Abstract
Objective
Periodontitis is a highly prevalent oral infectious disease and has been increasingly associated with H. pylori infection, gastric inflammation, and gastric cancer but little is known about epigenetic machinery underlying this potentially bidirectional association. The present study is aimed at identifying key deregulated miRNA, their associated genes, signaling pathways, and compounds linking periodontitis with H. pylori-associated peptic ulcer disease.
Methods
miRNA expression datasets for periodontitis-affected and H. pylori-associated peptic ulcer disease-affected tissues were sought from the GEO database. Differentially expressed miRNA (DEmiRNAs) were identified and the overlapping, shared-DEmiRNA between both datasets were determined. Shared-DEmiRNA-target networks construction and functional analyses were constructed using miRNet 2.0, including shared-DEmiRNA-gene, shared-DEmiRNA-transcription factor (TF), and shared-DEmiRNA-compound networks. Functional enrichment analysis for shared DEmiRNA-gene and shared DEmiRNA-TF networks was performed using the KEGG, Reactome, and Geno Ontology (GO) pathways.
Results
11 shared-DEmiRNAs were identified, among which 9 showed similar expression patterns in both diseases, and 7 were overexpressed. miRNA hsa-hsa-mir-155-5p and hsa-mir-29a-3p were top miRNA nodes in both gene and TF networks. The topmost candidate miRNA-deregulated genes were PTEN, CCND1, MDM2, TNRC6A, and SCD while topmost deregulated TFs included STAT3, HIF1A, EZH2, CEBPA, and RUNX1. Curcumin, 5-fluorouracil, and the gallotanin 1,2,6-Tri-O-galloyl-beta-D-glucopyranose emerged as the most relevant linkage compound targets. Functional analyses revealed multiple cancer-associated pathways, PI3K pathways, kinase binding, and transcription factor binding among as enriched by the network-associated genes and TFs.
Conclusion
Integrative analysis of deregulated miRNAs revealed candidate molecular mechanisms comprising of top miRNA, their gene, and TF targets linking H. pylori-infected peptic ulcer disease with periodontitis and highlighted compounds targeting both diseases. These findings provide basis for directing future experimental research.
1. Introduction
Periodontitis is a multifactorial infectious disease characterized by the inflammatory destruction of the supporting structures of the teeth in response to a dysbiotic dental plaque biofilm [1]. Periodontitis lesions represent chronic wounds that can impose significant systemic inflammatory burden and periodontal disease has been linked with a plethora of metabolic and other nonoral diseases [2]. Emerging evidence has highlighted an association of severe periodontal disease with peptic ulcer disease, chronic gastritis [3, 4], and also with gastric carcinogenesis [5]. Gastric inflammation and carcinogenesis are strongly associated with H. pylori infection [6–8]. Considering the serious global burden imposed by gastric cancer [9], the association of H. pylori infection and gastric inflammation with periodontal disease merits further mechanistic investigation. Several studies have identified the presence of a reservoir of H. pylori in the oral microflora of periodontitis patients [10–13]. In concurrence, the treatment of periodontitis has been found to improve gastric H. pylori elimination among peptic ulcer disease patients in multiple trials [14]. Others have suggested oral and gastric H. pylori infections may comprise a risk factor for periodontitis [13, 15]. Although it was earlier believed that oral detection of H. pylori did not represent viable bacteria but represented the genetic remnants derived from gastric colonization, viable H. pylori has been isolated from root canals [16]. In addition, live H. pylori bacterial could be cultured from saliva of gastric H. pylori-positive subjects [17]. Experimental data has shown that H. pylori infection and its virulence factor CagA can exert proinflammatory effects and promote the growth of periodontal pathogens [18].
Currently, very little is understood about the molecular mechanisms underlying the possibly syndemic association of periodontitis and H. pylori-associated gastric inflammatory conditions. MircoRNAs (miRNA) are small RNAs that play critical regulatory roles in controlling gene expression, thereby modulating cellular processes and playing key roles in mediating disease [19, 20]. The identification of disease-associated miRNA and their deregulation patterns can provide an insight into molecular pathogenesis events. In addition, such miRNA can serve as biomarkers or therapeutic drug target candidates [21]. Integrative bioinformatics has been applied successfully to decipher miRNA deregulation in oral diseases [22]. miRNAs have been identified as biomarkers of periodontal disease [23], and bioinformatic investigation has identified miR-125a-3p, miR-200a, and miR-142-3p as key deregulated miRNA in periodontitis [24]. In principle, deregulated miRNAs in periodontitis are not contratined to the tissue and can possibly enter circulation via exosomal carriage and thereby alter gene expression in remote sites [25]. In support, miR-146a has been implicated in the association of periodontitis with cardiovascular disease [26].
With this context, the present study is aimed at performing an integrated analysis of miRNA deregulation patterns in periodontitis and H. pylori-associated peptic ulcer disease to identify candidate linkage miRNAs, their associated genes, signaling pathways, and relevant compounds. The broader goal of this approach was to advance the understanding of molecular linkage mechanisms in context of these diseases and provide theoretical basis for driving future experimental research.
2. Materials and Methods
2.1. Microarray Data
miRNA expression datasets of the target conditions were sought from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The miRNA expression dataset for periodontitis, GSE54710 [27] (acquired on the Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray 030840 platform), and the dataset GSE32174, for H. pylori-associated duodenal ulcers [28] (acquired on the Illumina Human v1 microRNA expression beadchip) were identified for analysis. The GSE54710 dataset contains 200 samples, with 158 periodontitis-affected and 41 control tissue samples. The GSE32174 dataset contains samples from antral biopsies, including a total of 48 samples with 9 H. pylori-negative and 39 H. pylori-positive samples identified in the metadata.
2.2. Identification of DEmiRNA and Shared-DEmiRNA
The microarray data expression matrix and metadata were downloaded using the “GEOquery” v 2.32.0 package in R (https://www.r-project.org/). Next, differentially expressed miRNA was identified using the R package “limma” v.3.22.7 [29]. miRNA with expression levels above the median and in more than 2 samples was considered as “expressed” and utilized in the analysis. The assigned labels of the samples (periodontitis affected versus controls for GSE54710, and H. pylori-positive versus negative for GSE32174) were utilized for grouping, and DEmiRNA was identified using an FDR adjusted p value < 0.05. Probe IDs were mapped to miRBase miRNA IDs (version 8.). The two DEmiRNA lists were processed using the R package “Venndiagram” v 1.6.9 [30], and the shared-DEmiRNA was obtained. These were considered the shared DEmiRNA and were subjected to further analysis.
2.3. Shared-DEmiRNA-Target Network Construction and Functional Enrichment Analyses
DEmiRNA target networks were constructed using miRNet 2.0 (https://www.mirnet.ca/) [31]. For the shared-DEmiRNA-gene network, target genes were selected from the 3 databases (miRTarBase v8.0, TarBase v8.0, and miRecords). A “minimum network” was selected in order to reduce network complexity and retain the key features demonstrating network connectivity. It was computed using all the seed or query nodes. To construct a “minimum network,” pair-wise shortest paths between the seed nodes are determined, and any nodes not on the shortest paths are removed. A similar approach was used to construct a shared-DEmiRNA-transcription factor (TF) network, which utilizes TransmiR v2.0., and a shared-DEmiRNA-small molecule network, which is based upon data from SM2miR and PharmacomiR, each. Functional enrichment analysis was conducted with a hypergeometric test algorithm for shared DEmiRNA-gene and shared DEmiRNA-TF networks using the KEGG, Reactome, and Geno Ontology (GO) pathways with the query set as “all genes” in the 2 minimum networks and top 10 significantly enriched functions ranked by p value were tabulated.
3. Results
3.1. Identification of DEmiRNA and Shared-DEmiRNA
After filtering, 1200 miRNA were retained in the analysis for the GSE54710 dataset, and a total of 172 significant periodontitis-associated DEmiRNA were identified, whereas 425 miRNAs were retained for the GSE32174 dataset analysis and 82 significant H. pylori infection-associated DEmiRNA were identified (Supplementary Table-S1). 177 DEmiRNA were overexpressed, and 55 DEmiRNA were underexpressed as compared to controls in periodontitis-affected tissue. Similarly, 49 miRNAs were overexpressed in H. pylori-infected tissue, while 33 were underexpressed. The overlap between the two DEmiRNA lists was sought using Venn diagram, and a total of the 11 shared-DEmiRNA were identified (Figure 1).
Figure 1.
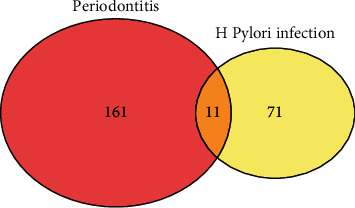
Venn diagram depicting the shared DEmiRNAs between periodontits-affected and H. pylori-infected tissues identified using the GEO datasets GSE54710 and GSE32174, respectively.
Corresponding expression patterns in the 2 conditions are depicted in Figure 2. Similar expression patterns were evident in case of 9 shared-DE miRNAs, where overexpression was noted in 7 and underexpression was seen in 2 shared-DEmiRNAs as compared to control tissues in both conditions. 2 shared-DEmiRNAs, hsa-miR-652 and has-miR-30a, showed opposing expression patterns in both the conditions, with underexpression as compared to control in case of H. pylori infection but overexpression in case of periodontitis-affected tissue.
Figure 2.
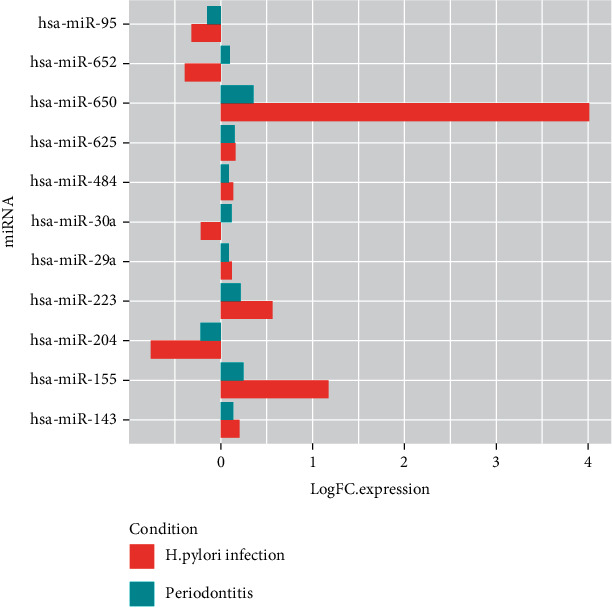
Bar plots depicting the log FC expression values of the shared-DEmiRNAs in periodontitis and H. pylori infection.
3.2. Shared-DEmiRNA-Target Networks and Functional Analyses
Shared-DEmiRNA-gene network: the shared-DEmiRNA-gene minimum network comprised of 105 genes and 27miRNA with 524 edges (Figure 3, Supplementary Table S2). The top DEmiRNA nodes with the highest degrees in the network were hsa-mir-484, hsa-mir-155-5p, hsa-mir-29a-3p, and hsa-mir-30a-5p. The top 5 gene nodes with the highest degrees in the network included PTEN, CCND1, MDM2, and TNRC6A. The top 10 enriched functions in each are listed in Table 1(a). The reactome analysis showed signaling by SCF-KIT, oncogene-induced senescence, pre-NOTCH transcription and translation, and multiple PI3K signaling-related pathways among the top enriched pathways. GO biological process analysis showed negative regulation of multiple processes such as cellular and RNA metabolic processes and transcription. Top enriched GO molecular functions included multiple binding associated functions including protein kinase, enzyme, TF, and chromatin binding. Top GO cellular components enriched included cytosol, nucleoplasm, and nuclear lumen.
Figure 3.
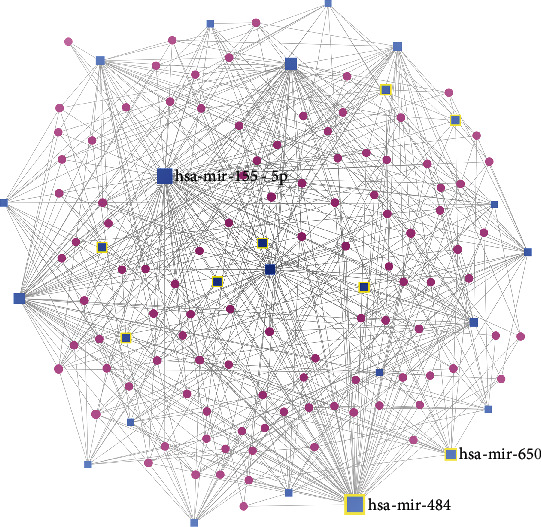
Shared DEmiRNA-genes minimum network visualized using the visual analytics platform “miRnet.” The seed nodes are highlighted. It comprised of 105 genes and 27miRNA with 524 edges.
Shared-DEmiRNA-TF network: the shared-DEmiRNA-TF minimum network comprised of 10 TFs and 7miRNA with 24 edges (Figure 4, Supplementary Table S3). The top 5 TFs included STAT3, HIF1A, EZH2, CEBPA, and RUNX1, while hsa-mir-29a and hsa-mir-155 were the topmost miRNA nodes. The top enriched KEGG, Reactome, and GO pathways are listed in Table 1(b). These included the reactome pathways of responses to stress, cellular senescence, and multiple NOTCH signaling-related pathways. GO BP pathway analysis showed the regulation of cell development, differentiation, and transcription as among those implicated. GO molecular functions enhanced among the TFs in the network included multiple transcription-related functions.
Figure 4.
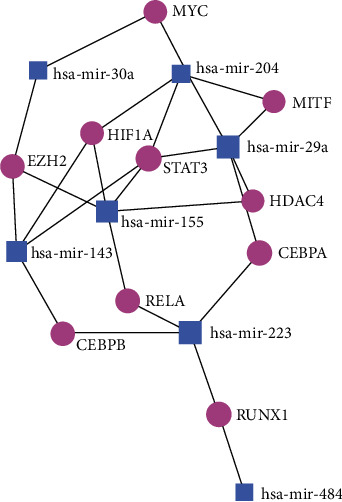
Shared-DEmiRNA-TF minimum network comprised of 10 TFs and 7 miRNA with 24 edges.
Shared DE-miRNA-small molecule network: the shared DE-miRNA-small molecule minimum network comprised of 7 compounds and 10 miRNAs with 21 edges (Figure 5, Supplementary Table S4). The 7 compounds in this network included 5-fluorouracil, curcumin, 1,2,6-Tri-O-galloyl-beta-D-glucopyranose, glucocorticoid, cisplatin, doxorubicin, and formaldehyde. The compound with the highest degree and betweeness was curcumin, followed by 5-fluorouracil.
Figure 5.
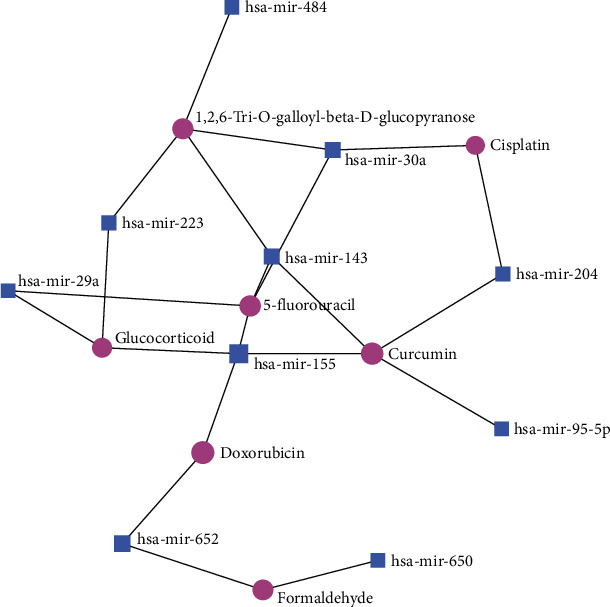
The shared DE-miRNA-small molecule minimum network comprised of 7 compounds and 10 miRNAs with 21 edges.
4. Discussion
The present bioinformatics study explored miRNA-mediated regulatory mechanisms implicated in the association of gastric H. pylori infection with periodontitis by identifying common deregulated miRNA in both conditions. Network analysis was applied to identify the key candidate genes and TFs that may act as linkage molecular mechanisms operating via miRNA-mediated deregulation. In addition, small molecules and compounds associated with the shared DEmiRNA were identified. Similar expression patterns of the shared DEmiRNA (Figure 1) were evident for most of the deregulated miRNA in both conditions, suggesting common deregulatory patterns in host immune mechanisms implicated in the two conditions. The shared DEmiRNA with the highest degrees included hsa-mir-484 and hsa-mir-155, followed by hsa-mir-29a, which were found overexpressed in both conditions, and hsa-mir-30a which showed contradictory expression patterns. Both miR 155 [32–34] and miR-484 [35, 36] have been widely implicated in inflammation and cancer. miRNA 29a is shown to be an important regulator of the intestinal barrier function [37] and also implicated as a cancer biomarker [38, 39]. miRNA is an important effector of the host-bacterial interaction, whereby pathogenic microbiota alter host miRNA production to enhance their survival [40]. Chronic gastric H. pylori infection is an important risk factor for gastric cancer [6–8], while severe chronic periodontitis has also been linked to gastric cancer [5, 41]. H. pylori infection has been associated with higher levels of key periodontal pathogens and was found to enhance the expression of IL-8 and Wnt5a, suggesting it may act as an aggravating factor for periodontal disease [18]. Overall, a bidirectional and syndemic link between H. pylori and periodontitis is likely, and the key deregulated miRNA identified in this study may contribute to the oncogenic risks for oral and gastric carcinogenesis arising from both conditions.
miRNA-mediated deregulation of gene expression is likely to be a key molecular mechanism linking periodontitis with systemic diseases [25]. The genes with the highest degrees in shared-DEminRNA-gene network included PTEN, CCND1, MDM2, SCD, and TNRC6A, and these may be important miRNA-deregulated mediators linking H. pylori infection and periodontitis. PTEN (Phosphatase and Tensin Homolog) signaling is involved in regulating multiple cellular processes, and its deregulation has been associated with cancer [42]. PTEN is a well-known tumor suppressor and is commonly inactivated in multiple cancers, playing key role in regulating the PI3K/AKT/mTOR pathway which effects cancer cell survival [43]. Multiple studies have associated the deregulation of PTEN with gastric carcinogenesis, leading to inhibition of its tumor suppressor function [44–46]. miRNA-mediated alteration of PTEN signaling has been shown as an important pathway in gastric cancer development [47], and its phosphorylation and inhibition by H. pylori infection have been demonstrated [48]. PTEN downregulation is also documented in periodontitis [49] and P. ginivalis, and the keystone periodontal pathogen was found to promote esophageal carcinoma cell by modulation of PTN signaling [50]. CCND1 has been associated with gastric cancer [51–53], and its polymorphisms have been hypothesized to mediate risk for periodontitis-linked oral cancer [54], suggesting similar phenomena in gastric cancer. MDM2 has been implicated in oxidative stress-mediated P53 gene upregulation and damage of gingival fibroblasts [55], and its upregulation is also associated with gastric H. pylori infection [56]. SCD or stearoyl CoA desaturase-1 is implicated in fatty acid synthesis and has been associated with metabolic disorders [57] and gastric cancer [58]. In a rat model of periodontitis, SCD-1 was found markedly upregulated in the liver and associated with metabolic aberration [59]. miRNA-mediated deregulation of TNRC6A has been implicated in gastric and colorectal cancers [60] although knowledge about its role in periodontitis remains scarce.
In the shared-DEmiRNA TF network, STAT3, EZH2, HIF1A, CEBPA, and RUNX1 showed the highest degrees. STAT3 (signal transducer and activator of transcription 3) activation is implicated in inflammation-associated carcinogenesis, and its phosphorylation by H. pylori has been documented [61]. STAT3 pathway activation is documented in periodontitis [62] and is also found to contribute to neuroinflammation [63]. EZH2 encodes for a methyltransferase enzyme that is involved in osteoclast regulation and is proposed as a target for periodontitis [64]. EZH2 was found downregulated in response to short-chain fatty acids produced by periodontal pathogens, which contributes to cancer development [65]. Its expression was, however, reported to be upregulated by H. pylori virulence factor cytotoxin-associated gene A [66] and increasing expression levels associated with the multistep gastric carcinogenesis process [67]. HIF1A, involved in cellular responses to oxidative stress, has been implicated in the immune-inflammatory response in periodontitis [68] and is also upregulated in H. pylori-mediated gastric inflammation [69] and carcinogenesis [70]. Consistent with our finding, CEBPA, an NF-κB-related associated gene, has been previously reported in a bioinformatics study as a key TF involved in H. pylori-mediated immune deregulation [71, 72] and validated as downregulated in infected gastric tissues [73]. In periodontitis, lower serum levels of CEBPA have been reported [74]. The RUNX genes are important regulators of cellular development, and reduced RUX1 expression in gastric cancer cell lines is documented [75]. RUNX1 is involved in alveolar osteocalstogenesis [76].
Functional enrichment analysis of the shared-DEmiRNA gene and TF networks showed several cancer-related KEGG pathways as enriched, supporting the emerging findings of increased gastric and esophageal carcinogenesis risk in periodontitis, evidenced in large sample prospective data [77]. Reactome analysis indicated multiple PI3K pathway alterations as shared linkages, which are implicated in H. pylori-mediated inflammation [78] and widely understood to function in carcinogenesis [79]. Multiple GO molecular functions related to kinase binding were enriched, which are pivotal to the regulation of innate immune responses and inflammation [80].
In the present study, small compounds targeted by the shared-DEmiRNA were also analyzed. 5-fluorouracil, curcumin, and 1,2,6-Tri-O-galloyl-beta-D-glucopyranose were identified as the top candidates. H. pylori CagA protein is found to reduce the sensitivity of gastric cancer cells to the antineoplastic agent 5-fluorouracil [81], which is also shown to exacerbate periodontitis progression [82]. However, the role of periodontitis in modulating resistance to 5-fluorouracil in gastric cancer remains unknown. Curcumin, a phytochemical, has shown anti-inflammatory effects in periodontitis [83] and also shows anti-infective efficacy against gastric H. pylori [84]. Taken together, these findings suggest curcumin might be a candidate therapeutic for cotreatment of both these conditions. 1,2,6-Tri-O-galloyl-β-d-glucopyranose (1,2,6-TGGP) is a gallotanin that has shown efficacy against multiple bacteria and diseases [85] and can be considered a viable candidate phytochemical to combat both H. pylori gastric infections and periodontitis. Future studies are warranted to investigate the identified molecular candidates in the context of the association of these diseases.
The major limitation of the present study is the lack of validating experimental data to verify to candidate molecular linkage mechanisms discovered through bioinformatics. The datasets used in the present study include small sample numbers and are from single centers, which can limit the robustness of the predictions. Similar studies using multiple large datasets with comprehensive metadata are essential. An important caveat is that deregulated exosomal or circulatory miRNA was not analyzed in the present investigation, and these may be more important in the pathogenic mechanisms linking periodontitis with systemic disease [25]. In addition, future studies may address deregulated, genes, or other noncoding RNAs as linkage mechanisms. Future studies should be designed to verify the miRNAs, their associated genes, TF, and functional pathways that constitute putative linkages between H. pylori-infected gastric diseases and periodontitis using validation experiments using clinical cohorts, animal, and cell models. Such data can permit the discovery of molecular targets for both conditions. Furthermore, as the association is likely to be bidirectional in nature, a comprehensive understanding the biological mechanisms involved via both experimental and clinical research is warranted.
5. Conclusions
Integrative analysis of common deregulated miRNAs in H. pylori-associated peptic ulcer and periodontitis-affected tissues revealed key candidate linkage molecular mechanisms comprising of miRNAs (including hsa-mir-155-5p, hsa-mir-484, and hsa-mir-29a-3p), genes (including PTEN, CCND1, MDM2, TNRC6A, and SCD), and TF targets (including STAT3, HIF1A, EZH2, CEBPA, and RUNX1) and highlighted the most relevant compounds (including curcumin, 5-fluorouracil, and 1,2,6-tri-O-galloyl-beta-D-glucopyranose) in this context. These findings provide a theoretical basis for directing future experimental studies.
Table 1.
(a) Top enriched pathways in the shared DEmiRNA-gene network
| KEGG pathway | p value |
| Glioma | 4.48E-06 |
| Neurotrophin signaling pathway | 4.51E-06 |
| Prostate cancer | 3.14E-05 |
| Pathways in cancer | 7.11E-05 |
| Melanoma | 7.23E-05 |
| Chronic myeloid leukemia | 0.000108 |
| Bladder cancer | 0.000225 |
| Cell cycle | 3.00E-04 |
| Cysteine and methionine metabolism | 0.000422 |
| p53 signaling pathway | 0.000713 |
| Reactome pathway | |
| Signaling by SCF-KIT | 1.43E-05 |
| Oncogene induced senescence | 1.70E-05 |
| Pre-NOTCH transcription and translation | 4.81E-05 |
| PI3K events in ERBB4 signaling | 7.45E-05 |
| PIP3 activates AKT signaling | 7.45E-05 |
| PI3K events in ERBB2 signaling | 7.45E-05 |
| PI-3K cascade:FGFR1 | 7.45E-05 |
| PI-3K cascade:FGFR2 | 7.45E-05 |
| PI-3K cascade:FGFR3 | 7.45E-05 |
| PI-3K cascade:FGFR4 | 7.45E-05 |
| GO-BP | |
| Negative regulation of metabolic process | 1.46E-09 |
| Negative regulation of cellular metabolic process | 1.96E-09 |
| Negative regulation of transcription from RNA polymerase II promoter | 2.02E-09 |
| Negative regulation of cellular biosynthetic process | 5.27E-09 |
| Negative regulation of biosynthetic process | 7.61E-09 |
| Negative regulation of transcription, DNA-dependent | 9.74E-08 |
| Negative regulation of transcription, DNA-dependent | 9.74E-08 |
| Negative regulation of RNA metabolic process | 1.93E-07 |
| Negative regulation of nucleobase-containing compound metabolic process | 3.08E-07 |
| Generation of neurons | 1.23E-06 |
| GO-MF | |
| Protein kinase binding | 4.93E-09 |
| Kinase binding | 2.20E-08 |
| Negative regulation of transcription, DNA-dependent | 3.35E-08 |
| Chromatin binding | 4.92E-07 |
| Transcription factor binding | 1.78E-06 |
| Enzyme binding | 4.41E-06 |
| Transcription from RNA polymerase II promoter | 8.05E-06 |
| Transcription corepressor activity | 8.85E-05 |
| SMAD binding | 1.00E-04 |
| Protein binding transcription factor activity | 0.000788 |
| GO-CC | |
| Cytosol | 4.70E-07 |
| Nucleoplasm | 2.96E-06 |
| Nuclear lumen | 5.22E-06 |
| Organelle lumen | 8.75E-06 |
| Nucleoplasm part | 9.20E-06 |
| Macromolecular complex | 1.19E-05 |
| Membrane-enclosed lumen | 1.35E-05 |
| Transcription factor complex | 1.65E-05 |
| Membrane-bounded vesicle | 3.32E-05 |
| Nuclear chromatin | 5.44E-05 |
(b) Top enriched pathways in shared DEmiRNA-TF network
| KEGG pathway | p value |
| Acute myeloid leukemia | 2.72E-08 |
| Pathways in cancer | 5.71E-06 |
| Chronic myeloid leukemia | 0.000286 |
| Epstein-Barr virus infection | 0.000549 |
| Transcriptional misregulation in cancer | 0.000553 |
| Adipocytokine signaling pathway | 0.00604 |
| Pancreatic cancer | 0.00721 |
| Small cell lung cancer | 0.00961 |
| Toxoplasmosis | 0.0128 |
| Jak-STAT signaling pathway | 0.0145 |
| Reactome pathway | |
| Cellular responses to stress | 6.47E-05 |
| Cellular senescence | 0.000306 |
| NOTCH1 intracellular domain regulates transcription | 0.000706 |
| Transcriptional regulation of white adipocyte differentiation | 0.00143 |
| Signaling by NOTCH1 | 0.00159 |
| Senescence-associated secretory phenotype (SASP) | 0.00228 |
| Signaling by NOTCH | 0.00323 |
| DEx/H-box helicases activate type I IFN and inflammatory cytokines production | 0.00724 |
| Binding of TCF/LEF:CTNNB1 to target gene promoters | 0.00724 |
| IkBA variant leads to EDA-ID | 0.00724 |
| GO-BP | |
| Regulation of cell differentiation | 3.46E-11 |
| Regulation of developmental process | 1.51E-09 |
| Cell proliferation | 1.72E-09 |
| Transcription from RNA polymerase II promoter | 1.97E-09 |
| Positive regulation of transcription from RNA polymerase II promoter | 3.80E-09 |
| Regulation of multicellular organismal process | 2.44E-08 |
| Regulation of transcription from RNA polymerase II promoter | 2.55E-08 |
| Positive regulation of transcription, DNA-dependent | 1.35E-07 |
| Positive regulation of transcription, DNA-dependent | 1.35E-07 |
| Positive regulation of RNA metabolic process | 2.13E-07 |
| GO-MF | |
| Transcription factor binding | 9.34E-11 |
| Transcription from RNA polymerase II promoter | 1.72E-09 |
| RNA polymerase II distal enhancer sequence-specific DNA binding transcription factor activity | 5.64E-09 |
| DNA binding | 6.20E-08 |
| Sequence-specific DNA binding | 8.61E-08 |
| Positive regulation of transcription, DNA-dependent | 1.22E-07 |
| Negative regulation of transcription, DNA-dependent | 6.69E-07 |
| Protein dimerization activity | 7.12E-07 |
| Protein heterodimerization activity | 8.13E-05 |
| Protein kinase binding | 8.30E-05 |
| GO-CC | |
| Nuclear lumen | 1.51E-05 |
| Nucleoplasm | 1.65E-05 |
| Nuclear part | 7.71E-05 |
| Organelle lumen | 8.69E-05 |
| Nucleoplasm part | 9.60E-05 |
| Membrane-enclosed lumen | 9.90E-05 |
| Nucleus | 0.000389 |
| Transcription factor complex | 0.000649 |
| Nuclear matrix | 0.00118 |
| Protein complex | 0.00294 |
Acknowledgments
The authors acknowledge the valuable inputs regarding the study from their colleagues and reviewers. We appreciate the funding by the Science and Technology Overcoming Research Obstacles Project of Quzhou City (Grant no.: 2020056), which was used for supporting the research project of Zhen Wang (Email: wang1121325@wmu.edu.cn).
Data Availability
The datasets utilized in the present study are publically available.
Conflicts of Interest
The authors declare no conflicts of interest.
Supplementary Materials
Table S1a: significant DEmiRNA in periodontitis (GSE54710). Table S1b: significant DEmiRNA in H. pylori-infected gastric tissue (GSE32174). Table S2a: shared-DEmiRNA-gene network interactions. Table S2b: KEGG pathway enrichment analysis of shared-DEmiRNA-gene network. Table S2c: reactome pathway enrichment analysis of shared-DEmiRNA-gene network. Table S2d: GO–biological pathways enrichment analysis of shared-DEmiRNA-gene network. Table S2e: GO-molecular functions enrichment analysis of shared-DEmiRNA-gene network. Table S2f: GO cellular component enrichment analysis of shared-DEmiRNA-gene network. Table S3a: shared-DEmiRNA-TF network interactions. Table S3b: KEGG pathway enrichment analysis of shared-DEmiRNA-TF network interactions. Table S3c: reactome pathway enrichment analysis of shared-DEmiRNA-TF network interactions. Table S3d: GO-BP pathway enrichment analysis in shared-DEmiRNA-TF network interactions. Table S3e: GO-molecular functions enrichment analysis in shared-DEmiRNA-TF network interactions. Table S3f: GO cellular component enrichment analysis in shared-DEmiRNA-TF network interactions. Table S4: shared-DEmiRNA-compound network interactions.
References
- 1.Eke P. I., Borgnakke W. S., Genco R. J. Recent epidemiologic trends in periodontitis in the USA. Periodontology . 2000;82(1):257–267. doi: 10.1111/prd.12323. [DOI] [PubMed] [Google Scholar]
- 2.Winning L., Linden G. J. Periodontitis and systemic disease: association or causality? Current Oral Health Reports . 2017;4(1):1–7. doi: 10.1007/s40496-017-0121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boylan M. R., Khalili H., Huang E. S., et al. A prospective study of periodontal disease and risk of gastric and duodenal ulcer in male health professionals. Clinical and Translational Gastroenterology . 2014;5(2):p. e49. doi: 10.1038/ctg.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H.-C., Chen T.-P., Wei C.-Y., Chang Y.-C. Association between peptic ulcer disease and periodontitis: a nationwide population-based case-control study in Taiwan. International Journal of Environmental Research and Public Health . 2018;15(5):p. 912. doi: 10.3390/ijerph15050912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chrysanthakopoulos N. A., Oikonomou A. A. A case-control study of the periodontal condition in gastric cancer patients. Stomatological Disease and Science . 2017;1(2):55–61. doi: 10.20517/2573-0002.2017.01. [DOI] [Google Scholar]
- 6.Polk D. B., Peek R. M. Helicobacter pylori: gastric cancer and beyond. Nature Reviews Cancer . 2010;10(6):403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qadri Q., Rasool R., Gulzar S. N., Naqash S., Shah Z. A. H. pylori infection, inflammation and gastric cancer. Journal of Gastrointestinal Cancer . 2014;45(2):126–132. doi: 10.1007/s12029-014-9583-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X.-Y., Zhang P.-Y., Aboul-Soud M. A. M. From inflammation to gastric cancer: role of Helicobacter pylori. Oncology Letters . 2017;13(2):543–548. doi: 10.3892/ol.2016.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thrift A. P., El-Serag H. B. Burden of gastric cancer. Clinical Gastroenterology and Hepatology . 2020;18(3):534–542. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umeda M., Kobayashi H., Takeuchi Y., et al. High prevalence of Helicobacter pylori detected by PCR in the oral cavities of periodontitis patients. Journal of Periodontology . 2003;74(1):129–134. doi: 10.1902/jop.2003.74.1.129. [DOI] [PubMed] [Google Scholar]
- 11.Gebara E. C. E., Pannuti C., Faria C. M., Chehter L., Mayer M. P. A., Lima L. A. P. A. Prevalence of Helicobacter pylori detected by polymerase chain reaction in the oral cavity of periodontitis patients. Oral Microbiology and Immunology . 2004;19(4):277–280. doi: 10.1111/j.1399-302X.2004.00153.x. [DOI] [PubMed] [Google Scholar]
- 12.Al Asqah M., Al Hamoudi N., Anil S., Al-hamoudi W. K. Is the presence of Helicobacter pylori in the dental plaque of patients with chronic periodontitis a risk factor for gastric infection? Canadian Journal of Gastroenterology . 2009;23(3, article 950527):177–179. doi: 10.1155/2009/950527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei X., Zhao H.-Q., Ma C., et al. The association between chronic periodontitis and oral Helicobacter pylori: a meta-analysis. PLoS One . 2019;14(12, article e0225247) doi: 10.1371/journal.pone.0225247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouziane A., Ahid S., Abouqal R., Ennibi O. Effect of periodontal therapy on prevention of gastric H elicobacter pylori recurrence: a systematic review and meta-analysis. Journal of Clinical Periodontology . 2012;39(12):1166–1173. doi: 10.1111/jcpe.12015. [DOI] [PubMed] [Google Scholar]
- 15.Adachi K., Notsu T., Mishiro T., Yoshikawa H., Kinoshita Y. Influence of Helicobacter pylori infection on periodontitis. Journal of Gastroenterology and Hepatology . 2019;34(1):120–123. doi: 10.1111/jgh.14358. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch C., Tegtmeyer N., Rohde M., Rowland M., Oyarzabal O. A., Backert S. Live Helicobacter pylori in the root canal of endodontic-infected deciduous teeth. Journal of Gastroenterology . 2012;47(8):936–940. doi: 10.1007/s00535-012-0618-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang X. M., Yee K. C., Hazeki-Taylor N., et al. Oral Helicobacter pylori, its relationship to successful eradication of gastric H. pylori and saliva culture confirmation. Journal of Physiology and Pharmacology . 2014;65(4):559–566. [PubMed] [Google Scholar]
- 18.Hu Z., Zhang Y., Li Z., et al. Effect of Helicobacter pylori infection on chronic periodontitis by the change of microecology and inflammation. Oncotarget . 2016;7(41):66700–66712. doi: 10.18632/oncotarget.11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavali S., Bruhn S., Tiemann K., et al. MicroRNAs act complementarily to regulate disease-related mRNA modules in human diseases. RNA . 2013;19(11):1552–1562. doi: 10.1261/rna.038414.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhayani M. K., Calin G. A., Lai S. Y. Functional relevance of miRNA∗ sequences in human disease. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis . 2012;731(1-2):14–19. doi: 10.1016/j.mrfmmm.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 21.Hanna J., Hossain G. S., Kocerha J. The potential for microRNA therapeutics and clinical research. Frontiers in Genetics . 2019;10:p. 478. doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning W., Jiang X., Sun Z., et al. Identification of the potential biomarkers involved in the human oral mucosal wound healing: a bioinformatic study. BioMed Research International . 2021;2021:16. doi: 10.1155/2021/6695245.6695245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmalz G., Li S., Burkhardt R., et al. MicroRNAs as salivary markers for periodontal diseases: a new diagnostic approach? BioMed Research International . 2016;2016:14. doi: 10.1155/2016/1027525.1027525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Liu X., Li H., et al. Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis. Journal of Periodontal Research . 2018;53(4):495–505. doi: 10.1111/jre.12539. [DOI] [PubMed] [Google Scholar]
- 25.Santonocito S., Polizzi A., Palazzo G., Isola G. The emerging role of microRNA in periodontitis: pathophysiology, clinical potential and future molecular perspectives. International Journal of Molecular Sciences . 2021;22(11):p. 5456. doi: 10.3390/ijms22115456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yagnik K., Mahendra J., Kurian V. M. The periodontal-cardiovascular alliance: evaluation of miRNA-146a in subgingival plaque samples of chronic periodontitis patients with and without coronary heart disease. Journal of Investigative and Clinical Dentistry . 2019;10(4, article e12442) doi: 10.1111/jicd.12442. [DOI] [PubMed] [Google Scholar]
- 27.Stoecklin-Wasmer C., Guarnieri P., Celenti R., Demmer R. T., Kebschull M., Papapanou P. N. MicroRNAs and their target genes in gingival tissues. Journal of Dental Research . 2012;91(10):934–940. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lario S., Ramírez-Lázaro M. J., Aransay A. M., et al. MicroRNA profiling in duodenal ulcer disease caused by Helicobacter pylori infection in a Western population. Clinical Microbiology and Infection . 2012;18(8):E273–E282. doi: 10.1111/j.1469-0691.2012.03849.x. [DOI] [PubMed] [Google Scholar]
- 29.Ritchie M. E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research . 2015;43(7):e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H., Boutros P. C. Venndiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics . 2011;12(1):1–7. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang L., Zhou G., Soufan O., Xia J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Research . 2020;48(W1):W244–W251. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S., Chen T., Zhong Z., Wang Y., Li Y., Zhao X. MicroRNA-155 silencing inhibits proliferation and migration and induces apoptosis by upregulating BACH1 in renal cancer cells. Molecular Medicine Reports . 2012;5(4):949–954. doi: 10.3892/mmr.2012.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tili E., Michaille J.-J., Wernicke D., et al. Mutator activity induced by microRNA-155 (miR-155) links inflammation and cancer. Proceedings of the National Academy of Sciences . 2011;108(12):4908–4913. doi: 10.1073/pnas.1101795108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgs G., Slack F. The multiple roles of microRNA-155 in oncogenesis. Journal of Clinical Bioinformatics . 2013;3(1):17–18. doi: 10.1186/2043-9113-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T., Ding Z.-L., Zheng Y.-L., Wang W. miR-484 promotes non-small-cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis. Biomedicine & Pharmacotherapy . 2017;96:153–164. doi: 10.1016/j.biopha.2017.09.102. [DOI] [PubMed] [Google Scholar]
- 36.Liu J., Li S. M. miR-484 suppressed proliferation, migration, invasion and induced apoptosis of gastric cancer via targeting CCL-18. International Journal of Experimental Pathology . 2020;101(6):203–214. doi: 10.1111/iep.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H., Xiao X., Shi Y., et al. Inhibition of miRNA-29a regulates intestinal barrier function in diarrhea-predominant irritable bowel syndrome by upregulating ZO-1 and CLDN1. Experimental and Therapeutic Medicine . 2020;20(6):p. 1. doi: 10.3892/etm.2020.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuntip N., Japrung D., Pongprayoon P. Modeling the adsorption of the miR-29a cancer biomarker on a graphene quantum dot. ACS Omega . 2021;6(33):21764–21772. doi: 10.1021/acsomega.1c03404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J.-y., Zhang Q., Wang D.-d., et al. MiR-29a: a potential therapeutic target and promising biomarker in tumors. Bioscience Reports . 2018;38(1) doi: 10.1042/BSR20171265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aguilar C., Mano M., Eulalio A. MicroRNAs at the host–bacteria interface: host defense or bacterial offense. Trends in Microbiology . 2019;27(3):206–218. doi: 10.1016/j.tim.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 41.Chou S.-H., Tung Y.-C., Wu L.-S., Chang C.-J., Kung S., Chu P.-H. Severity of chronic periodontitis and risk of gastrointestinal cancers: a population-based follow-up study from Taiwan. Medicine . 2018;97(27, article e11386) doi: 10.1097/MD.0000000000011386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keniry M., Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene . 2008;27(41):5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 43.Álvarez-Garcia V., Tawil Y., Wise H. M., Leslie N. R. Mechanisms of PTEN loss in cancer: It’s all about diversity. Seminars in Cancer Biology . 2019;59:66–79. doi: 10.1016/j.semcancer.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Xu W.-T., Yang Z., Lu N. H. Roles of PTEN (phosphatase and tensin homolog) in gastric cancer development and progression. Asian Pacific Journal of Cancer Prevention . 2014;15(1):17–24. doi: 10.7314/APJCP.2014.15.1.17. [DOI] [PubMed] [Google Scholar]
- 45.Sato K., Tamura G., Tsuchiya T., et al. Analysis of genetic and epigenetic alterations of the PTEN gene in gastric cancer. Virchows Archiv . 2002;440(2):160–165. doi: 10.1007/s004280100499. [DOI] [PubMed] [Google Scholar]
- 46.Yang Z., Yuan X.-G., Chen J., Luo S.-W., Luo Z.-J., Lu N. H. Reduced expression of PTEN and increased PTEN phosphorylation at residue Ser380 in gastric cancer tissues: a novel mechanism of PTEN inactivation. Clinics and Research in Hepatology and Gastroenterology . 2013;37(1):72–79. doi: 10.1016/j.clinre.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Hu M., Zhu S., Xiong S., Xue X., Zhou X. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer. Oncology Reports . 2019;41(3):1439–1454. doi: 10.3892/or.2019.6962. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z., Xie C., Xu W., et al. Phosphorylation and inactivation of PTEN at residues Ser380/Thr382/383 induced by Helicobacter pylori promotes gastric epithelial cell survival through PI3K/Akt pathway. Oncotarget . 2015;6(31):31916–31926. doi: 10.18632/oncotarget.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu C., Wei Z., Zhang D. PTEN inhibits inflammatory bone loss in ligature-induced periodontitis via IL1 and TNF-α. BioMed Research International . 2019;2019:9. doi: 10.1155/2019/6712591.6712591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang G., Wang H., Shi H., et al. Porphyromonas gingivalisPromotes the proliferation and migration of esophageal squamous cell carcinoma through the miR-194/GRHL3/PTEN/Akt axis. ACS infectious diseases . 2020;6(5):871–881. doi: 10.1021/acsinfecdis.0c00007. [DOI] [PubMed] [Google Scholar]
- 51.Nie M., Wang Y., Yu Z., et al. AURKB promotes gastric cancer progression via activation of CCND1 expression. Aging (Albany NY) . 2020;12(2):1304–1321. doi: 10.18632/aging.102684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuo H.-W., Huang C.-Y., Chun-Kai F., et al. The significant association of CCND1 genotypes with gastric cancer in Taiwan. Anticancer Research . 2014;34(9):4963–4968. [PubMed] [Google Scholar]
- 53.Bizari L., Borim A. A., Leite K. R. M., et al. Alterations of the CCND1 and HER-2/neu (ERBB2) proteins in esophageal and gastric cancers. Cancer Genetics and Cytogenetics . 2006;165(1):41–50. doi: 10.1016/j.cancergencyto.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 54.Khan M., Khan S., Mandal R. K., et al. Cell cycle regulatory CCND1 G870A gene polymorphism and periodontitis-induced oral cancer: a risk analysis. Revista Romana de Medicina de Laborator . 2021;29(4):349–363. [Google Scholar]
- 55.Kiyoshima T., Enoki N., Kobayashi I., et al. Oxidative stress caused by a low concentration of hydrogen peroxide induces senescence-like changes in mouse gingival fibroblasts. International Journal of Molecular Medicine . 2012;30(5):1007–1012. doi: 10.3892/ijmm.2012.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kodama M., Fujioka T., Murakami K., et al. Eradication of Helicobacter pylori reduced the immunohistochemical detection of p53 and MDM2 in gastric mucosa. Journal of Gastroenterology and Hepatology . 2005;20(6):941–946. doi: 10.1111/j.1440-1746.2005.03880.x. [DOI] [PubMed] [Google Scholar]
- 57.Rahman S. M., Dobrzyn A., Lee S.-H., Dobrzyn P., Miyazaki M., Ntambi J. M. Stearoyl-CoA desaturase 1 deficiency increases insulin signaling and glycogen accumulation in brown adipose tissue. American Journal of Physiology-Endocrinology and Metabolism . 2005;288(2):E381–E387. doi: 10.1152/ajpendo.00314.2004. [DOI] [PubMed] [Google Scholar]
- 58.Sun Q., Xiaojuan Y., Peng C., et al. Activation of SREBP-1c alters lipogenesis and promotes tumor growth and metastasis in gastric cancer. Biomedicine & Pharmacotherapy . 2020;128, article 110274 doi: 10.1016/j.biopha.2020.110274. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka H., Nakai K., Murakami F., et al. Ligature-induced periodontitis increased insulin resistance and triglyceride levels in Wistar rats. Journal of Hard Tissue Biology . 2017;26(3):261–267. [Google Scholar]
- 60.Kim M. S., Oh J. E., Kim Y. R., et al. Somatic mutations and losses of expression of microRNA regulation-related genes AGO2 and TNRC6A in gastric and colorectal cancers. The Journal of Pathology . 2010;221(2):139–146. doi: 10.1002/path.2683. [DOI] [PubMed] [Google Scholar]
- 61.Piao J.-Y., Kim S.-J., Kim D.-H., et al. Helicobacter pylori infection induces STAT3 phosphorylation on Ser727 and autophagy in human gastric epithelial cells and mouse stomach. Scientific Reports . 2020;10(1):1–13. doi: 10.1038/s41598-020-72594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ambili R., Janam P., Babu P. S. S., et al. Differential expression of transcription factors NF-κB and STAT3 in periodontal ligament fibroblasts and gingiva of healthy and diseased individuals. Archives of Oral Biology . 2017;82:19–26. doi: 10.1016/j.archoralbio.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 63.Hu Y., Xu Z., Zhang J., et al. Activated STAT3 signaling pathway by ligature-induced periodontitis could contribute to neuroinflammation and cognitive impairment in rats. Journal of Neuroinflammation . 2021;18(1):1–17. doi: 10.1186/s12974-021-02071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L., He Y., Ning W. Role of enhancer of zeste homolog 2 in osteoclast formation and periodontitis development by downregulating microRNA-101-regulated VCAM-1. Journal of Tissue Engineering and Regenerative Medicine . 2021;15(6):534–545. doi: 10.1002/term.3187. [DOI] [PubMed] [Google Scholar]
- 65.Yu X., Shahir A.-M., Sha J., et al. Short-chain fatty acids from periodontal pathogens suppress histone deacetylases, EZH2, and SUV39H1 to promote Kaposi's sarcoma-associated herpesvirus replication. Journal of Virology . 2014;88(8):4466–4479. doi: 10.1128/JVI.03326-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hayashi Y., Tsujii M., Wang J., et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut . 2013;62(11):1536–1546. doi: 10.1136/gutjnl-2011-301625. [DOI] [PubMed] [Google Scholar]
- 67.Cai G. H., Wang K., Miao Q., Peng Y. S., Chen X. Y. Expression of polycomb protein EZH2 in multi-stage tissues of gastric carcinogenesis. Journal of Digestive Diseases . 2010;11(2):88–93. doi: 10.1111/j.1751-2980.2010.00420.x. [DOI] [PubMed] [Google Scholar]
- 68.Ning W., Acharya A., Sun Z., et al. Deep learning reveals key immunosuppression genes and distinct immunotypes in periodontitis. Frontiers in Genetics . 2021;12, article 648329 doi: 10.3389/fgene.2021.648329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matak P., Heinis M., Mathieu J. R. R., et al. Myeloid HIF-1 is protective in Helicobacter pylori–mediated gastritis. The Journal of Immunology . 2015;194(7):3259–3266. doi: 10.4049/jimmunol.1401260. [DOI] [PubMed] [Google Scholar]
- 70.Bhattacharyya A., Chattopadhyay R., Hall E. H., Mebrahtu S. T., Ernst P. B., Crowe S. E. Mechanism of hypoxia-inducible factor 1α-mediated Mcl1 regulation in Helicobacter pylori-infected human gastric epithelium. American Journal of Physiology-Gastrointestinal and Liver Physiology . 2010;299(5):G1177–G1186. doi: 10.1152/ajpgi.00372.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bai Y., Li W., Guangyu X., Cui G. A bioinformatics approach revealed the transcription factors of Helicobacter pylori pathogenic genes and their regulatory network nodes. Electronic Journal of Biotechnology . 2020;45:53–59. doi: 10.1016/j.ejbt.2020.04.002. [DOI] [Google Scholar]
- 72.Hu T.-Z., Huang L.-H., Xu C. X., et al. Expressional profiles of transcription factors in the progression of Helicobacter pylori-associated gastric carcinoma based on protein/DNA array analysis. Medical Oncology . 2015;32(12):1–11. doi: 10.1007/s12032-015-0711-y. [DOI] [PubMed] [Google Scholar]
- 73.Afrough H., Ghafouri-Fard S., Yousefi H., Pakzad P., Oskooei V. K., Taheri M. DICER-AS1 lncRNA: a putative culprit in the pathogenesis of gastric cancer. Experimental and Molecular Pathology . 2020;116, article 104490 doi: 10.1016/j.yexmp.2020.104490. [DOI] [PubMed] [Google Scholar]
- 74.Ghafouri-Fard S., Gholami L., Nazer N., et al. Assessment of expression of NF-κB-related genes in periodontitis. Gene Reports . 2021;26, article 101454 doi: 10.1016/j.genrep.2021.101454. [DOI] [Google Scholar]
- 75.Sakakura C., Hagiwara A., Miyagawa K., et al. Frequent downregulation of the runt domain transcription factors RUNX1, RUNX3 and their cofactor CBFB in gastric cancer. International Journal of Cancer . 2005;113(2):221–228. doi: 10.1002/ijc.20551. [DOI] [PubMed] [Google Scholar]
- 76.Zhang X., Li Z., Zhao Z., Chen Y., Sun Y., Cai Q. Runx1/miR-26a/Jagged1 signaling axis controls osteoclastogenesis and alleviates orthodontically induced inflammatory root resorption. International Immunopharmacology . 2021;100, article 107991 doi: 10.1016/j.intimp.2021.107991. [DOI] [PubMed] [Google Scholar]
- 77.Lo C.-H., Kwon S., Wang L., et al. Periodontal disease, tooth loss, and risk of oesophageal and gastric adenocarcinoma: a prospective study. Gut . 2021;70(3):620–621. doi: 10.1136/gutjnl-2020-321949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li N., Tang B., Jia Y.-p., et al., editors; Wang K., Zhang W. J., Guo G., Wang T. J. Helicobacter pylori CagA protein negatively regulates autophagy and promotes inflammatory response via c-met-PI3K/Akt-mTOR signaling pathway. Frontiers in Cellular and Infection Microbiology . 2017;7:p. 417. doi: 10.3389/fcimb.2017.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuan T. L., Cantley L. C. PI3K pathway alterations in cancer: variations on a theme. Oncogene . 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zarrin A. A., Bao K., Lupardus P., Vucic D. Kinase inhibition in autoimmunity and inflammation. Nature Reviews Drug Discovery . 2021;20(1):39–63. doi: 10.1038/s41573-020-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao S., Song D., Liu Y., Yan H., Chen X. Helicobacter pylori CagA Protein Attenuates 5-Fu Sensitivity of Gastric Cancer Cells Through Upregulating Cellular Glucose Metabolism. Oncotargets and Therapy . 2020;Volume 13:6339–6349. doi: 10.2147/OTT.S230875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Novaes V. C. N., Ervolino E., Fernandes G. L., et al. Influence of the treatment with the antineoplastic agents 5-fluorouracil and cisplatin on the severity of experimental periodontitis in rats. Supportive Care in Cancer . 2021:1–14. doi: 10.1007/s00520-021-06586-y. [DOI] [PubMed] [Google Scholar]
- 83.Zhou T., Chen D., Li Q., Sun X., Song Y., Wang C. Curcumin inhibits inflammatory response and bone loss during experimental periodontitis in rats. Acta Odontologica Scandinavica . 2013;71(2):349–356. doi: 10.3109/00016357.2012.682092. [DOI] [PubMed] [Google Scholar]
- 84.Mahady G. B., Pendland S. L., Yun G., Lu Z. Z. Turmeric (Curcuma longa) and curcumin inhibit the growth of Helicobacter pylori, a group 1 carcinogen. Anticancer Research . 2002;22(6C):4179–4181. [PubMed] [Google Scholar]
- 85.Bag A., Bhattacharyya S. K., Chattopadhyay R. R. Isolation and identification of a gallotannin 1,2,6-tri-O-galloyl-β-d-glucopyranose from hydroalcoholic extract ofTerminalia chebulafruits effective against multidrug-resistant uropathogens. Journal of Applied Microbiology . 2013;115(2):390–397. doi: 10.1111/jam.12256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1a: significant DEmiRNA in periodontitis (GSE54710). Table S1b: significant DEmiRNA in H. pylori-infected gastric tissue (GSE32174). Table S2a: shared-DEmiRNA-gene network interactions. Table S2b: KEGG pathway enrichment analysis of shared-DEmiRNA-gene network. Table S2c: reactome pathway enrichment analysis of shared-DEmiRNA-gene network. Table S2d: GO–biological pathways enrichment analysis of shared-DEmiRNA-gene network. Table S2e: GO-molecular functions enrichment analysis of shared-DEmiRNA-gene network. Table S2f: GO cellular component enrichment analysis of shared-DEmiRNA-gene network. Table S3a: shared-DEmiRNA-TF network interactions. Table S3b: KEGG pathway enrichment analysis of shared-DEmiRNA-TF network interactions. Table S3c: reactome pathway enrichment analysis of shared-DEmiRNA-TF network interactions. Table S3d: GO-BP pathway enrichment analysis in shared-DEmiRNA-TF network interactions. Table S3e: GO-molecular functions enrichment analysis in shared-DEmiRNA-TF network interactions. Table S3f: GO cellular component enrichment analysis in shared-DEmiRNA-TF network interactions. Table S4: shared-DEmiRNA-compound network interactions.
Data Availability Statement
The datasets utilized in the present study are publically available.


