SUMMARY
Visual arrestin (Arr1) terminates rhodopsin signaling by blocking its interaction with transducin. To do this, Arr1 translocates from the inner to the outer segment of photoreceptors upon light stimulation. Mounting evidence indicates that inositol phosphates (InsPs) affect Arr1 activity, but the Arr1-InsP molecular interaction remains poorly defined. We report the structure of bovine Arr1 in a ligand-free state featuring a near-complete model of the previously unresolved C-tail, which plays a crucial role in regulating Arr1 activity. InsPs bind to the N-domain basic patch thus displacing the C-tail, suggesting that they prime Arr1 for interaction with rhodopsin and help direct Arr1 translocation. These structures exhibit intact polar cores, suggesting that C-tail removal by InsP-binding is insufficient to activate Arr1. These results show how Arr1 activity can be controlled by endogenous InsPs in molecular detail.
Keywords: arrestin; myo-D-inositol hexakisphosphate (InsP6); (1,4,5) myo-D-inositol triphosphate (InsP3); retina; phototransduction; G protein-coupled receptor (GPCR)
INTRODUCTION
Arrestins (Arr1–4 in vertebrates) are proteins involved in terminating G protein signaling. This signal cessation is accomplished through the recognition of activated and phosphorylated G protein-coupled receptor (GPCRs) (Kühn et al., 1984; Palczewski et al., 1989; Wilden et al., 1986). Besides their impact on G protein signaling, arrestins have been found to bind many other intracellular targets and initiate alternative signaling pathways (Ahmed et al., 2011; Coffa et al., 2011; Hanson et al., 2007; Nair et al., 2004; Song et al., 2006; Wu et al., 2006; Moore et al., 2007; Shukla et al., 2011; Moaven et al., 2013). In the context of phototransduction, activated rhodopsin (Rh*) is phosphorylated on C-terminal Ser/Thr residues (Rh*P) by rhodopsin kinase, which enhances its affinity for Arr1 (Palczewski, 2006; Palczewski et al., 1989; Vishnivetskiy et al., 2007). The resulting nanomolar affinity interaction, which has been structurally characterized, prevents further association of the phototransduction G protein, transducin (Palczewski et al., 1989; Palczewski, 1994; Deming et al., 2015; Zhou et al., 2017).
The process of quenching rhodopsin signaling involves a dramatic translocation of Arr1 from the inner segment to the outer segment that occurs upon light activation of photoreceptors (Broekhuyse et al., 1985; Philp et al., 1987). Translocation of Arr1 in Xenopus laevis and mice was shown to be dependent on ATP (Orisme et al., 2010). However, the energy demands for a purely active-transport mechanism of transfer of Arr1 to the OS are prohibitively high (Gurevich et al., 2011). Therefore, it is more likely that Arr1 is either gated or involved in an interaction-restriction diffusion process (Gurevich et al., 2011; Orisme et al., 2010). These hypotheses are bolstered by findings that show Arr1 translocation is superstoichiometric to the amount of bleached rhodopsin (Strissel et al., 2006). Steric volume exclusion is also a potentially important mechanism for the unequal distribution of Arr1 and its multimeric forms in the cytoplasm of photoreceptors, suggesting that Arr1 dissociation from its tetrameric to monomeric forms may be critical for controlling translocation (Najafi et al., 2012; Malhotra et al., 2021). Although recent work on putative oligomerization-defective Arr1 mutants revealed normal translocation activity (Samaranayake et al., 2020), the in vitro assessment of oligomerization for these mutants was not conducted at physiological concentrations of Arr1 (Kim et al., 2011). Thus, steric volume occlusion remains a viable mechanism for Arr1 translocation. How Arr1 tetramer dissociation and translocation could be orchestrated in a light-dependent manner is still a matter of debate.
It has been hypothesized that the activity of phospholipase C (PLC) and/or protein kinase C (PKC) is responsible for Arr1 translocation from IS to OS (Orisme et al., 2010). The evidence for PLC activation as the cause of Arr1 translocation becomes even more important in the context of previously established work that demonstrated a light-dependent change in D-myo-inositol phosphate (InsP) concentrations in vertebrate photoreceptors (Ghalayini and Anderson, 1984). The reason for the light-dependent decrease in phosphoinositol 4,5-bisphosphate (PtdInsP2, also referred to as PIP2) has not been fully explained, and has been debated (Berridge and Irvine, 1984; Brown et al., 1984; Das et al., 1986, 1987; Fein et al., 1984; Ghalayini and Anderson, 1984; Hayashi and Amakawa, 1985; Millar et al., 1988; Vandenberg and Montal, 1984). InsPs have been shown to bind Arr1 with low micromolar affinities, which increase with increasing number of phosphates and decline sharply with less than 3 phosphates (D-myo-inositol-1-monophosphate and D-myo-inositol-1,4-bisphosphate) (Palczewski et al., 1991a).
Prior structural work on arrestins has suggested a mechanism of arrestin activation through distinct conformational changes that enable the protein to bind GPCRs and initiate signaling pathways. Arrestins fold into two domains that interact minimally. However, the interaction includes two critical areas that have been identified in the context of activation. The C-tail of Arr1 makes a regulatory interaction with the concave side of the N-domain, keeping the protein from being constitutively active (as in the p44 splice mutant whose C-tail is missing). The discovery and characterization of the p44 splice variant of Arr1 identified that the C-terminus of Arr1 is critical to regulating its cellular localization and ability to readily bind Rh*P (Palczewski et al., 1994; Smith et al., 1994; Langlois et al., 1996; Palczewski and Smith, 1996; Pulvermüller et al., 1997; Ridge et al., 2003). The other element of inter-domain interaction is the polar core, which is a 5-residue interaction involving the gate loop (residues 296–305, known as the “lariat loop”) and the C-tail, as well as other residues. The polar core is disrupted in arrestin structures that are said to adopt an active conformation. One example of this is the Rh*P-Arr1 fusion protein where the binding of Arr1 to Rh*P results in disruption of the polar core accompanied by a shift in the gate loop (Zhou et al., 2017).
All vertebrate arrestins have at least one structure reported, including basal conformations, a C-tail-truncated splice variant of Arr1 (p44), and a fusion protein of Arr1 bound to Rh*P (Granzin et al., 1998; Hirsch et al., 1999; Kim et al., 2013; Sutton et al., 2005; Zhou et al., 2017). Arr2 and Arr3 are expressed ubiquitously throughout the body and have been crystallized with bound D-myo-inositol-1,2,3,4,5,6-hexakisphosphate (InsP6) (Milano et al., 2006; Chen et al., 2017). In both cases, InsP6 showed two-protomer binding, with contacts primarily in the C-domain of one of the protomers, but two of the residues in that domain involved in InsP6 binding are not conserved in the Arr1 sequence. In the case of Arr3, the bound InsP6 appeared to destabilize the polar core and cause activation-like changes to the protein similar to the fusion protein structure of Rh*P-Arr1 (Zhou et al., 2017). Arr2 did not show such activation, and instead seemed to be in a pre-activated or ‘primed’ state where the C-tail was removed from the N-domain contemporaneous with an intact polar core.
Unlike the self-association that occurs for Arr2/3 in the presence of InsP6, Arr1 is converted from a tetrameric to a monomeric state in the presence of sufficient InsP6 (Hanson et al., 2008a). It is thought that cytoplasmic concentrations of InsP6 or other InsPs are sufficient to have an impact on Arr2/3 activity in vivo, suggesting the same could be true for Arr1 (Milano et al., 2006). Given the evidence for light-dependent InsP production, specifically related to PLC activity (which releases InsP3 from PIP2), InsPs could play an important role in Arr1 translocation. An NMR study of Arr1 bound to InsP6 or heparin suggested that both ligands bound to the N-domain and displaced the regulatory C-tail, as suggested also by earlier limited proteolysis experiments (Palczewski et al., 1991b; Zhuang et al., 2010). A recent report indicated that R171, in the basic groove of the N-domain, plays a crucial role in coordinating the C-tail release mechanism, owing to its interaction with the phosphorylated Rh*P C-tail in the fusion protein structure (Haider et al., 2019; Zhou et al., 2017).
Precise modeling of the C-tail and potential InsPs in the N-domain would help elucidate how the self-affinity of the C-tail for the N-terminal basic groove regulates Arr1 activity and localization. This C-tail interaction with the N-domain is thought to keep full length Arr1 from being constitutively active and monomeric, and to prevent its spreading across the IS and OS in dark conditions like the p44 splice variant (Kim et al., 2013). The detail of this molecular interaction is crucial to our understanding of Arr1 translocation to the OS. In the current study we used X-ray crystallography to resolve the Arr1 C-tail/N-domain interaction and elucidate structures of Arr1 bound to InsP3, InsP6 and higher-order InsP compounds, showing that the binding site for these ligands partially overlaps with that of the regulatory C-tail. These structures provide insights into how Arr1 can undergo structural changes that prime it for translocation and Rh*P deactivation.
RESULTS
Crystallization of fully inactive and InsP-bound Arr1
Limited proteolysis studies predicted that the binding of Arr1 to ligands or to its native receptor rhodopsin causes a destabilization of the C-terminus, leading to its removal from the N-domain (Palczewski et al., 1991b). At the outset of the current work, it remained to be determined if InsP binding to Arr1 causes activation, as in the case of Arr3, or if it is already ‘primed’ by removal of the C-tail without disruption of the polar core, as in the case of Arr2 (Chen et al., 2017; Milano et al., 2006). To test these hypotheses, we crystallized native, bovine Arr1 in basal and InsP-bound states, including InsP6 and InsP3, and determined their structures to 2.5, 2.6, and 2.4 Å resolutions, respectively (Figure 1, Table 1). Prior structural studies of basal Arr1 have used crystallization conditions of relatively high ionic strength (Granzin et al., 2012; Hirsch et al., 1999). We searched for new crystallization conditions that would enable resolution of the critical C-tail regulatory region of Arr1, which has remained absent from all prior models. Our basal Arr1 crystallized over 2–4 days in relatively low ionic strength conditions (Figures 1, 2, Methods) in a form isomorphous to prior Arr1 structures in space group P21212. InsP6 co-crystallized with Arr1 in the P21212 space group, and InsP3 was soaked into a basal crystal that also had a P21212 space group. To help rule out the possibility of InsP6 and InsP3 binding positions being dependent on crystallization conditions and crystal packing, we also determined InsP6 and InsP3-bound Arr1 structures through co-crystallization in an alternative space group (C2221) (Table 1). Both ligands bound in the same pocket of Arr1 regardless of space group and co-crystallization/soaking conditions.
Figure 1. Basal and primed conformations of Arr1.

(A) Full-length, native, bovine Arr1 structure with stabilized C-terminus (novel residues of extreme C-terminus in green). (B) Arr1 in primed (C-tail released) conformation caused by InsP6 binding to the N-domain. (C) Arr1 in a primed conformation caused by InsP3 binding to the N-domain.
Table 1.
Summary of crystallographic data collection and refinement statistics for basal, InsP3 and InsP6
| Data Collection | Basal Arr1 | Arr1-InsP6 | Arr1-InsP3 | Arr1-InsP6 | Arr1-InsP3 |
|---|---|---|---|---|---|
| X-ray source | NECAT - ID-C | NECAT - ID-C | SSRL 12–2 | PAL 7A SB I | PAL 7A SB I |
| Wavelength (Å) | 0.97920 | 0.97920 | 0.979460 | 1.000040 | 1.000020 |
| Space group | P21212 | P21212 | P21212 | C2221 | C2221 |
| Unit cell dimensions (Å) | a=167.18, b=187.00, c=90.34 |
a=167.21, b=187.76, c=90.38 |
a=168.03, b=187.64, c=90.62 |
a= 167.59, b= 190.68, c= 190.52 |
a= 167.57, b= 187.70, c= 190.82 |
| Resolution (Å) | 50.0–2.5 (2.65–2.50) | 50.0–2.60 (2.76–2.60) | 50.0–2.40 (2.54–2.40) | 50.0–3.10 (3.28–3.10) | 47.7–3.00 (3.16–3.00) |
| Unique reflections | 98,577 (15,684) | 88,202 (14,017) | 112,146 (17,835) | 105,812 (16,859) | 59,857 (8,712) |
| Multiplicity | 6.5 (6.6) | 13.1 (14.0) | 6.8 (6.9) | 2.5 (2.5) | 7.3 (7.5) |
| Completeness (%) | 99.9 (99.8) | 99.8 (99.4) | 99.6 (99.1) | 98.6 (97.8) | 99.2 (100.0) |
| <I/σI> | 10.93 (0.80) | 13.92 (0.96) | 16.35 (0.74) | 6.56 (1.93) | 19.8 (4.5) |
| Rsym (%) | 11.0 (172.1) | 13.6 (223.3) | 8.6 (262.6) | 14.1 (43.5) | 7.5 (46.9) |
| CC1/2 (%) | 99.8 (52.0) | 99.9 (62.5) | 99.9 (31.3) | 98.4 (82.4) | 99.8 (91.8) |
| Wilson B factor (Å2) | 72.4 | 77.3 | 72.9 | 52.7 | 61.9 |
| Refinement | |||||
| Resolution (Å) | 49.04–2.50 | 49.20–2.60 | 49.14–2.40 | 46.20–3.10 | 46.93–3.00 |
| Number of reflections | 93,643 (4,928) | 83,786 (4,410) | 106,534 (5,602) | 55,209 (2,757) |
59,591 (2,993) |
| Rwork/Rfree (%) | 20.8/23.3 | 20.5/23.8 | 21.0/24.9 | 21.5/24.1 | 20.6/23.9 |
| Number of atoms | 11,919 | 11,763 | 11,771 | 11,468 | 11,598 |
| Protein | 11,759 | 11,444 | 11,448 | 11,360 | 11,475 |
| InsP | N/A | 144 | 96 | 108 | 96 |
| Water | 160 | 175 | 206 | N/A | 2 |
| <B-factor> (Å2) | 78.2 | 86.2 | 80.0 | 71.6 | 73.3 |
| Protein | 78.4 | 85.4 | 80.3 | 70.9 | 73.1 |
| InsP | N/A | 169.4 | 92.9 | 150.3 | 105.9 |
| Water | 61.5 | 65.8 | 62.2 | N/A | 44.4 |
| RMS deviation from ideal | |||||
| Bond lengths (Å) | 0.008 | 0.005 | 0.003 | 0.002 | 0.003 |
| Bond angles (°) | 1.52 | 1.35 | 1.26 | 0.56 | 0.56 |
| Ramachandran plot (% favored/# outliers) | 97.1/0 | 95.5/0 | 95.1/0 | 94.9/4 | 95.6/1 |
| Molprobity score (%) | 99 | 100 | 100 | 100 | 100 |
| PDB accession code | 7JSM | 7JTB | 7JXA | 7F1W | 7F1X |
Figure 2. Basal Arr1 has C-tail spanning the N-domain.
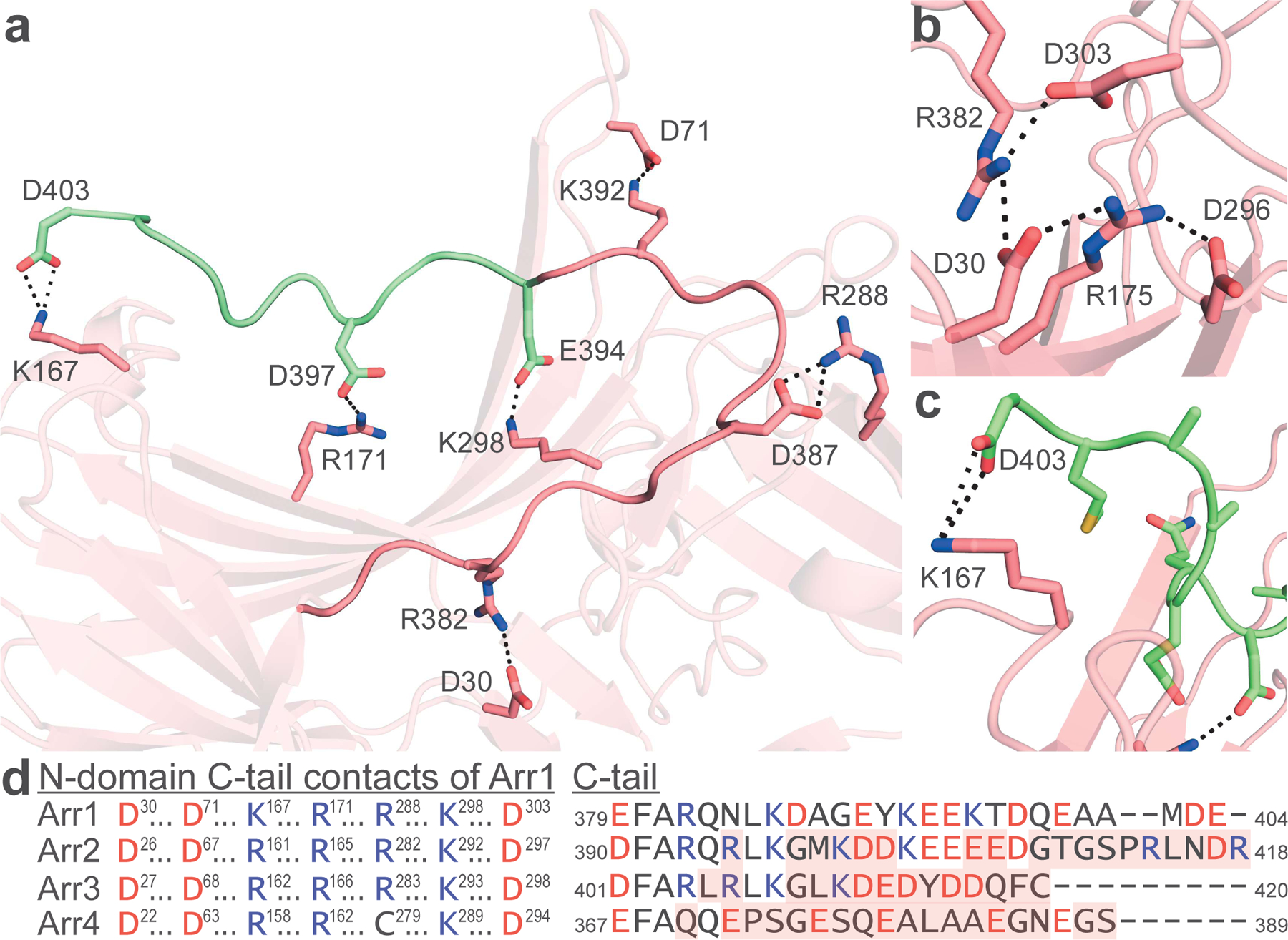
(A) Arr1 C-tail that was resolved previously (dark pink) shows stabilizing interactions with the N-domain. The extreme C-terminus is now resolved (green) and shows several interactions stabilizing its association with the N-domain. (B) Basal Arr1 shows an intact polar core. (C) The C-latch of basal Arr1 is an interaction between the penultimate residue (D403) and K167. All noted interactions in a-c are 4.0 Å or less. (D) The N-domain contacts with the C-tail are highly conserved across all arrestins. Shown is the full-sequence-aligned comparison of all four bovine arrestins highlighted at residues responsible for salt-bridge interaction with the C-tail (left); aligned C-tail residues of each bovine arrestin are shown (right). Acidic residues are shown in red, basic residues in blue. Mismatch from Arr1 is noted with red shading in the C-tail alignment. N-domain and C-tail interaction pairs noted with like symbols.
Structure of basal Arr1 reveals residues responsible for C-tail stabilization
The basal structure showed increased C-tail density in two of the four protomers as compared to prior basal structures, allowing the region to be modeled up to the penultimate residue (Figure 2 and Figure S1). The C-tail contains one recognizable structural element, an inverse gamma turn. Six salt bridges were found to bind the C-tail, and the N-domain residues of each pair are highly conserved among vertebrate Arr paralogs (Figure 2A and 2D). The density map shows the C-tail crossing directly over the concave, basic patch of the N-domain. The basal structure is otherwise largely similar to the prior basal structures and has an intact polar core (Figure 2B). Of particular note are the terminal residues of the C-tail stabilized in the same region (here termed ‘C-latch’) where the Rh*P phosphorylated C-tail was shown to begin a beta-sheet interaction with Arr1 in the Rh*P-Arr1 fusion protein structure (Figure 2C) (Zhou et al., 2017). The C-latch orients K167 in the basal structure, forming an interaction with D403. Nearby, R18 makes a hydrogen bond contact with the backbone of A400, further stabilizing the basal C-tail in the C-latch. The structure of the active conformation Rh*P-Arr1 fusion protein shows a different orientation of R18 from that of the basal structure.
Inositol phosphates prime Arr1 by removing the regulatory C-tail
To gain insight into the influence of InsPs on the activation state of Arr-1, we solved structures of Arr1 bound to InsP6 and InsP3 (Figure 3). The binding site was found to be in the concave portion of the N-domain as had been predicted by NMR (Figure 3A) (Zhuang et al., 2010). In the case of InsP6, Arr1 uses 6 basic residues to bind all 6 phosphate groups, with some phosphates showing interactions with multiple residues (Figure 3B). The C-tail is removed from the top of the N-domain, bringing the last modeled residue to L385.
Figure 3. InsPs stably bind to the N-domain of Arr1.
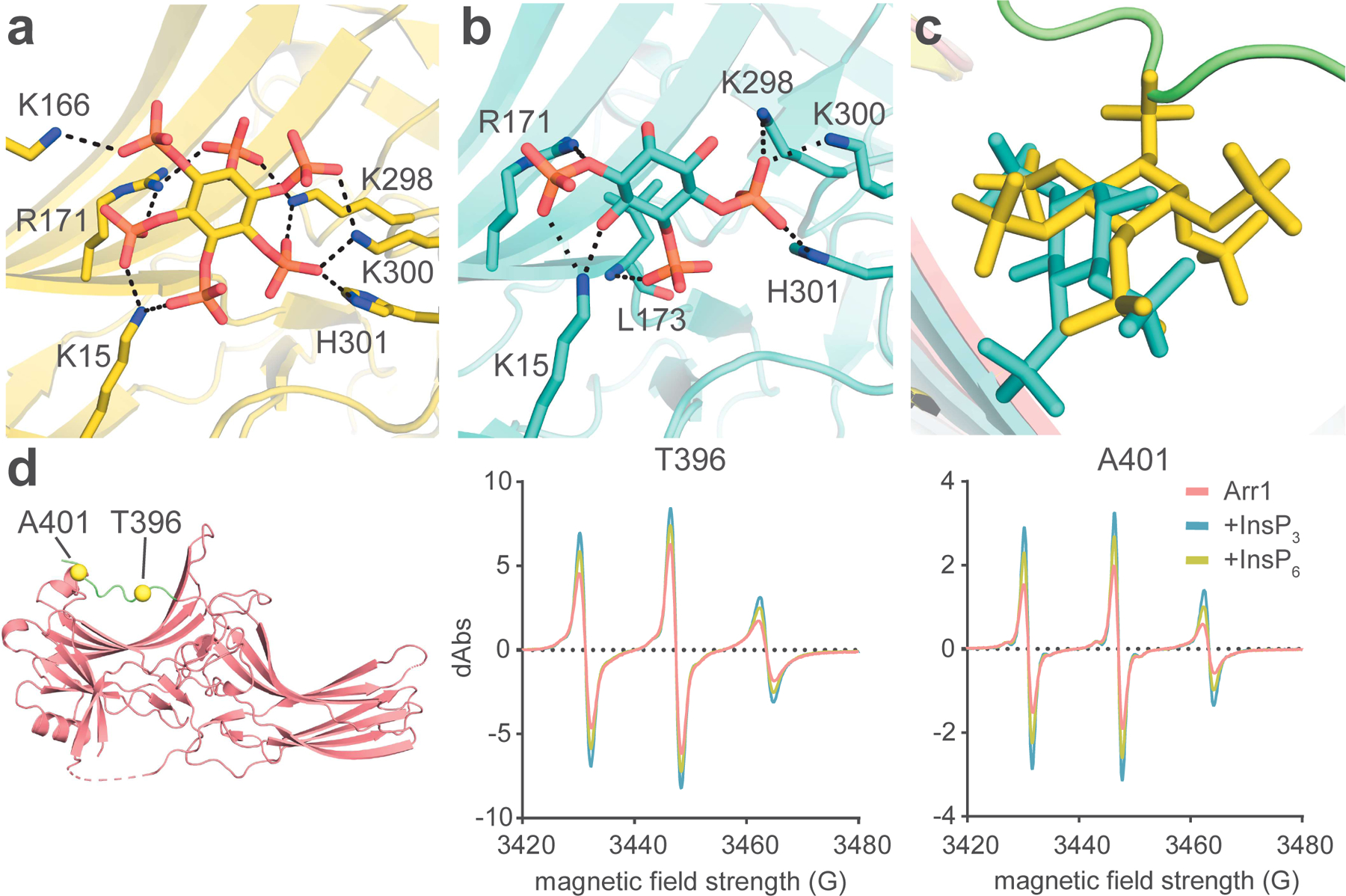
(A) InsP6 binds to the concave, basic region of the N-domain. (B) InsP3 binds to the concave, basic region of the N-domain. All noted interactions are 4.0 Å or less. (C) Comparison of the relative location of the basal C-tail, InsP6, and InsP3. (D) Arr1 structure showing the positions of A401 and T396 (left); cwEPR data of Arr1 at the two positions on the C-tail, T396 (middle) and A401 (right). Measurements from both T396 and A401 show increased flexibility upon InsP-binding.
InsP3 bound to the same region as InsP6, although with fewer points of interaction due to it having half the phosphate groups of InsP6 (Figure 3A and 3B). The all-atom RMSD between the InsP3- and InsP6-Arr1 structures is 0.955 Å. InsP3 was located deeper in the binding pocket than InsP6, making a hydrogen bond with the backbone amide hydrogen of L173 (Figure 3B and 3C). Three of the same residues that bound InsP6 from the gate loop of Arr1 were responsible for binding the C4-phosphate on InsP3 (K298, K300, and H301). K15 also engaged with InsP3, interacting with the C1-phosphate and potentially hydrogen bonding with the C6-hydroxyl group of the inositol ring. R171 also has a possible hydrogen bond interaction with the phosphoester oxygen of the C1-phosphate. The C-tail in the Arr1-InsP3 structure was displaced from the N-domain to the same degree as in the InsP6 structure. Because the C-tail was removed just as completely with half the phosphates present, it is likely that the residues shown to bind InsP3 contain critical contacts for C-tail removal.
Comparing the residues that bound to both InsP3 and the C-tail in our basal structure reveals that R171 and K298 both have interactions, suggesting that disruptions of the C-tail contacts with the N-domain at those two residues by InsP3 binding may initiate C-tail displacement. To test the relative ability of InsP3 to remove the C-tail as compared to InsP6, we used continuous wave electron paramagnetic resonance (cwEPR) to measure changes in the flexibility of the C-tail in the absence and presence of the InsPs (Figure 3D). We used two residues on the C-tail for this evaluation, one at the extreme end of the tail (A401) and the other located over the N-domain binding site (T396). The spectra of the spin labels at both sites showed a sharpening of their peaks upon binding to InsPs, indicating increased flexibility within the local environment, consistent with release of the C-tail from its bound state. While the basal protein at both points of the C-tail was already somewhat flexible in the apo-Arr1 sample, the flexibility increased at both sites upon addition of the InsPs, with InsP3 imparting slightly more flexibility than InsP6. This result highlights the ability of InsP3 to bind and disrupt the C-tail interaction more than higher InsPs when measured in solution. The different functional effect is likely caused by the different binding orientation seen in the Arr1-InsP3 crystal structure (Figure 3C). The ability of InsP3 to remove the C-tail was predicted based on the limited proteolysis experiments done previously with InsP6 and heparin and prior binding experiments with InsP and Arr1, but it is now confirmed directly via cwEPR (Palczewski et al., 1991a, 1991b). The activation of the phosphate sensor of Arr1 by InsP3 clarifies how Arr1 can be so sensitive to the Rh*P C-terminus, as it takes only a few phosphate groups to completely displace the C-tail. Prior structural and biochemical work comparing Arr3/4 to Arr1 suggested that R18 of Arr1 is a phosphate sensor, and that the proline substitutions in Arr3/4 ablate phosphate-sensing activity (Sutton et al., 2005). While we cannot confirm R18 binding an InsPs in our structures, it is entirely possible that the C-tail could interact with R18. There is already a long-distance hydrogen bond between the C-tail and R18 in our model, and the unresolved final residue, E404, could be an additional point of connection.
InsPs prime Arr1, but do not activate it
Prior studies of nonvisual arrestins showed that InsPs can ‘activate’ Arr3 by disrupting the polar core and causing a 15–20° twist in the protein, as seen in receptor-bound structures of arrestins (Zhou et al., 2017; Chen et al., 2017; Zhuang et al., 2010). Arr3 bound each pair of InsP6 molecule between two protomers (six InsP6 molecules for three protomers in a triangle formation), with the majority of the interaction with InsP6_1 (nomenclature matching original publication) from the C-domain of one protomer and 1–2 bonds from the N-domain (Figure 4A). Conversely, the Arr2-InsP6 complex did not have a stabilized C-tail and there was no polar core disruption, leading it to be classified as ‘pre-activated’ or ‘primed’ (Figure 4B). Arr1 bound InsP3 or InsP6 via the N-domain in a monomeric fashion without disrupting the polar core (Figure 4A and 4B). There is no large inter-domain twist in any of the InsP-bound structures as compared to the p44 mutant or the rhodopsin-bound Arr1 (Figure S2). Because of these features, we find the Arr1-InsP complexes to be primed, not activated.
Figure 4. Structure of Arr1 bound to InsP6 – comparisons with Arr2- and Arr3-InsP6 complexes.
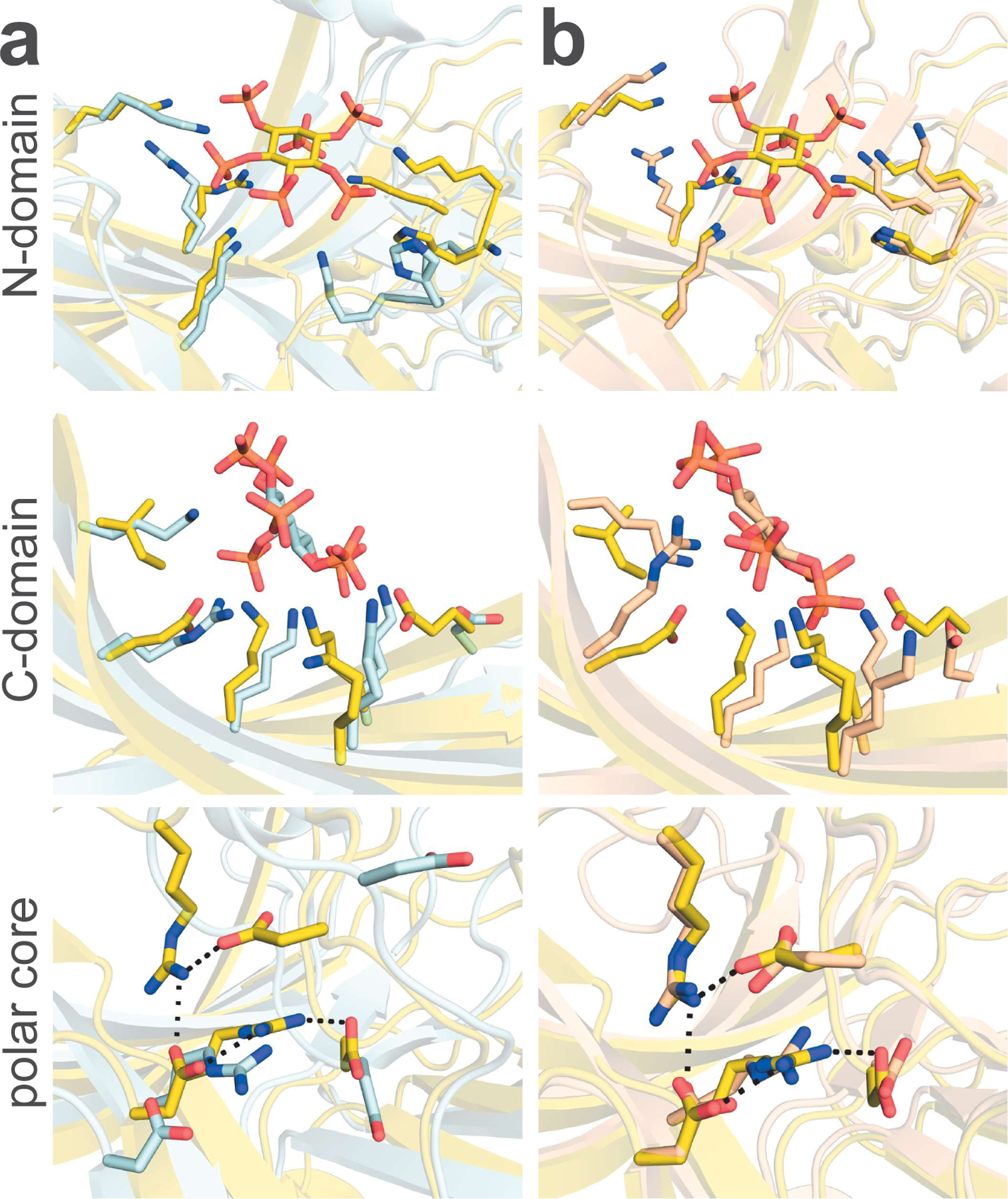
(A) Arr1 (yellow) compared to Arr3 (light blue). Arr1 binds InsP6 in the N-domain, recruiting R171 and the lariat loop in ways distinct from the conformations seen for the same residues in Arr3. The C-domain of Arr3 binds InsP6 with many points of contact, while Arr1 has two substitutions (KI → and → RE) that make the binding site less favorable for InsP6. The active state of Arr3 also forces a disruption of the polar core, whereas Arr1 keeps its polar core intact. (B) Arr1 shows a change in the orientation of R171 relative to the Arr2 structure (light orange), but both Arr1 and Arr2 maintain the position of the lariat loop and polar core. The same substitutions from Arr3 to Arr1 (KI → and → RE) in the C-domain binding pocket are conserved from Arr2 to Arr1.
N-domain of Arr1-InsP6 is primed relative to the active conformation
The N-domain binding pocket of the Arr1-InsP3 complex is not consistent with the active structure of the Rh*P-Arr1 fusion protein. Three key differences differentiate the two states, and they indicate how Arr1 transitions from the primed to active state during receptor binding (Figure 5). First, R171 is in the basal position in the Arr1-InsP3 structure, and it directly participates in holding InsP3 in the binding pocket (Figure 3B). In the Rh*P-Arr1 fusion protein structure, R171 is turned away from its basal position toward pT336 on Rh*P. Second, the lariat loop of Arr1 is found in a mainly basal position, with K298 and K300 binding to several phosphates of the InsP6 molecule. In the fusion protein structure, the lariat loop spans the N-domain binding cavity, bound to the C-terminal pS338 of Rh*P. Third, the C-tail is largely displaced in the Arr1-InsP6 structure, but it remains in the N-domain up to L385, passing by the lariat loop. The receptor-bound Arr1 structure shows a complete displacement of the C-tail, leaving room for the lariat loop to move, further disrupting the polar core.
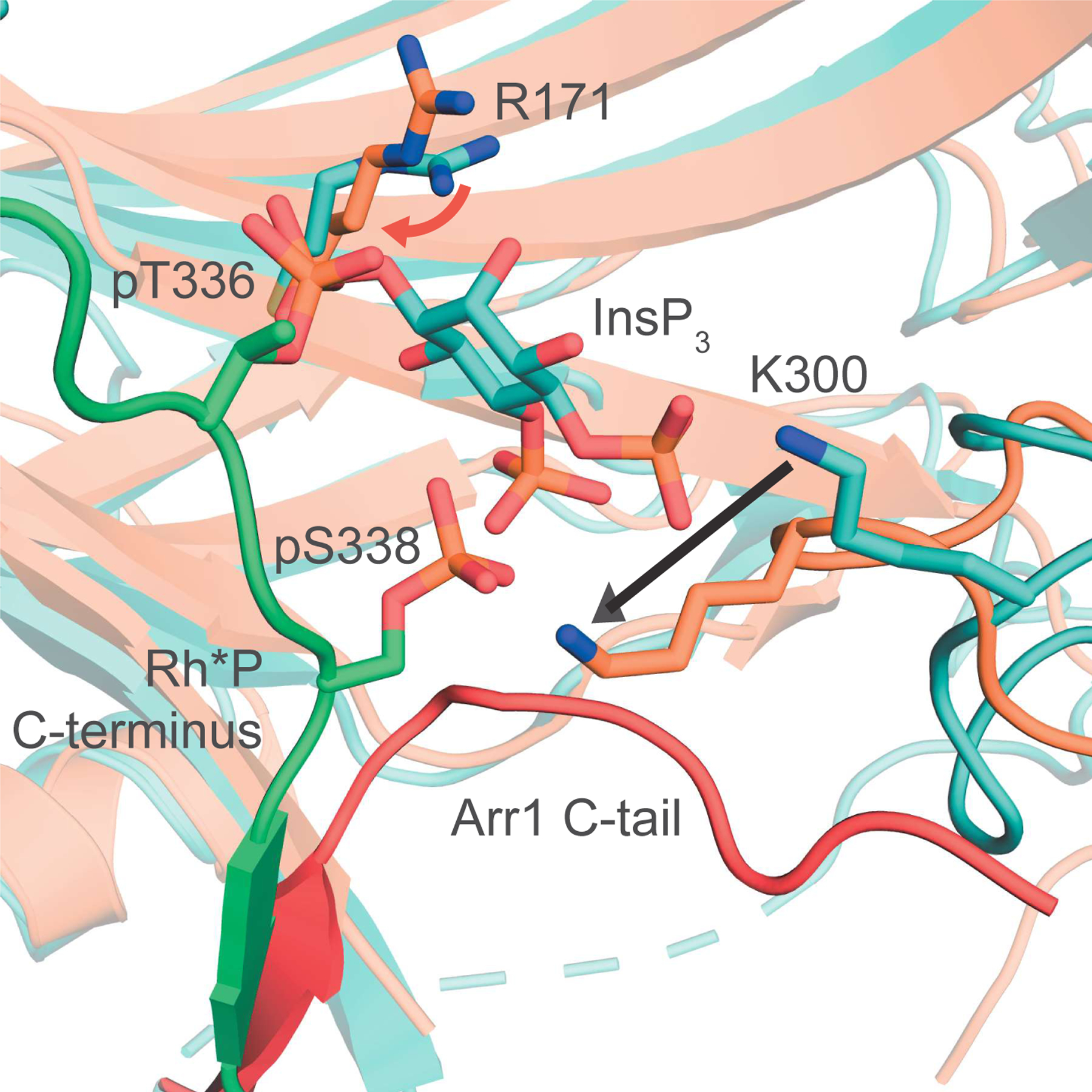
Figure 5. Structure of Arr1-InsP3 in primed conformation – comparison with structure of active, Rh*P-Arr1 fusion protein.

The N-domain of the Arr1-InsP3 complex (teal) differs from active Arr1 (orange, PDB ID: 5W0P), in three main ways. First, R171 in the active structure points more toward pT336 than the center of the phosphate sensor (red arrow). Second, the C-tail of the active protein is completely displaced, being replaced in the 3-element interaction by the C-terminus of Rh*P (Arr1-InsP3 C-tail in red, missing in active structure). Third, the lariat loop containing K300 leaves the basal-like state of the Arr1-InsP3 structure to stretch across the N-domain, binding pS338 (indicated by black arrow).
InsPs larger than InsP3 are found in the retina and bind Arr1 with high affinity
High density polyacrylamide gel electrophoresis (PAGE) analysis shows the presence of InsP5 and InsP6 in bovine retina homogenates (Figure 6A). Roughly a quarter of the sample extracted from 25 bovine retinas was sufficient to give a very dense band of InsP6, whose identity was confirmed by comparison to an authentic standard (Figure S3). Although we do not see bands for 5-diphosphoinositol pentakisphosphate (5PP-InsP5) and bis-1,5-diphosphoinositol tetrakisphosphate (1,5(PP)2-InsP4), we cannot conclude that they were never in the retina samples before the beginning of the extraction process, or that they perhaps have concentrations below the limit of detection by PAGE.
Figure 6. InsPs are present in the vertebrate retina and bind Arr1.
(A) PAGE analysis shows the presence of InsP5 and InsP6 in bovine retina homogenates. Amounts of standards (nmol) are noted at the top. 25% of the sample extracted from 25 bovine retinas was loaded in the “retina” lane. (B-C) Single cell RNA-sequencing (scRNA-seq) heat map depicting absolute (unscaled, B) and relative (scaled as a fraction of the absolute expression value in the retinal cell type with maximal expression for each gene, C) expression values of PP-InsP enzymes by cell type in wild-type (WT) murine retina (Hoang et al., 2020). MC, Müller cell; BC, bipolar cell; AC, amacrine cell; HC, horizontal cell; As, astrocyte; EC, endothelial cell; RPE, retinal pigment epithelium. (D) In situ hybridization analyses of WT murine retinal cryo-sections showing Ppip5k1 and Ip6k2 message throughout the retina with strongest signals in the photoreceptor outer nuclear layer (ONL) (n = 3, representative image shown). The section is co-stained with peanut agglutinin (PNA) and DAPI to demarcate cone photoreceptors and nuclei, respectively. INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar, 50 μm. (E-F) Affinity measurements of natural (E) and synthetic (F) InsPs for Arr1 performed using a fluorescence quenching assay (n=3, mean ± SD). The data in E and F were corrected to account for ligand depletion and the curves based on a one-site binding model.
The inositol pyrophosphates (PP-InsPs) are particularly sensitive to the perchloric acid extraction used in this study (Wilson and Saiardi, 2018). To address the possible presence of PP-InsPs in the retina, we generated heat maps showing absolute (unscaled) and relative expression values of PP-InsP kinases according to cell type in wild-type (WT) murine retina, from a previously published single cell RNA-sequencing (scRNA-seq) data (Figure 6B and 6C) (Hoang et al., 2020). Ip6k2 and Ppip5k1, known to phosphorylate InsP6 and 5PP-InsP5, respectively, were the most abundant transcripts found. To confirm the expression of these transcripts in photoreceptors, combined fluorescence in situ hybridization and immunohistochemical analyses of WT murine retinal cryo-sections were performed (Figure 6D). Ip6k2 and Ppip5k1 expression was observed throughout the retina, with the strongest signals observed in the photoreceptor-containing outer nuclear layer (ONL), consistent with our scRNA-seq analysis. We assessed the affinity of each natural InsP for Arr1 using an intrinsic quenching assay. InsP3 has somewhat lower affinity for Arr1 than the more highly phosphorylated species, which were all slightly lower than 1 micromolar in affinity (Figure 6E, Table S1). Because of the relatively labile nature of the natural PP-InsPs, we also tested the affinity of synthetic, bisphosphonate versions of the PP-InsPs (5PCP-InsP5 and 1,5(PCP)2-InsP4; PCP-InsPs collectively) (Figure 6F). The PCP-InsPs exhibited weaker interactions with Arr1 than their natural counterparts (Table S1).
Structures of Arr1 in complex with higher-order InsPs confirm the versatility of anionic ligand binding within the N-domain.
We were able to crystallize the PP-InsPs and PCP-InsPs with Arr1 using the same soaking method used for InsP3 (Figure 7A-D and S4, Table 2). The structures were similar to the other InsP-bound-Arr1 structures and each other, with hardly any inter-domain movement (Figure S2). All four ligands utilize the additional 5-position phosphate to interact with H301, and they all appear to reach slightly deeper in the binding region of the N-domain than InsP6 (Figure 7E). The added 1-position phosphate did not seem to have a conserved interaction with any of the residues in the N-domain, but the solvent accessible nature of the exposed N-domain binding region leaves many possible ways for PP-InsPs to bind to Arr1.
Figure 7. Higher-order InsPs bind to the same region as InsP6.
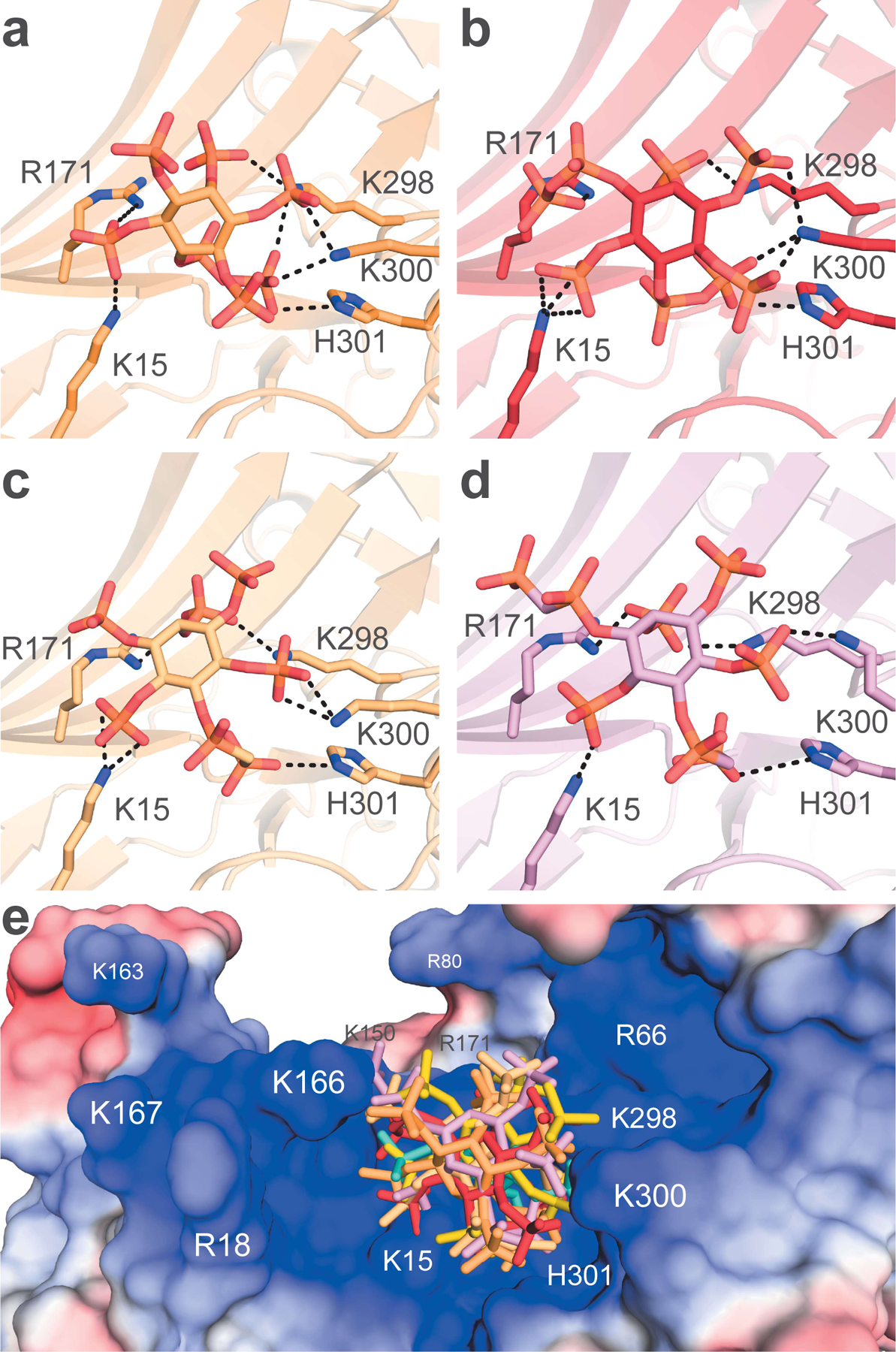
(A-D) 5PP-InsP5, 1,5(PP)2-InsP4, 5PCP-InsP5, 1,5(PCP)2-InsP4, respectively, are shown to bind to the N-domain. (E) All InsPs in this study are shown to bind to the positively-charged pit of the N-domain.
Table 2.
Summary of crystallographic data collection and refinement statistics for PP-InsPs and PCP-InsPs
| Data Collection | 5PP-InsP5 | 5PCP-InsP5 | 1,5(PP)2-InsP4 | 1,5(PCP)2-InsP4 |
|---|---|---|---|---|
| X-ray source | SSRL 12–2 | NSLS II - FMX | SSRL 12–2 | SSRL 12–2 |
| Wavelength (Å) | 0.97946 | 0.979358 | 0.97946 | 0.97946 |
| Space group | P21212 | P21212 | P21212 | P21212 |
| Unit cell dimensions (Å) | a= 169.36, b= 187.00, c= 90.79 |
a= 169.36, b= 187.00, c= 90.79 |
a= 168.90, b= 187.09, c= 90.39 |
a= 168.47, b= 187.23, c= 90.74 |
| Resolution (Å) | 50.0–2.6 (2.60–2.76) | 50.0–2.8 (2.80–2.97) | 50.0–3.0 (3.00–3.18) | 50.0–2.7 (2.66–2.82) |
| Unique reflections | 88,724 (14,073) | 71,781 (11,401) | 58,102 (9,152) | 83,087 (13,219) |
| Multiplicity | 6.70 (6.77) | 8.37 (8.27) | 13.3 (13.0) | 13.6 (13.9) |
| Completeness (%) | 99.8 (99.6) | 99.9 (99.6) | 99.7 (98.7) | 99.9 (99.8) |
| <I/σI> | 18.16 (0.98) | 14.95 (1.12) | 12.71 (0.69) | 16.07 (0.89) |
| Rsym (%) | 8.1 (177.2) | 12.4 (234.0) | 20.4 (362.3) | 14.4 (277.0) |
| CC1/2 (%) | 99.9 (61.1) | 99.9 (60.5) | 99.9 (59.6) | 99.9 (53.2) |
| Wilson B factor (Å2) | 78.6 | 80.5 | 87.9 | 77.3 |
| Refinement | ||||
| Resolution (Å) | 49.14–2.60 | 48.47–2.80 | 49.16–3.00 | 49.23–2.66 |
| Number of reflections | 84,114 (4,431) | 68,199 (3,573) | 53,859 (2,775) | 78,647 (4,133) |
| Rwork/Rfree (%) | 22.2/24.8 | 21.5/24.6 | 21.0/24.5 | 22.0/24.9 |
| Number of atoms | 11,759 | 11,548 | 11,777 | 11,750 |
| Protein | 11,500 | 11,280 | 11,504 | 11,514 |
| InsP | 200 | 240 | 264 | 220 |
| Water | 53 | 28 | 9 | 16 |
| <B-factor> (Å2) | 93.7 | 101.5 | 113.3 | 93.6 |
| Protein | 92.0 | 99.7 | 111.1 | 91.7 |
| InsP | 201.1 | 192.7 | 214.5 | 197.5 |
| Water | 64.2 | 76.4 | 69.5 | 60.9 |
| RMS deviation from ideal | ||||
| Bond lengths (Å) | 0.003 | 0.004 | 0.004 | 0.004 |
| Bond angles (°) | 0.56 | 1.23 | 1.24 | 1.24 |
| Ramachandran plot (% favored/# outliers) | 96.4/0 | 97.7/0 | 94.9/0 | 95.8/0 |
| Molprobity score (%) | 100 | 100 | 100 | 100 |
| PDB accession code | 7MP0 | 7MOR | 7MP2 | 7MP1 |
DISCUSSION
The structural and complementary biochemical data on Arr1 herein fill gaps in our knowledge regarding the conformational changes needed for full receptor engagement and termination of GPCR signaling (Figure 8). The basal C-tail of Arr1 binds the N-domain via at least 6 salt bridges, creating a binding pocket or tunnel to attract negative charges (Figure 8A). Of the six salt-bridge interactions binding the C-tail to the N-domain, five were not observed in previously reported structures. While not the only possible form that C-tail-binding can take as it vibrates in solution, we believe we have identified specific interactions responsible for C-tail regulation of Rh*P binding. When the phosphate sensor region interacts with InsPs or other phosphorylated ligands, it releases the portion of the C-tail spanning the N-domain cavity, likely starting with E394 and D397, as their respective binding partners, K298 and R171, engage with the ligand phosphates (Figure 8B). As the ligand is further stabilized by residues in the N-domain, the C-tail is further displaced, and the C-latch is released concomitant with solvation of the C-tail to residue L385. As in the case of the truncated p44 splice variant and active structures of Arr1, the C-tail must detach further, disengaging one of the components of the 3-element interaction (Kim et al., 2013). As the Arr1 C-tail is leaving the N-domain, the phosphorylated C-terminus of Rh*P can simultaneously form a more stable contact with the N-terminus of Arr1, and, in so doing, drag the lariat loop of Arr1 away from the center of the protein, thereby disrupting the polar core and initiating full Arr1 activation (Figure 8C). We believe that the position of the lariat loop in the structure of the Rh*P-Arr1 fusion protein is not seen in the p44 splice variant of Arr1 because there is no phosphorylated C-terminus to drag the loop across the N-domain, leaving the conformation adopted by the p44 mutant between our InsP-primed structures and the active, Rh*P-Arr1 fusion structure. The InsP-bound structure, therefore, serves both as a model of the transitional state in the Rh*P-Arr1 desensitization process, as well as a conformation achieved when InsPs bind Arr1.
Figure 8. InsPs displace Arr1 C-tail, priming the protein and modeling how the Rh*P C-terminus can prime and then activate Arr1.
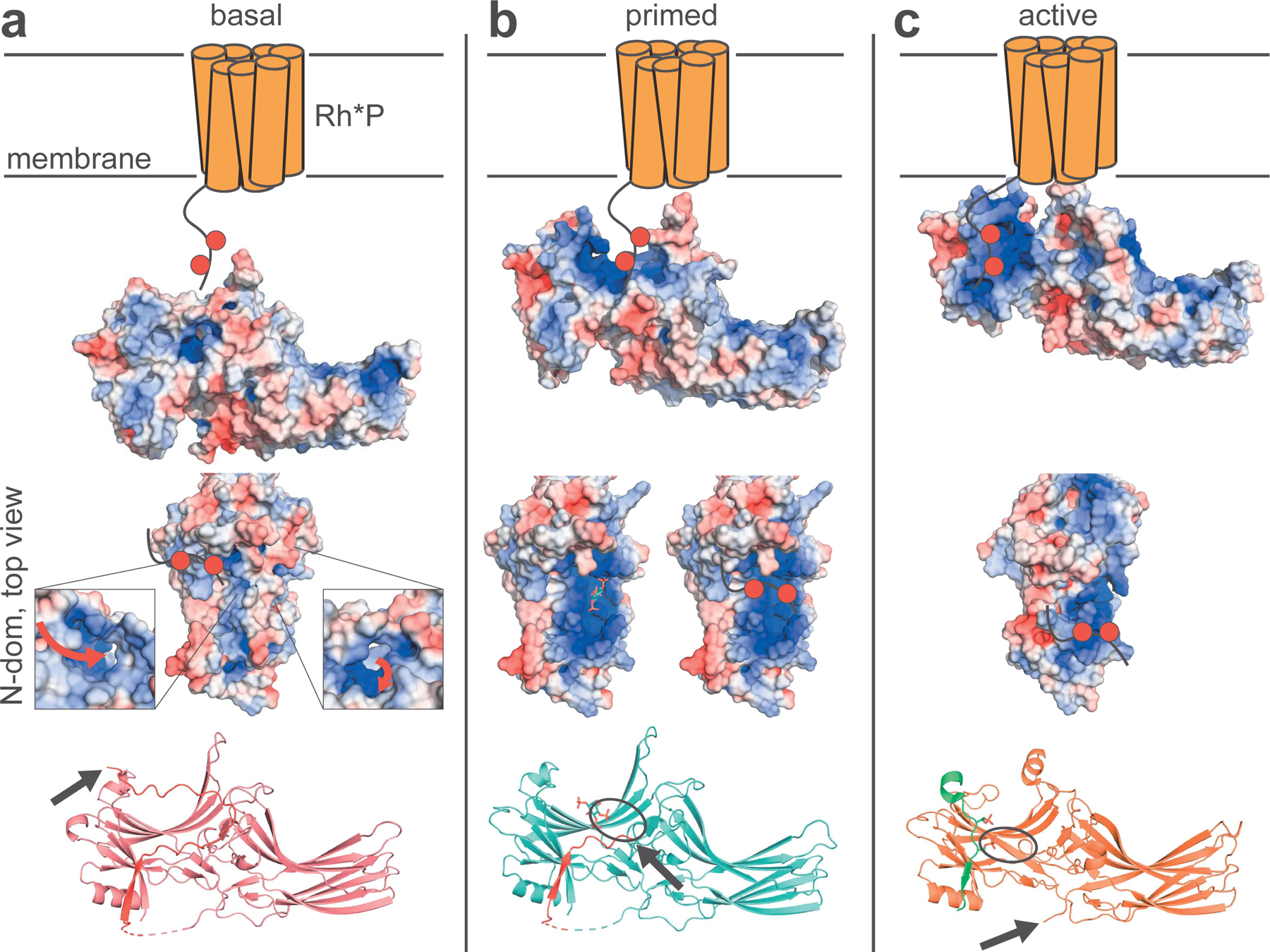
(A) The C-tail of basal Arr1 creates a binding pocket or tunnel to attract negative charges near the mid-point of the protein. When the phosphate sensor interacts with InsPs or Rh*P, it releases the C-tail spanning the N-domain cavity (orange receptor is Rh*P; red circles on the C-terminus of Rh*P represent phosphate groups; red arrows show the possible path of phosphate binding; the black arrow denotes the end of the Arr1-C-tail, which is highlighted in red). (B) The C-tail is further displaced by the ligand, opening up the basic patch across the N-domain (interactions between phosphates and the lariat loop are denoted by black ovals). (C) After complete C-tail displacement, the phosphorylated C-terminus of Rh*P can traverse the basic patch toward the N-terminus of Arr1 to form a stable contact, dragging the lariat loop of Arr1 away from the center of the protein and disrupting the polar core, thereby causing activation (Rh*P C-terminus highlighted in green).
Highly phosphorylated InsPs such as 5PP-InsP5 and 1,5(PP)2-InsP4 could also play a functional role in photoreceptors. To our knowledge, InsP6 has not been directly reported in the retina, although concentrations of InsP6 in many other cell types have been established (Shears, 2001). We also found relatively high levels of photoreceptor transcripts for the kinases responsible for conversion of InsP6 into the pyrophosphates 5PP-InsP5 and 1,5(PP)2-InsP4, which bind arrestin similarly to InsP6. These data suggest a hitherto unexplored role for these highly charge-dense InsP species in photoreceptors.
Taken together, our more complete C-tail- and InsP-bound structures have inspired a working molecular model that can explain how phosphate sensor binding leads to C-tail displacement and/or Rh*P binding (Figure 8). As the InsPs (or other phosphorylated ligands, e.g., Rh*P) enter the concave cavity of the N-domain, the phosphate groups displace the C-tail by competing for binding to R171 and K298. At this stage only D403 of the C-latch (bound to K167) is left to hold the C-tail in place in the region spanning the N-domain. As the InsP becomes more stabilized on Arr1 by interacting further with any combination of K300, K15, H301, and K166, the binding pocket begins to adjust conformationally to accommodate the negative charges, forcing the negatively charged C-tail to move further away from the N-domain. The partial dissociation of the C-tail affects interactions in the C-latch, causing it to release D403 and near-fully liberate the C-tail. The partial separation of the C-tail enables solvation to compete with the remaining C-tail interactions (K392-D71 and D387-R288) and transition to the primed, InsP-bound conformation.
At this stage, the C-tail displacement could have a key impact on arrestin oligomerization. The solution tetramer of Arr1 is thought to have a N-N, C-C, N-N organization, resembling a “boat” (Hanson et al., 2008b). After arranging our basal structure in the proposed solution tetramer configuration, we believe that the homotetramer could be disrupted if the C-tail of adjacent N-domains become more dynamic (Figure S5). Based on our data indicating that InsP3 potently affects C-tail binding to the N-domain, as well as the high-resolution structure which confirms deep penetration of the ligand into the N-domain cavity, it is plausible that any InsPs in proximity of Arr1 will bind the Arr1 N-domain in a C-tail-destabilizing manner, thereby disrupting the likely solution tetramer N-N contacts. This hypothesis is supported by our data on the affinity of all tested InsPs for Arr1 near or below 1 µM, lower than the reported KD of bovine Arr1 dimer and tetramer formation (KDdimer = 36.8 µM, KDtetramer = 7.5 µM) (Imamoto et al., 2003).
The relative InsP affinities must also be considered in the context of the concentrations of the Arr1 monomer/dimer/tetramers and InsPs. The concentrations of total Arr1 are reportedly in the low millimolar range. InsPs are not well quantified in photoreceptors, but InsP6 has been quantified across a variety of mammalian tissues with concentrations averaging between 15–100 µM (Shears, 2001). Assuming the majority of Arr1 is in the tetrameric form given the cooperativity of their formation over dimers, the tetramers are roughly 2.5- to 15-fold higher in concentration than InsP6, a range over which Arr1 is likely to be sensitive to InsP concentrations. Foundational work on InsP6 homeostasis showed that up to 50% of InsP6 is converted to 5PP-InsP5 each hour, which, in the context of the relatively low levels of 5PP-InsP5, means that the PP-InsPs turn over rapidly (Glennon and Shears, 1993; Menniti et al., 1993). As has been noted elsewhere, the energy expenditure required to maintain higher-order InsP flux suggests a usefulness that is not yet fully understood (Saiardi, 2012). Precise measurements of InsPs concentrations and turnover in the retina should be pursued, with special attention to possible fluctuations before and after light exposure.
To enable historical comparison to prior binding experiments between InsPs and Arr1, we omitted Mg2+ from the current binding assay (Palczewski et al., 1991b; Wilson and Copeland, 1997). Research into the physiological state of InsP6 and PP-InsPs in the cell since those early InsP-Arr1 reports have found Mg2+ as essential for differentiating between specific and non-specific binding interactions when comparing high-order InsPs (e.g., InsP6 vs. 5PP-InsP5 vs. 1,5(PP)2-InsP4) (Shears, 2001). This is likely not an issue in the case of InsP3, however, so the lower relative affinity measured for InsP3 compared to the higher InsPs is likely less/not significant in physiological conditions.
There are no prior PDB structures of primed Arr1 bound a non-peptide ligand such as InsPs. Comparison of our primed structures to the basal structure shows that none is in the ‘active’ conformation; they show neither the finger-loop contortion nor the polar core disruption seen in the Arr3-InsP6 complex structure. Because only the C-tail is perturbed in our InsP-bound structures, they more closely resemble the primed but un-activated Arr2-InsP6 complex.
Remarkably, our Arr1-InsP structures are distinct from prior Arr2/3-InsP6 structures; the Arr1 structure shows single-domain binding of all InsPs, whereas in the structures of both Arr2 and Arr3, InsP6 is bound by 2 protomers. Two differences between Arr2/3 and Arr1 regarding the C-domain InsP6-binding residues are a basic-to-neutral (K251/I256) and basic-to-acidic substitution (R237/E242) (Figure 4). Because of these differences, and the ability of InsP6 to dissociate Arr1 oligomers (inverse to the behavior of nonvisual arrestins), we surmise that the weak interactions between InsP6 and the Arr1 C-domain in our structures are likely to be crystal artefacts.
The potential evolutionary path to repurposing an arrestin protein for the specialized function of termination of phototransduction is fascinating to consider. Mutations in the C-domain of Arr1 that reduce that domain’s affinity for InsPs apparently confer an alternative relationship of the protein to InsPs, promoting monomers over self-association. The relative loss in affinity of the C-domain of Arr1 for InsPs shifted the binding site to the N-domain, where the crucial C-tail interaction regulates Arr1’s ability to access the OS. The unique biology of the retinal photoreceptor systems necessitates a robust, but strictly regulated, level of arrestin-mediated signal termination. Arr1 utilizes conserved N-domain contacts to bind the C-tail regulatory domain, thereby possibly making multimerization more likely. Arr1 has the added feature of being targeted by phosphate groups first in the N-domain, enabling InsP regulation of the multimer/monomer equilibrium and consequently the diffusion of Arr1 in the photoreceptors.
Our structural data raise the question of the role of the higher InsPs and PP-InsPs in the photoreceptors, as well as if these molecules have specific effects on Arr1 upon binding. Given that the higher InsPs and PP-InsPs target the same polybasic binding site and considering that conformational changes of Arr1 are minimal upon binding to InsPs and PP-InsPs, it appears unlikely that distinct conformational changes will occur in the InsP/PP-InsP complexes in solution. More broadly, genetic studies have revealed complex functions for PP-InsPs in signal transduction (Brown et al., 2016). The underlying molecular mechanisms explaining how PP-InsPs mediate specific cellular responses, however, are not clear. Allosteric regulation of proteins is one signaling mechanism for PP-InsPs, something well demonstrated by the yeast VTC complex (Wild et al., 2016). In other cell types, PP-InsPs promote protein-protein interaction by recruitment of IP6Ks, which foster local production of 5PP-InsP5 (Chin et al., 2020; Rojas et al., 2019). Another mode of PP-InsP signal transduction is associated with their similarity to membrane bound PIPs. Many PP-InsP binding proteins contain PIP binding domains, such as PH and C2 domains (Luo et al., 2003; Chakraborty et al., 2010; Gokhale et al., 2013; Furkert et al., 2020). Recent studies have shown that PP-InsPs can effectively compete with PIPs and can delocalize proteins from the membrane, thereby regulating membrane-cytosol communication (Gokhale et al., 2013; Lorenzo-Orts et al., 2020; Pavlovic et al., 2016).
It is established that Arr1 translocation is neither reliant upon rhodopsin phosphorylation nor transducin signaling in vivo, but depends on non-classical rhodopsin signaling (unless there are additional light-dependent pathways yet undiscovered) (Mendez et al., 2003). There are remaining questions regarding recruitment of Arr1 to the OS. Arr1 has been shown to mislocalize to the OS in dark-adapted Rpe65−/− mice, suggesting that apo-opsin cycles through conformations approximating active rhodopsin that attract Arr1, either directly or through an uncharacterized pathway. It has been speculated that Gαq and/or Gα11 play a role in transmitting the signal from light-activated rhodopsin. Gαq and Gα11 are known to activate PLC and PKC signaling pathways, both of which are sufficient to cause Arr1 translocation in vivo, indicating that they affect Arr1 stores in the photoreceptor IS (Orisme et al., 2010). Decades before the PLC-PKC-translocation relationship was discovered, several groups reported changes in PIP2 and its PLC-cleavage products upon brief illumination of the retina and isolated rod OS (Brown et al., 1984; Ghalayini and Anderson, 1984; Vandenberg and Montal, 1984; Hayashi and Amakawa, 1985), although there may be a role for phosphatases in the observed PIP2 depletion (Millar et al., 1988). The cumulative results show that PIP2 is cleaved in a rapid and light-dependent manner in vertebrate photoreceptors, which likely releases InsP3 and diacylglycerol (DAG). Our work has provided high-resolution details as to how InsPs bind and disengage the autoregulation of Arr1.
STAR METHODS
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Philip Kiser (pkiser@uci.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper analyzes existing, publicly available data. The public location of the dataset is listed in the key resources table.
This paper does not report original code.
The atomic coordinates and structure factors of all structures have been deposited in the Protein Data Bank under accession codes 7F1W, 7F1X, 7JSM, 7JTB, 7JXA, 7MOR, 7MP0, 7MP1, and 7MP2. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Animals
Female WT C57BL/6J mice approximately one yr of age were purchased from The Jackson Laboratory. Animals were housed in a standard 12-hour light/12-hour dark cycle environment, fed standard soy protein–free rodent chow diet (Envigo Teklad 2020X, Indianapolis, IN), provided water ad libitum, and housed in plastic cages with standard corncob rodent bedding and 6-gram nestlets (Ancare, Bellmore, NY). They were euthanized in a CO2 chamber prior to enucleation. Animal procedures were approved by the Institutional Animal Care and Use Committees at the VA Long Beach Healthcare System and the University of California Irvine. All experimental protocols were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals, the recommendations of the American Veterinary Medical Association Panel on Euthanasia, and the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Visual Research.
Microbe strains
E. coli BL21 DE3 cells from Agilent were transformed with expression plasmids and were grown at 37 °C in lysogeny broth media supplemented with ampicillin until OD600 reached 0.6. Expression was induced with 25 μM IPTG for 18 hours at 16 °C.
Native tissue
Native bovine retinas were sourced from local slaughterhouses. Eyes were collected and transported on ice, then dissected on ice under dim red light in a dark room. Retinas were then stored at −80 °C until ROS purification.
Method Details
Purification of Arr1
The purification of Arr1 from bovine retinas was done largely according to the protocol by Buczylko and Palczewski (Buczyłko and Palczewski, 1993). Sixty bovine retinas (American Beef Packers, Chino, CA) were homogenized under dim red light with 20 loose and 20 tight pumps in a 40 ml Wheaton glass homogenizer on ice and then centrifuged at 28,969 × g in a JA-17 rotor for 35 min. The supernatant was diluted with Homogenization Buffer (10 mM HEPES, pH 7.5; 1 EDTA-free cOmplete protease inhibitor tablet (Roche, Basel, Switzerland) per 50 ml) to 4× volume and loaded on a self-packed, ~30 ml Whatman DE52 cellulose (Maidstone, UK) XK16 column (GE Life Sciences, Marlborough, MA) which was equilibrated with Homogenization Buffer, at 0.4 ml/min using a Bio-Rad NGC Quest FPLC (Hercules, CA). The column was washed with 150 ml of Wash Buffer (10 mM HEPES, pH 7.5; 15 mM NaCl) at 0.4 ml/min. Elution was performed with a linear gradient (15–150 mM NaCl) and collected in 1.5 ml fractions at 0.25 ml/min over 240 ml. Anion exchange fractions roughly between 25% and 50% of the elution gradient were pooled and loaded onto a self-packed heparin Sepharose column (BioVision Research Products, Mountain View, CA; 7 ml of loose resin packed in a XK-16 column) which had been equilibrated with Heparin Equilibration Buffer (10 mM HEPES, pH 7.5; 100 mM NaCl) at 0.3 ml/min. Heparin Wash Buffer (10 mM HEPES, pH 7.5; 150 mM NaCl) was applied to the column at 0.3 ml/min for a total of 100 ml. Arr1 was selectively eluted from the column by a linear gradient of 0–8 mM InsP6 (in Heparin Wash Buffer) at 0.2 ml/min and collected in 1.5 ml fractions. To remove the InsP6 bound to Arr1, heparin column elution fractions containing Arr1 were pooled and dialyzed for 12 hrs with 3 buffer changes of 1 L (4 °C, with vigorous stirring) of Heparin Equilibration Buffer. Material from dialysis was loaded onto a regenerated heparin column (same 7 ml of loose resin packed on a XK16 column, washed with 2M NaCl and equilibrated with Heparin Equilibration Buffer) at 0.3 ml/min. The column was washed with 100 ml of Heparin Wash Buffer at 0.3 ml/min, followed by a Second Heparin Elution Buffer (10 mM HEPES, pH 7.5; 400 mM NaCl) at 0.2 ml/min, collected in 1.0 ml fractions. All steps were carried out on ice or at 4 °C.
Tyrosine fluorescence quenching assay of Arr1
To assess InsP binding to Arr1, an intrinsic tyrosine fluorescence quenching assay was performed (Arr1 has 14 Tyr residues/monomer), as done previously (Wilson and Copeland, 1997). 300 nM Arr1 in 500 µL of Heparin Equilibration Buffer was mixed in a quartz cuvette (PerkinElmer, Waltham, MA) with mini stir bar mixing on low and temperature maintained at 20 °C. A PerkinElmer LS55, coupled to an Isotemp 3016S (Fisher Scientific, Pittsburgh, PA) to regulate temperature, was used to measure fluorescence of samples after serial addition of InsP stocks diluted in the same buffer as the Arr1. Device settings for the scans were Ex/Em range of wavelengths of 275/290–350 nm. A slit width of 7 nm was used with a scan rate of 100 nm per min. Every scan was repeated 3 times, and the emission peak at 305 nm was used for the tyrosine emission wavelength. Percent quench values were adjusted for increasing dilution with additional ligand added 5 µL at a time. Curves were fit to the means of the replicates with one site binding accounting for ligand depletion.
Crystallization conditions – P21212 space group crystals
All crystals were formed in hanging drops at 8 °C over 2–7 days. Basal state Arr1 crystals were formed in 2 µl of 10–11 mg/ml of purified Arr1 and 2 µl of well solution, consisting of 100 mM bis-tris propane (BTP, pH 7.0), 35% 2-ethoxyethanolamine, and 1 mM MgCl2. InsP6-Arr1 complexes co-crystallized in the same buffer and volume drops as basal, with the addition of 0.9 mM InsP6 to the well solution. To generate Arr1-InsP3 crystals, InsP3 solution was added to the drop of basal crystals grown in 100 mM bis-tris propane (BTP, pH 7.0), 35% 2-ethoxyethanolamine, and 2.5 mM NaCl using a 2.93 mM final concentration of InsP3. The soaked crystals were incubated overnight, then harvested. The crystals formed as either square or triangular plates roughly 0.4 mm wide and 0.05 mm thick. The crystals were flash-cooled in liquid nitrogen for X-ray data collection.
Crystallization conditions – C2221 space group crystals
Native arrestin was purified basically as described elsewhere from bovine retinae that were supplied by a local slaughterhouse (Chuklim, Republic of Korea) (Buczyłko and Palczewski, 1993). 50 bovine retinas in 10 mM HEPES Buffer (pH 7.5) containing 1 mM benzamidine and SigmaFast protease inhibitor cocktail were homogenized with a glass-glass homogenizer (Sigma Aldrich, St. Louis, MO). The lysate was then separated from the cell debris by centrifugation (4°C, 20,000 rpm, Hanil A85–6 rotor, 20 min). The supernatant was applied onto a DEAE-cellulose column equilibrated with Buffer A (10 mM HEPES, pH 7.5). The column was washed with Buffer A and elution was performed with 150 mM NaCl and 0.5 mM benzamidine in Buffer A. The extract was loaded onto a 5-ml HiTrap Heparin column (Cytiva, Marlborough, MA) equilibrated with Buffer A. After washing the column with Buffer A, proteins were eluted with a 0–10 mM phytic acid gradient in Buffer A. Eluted arrestin was dialyzed against Buffer A, loaded onto a HiTrap Heparin column and eluted with a 0–1 M NaCl gradient in Buffer A. Arrestin eluted at a concentration of 300 to 400 mM NaCl was dialysed thoroughly against 100 mM NaCl in Buffer A. Purified arrestin was concentrated to 20 mg/ml using Amicon filters (30 kDa cutoff). For crystallization, concentrated arrestin (2 µl) was mixed with reservoir solution (2 µl) consisting of crystallization buffer as described previously (Wilden et al., 1997). Arrestin crystals grew within a week using the hanging-drop method. Crystals of arrestin bound to InsP3 or InsP6 were obtained by co-crystallization in a 1:8 (arrestin:inositolphosphate) molar ratio. For X-ray diffraction data collection, single crystals with InsP6 or InsP3 were harvested using cryo-loops and directly frozen in liquid nitrogen without addition of any cryoprotectant.
Diffraction data collection, structure solution and refinement
Soaked crystal data was collected and processed as follows. Diffraction data were collected at the APS NECAT IDC beamline. Data reduction was carried out with XDS (Kabsch, 2010). The basal and InsP6 Arr1 structures were solved by direct refinement using a previously determined Arr1 structure as a search model (PDB accession code 3UGX) (Murshudov et al., 2011). The Arr1-InsP3 structure was solved by molecular replacement using chain D of the Arr1-InsP6 structure as a starting model. Structural models were iteratively improved by manual real space refinement in Coot and reciprocal space refinement in REFMAC (Emsley and Cowtan, 2004; Murshudov et al., 2011). The models were periodically checked for geometric soundness with MOLPROBITY and the wwPDB validation server (Chen et al., 2010; Read et al., 2011).
Data for co-crystallized samples were collected and processed as follows. X-ray diffraction data were collected at 100 K at beamline 7A of the Pohang Accelerator Laboratory (PAL 7A SB I; Pohang, Republic of Korea) using a CCD detector (ADSC Quantum 270) and an X-ray wavelength of 1.0 Å. All images were indexed, integrated and scaled using the XDS program package and the CCP4 program (Collaborative Computational Project, Number 4, 1994; Evans, 2006; Kabsch, 2010). The InsP6- and InsP3-bound arrestin crystals belong to the orthorhombic space group C2221 with unit-cell parameters a = 167.85 Å, b = 190.46 Å, c = 190.47 Å, α = β = γ = 90° in case of arrestin complexed with InsP6 and C2221 with unit-cell parameters a = 167.57Å, b = 187.70 Å, c = 190.82Å, α = β = γ = 90° in case of arrestin complexed with InsP3. Initial phases for arrestin were obtained by a conventional molecular-replacement protocol (rotation, translation and rigid-body fitting) using PHASER and the structure of bovine visual arrestin (PDB entry 1CF1) as an initial search model (Hirsch et al., 1999; McCoy et al., 2007). The model was fitted more appropriately by simulated annealing in CNS (Brünger et al., 1998; Brunger, 2007). Manual rebuilding of the model for arrestin complexed with inositol phosphates and electron-density interpretation were performed after each refinement cycle using Coot (Emsley and Cowtan, 2004). Restrained, individual B factors were refined and the crystal structure was finalized using Phenix.refine software (Adams et al., 2010). Structure validation was performed with PROCHECK and OneDep system (Hodsdon et al., 1996; Read et al., 2011). Some of the software used for the building and refinement of the structures was from the SBGrid software consortium (Morin et al., 2013).
Structure analysis
Electrostatic surfaces were calculated using the adaptive Poisson-Boltzmann solver (APBS) electrostatics plugin available in PyMOL (Baker et al., 2001). Figures displaying three-dimensional structures were rendered using PyMOL (Schrödinger, LLC, New York, NY).
PAGE of InsP compounds extracted from bovine retina
InsPs (and other phosphate-containing compounds) were extracted from bovine retinas using a modification of a prior protocol (Wilson and Saiardi, 2018). Elution fractions from the extraction of InsPs from 25 fresh bovine retinas (14.02 g wet weight) was concentrated to roughly 20 µL, and 5 µL were loaded in the lane indicated in Figure 5a. Standard compounds were either purified by the authors as described previously (5PP-InsP5 and 1,5(PP)2-InsP4) or purchased (InsP6 and ATP were from Sigma Aldrich, and InsP5 and GTP were from Cayman Chemical, Ann Arbor, Michigan) (Puschmann et al., 2019). The 33.3% bis-acrylamide gel and staining procedures used to identify the components of the purified, phosphate-rich samples were adapted from a prior work to run on a shorter, 16 cm gel, and included 0.1% bromphenol blue in place of Orange G as the dye-front indicator (Losito et al., 2009). The gel was cast at room temperature, then pre-run for 2 hrs at 300 V at 4 °C. Once samples were loaded, the gel ran 24 hr at 300 V at 4 °C until the lowest visible dye front was 8 cm from the bottom of the gel. The gel was stained for 30 min. in a solution of 20% methanol, 2% glycerol, and 0.05 % (w/v) Toluidine Blue O (Polysciences, Inc., Warrington, PA). The gel was then destained in 20% methanol and 2% glycerol for 2 hr with mild agitation and several exchanges of buffer. Images were taken on a light box and the color balance was warmed using ImageJ by increasing the yellow and red channels (to make the metachromatic shift normally evident to the eye also visible in the photo) (Schneider et al., 2012).
In situ hybridization of murine retinas
WT C57BL/6J mice approximately one yr of age (The Jackson Laboratory, Bar Harbor, ME) were euthanized in a CO2 chamber prior to enucleation. Eyes were fixed in Hartman’s Fixative Solution (Sigma-Aldrich) overnight at room temperature (RT) and transferred to 30% sucrose in PBS for overnight incubation at 4 °C, prior to embedding in OCT compound for cryosectioning. Slides were processed for in-situ hybridization using the RNAscope Multiplex Fluorescent Kit, according to manufacturer instructions using probes Mm-Ppip5k1and Mm-Ip6k2-C2 (Advanced Cell Diagnostics, Newark, CA). Upon completion of the RNAscope assay, slides were incubated in a blocking buffer containing 5% FBS, 1% BSA, and 0.2% Triton X-100 in PBS for 1 hr at RT for immunohistochemistry. Slides were then incubated with biotinylated PNA (1:250; Vector Labs, Burlingame, CA) in blocking buffer overnight at 4 °C, followed by a 0.5 hr incubation with Alexa Fluor 488 streptavidin (1:250, Invitrogen, Carlsbad, CA) in blocking buffer at RT. Fluorescence microscopy images were obtained on a Keyence BZ-X810 fluorescent microscope (Keyence, Itasca, IL).
Purification and spin-labelling of arrestin cysteine mutants
For bovine arrestin-1, E. coli codon-optimized genes for cysteine-free base mutants (C63V/C128S/C143A, containing two additional mutations, F85A/F197A, which reduce arrestin self-association) were synthesized by GenScript (Piscataway, NJ) and cloned into vector pET-15a between NcoI and XhoI sites (Vishnivetskiy et al., 2013). All cysteine mutations for site-directed spin labeling were introduced by PCR using QuikChange mutagenesis and confirmed by DNA sequencing. E. coli BL21 (DE3, Agilent, Santa Clara, CA) transformed with the arrestin expression plasmids were grown at 37 °C in lysogeny broth media supplemented with ampicillin until OD600 reached 0.6. Then expression was induced with 25 μM IPTG for 18 hours at 16 °C. Harvested cell pellets were resuspended in lysis buffer (10 mM Tris pH 7.0, 100 mM NaCl, 2 mM EDTA, 1 mM DTT) containing a SigmaFast protease inhibitor cocktail tablet (Sigma-Aldrich). Cells were lysed by 4 passes through an EmulsiFlex-C3 homogenizer (Avestin, Ottawa, Canada) at 15,000 psi, and centrifuged at 100,000 g at 4°C for 1 hour. The supernatant was loaded onto a 20 ml Heparin-FF column (Cytiva) and elution was performed with a linear 0.1–1 M NaCl gradient in lysis buffer.
Fractions containing arrestin were dialyzed against 10 mM Tris pH 8.3, 2 mM EDTA, 1 mM DTT and then loaded onto SP-HP and Q-HP columns (Cytiva) connected in a tandem setup. The SP column was removed after sample loading and elution from the Q-HP column was performed with a linear 20–500 mM NaCl gradient in dialysis buffer. Fractions containing pure arrestin were pooled, concentrated and stored at −80 °C.
All single cysteine constructs of arrestin were labelled with a 10-fold excess of 1-oxyl-2,2,5,5-tetramethyl-∆3-pyrroline-3-methyl methanethiosulfonate (Toronto Research Chemicals, North York, Canada) at 4 °C overnight to generate the R1 spin label side chain. Excess spin label was removed using a PD-10 desalting column (Cytiva). Spin labelled arrestin samples were concentrated to 100 µM for cwEPR measurements.
CW-EPR measurements
X-band cwEPR data of the spin-labeled arrestin mutants (T396C and A401C) at different buffer compositions were acquired using a BRUKER ELEXSYS E500 spectrometer equipped with a BRUKER EP 041 MR microwave bridge (BRUKER Corporation, Billerica, MA). Samples were loaded into 0.6 mm inner diameter and 0.84 mm outer diameter capillaries and inserted into a high-sensitivity ER 4123D dielectric resonator (BRUKER Corporation) for cwEPR measurements. The field sweep for data collection was 100-G, modulation amplitude was 2 G, and the incident microwave power was 0.5 mW. Data sets were averaged for 30 scans. Each cwEPR measurement was acquired at room temperature.
Quantification and statistical analysis
Curves from Tyr-quenching assays were fit to the means of the replicates with one site binding accounting for ligand depletion using Graphpad Prism’s linear regression function. Data shown in Supplementary Table 1 and Figure 6 e and f are the mean and standard deviation for each sample. All crystal statistics can be found in Tables 1 and 2.
Supplementary Material
ACKNOWLEDGEMENTS:
We thank Tim Dinh and Dr. Huajun Yan for isolation of bovine retinas. We thank Peter Wong for helping purify Arr1. This research was supported in part by grants from the National Institutes of Health (EY009339, EY027283 and EY024864 to K.P., training grants F30EY031566 to J.L., T32EY007157 and T32GM007250 and core grants P30EY011373 and P30EY025585), the Department of Veterans Affairs (I01BX004939 to P.D.K) ), the Canada Excellence Research Chairs program (to O.P.E.), the Canadian Institutes of Health Research (PJT-159464 to O.P.E.), the Natural Sciences and Engineering Research Council of Canada (RGPIN-2017–06862 to O.P.E.), and a Mitacs Accelerate grant (IT17088) to J.R. and O.P.E. O.P.E. holds the Anne and Max Tanenbaum Chair in Neuroscience at the University of Toronto. The authors acknowledge support from an RPB unrestricted grant to the Department of Ophthalmology at UCI. Data were measured at beamline 17-ID-2 (FMX) of the NSLS-II, which is supported by NIH grant GM111244 and the DOE Office of Biological and Environmental Research KP1605010. The NSLS-II is supported by the DOE Office of Science, Office of Basic Energy Sciences Program under contract number DE-SC0012704 (KC0401040). Use of beamline 12–2 at the SSRL, SLAC National Accelerator Laboratory, is supported by the U.S. DOE Office of Science under Contract No. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health (P41GM103393). This work is based upon research conducted at the NE-CAT beamlines, which are funded by the NIH (P30 GM124165). This research used resources of the APS, a U.S. DOE Office of Science User Facility operated by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science and ICT(NRF-2017M3A9F6029733), and Basic Science Research Program of the NRF funded by the Ministry of Science (NRF-2021R1I1A3060013). The contents of this publication do not necessarily represent the official views of any funding agency.
ABBREVIATIONS:
- 1,5(PP)2-InsP4
bis-1,5-diphosphoinositol tetrakisphosphate
- 1,5(PCP)2-InsP4
bis-1,5-bisphosphonate tetrakisphosphate
- 5PP-InsP5
5-diphosphoinositol pentakisphosphate
- 5PCP-InsP5
5-bisphosphonate pentakisphosphate
- AC
amacrine cell
- As
astrocyte
- Arr
arrestin
- ATP
adenosine triphosphate
- BC
bipolar cell
- BTP
bis-tris propane
- cwEPR
continuous wave electron paramagnetic resonance
- DAG
diacylglycerol
- DAPI
4′,6-diamidino-2-phenylindole
- DEER
double electron-electron resonance
- EC
endothelial cell
- EDTA
ethylenediaminetetraacetic acid
- EPR
electron paramagnetic resonance
- GCL
ganglion cell layer
- GPCR
G protein-coupled receptor
- HC
horizontal cell
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- INL
inner nuclear layer
- InsP
myo-D-inositol phosphate
- InsP3
(1,4,5) myo-D-inositol triphosphate
- InsP6
myo-D-inositol hexakisphosphate
- IS
inner segement
- MT
microtubules
- MC
Müller cell
- NMR
nuclear magnetic resonance
- ONL
outer nuclear layer
- p44
constitutively active 44 kDa splice variant of arrestin 1
- PAGE
polyacrylamide gel electrophoresis
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PLC
phospholipase C
- PKC
phosphokinase C
- PNA
peanut agglutinin
- scRNA-seq
single cell RNA-sequencing
- SPR
surface plasmon resonance
- Rh*
activated rhodopsin
- Rh*P
activated and phosphorylated rhodopsin
- RK
rhodopsin kinase
- Rpe65
retinal pigment epithelium-specific 65 kDa protein
- WT
wild-type
Footnotes
DECLARATION OF INTERESTS: The authors have no competing interests to declare.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, and Gurevich EV (2011). Ubiquitin Ligase Parkin Promotes Mdm2–Arrestin Interaction but Inhibits Arrestin Ubiquitination. Biochemistry 50, 3749–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA (2001). Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, and Irvine RF (1984). Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature 312, 315–321. [DOI] [PubMed] [Google Scholar]
- Broekhuyse RM, Tolhuizen EFJ, Janssen APM, and Winkens HJ (1985). Light induced shift and binding of S-antigen in retinal rods. Curr. Eye Res 4, 613–618. [DOI] [PubMed] [Google Scholar]
- Brown JE, Rubin LJ, Ghalayini AJ, Tarver AP, Irvine RF, Berridge MJ, and Anderson RE (1984). myo -inositol polyphosphate may be a messenger for visual excitation in Limulus photoreceptors. Nature 311, 160–163. [DOI] [PubMed] [Google Scholar]
- Brown NW, Marmelstein AM, and Fiedler D. (2016). Chemical tools for interrogating inositol pyrophosphate structure and function. Chem. Soc. Rev 45, 6311–6326. [DOI] [PubMed] [Google Scholar]
- Brunger AT (2007). Version 1.2 of the Crystallography and NMR system. Nat. Protoc 2, 2728–2733. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Buczyłko J, and Palczewski K. (1993). 15 - Purification of Arrestin from Bovine Retinas. In Methods in Neurosciences, Hargrave PA, ed. (Academic Press; ), pp. 226–236. [Google Scholar]
- Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, Maag D, Kim S, Huang AS, Dailey MJ, et al. (2010). Inositol Pyrophosphates Inhibit Akt Signaling, Thereby Regulating Insulin Sensitivity and Weight Gain. Cell 143, 897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Perry NA, Vishnivetskiy SA, Berndt S, Gilbert NC, Zhuo Y, Singh PK, Tholen J, Ohi MD, Gurevich EV, et al. (2017). Structural basis of arrestin-3 activation and signaling. Nat. Commun 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AC, Gao Z, Riley AM, Furkert D, Wittwer C, Dutta A, Rojas T, Semenza ER, Felder RA, Pluznick JL, et al. (2020). The inositol pyrophosphate 5-InsP7 drives sodium-potassium pump degradation by relieving an autoinhibitory domain of PI3K p85α. Sci. Adv 6, eabb8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffa S, Breitman M, Spiller BW, and Gurevich VV (2011). A Single Mutation in Arrestin-2 Prevents ERK1/2 Activation by Reducing c-Raf1 Binding. Biochemistry 50, 6951–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Das ND, Yoshioka T, Samuelson D, and Shichi H. (1986). Immunocytochemical Localization of Phosphatidylinositol-4, 5-Bisphosphate in Dark-and Light-Adapted Rat Retinas. Cell Struct. Funct 11, 53–63. [DOI] [PubMed] [Google Scholar]
- Das ND, Yoshioka T, Samuelson D, Cohen RJ, and Shichi H. (1987). Immunochemical Evidence for the Light-Regulated Modulation of Phosphatidylinositol-4, 5-Bisphosphate in Rat Photoreceptor Cells. Cell Struct. Funct 12, 471–481. [DOI] [PubMed] [Google Scholar]
- Deming JD, Pak JS, Shin J, Brown BM, Kim MK, Aung MH, Lee E-J, Pardue MT, and Craft CM (2015). Arrestin 1 and Cone Arrestin 4 Have Unique Roles in Visual Function in an All-Cone Mouse Retina. Invest. Ophthalmol. Vis. Sci 56, 7618–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Evans P. (2006). Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr 62, 72–82. [DOI] [PubMed] [Google Scholar]
- Fein A, Payne R, Wesley Corson D, Berridge MJ, and Irvine RF (1984). Photoreceptor excitation and adaptation by inositol 1,4,5-trisphosphate. Nature 311, 157–160. [DOI] [PubMed] [Google Scholar]
- Furkert D, Hostachy S, Nadler-Holly M, and Fiedler D. (2020). Triplexed Affinity Reagents to Sample the Mammalian Inositol Pyrophosphate Interactome. Cell Chem. Biol 27, 1097–1108.e4. [DOI] [PubMed] [Google Scholar]
- Ghalayini A, and Anderson RE (1984). Phosphatidylinositol 4,5-bisphosphate: Light-mediated breakdown in the vertebrate retina. Biochem. Biophys. Res. Commun 124, 503–506. [DOI] [PubMed] [Google Scholar]
- Glennon MC, and Shears SB (1993). Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem. J 293, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Janoshazi AK, Weaver JD, and Shears SB (2013). PPIP5K1 modulates ligand competition between diphosphoinositol polyphosphates and PtdIns(3,4,5)P3 for polyphosphoinositide-binding domains. Biochem. J 453, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, and Büldt G. (1998). X-ray crystal structure of arrestin from bovine rod outer segments. Nature 391, 918–921. [DOI] [PubMed] [Google Scholar]
- Granzin J, Cousin A, Weirauch M, Schlesinger R, Büldt G, and Batra-Safferling R. (2012). Crystal Structure of p44, a Constitutively Active Splice Variant of Visual Arrestin. J. Mol. Biol 416, 611–618. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, and Gurevich EV (2011). The functional cycle of visual arrestins in photoreceptor cells. Prog. Retin. Eye Res 30, 405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider RS, Wilhelm F, Rizk A, Mutt E, Deupi X, Peterhans C, Mühle J, Berger P, Schertler GFX, Standfuss J., et al. (2019). Arrestin-1 engineering facilitates complex stabilization with native rhodopsin. Sci. Rep 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song X, Nair KS, Slepak VZ, Klug CS, and Gurevich VV (2007). Arrestin Mobilizes Signaling Proteins to the Cytoskeleton and Redirects their Activity. J. Mol. Biol 368, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Vishnivetskiy SA, Hubbell WL, and Gurevich VV (2008a). Opposing Effects of Inositol Hexakisphosphate on Rod Arrestin and Arrestin2 Self-Association. Biochemistry 47, 1070–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Dawson ES, Francis DJ, Eps NV, Klug CS, Hubbell WL, Meiler J, and Gurevich VV (2008b). A Model for the Solution Structure of the Rod Arrestin Tetramer. Structure 16, 924–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, and Amakawa T. (1985). Light-mediated breakdown of phosphatidylinositol-4,5-bisphosphate in isolated rod outer segments of frog photoreceptor. Biochem. Biophys. Res. Commun 128, 954–959. [DOI] [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, and Sigler PB (1999). A Model for Arrestin’s Regulation: The 2.8 Å Crystal Structure of Visual Arrestin. Cell 97, 257–269. [DOI] [PubMed] [Google Scholar]
- Hoang T, Wang J, Boyd P, Wang F, Santiago C, Jiang L, Yoo S, Lahne M, Todd LJ, Jia M., et al. (2020). Gene regulatory networks controlling vertebrate retinal regeneration. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodsdon ME, Ponder JW, and Cistola DP (1996). The NMR solution structure of intestinal fatty acid-binding protein complexed with palmitate: application of a novel distance geometry algorithm. J. Mol. Biol 264, 585–602. [DOI] [PubMed] [Google Scholar]
- Imamoto Y, Tamura C, Kamikubo H, and Kataoka M. (2003). Concentration-Dependent Tetramerization of Bovine Visual Arrestin. Biophys. J 85, 1186–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Hanson SM, Vishnivetskiy SA, Song X, Cleghorn WM, Hubbell WL, and Gurevich VV (2011). Robust Self-Association Is a Common Feature of Mammalian Visual Arrestin-1. Biochemistry 50, 2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe H-W, and Sommer ME (2013). Crystal structure of pre-activated arrestin p44. Nature 497, 142–146. [DOI] [PubMed] [Google Scholar]
- Kühn H, Hall SW, and Wilden U. (1984). Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett 176, 473–478. [DOI] [PubMed] [Google Scholar]
- Langlois G, Chen CK, Palczewski K, Hurley JB, and Vuong TM (1996). Responses of the phototransduction cascade to dim light. Proc. Natl. Acad. Sci 93, 4677–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Orts L, Couto D, and Hothorn M. (2020). Identity and functions of inorganic and inositol polyphosphates in plants. New Phytol 225, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losito O, Szijgyarto Z, Resnick AC, and Saiardi A. (2009). Inositol Pyrophosphates and Their Unique Metabolic Complexity: Analysis by Gel Electrophoresis. PLOS ONE 4, e5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, and Snyder SH (2003). Inositol Pyrophosphates Mediate Chemotaxis in Dictyostelium via Pleckstrin Homology Domain-PtdIns(3,4,5)P3 Interactions. Cell 114, 559–572. [DOI] [PubMed] [Google Scholar]
- Malhotra H, Barnes CL, and Calvert PD (2021). Functional compartmentalization of photoreceptor neurons. Pflüg. Arch. - Eur. J. Physiol [DOI] [PMC free article] [PubMed]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez A, Lem J, Simon M, and Chen J. (2003). Light-Dependent Translocation of Arrestin in the Absence of Rhodopsin Phosphorylation and Transducin Signaling. J. Neurosci 23, 3124–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menniti FS, Miller RN, Putney JW, and Shears SB (1993). Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem 268, 3850–3856. [PubMed] [Google Scholar]
- Milano SK, Kim Y-M, Stefano FP, Benovic JL, and Brenner C. (2006). Nonvisual Arrestin Oligomerization and Cellular Localization Are Regulated by Inositol Hexakisphosphate Binding. J. Biol. Chem 281, 9812–9823. [DOI] [PubMed] [Google Scholar]
- Millar FA, Fisher SC, Muir CA, Edwards E, and Hawthorne JN (1988). Polyphosphoinositide hydrolysis in response to light stimulation of rat and chick retina and retinal rod outer segments. Biochim. Biophys. Acta BBA - Mol. Cell Res 970, 205–211. [DOI] [PubMed] [Google Scholar]
- Moaven H, Koike Y, Jao CC, Gurevich VV, Langen R, and Chen J. (2013). Visual arrestin interaction with clathrin adaptor AP-2 regulates photoreceptor survival in the vertebrate retina. Proc. Natl. Acad. Sci 110, 9463–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CAC, Milano SK, and Benovic JL (2007). Regulation of Receptor Trafficking by GRKs and Arrestins. Annu. Rev. Physiol 69, 451–482. [DOI] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, and Sliz P. (2013). Collaboration gets the most out of software. ELife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, and Vagin AA (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Gurevich VV, and Slepak VZ (2004). Direct Binding of Visual Arrestin to Microtubules Determines the Differential Subcellular Localization of Its Splice Variants in Rod Photoreceptors. J. Biol. Chem 279, 41240–41248. [DOI] [PubMed] [Google Scholar]
- Najafi M, Maza NA, and Calvert PD (2012). Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc. Natl. Acad. Sci 109, 203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orisme W, Li J, Goldmann T, Bolch S, Wolfrum U, and Smith WC (2010). Light-dependent translocation of arrestin in rod photoreceptors is signaled through a phospholipase C cascade and requires ATP. Cell. Signal 22, 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. (1994). Structure and functions of arrestins. Protein Sci. Publ. Protein Soc 3, 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. (2006). G Protein–Coupled Receptor Rhodopsin. Annu. Rev. Biochem 75, 743–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, and Smith WC (1996). Splice Variants of Arrestins. Exp. Eye Res 63, 599–602. [DOI] [PubMed] [Google Scholar]
- Palczewski K, McDowell JH, Jakes S, Ingebritsen TS, and Hargrave PA (1989). Regulation of rhodopsin dephosphorylation by arrestin. J. Biol. Chem 264, 15770–15773. [PubMed] [Google Scholar]
- Palczewski K, Pulvermüller A, Buczylko J, Gutmann C, and Hofmann KP (1991a). Binding of inositol phosphates to arrestin. FEBS Lett 295, 195–199. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Buczyłko J, Imami NR, McDowell JH, and Hargrave PA (1991b). Role of the carboxyl-terminal region of arrestin in binding to phosphorylated rhodopsin. J. Biol. Chem 266, 15334–15339. [PubMed] [Google Scholar]
- Palczewski K, Buczylko J, Ohguro H, Annan RS, Carr SA, Crabb JW, Kaplan MW, Johnson RS, and Walsh KA (1994). Characterization of a truncated form of arrestin isolated from bovine rod outer segments: A novel truncated form of arrestin. Protein Sci 3, 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic I, Thakor DT, Vargas JR, McKinlay CJ, Hauke S, Anstaett P, Camuña RC, Bigler L, Gasser G, Schultz C., et al. (2016). Cellular delivery and photochemical release of a caged inositol-pyrophosphate induces PH-domain translocation in cellulo. Nat. Commun 7, 10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philp NJ, Chang W, and Long K. (1987). Light-stimulated protein movement in rod photoreceptor cells of the rat retina. FEBS Lett 225, 127–132. [DOI] [PubMed] [Google Scholar]
- Pulvermüller A, Maretzki D, Rudnicka-Nawrot M, Smith WC, Palczewski K, and Hofmann KP (1997). Functional Differences in the Interaction of Arrestin and Its Splice Variant, p44, with Rhodopsin. Biochemistry 36, 9253–9260. [DOI] [PubMed] [Google Scholar]
- Puschmann R, Harmel RK, and Fiedler D. (2019). Scalable Chemoenzymatic Synthesis of Inositol Pyrophosphates. Biochemistry 58, 3927–3932. [DOI] [PubMed] [Google Scholar]
- Read RJ, Adams PD, Arendall WB, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lütteke T, Otwinowski Z., et al. (2011). A New Generation of Crystallographic Validation Tools for the Protein Data Bank. Structure 19, 1395–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge KD, Abdulaev NG, Sousa M, and Palczewski K. (2003). Phototransduction: crystal clear. Trends Biochem. Sci. 28, 479–487. [DOI] [PubMed] [Google Scholar]
- Rojas T, Cheng W, Gao Z, Liu X, Wang Y, Malla AP, Chin AC, Romer LH, Snyder SH, and Fu C. (2019). Inositol hexakisphosphate kinase 3 promotes focal adhesion turnover via interactions with dynein intermediate chain 2. Proc. Natl. Acad. Sci 116, 3278–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiardi A. (2012). Cell Signalling by Inositol Pyrophosphates. In Phosphoinositides II: The Diverse Biological Functions, Balla T, Wymann M, and York JD, eds. (Dordrecht: Springer Netherlands; ), pp. 413–443. [Google Scholar]
- Samaranayake S, Vishnivetskiy SA, Shores CR, Thibeault KC, Kook S, Chen J, Burns ME, Gurevich EV, and Gurevich VV (2020). Biological Role of Arrestin-1 Oligomerization. J. Neurosci 40, 8055–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shears SB (2001). Assessing the omnipotence of inositol hexakisphosphate. Cell. Signal 13, 151–158. [DOI] [PubMed] [Google Scholar]
- Shukla AK, Xiao K, and Lefkowitz RJ (2011). Emerging paradigms of β-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem. Sci 36, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WC, Milam AH, Dugger D, Arendt A, Hargrave PA, and Palczewski K. (1994). A splice variant of arrestin. Molecular cloning and localization in bovine retina. J. Biol. Chem 269, 15407–15410. [PubMed] [Google Scholar]
- Song X, Raman D, Gurevich EV, Vishnivetskiy SA, and Gurevich VV (2006). Visual and Both Non-visual Arrestins in Their “Inactive” Conformation Bind JNK3 and Mdm2 and Relocalize Them from the Nucleus to the Cytoplasm. J. Biol. Chem 281, 21491–21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strissel KJ, Sokolov M, Trieu LH, and Arshavsky VY (2006). Arrestin Translocation Is Induced at a Critical Threshold of Visual Signaling and Is Superstoichiometric to Bleached Rhodopsin. J. Neurosci 26, 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, and Gurevich VV (2005). Crystal Structure of Cone Arrestin at 2.3Å: Evolution of Receptor Specificity. J. Mol. Biol 354, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Vandenberg CA, and Montal M. (1984). Light-regulated biochemical events in invertebrate photoreceptors. 2. Light-regulated phosphorylation of rhodopsin and phosphoinositides in squid photoreceptor membranes. Biochemistry 23, 2347–2352. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, and Gurevich VV (2007). Regulation of Arrestin Binding by Rhodopsin Phosphorylation Level. J. Biol. Chem 282, 32075–32083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Chen Q, Palazzo MC, Brooks EK, Altenbach C, Iverson TM, Hubbell WL, and Gurevich VV (2013). Engineering visual arrestin-1 with special functional characteristics. J. Biol. Chem 288, 3394–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung J-Y, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M., et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. [DOI] [PubMed] [Google Scholar]
- Wilden U, Hall SW, and Kühn H. (1986). Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc. Natl. Acad. Sci 83, 1174–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden U, Choe HW, Krafft B, and Granzin J. (1997). Crystallization and preliminary X-ray analysis of arrestin from bovine rod outer segment. FEBS Lett 415, 268–270. [DOI] [PubMed] [Google Scholar]
- Wilson CJ, and Copeland RA (1997). Spectroscopic Characterization of Arrestin Interactions with Competitive Ligands: Study of Heparin and Phytic Acid Binding. J. Protein Chem 16, 755–763. [DOI] [PubMed] [Google Scholar]
- Wilson MS, and Saiardi A. (2018). Inositol Phosphates Purification Using Titanium Dioxide Beads. Bio-Protoc 8, e2959–e2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, and Gurevich VV (2006). Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J. Mol. Biol 364, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, et al. (2017). Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell 170, 457–469.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang T, Vishnivetskiy SA, Gurevich VV, and Sanders CR (2010). Elucidation of Inositol Hexaphosphate and Heparin Interaction Sites and Conformational Changes in Arrestin-1 by Solution Nuclear Magnetic Resonance. Biochemistry 49, 10473–10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper analyzes existing, publicly available data. The public location of the dataset is listed in the key resources table.
This paper does not report original code.
The atomic coordinates and structure factors of all structures have been deposited in the Protein Data Bank under accession codes 7F1W, 7F1X, 7JSM, 7JTB, 7JXA, 7MOR, 7MP0, 7MP1, and 7MP2. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


