleus, pedunculotegmental nucleus, and isRt. It also consisted of PBG laterally, which lies anteriorly to the inferior colliculus.
FIG. 3.
The probabilistic (n = 12) atlas label in MNI space of the MiTg-PBG is shown (red-to-yellow) with neighboring nuclei (white arrows) such as IC (cool), CnF (blue), PTg (green), isRt (red), and white matter spth. As a background image, we used the FA map in the MNI space. Very good (i.e., up to 100%) spatial agreement of labels across subjects was observed, indicating the feasibility of delineating the probabilistic label of this complex of nuclei. IC, inferior colliculus; spth, spinothalamic tract.
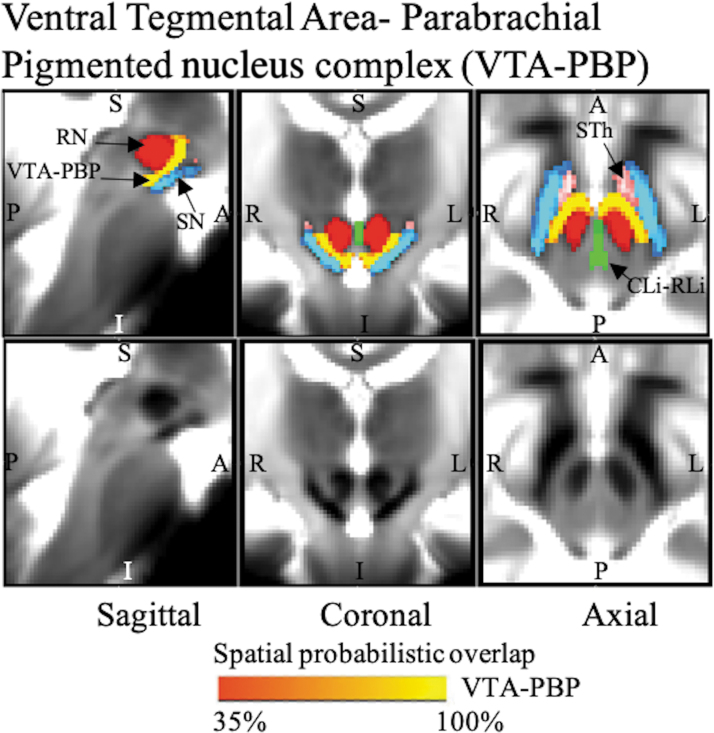
VTA-PBP (Fig. 4) was seen as a horseshoe-shaped nucleus complex visible as a hyperintense region on T2-weighted maps lying between the hypointense regions of the substantia nigra and red nucleus with its hilus pointing dorsally. Specifically, as observed in our images, it lies orally to the decussation of the superior cerebellar peduncle, commencing with the red nucleus. It was seen medially to the substantia nigra, laterally to the paranigral nucleus, and laterally/anteriorly to the oroventral extension of the CLi-RLi. A bridge of cells passing ventral to the linear raphe (namely the interfascicular nucleus/interpeduncular nucleus, dorsomedial subnucleus/interpeduncular nucleus, and caudal subnucleus/interpeduncular fossa/parapeduncular nucleus/parainterfascicular nucleus/paranigral nucleus of ventral tegmental area) and dorsal to the midline portion of the paranigral nucleus connects the VTA-PBP with the corresponding nucleus of the opposite side.
FIG. 4.
The probabilistic (n = 12) atlas label in the MNI space of VTA-PBP is shown (red-to-yellow) with neighboring nuclei (black arrows), such as STh (pink), SN (blue-light blue), RN (red), and CLi-RLi (green). As a background image, we used the T2w map in the MNI space. Very good (i.e., up to 100%) spatial agreement of labels across subjects was observed, indicating the feasibility of delineating the probabilistic label of this nucleus complex. Of note, we found good contrast for these nuclei, sandwiched between RN and SN, showing a horseshoe-shaped structure in the coronal and axial sections, which matched the description of these nuclei from literature (Olszewski and Baxter, 1954; Paxinos et al., 2012). CLi-RLi, caudal–rostral linear nucleus of the raphe; RN, red nucleus; SN, substantia nigra; STh, subthalamic nucleus; VTA-PBP, ventral tegmental area–parabrachial pigmented nucleus complex.
CLi-RLi was seen as a thin stripe of gray matter nuclei lying medially, connecting the VTA-PBP and central tegmental tract (Fig. 5).
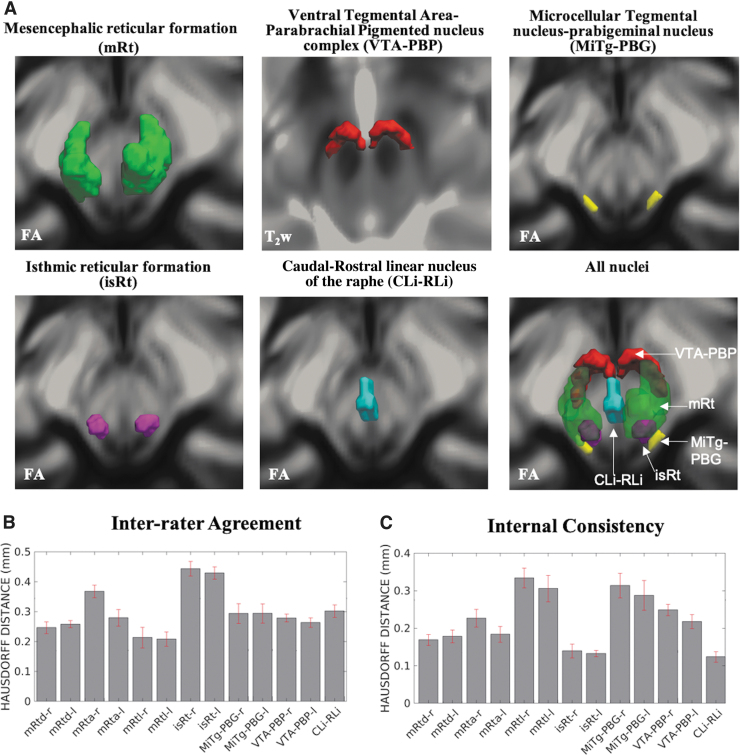
We show 3D rendered images of nucleus labels in Figure 6.
FIG. 6.
(A) 3D rendering of our nucleus labels of the entire mRt, isRt, MiTg-PBG, VTA-PBP, and CLi-RLi in the MNI space and of all the nuclei. As a background image, we used the FA map in the MNI space, except for VTA-PBP, for which we used the T2w image in the MNI space. The 3D rendering was achieved by the use of ITK-SNAP, v. 3.8.0 (Yushkevich et al., 2006), and ParaView, v. 5.9.0 (Ayachit, 2015); images were mildly rotated in the left/right direction to also display the nucleus label depth. (B, C) Atlas validation. (B) The inter-rater agreement of nucleus segmentation (bar/error bar = mean/SE modified Hausdorff distance across 12 subjects). (C) The internal consistency of nucleus labels across subjects (bar/error bar = mean/SE modified Hausdorff distance across 12 subjects). The labels of the mRtd-r/l, mRta-r/l, mRtl-r/l, isRt-r/l, MiTg-PBG-r/l, VTA-PBP-r/l, and CLi-RLi displayed good spatial overlap across raters and subjects (the modified Hausdorff distance was smaller than the spatial imaging resolution, p < 0.05), thus validating the probabilistic nucleus atlas. 3D, three-dimensional; SE, standard error.
For each nucleus, the results of atlas validation in the form of the average modified Hausdorff distance (of the inter-rater agreement and internal consistency) are shown in Figure 6. All the nuclei showed an inter-rater agreement below 0.5 and internal consistency below 0.4. The p-values of the unpaired one-tailed t-test for modified Hausdorff distance for inter-rater agreement and internal consistency were found to be below (p < 10−8) the linear spatial imaging resolution (1.1 mm), thus validating the generated probabilistic atlas labels.
The volume (mean ± SE across subjects) of each final label in native space and nuclei volumes from the literature (Paxinos et al., 2012) are shown in Table 1. The volumes of our labels did not differ from Paxinos' volumes (p < 0.05) (Paxinos et al., 2012). In Table 1, we report details of volume computation of mRt, isRt, MiTg-PBG, VTA-PBP, and CLi-RLi nuclei and of nucleus subregions, as obtained from the Paxinos atlas and our current study.
Discussion
Based on high-resolution imaging data of living humans at 7 Tesla, we demonstrated the feasibility of delineating five major brainstem nuclei, viz mRt (and subregions mRtd, mRta, and mRl), isRt, MiTg-PBG, VTA-PBP, and CLi-RLi. The spatial overlap of these delineated nuclei was computed across subjects, generating a probabilistic atlas of these nuclei in the stereotaxic neuroimaging space. The current atlas was further validated by inter-rater agreement, internal consistency, and volumes compared with that reported in literature (Paxinos et al., 2012). The authors describe here the strength and potential use of this atlas, limitations, conclusions, and future directions.
Feasibility of atlas generation and requirements for its use
We delineated five brainstem nuclei based on high-contrast FA maps and T2-weighted images obtained at 7 Tesla of living humans. 7 Tesla MRI images provided better image sensitivity (∼2.2-fold increase) and resolution compared with 3 Tesla for nucleus delineation. The increased field strength was further supplemented by the use of a custom-built volume transmit coil and 32-channel receive coil (Keil et al., 2010). This coil helped with improved sensitivity for the brainstem, as opposed to commercially available coils, due to (1) structurally complementing the human skull, thus facilitating better signal reception from the brainstem; and (2) efficient and customized arrangement of the coil elements for more efficient flip angle calibration even with same number of channels.
To achieve superior images at 7 Tesla, authors adopted to use (1) a monopolar scheme (given the lower gray and white matter T2 values at higher magnetic field), thus minimizing the echo time; (2) optimized radiofrequency transmit gain using the actual flip angle imaging pulse sequence (Yarnykh, 2007), in our region of interest (i.e., the brainstem), which displayed strong radiofrequency inhomogeneities at 7 Tesla; and (3) semiautomatic B0 shimming.
For wider acceptance and usage of this atlas in research and the clinic, precise coregistration to conventional MRI space would be warranted. Positively, coregistration of subcortical structures and the brainstem is less trivial than that of cortical regions, which present extensive folding (Fischl et al., 2008). Moreover, with advancement in registration methods and tools (Avants et al., 2011; Klein et al., 2010) and additional diffusion-based contrast and T2-weighted templates in the MNI space (Varentsova et al., 2014), including the original T1-weighted template, authors predict superior accuracy of brainstem coregistration. Furthermore, outcomes of brainstem coregistration of single-subject images to target stereotaxic images have shown precise results (e.g., for single-subject FA maps to the IIT FA map, as used here).
Authors have further validated the use of a similar atlas in studies of functional connectomes (Bianciardi et al., 2016), thus providing an example of feasibility of accurate coregistration of these small nuclei across subjects to a common template.
Nucleus function and atlas contribution to future knowledge
mRt is defined as an area in the midbrain tegmentum possibly involved in saccade triggering, feedback control of saccadic activity, and feed forward control of saccade-related activity (Cohen et al., 1985). It receives an extensive topographically organized input from the superior colliculus and has projections to the paramedian pontine reticular formation (Chen and May, 2000). The whole mRt is majorly studied in two divisions of rostral and caudal mRt (central mRt), where caudal mRt is further subdivided into dorsal and ventral parts. Based on high-resolution multicontrast MRI, we identified three subdivisions of mRt as dorsal, anterior, and lateral mRt subdivisions. We speculate that our current study will help in the spatiotemporal investigation of collicular saccadic signals by providing a structural distinction of these nuclei and subnuclei in living humans.
isRt, a relatively understudied nucleus, has been shown in sea lampreys (Barreiro-Iglesias et al., 2008; Villar-Cerviño et al., 2009) and humans (Paxinos et al., 2012) as a region sandwiched between more studied regions of cuneiform and pedunculotegmental nuclei. Due to its smaller volume and proximity to other nuclei, there is sparse literature elucidating its location and function. The current study delineates this nucleus and speculates further study based on the current atlas.
The anatomical location of the relatively understudied region of MiTg-PBG has been recently explored in rodents (Watson et al., 2017); however, there is a paucity of studies specifically reporting its functional significance. Under the light of past and current delineation from this group, where its neighboring regions have been distinctly reported, authors anticipate the use of the current atlas in developing a holistic understanding of these nuclei.
The midbrain dopamine system has been extensively studied to understand behavioral disorders, such as substance use disorder, schizophrenia, and Parkinson's disease. This system is highly heterogeneous where, among other regions, VTA and linear raphe neurons overlap in neurochemical characteristics, leading to lack of successful treatments for these conditions. Other overlapping factors may be their afferent and efferent projections, electrophysiological patterns, neurotransmitter release, and responsiveness to appetitive or aversive stimuli (Faget et al., 2016). With the current probabilistic atlas of VTA-PBP and CLi-RLi, authors speculate that future functional studies might benefit from structural and functional distinction and help in better understanding and treatment of such disease conditions.
Areas for improvement
The number of subjects in this study was limited to 12; yet, as for this study, a similar sample size in previous brain/brainstem template work (Bianciardi et al., 2015, 2018; Croxson et al., 2018; García-Gomar et al., 2019; Singh et al., 2019) was sufficient to achieve significant results. There was no significant difference in probabilistic label volumes of the current study compared with Paxinos histology-based atlas drawings (Paxinos et al., 2012). However, authors acknowledge that in vivo validation with ex vivo histology would have been a step ahead and provided additional anatomical validation.
Summary, Conclusion, and Future Directions
In summary, we foresee that the generated probabilistic template of mRt (mRtd, mRta, and mRl), isRt, MiTg-PBG, VTA-PBP, and CLi-RLi in the stereotaxic space—representative of younger human adults—might be a useful tool for improving localization of brainstem nuclei in MRI of living humans. The current probabilistic atlas based on MRI contrast encodes the uncertainty of boundary locations across subjects, which enables its applicability with included chances of error. The developed in vivo atlas of five brainstem nuclei is a new, user-friendly, 3D, probabilistic, and deformable tool that can be used to localize these structures in both conventional (e.g., 3 Tesla) and advanced MRI images, circumventing the challenging task of extrapolating locations of small nuclei from histology-based atlases.
Public release of the current atlas will enable potential users to adopt it and help in unraveling the brainstem structure-to-function link, in health and disease, with the potential of improving clinical outcomes.
Authors' Contributions
M.B. designed the research, performed the experiments, and secured the funding. M.B. preprocessed and analyzed the in vivo data. K.S. and M.B. manually labeled the brainstem nuclei. K.S., M.G.G.-G., and M.B. wrote the manuscript. All authors gave feedback along the process.
Author Disclosure Statement
Authors have no conflicts of interest to disclose.
Funding Information
This work was supported by the following sources of funding: the National Institute on Deafness and other Communication Disorders, R21 DC015888; National Institutes of Health (NIH) National Institute of Biomedical Imaging and Bioengineering, K01 EB019474 and P41 EB015896, National Institute on Aging, R01 AG063982, and National Cancer Institute, U01 CA193632; Massachusetts General Hospital Claflin Distinguished Scholar Award; and Harvard University Mind/Brain/Behavior Faculty Award. This work was also, in part, made possible by the resources provided by Shared Instrumentation Grants, 1S10RR023401, 1S10RR019307, and 1S10RR023043.
References
- Avants BB, Tustison NJ, Song G, et al. 2011. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54:2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayachit U. 2015. The Paraview Guide: A Parallel Visualization Application. NY, USA: Kitware, Inc. [Google Scholar]
- Barreiro-Iglesias A, Villar-Cerviño V, Anadón R, et al. 2008. Development and organization of the descending serotonergic brainstem-spinal projections in the sea lamprey. J Chem Neuroanat 36:77–84. [DOI] [PubMed] [Google Scholar]
- Bianciardi M, Strong C, Toschi N, et al. 2018. A probabilistic template of human mesopontine tegmental nuclei from in vivo 7T MRI. Neuroimage 170:222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciardi M, Toschi N, Edlow BL, et al. 2015. Toward an in vivo neuroimaging template of human brainstem nuclei of the ascending arousal, autonomic, and motor systems. Brain Connect 5:597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciardi M, Toschi N, Eichner C, et al. 2016. In vivo functional connectome of human brainstem nuclei of the ascending arousal, autonomic, and motor systems by high spatial resolution 7-Tesla fMRI. MAGMA 29:451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, May PJ. 2000. The feedback circuit connecting the superior colliculus and central mesencephalic reticular formation: a direct morphological demonstration. Exp Brain Res 131:10–21. [DOI] [PubMed] [Google Scholar]
- Cohen B, Matsuo V, Fradin J, et al. 1985. Horizontal saccades induced by stimulation of the central mesencephalic reticular formation. Exp Brain Res 57:605–616. [DOI] [PubMed] [Google Scholar]
- Croxson PL, Forkel SJ, Cerliani L, et al. 2018. Structural variability across the primate brain: a cross-species comparison. Cereb Cortex 28:3829–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson MP, Jain AK. A modified Hausdorff distance for object matching. In Proceedings of 12th International Conference on Pattern Recognition, Jerusalem, Israel, 1994, pp. 566–568. [Google Scholar]
- Faget L, Osakada F, Duan J, et al. 2016. Afferent inputs to neurotransmitter-defined cell types in the ventral tegmental area. Cell Rep 15:2796–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Rajendran N, Busa E, et al. 2008. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex 18:1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gomar MG, Strong C, Toschi N, et al. 2019. In vivo probabilistic structural atlas of the inferior and superior colliculi, medial and lateral geniculate nuclei and superior olivary complex in humans based on 7 Tesla MRI. Front Neurosci 13:764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil B, Triantafyllou C, Hamm M, et al. Design optimization of a 32-channel head coil at 7 T. In Proceedings of the Annual Meeting of the International Society for Magnetic Resonance in Medicine; Stockholm, 2010, p. 1493. [Google Scholar]
- Klein A, Ghosh SS, Avants B, et al. 2010. Evaluation of volume-based and surface-based brain image registration methods. Neuroimage 51:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moruzzi G, Magoun HW. 1949. Brain stem reticular formation and activation of the EEG. Electroencephalogr Clin Neurophysiol 1:455–473. [PubMed] [Google Scholar]
- Naidich TP, Duvernoy HM, Delman BN, et al. 2009. Duvernoy's Atlas of the Human Brain Stem and Cerebellum: High-Field MRI, Surface Anatomy, Internal Structure, Vascularization and 3 D Sectional Anatomy. Vienna, Austria: Springer Science & Business Media. [Google Scholar]
- Olszewski J, Baxter D. 1954. Cytoarchitecture of the human brain stem. www.cabdirect.org/cabdirect/abstract/19562201844 Last accessed Apr. 19, 2019.
- Pauli WM, Nili AN, Tyszka JM. 2018. A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Sci Data 5:180063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Huang X. 1995. Atlas of the Human Brain Stem. San Diego: Academic Press. [Google Scholar]
- Paxinos G, Xu-Feng H, Sengul G, et al. 2012. Chapter 8—organization of brainstem nuclei. In: Mai JK, Paxinos G (eds.) The Human Nervous System, 3rd ed. San Diego, CA: Academic Press; pp. 260–327. [Google Scholar]
- Reese TG, Heid O, Weisskoff RM, et al. 2003. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn Reson Med 49:177–182. [DOI] [PubMed] [Google Scholar]
- Singh K, Indovina I, Augustinack JC, et al. 2019. Probabilistic template of the lateral parabrachial nucleus, medial parabrachial nucleus, vestibular nuclei complex, and medullary viscero-sensory-motor nuclei complex in living humans from 7 Tesla MRI. Front Neurosci 13:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stejskal EO, Tanner JE. 1965. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J Chem Phys 42:288–292. [Google Scholar]
- Trutti AC, Mulder MJ, Hommel B, et al. 2019. Functional neuroanatomical review of the ventral tegmental area. Neuroimage 191:258–268. [DOI] [PubMed] [Google Scholar]
- Varentsova A, Zhang S, Arfanakis K. 2014. Development of a high angular resolution diffusion imaging human brain template. Neuroimage 91:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Cerviño V, Barreiro-Iglesias A, Anadón R, et al. 2009. Development of glycine immunoreactivity in the brain of the sea lamprey: comparison with gamma-aminobutyric acid immunoreactivity. J Comp Neurol 512:747–767. [DOI] [PubMed] [Google Scholar]
- Watson C, Shimogori T, Puelles L. 2017. Mouse Fgf8-Cre-LacZ lineage analysis defines the territory of the postnatal mammalian isthmus. J Comp Neurol 525:2782–2799. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Jannin P, D'Albis T, et al. 2014. Investigation of morphometric variability of subthalamic nucleus, red nucleus, and substantia nigra in advanced Parkinson's disease patients using automatic segmentation and PCA-based analysis. Hum Brain Mapp 35:4330–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnykh VL. 2007. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn Reson Med 57:192–200. [DOI] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, et al. 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128. [DOI] [PubMed] [Google Scholar]