Abstract
Resistant starch (RS) in the diet reaches the large intestine and is fermented by the gut microbiota, providing beneficial effects on human health. The human gut bacterium FMB-CY1 was isolated and identified as a new species closest to Ruminococcus bromii. Ruminococcus sp. FMB-CY1 completely degraded RS including commercial RS types 2, 3, and 4, and generated glucose and maltose; however, it did not assimilate glucose. Genome analysis revealed 15 amylolytic enzymes (Amy) present in FMB-CY1. The evolutionary trees revealed that the Amys were well divided each other. All Amys (4, 9, 10, 12, and 16) containing cohesin and/or dockerin and scaffolding proteins known to be involved in constituting the amylosome, were identified. A new species of Ruminococcus, strain FMB-CY1, was considered to have the ability to form amylosomes for the degradation of RS. This new RS-degrading Ruminococcus species provides insights into the mechanism(s) underlying RS degradation in the human gut.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10068-021-01027-2.
Keywords: Amylosome, Gut microbiota, Resistant starch, Ruminococcus, Starch granule
Introduction
Starch consists of simple glucose molecules linked by α-1,4 and α-1,6-glycosidic bonds. It is composed of amylose and amylopectin and is considered a major component of the human diet, accounting for over 25% of a typical individual’s daily calories (Bello‐Perez et al., 2020). Within the digestive tract of humans, there are a series of starch-degrading enzymes, including amylase, isoamylase, and amyloglucosidase. These enzymes help in degrading and assimilating starch in the digestive tract for use as an energy source (Bello‐Perez et al., 2020). However, a type of starch called resistant starch (RS) is known to be resistant to human digestive enzymes. Without being degraded in the early digestive tract, RS is able to reach the colon and is used as a source of energy by gut microbiota (Fuentes‐Zaragoza et al., 2011). According to their physical structures, RS is generally divided into five groups: RS1, physically inaccessible; RS2, native granular structure; RS3, retrogradation; RS4, chemically modified; and RS5, amylose–lipid complexes (Raigond et al., 2015). Recently, numerous studies have suggested that the intake of RS has many positive roles in improving human health. For instance, RS is known to lower the risk of insulin resistance and leptin resistance by preventing rapid glucose release due to its lower digestibility (Maziarz et al., 2017; Sandberg et al., 2017; Shaikh et al., 2019). In addition, RS is fermented by intrinsic gut microbiota in the large intestine and is transformed into short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate. These small molecules are known to play a role in maintaining human health by regulating luminal pH, providing fuel for epithelial cells, and influencing mucosal immune function (Blaak et al., 2020).
Although many amylolytic gut bacteria, including Bacteroides spp., Eubacterium spp., and Bifidobacterium spp., have been reported, the number of bacteria that can degrade RS remains scarce. Ruminococcus bromii and B. adolescentis (and other related Bifidobacterium species) are the two main species with RS-degrading capability found in the human gut microbiota (DeMartino and Cockburn, 2020). R. bromii, a strict anaerobe, is important for the gut microbial community related to RS. One study found that the abundance of R. bromii increased markedly in fecal samples from human volunteers who switched to diets high in RS (Flint, 2012; La Reau et al., 2016). Furthermore, R. bromii has been defined as a keystone species because of its excellent RS degradation ability and its ability to feed other gut microbiota via RS fermentation products in the large intestine (Ze et al., 2012). Ze et al. (2015) also reported a distinctive degradation mechanism of R. bromii L2-63, in which extracellular protein complexes called amylosomes were involved. Amylosomes are unique multienzyme complexes similar to cellulosomes and are found in various cellulolytic microorganisms. In amylosome complexes, several extracellular amylases are assembled through interactions between dockerin and cohesin modules present in enzymes and structural proteins, similar to cellulosomes. In contrast, Jung et al. (2018, 2019) identified two strong RS-degrading bacteria, B. choerinum FMB-1, from the rumen fluids of Korean native cattle (Bos taurus coreanae) and B. adolescentis P2P3 from human feces. Interestingly, in these strains, the formation and precipitation of insoluble high-amylose corn starch (HACS) granule clusters were observed before the HACS granules were decomposed by these bacteria. This unusual characteristic may be explained by cell surface-anchored protein complexes in which starch-binding domains and RS-degrading enzymes are involved. This mechanism is strikingly different from that of the amylosome of R. bromii L2-63 (Ze et al., 2015).
To date, only two species of gut bacteria, so-called R. bromii and B. adolescentis, have been found to be involved in primary RS degradation in the human gut; however, it is likely that others exist (DeMartino and Cockburn, 2020). In the present study, we isolated unidentified strains that are capable of utilizing RS as a primary degrader from human fecal samples. As a result, the new Ruminococcus species strain FMB-CY1, which was able to degrade HACS superior to other previously reported gut bacteria, was isolated (Jung et al., 2019; Ze et al., 2012). Whole-genome sequence analysis of Ruminococcus sp. FMB-CY1 was performed, and the uniqueness of its gene content in relation to RS degradation was investigated.
Materials and methods
Starches substrates and other reagent
As resistant starch (RS) substrates, S4126 and S4180 (Sigma-Aldrich, St Louis, MO, USA) were employed. S4126 is an unmodified regular corn starch containing approximately 73% amylopectin and 27% amylose, whereas S4180 is an unmodified high amylose corn starch (HACS) classified as RS2 containing 70% amylose. The food-grade commercial RS, Hi-Maize (HM) 260, HM 958, Novelose 330, Versafibe (VF) 1490, and VF 2470 were kindly provided by Ingredion (Westchester, IL, USA). In the case of commercial RS2 type starches (HM 260 and 958), HM 260 was prepared from Hylon VII, which is a commercial HACS (70% amylose) (Le Leu et al., 2009) whereas Hi-maize 958 is unmodified food-grade RS2 containing 61.8% of RS (Le Leu et al., 2005). RS3 type starch (NV 330) is a retrograded starch generated from hydrolyzed high-amylose corn starch products. VF 1490 and 2470 (RS4 type starch) were derived from potato starch and HACS, respectively. VF 1490 is a distarch phosphate modified using phosphorus oxychloride, and VF 2470 is produced by hydrolysis and heat treatment of high-amylose maize starch.
Mono-, di-, and tri-saccharides (glucose, fructose, maltose, sucrose, cellobiose, maltotriose, and panose) and polysaccharides (soluble starch, pectin, pullulan, glycogen, and carboxymethyl cellulose) were purchased from Sigma-Aldrich and Megazyme (Bray, Ireland). The medium, chopped meat (CM) broth, was obtained from MBcell (Seoul, Korea), and all other medium supplements for human fecal bacteria were purchased from Sigma-Aldrich. All other chemicals used in this study were of analytical grade.
Growth media for human fecal bacteria
CM broth containing 0.1% (v/v) trace mineral and vitamin solutions (CMTV) was used to isolate and incubate anaerobic human fecal bacteria. The CM broth consisted of (per 100 mL) peptone (3.0 g), yeast extract (0.5 g), dipotassium phosphate (0.5 g), l-cysteine (0.05 g), and resazurin (0.1 mg). The trace mineral solution (per liter) consisted of nitrilotriacetic acid (1.5 g), MgSO4·7H2O (3 g), MnSO4 (0.5 g), NaCl (1 g), FeSO4·7H2O (0.1 g), CoSO4·7H2O (0.18 g), CaCl2·2H2O (0.1 g), ZnSO4·7H2O (0.18 g), CuSO4·5H2O (0.01 g), KAl(SO4)2·12H2O (0.02 g), H3BO3 (0.01 g), Na2MoO4·2H2O (0.01 g), and NiCl2·6H2O (0.025 g). The vitamin solution (per liter) consisted of biotin (2 mg), folic acid (2 mg), pyridoxine–HCl (10 mg), thiamine-HCl·2H2O (5 mg), riboflavin (5 mg), nicotinic acid (5 mg), d-pantothenic acid (5 mg), vitamin B12 (0.1 mg), 4-aminobenzoic acid (5 mg), and lipoic acid (5 mg). The CMTV medium was dissolved in serum vials and autoclaved. Starch substrates were sterilized separately from the medium to prevent gelatinization. Then, sterilized starch substrates were mixed with a sterilized medium, followed by flushing with 99.5% CO2 gas using a 0.2-μm filter.
Isolation of RS-degrading human fecal bacteria
Fecal samples were obtained from a healthy Korean male adult (age 53 years) with no history of gastrointestinal diseases and who had not taken any antibiotics in the previous year. The donors consumed a regular diet for four weeks. The human fecal sample was quickly placed in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI, USA) filled with 5% CO2 and 95% N2 gas within five minutes. The human fecal sample was diluted ten-fold (5 g/50 mL) with a sterile PBS solution (pH 7.4), followed by homogenization for inoculation.
RS-degrading human fecal bacteria were isolated in four steps: (1) Preparation of agar medium: Modified CMTV medium containing 0.5% soluble starch was autoclaved and mixed with sterile S4126. The mixed medium was poured with gentle pipetting to prevent bubble formation and starch precipitation. (2) Spreading of fecal slurry: 100 μL of prepared fecal slurry was spread on CMTV agar medium and incubated under anaerobic conditions at 37 °C for 5–6 days. (3) Selection of raw corn starch-degrading strains: Colonies exhibiting a clear zone around the colony, that is, decomposing raw corn starch, were selected. (4) Identification of RS-degradation: Selected strains were inoculated in CMTV broth containing 0.5% S4180. After incubating for 3 days at 37 °C with 30 rotations per minute, residual RS was measured to confirm the degradation of RS.
Identification and phylogenetic analysis of RS-degrading human fecal bacteria
Genomic DNA was extracted using a stool DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. 16S rRNA gene amplification was performed using universal primers (27F and 1492R) and KOD polymerase (Toyobo, Osaka, Japan). The PCR conditions included an initial denaturation at 95 °C for 5 min; 30 cycles consisting of 30 s at 95 °C, 30 s at 55 °C, and 1.5 min at 72 °C; and a final extension at 72 °C for 10 min. The amplified fragments were sequenced and identified using the EzBioCloud database ver. 2021.04.13 (https://www.ezbiocloud.net/). The phylogenetic tree was constructed using MEGA X software (Kumar et al., 2018). The 16S rRNA gene sequence of Ruminococcus sp. FMB-CY1 was deposited in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession number MZ573208. The average nucleotide identity (ANI) values were computed using OrthoANI with the original ANI option (Lee et al., 2016). Ruminococcus sp. FMB-CY1 isolated in this study was deposited in the Korean Agricultural Culture Collection (KACC, Wanju, Korea) under collection number KACC 22405.
RS degradation ability and utilization of other carbohydrates
Ruminococcus sp. FMB CY1 were anaerobically cultivated in serum vials containing CMTV broth prepared as in the 'Growth media' section. As a seed culture, Ruminococcus sp. FMB-CY1 was anaerobically pre-cultured in 20 mL of CMTV broth containing 0.5% maltose at 37 °C until an optical density at 600 nm of 0.6 was reached. The seed culture (200 μL) was inoculated into 20 mL of CMTV broth containing 0.5% (w/v) S4180. The inoculation and subculture process were performed in anaerobic chamber (Coy Laboratory Products) filled with 5% CO2 and 95% N2 gas. After incubating at 37 °C with 30 rotations per minute, residual RS and released reducing sugars were measured every 4 h to confirm the degradation of RS granules. The seed culture (200 μL) was inoculated into 20 mL of CMTV broth containing 0.3% (w/v) other carbohydrate substrates (mono-, di-, tri-, and polysaccharides) to confirm the utilization of other carbohydrates. After incubation under the same conditions, growth was measured spectrophotometrically at an optical density (OD) of 600 nm.
Scanning electron microscopy (SEM)
The culture samples of RS and human fecal bacteria were cautiously dried with nitrogen gas and placed on a carbon tape over a microscope slide to be coated with gold under vacuum. The images were obtained using a Tabletop Microscope TM 3000 (Hitachi, Japan). The particle size was determined from a micrograph with a scale bar of 30 μm (3000×).
Quantification of residual starch and reducing sugars
RS utilization was measured using all residual carbohydrates via the phenol–sulfuric acid method (Masuko et al., 2005). Briefly, a culture sample (100 μL) was mixed with a 5% phenol solution (200 μL). Eight-hundred microliters of 99% sulfuric acid was added, and the reaction was gently mixed and incubated at 30 °C for 20 min. In contrast, the RS degradation ability was measured using only the remaining insoluble starch. First, insoluble starch in the culture was isolated by centrifugation. Next, the pellet was washed twice and resuspended in distilled water. Then, 100 μL of the suspension solution was assigned to the sample. After the addition of 5% phenol solution (200 μL), the sample was treated as described above.
Reducing sugars in the culture was measured using the dinitrosalicylic acid (DNS) method (Miller, 1959). Briefly, 100 μL of cell-free supernatant was mixed with 300 μL of DNS solution, followed by boiling for 5 min. The absorbance of the phenol–sulfuric acid and DNS reactant was measured at 555 nm using an iMark microplate reader (BioRad, Hercules, CA, USA). The experiments were repeated in triplicate.
Thin layer chromatography (TLC) analysis
TLC silica gel 60G F25425 glass plates (Merck Millipore, Billerica, MA, USA) were activated for 5 min at 110 °C. One microliter of culture supernatant was loaded twice onto TLC plates and developed with developing solution composed of 1-butanol: acetic acid: water at a 5:2:3 (v/v/v) ratio. The plate was dried and developed rapidly by dipping in a solution containing 0.3% (w/v) 1-naphthol and 5% (v/v) sulfuric acid in methanol. The plate was dried and placed in an oven at 110 °C for 5 min.
Genome sequencing and gene analysis
The genomic DNA of Ruminococcus sp. FMB-CY1 was extracted using a QIAamp DNA Mini Kit (Qiagen). The extracted genomic DNA was quantified using a NanoDrop 2000 UV–Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and a Qubit 2.0 fluorometer (Thermo Fisher Scientific). The whole genome of the strain was sequenced using a PacBio RS II (Pacific Biosciences, Menlo Park, CA, USA) sequencing platform. The sequenced reads were assembled using HGAP 3.0 (Chin et al., 2013) with a 2-Mb expected genome size. Chromosome topology was drawn using DNAPlotter (Carver et al., 2008). Annotation of the protein-coding genes was performed using the NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016).
Functional classification of the protein-coding genes of Ruminococcus sp. FMB-CY1 was predicted using BlastP search with two criteria: an e-value of 1e − 5 and a minimum coverage of 50% from the NCBI Clusters of Orthologous Groups (COG) database. Each gene was classified into COG categories, and unassigned genes were described as “not assigned.” Putative antimicrobial resistance genes were predicted using the MEGARes database (Doster et al., 2019) with BlastN. The Blast hits were screened using the following criteria: > 80% identity, > 50% coverage, and a 40 bp minimum alignment length. The genome sequence was uploaded to the NCBI Genome Database under accession number CP084034 (https://www.ncbi.nlm.nih.gov/genome/).
Carbohydrate-active enzymes were detected using dbCAN 2 with HMMER, DIAMOND, and Hotpep databases (Zhang et al., 2018), and amylolytic enzymes and proteins were organized. The prediction of signal peptides (Sec- and Tat-secretion) of enzymes and proteins was performed using the CBS-SignalP 5.0 server (Armenteros et al., 2019). The conserved domains of enzymes and proteins were defined using the NCBI-CD search (Marchler-Bauer et al., 2017). The corresponding proteins against R. bromii species (ATCC 27255 and L2-63) were analyzed by alignment using NCBI BlastP (Johnson et al., 2008). Multiple alignment of the corresponding proteins was performed using EBI-Clustal Omega (Sievers et al., 2011). Evolutionary trees were constructed using MEGA X software (Kumar et al., 2018).
Results and discussion
Isolation and identification of RS-degrading Ruminococcus sp. FMB-CY1
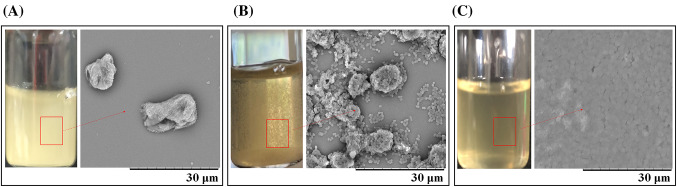
An RS-degrading obligate anaerobic gut bacterium, strain FMB-CY1, was isolated from the human fecal microbiota of a healthy 53-years old male. The strain FMB-CY1 showed a clear zone around the colony on the raw corn starch-containing agar plate (data not shown) and the aggregation of RS granules in CMTV broth, indicating that this strain had the ability to decompose raw starch and RS granules. The granules remained intact at the initial stage (Fig. 1a). However, after 14 h, the RS granules aggregated to form a granule cluster (Fig. 1b). These granule clusters gradually disappeared until 36 h, as all the RS clusters were degraded (Fig. 1c). The aggregation of RS granules by an RS-degrading bacterium was previously observed in B. adolescentis P2P3 (Jung et al., 2020).
Fig. 1.
Scanning electron microscopy during degradation of RS granules by Ruminococcus sp. FMB-CY1. (A) Dispersed RS granule at 0 h of incubation. (B) Granule cluster formation at 14 h of incubation. (C) Solely degraded RS at 36 h of incubation
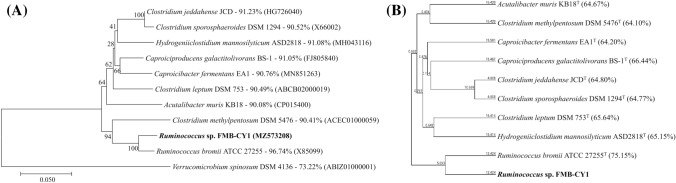
As a result of identification (Fig. 2a), the 16S rRNA sequence of the strain FMB-CY1 was mostly closest to R. bromii ATCC 27255 T (96.74%), followed by Clostridium jeddahense JCDT (91.23%), Hydrogeniiclostidium mannosilyticum ASD2818T (91.08%), Caproiciproducens galactitolivorans BS-1 T (91.05%), Ca. fermentans EA1T (90.76%) and Cl. sporosphaeroides DSM 1294 T (90.52%), and R. champanellensis 18P13T (89.85%). Based on the phylogenetic tree, the strain FMB-CY1 was bound to a clade with R. bromii ATCC 27255 T. In terms of the average nucleotide identity (ANI) values (Fig. 2b), strain FMB-CY1 showed a high similarity to R. bromii ATCC 27255 T (75.15%), but was far below 94%. Therefore, strain FMB-CY1 is proposed as a new species with a high possibility of belonging to the genus Ruminococcus. Therefore, the isolated strain FMB-CY1 was designated as Ruminococcus sp. FMB-CY1.
Fig. 2.
Identification of the human gut bacterium Ruminococcus sp. FMB-CY1. (A) Phylogenetic tree of 16S rRNA gene sequences of strain FMB-CY1. The type strains of closely related species based on the neighbor-joining algorithm applied with bootstrap replications of 1000 datasets. Scale bar, 0.05 accumulated changes per nucleotide. (B) Average nucleotide identity tree of whole genome of strain FMB-CY1. The type strains based on original ANI method chopped to 1020 bp. The numbers on the tree represent the distance between species
RS degradation ability of Ruminococcus sp. FMB-CY1
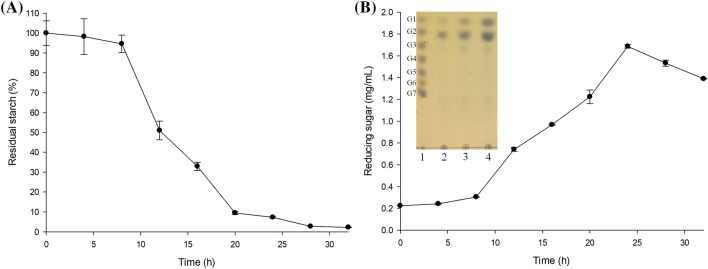
The degradation of RS Type 2 starch (non-gelatinized high-amylose corn starch; S4180) and the resulting reducing sugar products were measured over time by incubating Ruminococcus sp. FMB-CY1 (Fig. 3). S4180 was degraded slowly until 8 h after inoculation, then rapidly decomposed until 20 h, and almost all S4180 disappeared within 30 h (Fig. 3a). The amount of reducing sugars increased until 24 h when RS degradation was nearly complete, and thereafter slightly decreased (Fig. 3b). It was thought that the assimilation rate of the sugars released from S4180 by bacteria overcame the degradation rate of RS at 24 h. In addition, an increase in sugars, such as glucose, maltose, and maltotriose, during the growth of Ruminococcus sp. FMB-CY1 expression was confirmed by TLC analysis (Fig. 3b). Similar to S4180, many commercial RS Types 2, 3, and 4, such as HM 260, NV 330, and VF 2478, were also completely degraded, except for VF1490, which was processed from potato starch (Fig. S1). Only 70% of VF1490 was degraded by Ruminococcus sp. FMB-CY1. Hence, raw potato starch is considered to be more difficult to decompose by this strain than other RS.
Fig. 3.
RS degradation ability and reducing sugar release incubating Ruminococcus sp. FMB-CY1. The time course profiles of (A) the quantification of residual insoluble starch during incubation criteria on 0 h as control, and (B) the quantification of reducing sugars releasing from RS granule decomposition and TLC analysis of supernatant of culture medium. Lane 1 to 5: G1-G7 standard, incubated to 0 h, 24, and 48 h, respectively
Utilization of various carbohydrates by Ruminococcus sp. FMB-CY1
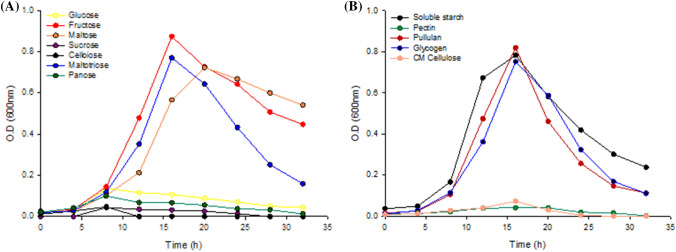
The growth of Ruminococcus sp. FMB-CY1 was measured to evaluate the carbon source utilization ability using several carbohydrates as the sole substrate. With mono-, di-, and tri-saccharides (Fig. 4a), Ruminococcus sp. FMB-CY1 grew well under maltose, fructose, and maltotriose. However, glucose, sucrose, cellobiose, and panose could not be used as the sole carbon sources. In the case of polysaccharides (Fig. 4b), soluble starch, pullulan, and glycogen were considered as easily accessible carbon sources for this bacterium. However, pectin and CM cellulose were not assimilated by Ruminococcus sp. FMB-CY1. Although the strain FMB-CY1 has high resistant starch degradation activity, it did not utilize glucose as a single carbon source, but instead used fructose and maltose preferentially (Bren et al., 2016). This observation suggested that there is no functional glucose transport/uptake system in Ruminococcus sp. FMB-CY1. Previously, three R. bromii strains (L2-63, 5AMG, and ATCC 27255) isolated from human feces were also found to grow on fructose, galactose, and maltose, but not glucose (Mukhopadhya et al., 2018). The fructose-specific phosphotransferase gene cluster was inherent in these strains, and it has been suggested that maltose is metabolized by a phosphorylase/glucanotransferase cycle similar to that of E. coli (Boos and Shuman, 1998; Mukhopadhya et al., 2018). Likewise, the fructose-specific phosphotransferase gene cluster and ABC-type maltose transport system were present in Ruminococcus sp. FMB-CY1 (data not shown). In addition, the inability to absorb glucose by Ruminococcus sp. FMB-CY1 may be beneficial for promoting the growth of other commensal gut bacteria.
Fig. 4.
Growth of Ruminococcus sp. FMB-CY1 on various carbohydrate sources. The growth curves on CMTV broth containing 0.3% of the (A) mono-, di-, tri-saccharides, and (B) polysaccharides
In addition, the number of bacteria rapidly decreased without a stationary phase after growth up to OD 0.8–0.9 for all carbon source conditions. This is an interesting result because the stationary phase is maintained for a certain period of time in most bacteria. This characteristic of rapid death is believed to be related to the autolytic activity of Ruminococcus sp. FMB-CY1 (Molinero et al., 2021). This indicates that the autolysis mechanism may be turned on when the cell density reaches a certain point.
Genome analysis of Ruminococcus sp. FMB-CY1
The complete genome of Ruminococcus sp. FMB-CY1 was composed of a 2,375,995-bp single chromosome with a 38.98% GC content (Fig. S2 and Table S1). A total of 2324 genes were identified, including 2245 protein-coding genes, 79 RNA genes, and 35 pseudogenes. The genome contained five, four, and four sets of full-length 5S, 16S, and 23S rRNA genes, 61 tRNA genes, and five non-coding RNA genes. One CRISPR array was identified at position 378,712–384,474 of the genome. Ruminococcus sp. FMB-CY1 is the first species with a complete genome sequence compared to R. bromii as the most related species. As shown in Table S1, the other genome sequences (ATCC 27225 and L2-63) were not assembled completely, and 74 and 40 contigs were reported, respectively. Genome sizes of R. bromii ATCC 27225 and R. bromii L2-63 were 2.15 Mb, and 2.24 Mb, respectively, similar to that of FMB-CY1. Likewise, the GC content of these strains was almost similar to that of FMB-CY1 by 40.7% and 41.4%, respectively.
Among the 2324 protein-coding genes of strain FMB-CY1, 1933 genes were assigned as defined proteins, whereas 391 genes were designated as hypothetical proteins (Table S2). The percentage of proteins with unknown functions, including “general function prediction only (R),” “function unknown (S),” and “not assigned,” was 27.3%. One antimicrobial resistance gene (FMB-CY1_001903) was found in the FMB-CY1 genome (Table S3). The biological mechanism of FMB-CY1_001903 was annotated as an aminoglycoside O-nucleotidyltransferase based on the definition of the MEGARes. Bacteria typically develop their antibiotic resistance mechanisms by gene(s) encoded in chromosome or plasmid. Antibiotic resistance can be expressed by a single gene or multiple genes requiring complex relationship. In fact, the gene corresponding to an aminoglycoside O-nucleotidyltransferase was not found by Blast alignment against the genomes of R. bromii strains ATCC 27255 and L2-63.
Putative amylolytic enzymes in Ruminococcus sp. FMB-CY1
When raw granular starch is consumed by bacteria, starch granules cannot be transported into the cell because of their size and structure. Starch granules are resistant to common starch-hydrolyzing enzymes; hence, specific granule-degrading enzymes are required. The enzymes and proteins of Ruminococcus sp. FMB-CY1, which is considered to directly or indirectly affect RS degradation, was determined. Eighteen genes encoding amylolytic enzymes active on α-glucan substrates and two genes possibly translating to the starch-binding protein (FMBCY1_000250 and FMBCY1_000714) were detected (Table 1). Most enzymes belonged to the glycosyl hydrolase (GH) family 13 and were predicted to function as cyclomaltodextrinases, 1,4-α-glucan branching enzymes, pullulanases, and α-amylases. In addition, one glycogen phosphorylase and one 4-α-glucanotransferase belonging to the glycosyltransferase (GT) 35 and GH77 family, respectively, were identified. Interestingly, most of the aforementioned enzymes and proteins had a signal peptide, and as such were considered to be localized extracellularly. Moreover, all proteins with signal peptides also possessed carbohydrate-binding modules (CBM) in the protein, except for one (FMBCY1_001956). In fact, CBM is known to be involved in binding to polysaccharides and carrying appended catalytic domains into intimate contact with target substrates (Blake et al., 2006). Therefore, these enzymes and proteins may be secreted outside the cell by signal peptides, playing a crucial role in decomposing relatively large insoluble starch granules at the cell surface.
Table 1.
Enzymes and proteins involved in the degradation of starch substrates of Ruminococcus sp. FMB-CY1
| Protein ID | Presumed function | Length (aa) | Signal peptide | Carbohydrate binding modules | Anchoring/binding region | Enzyme Class | Corresponding protein (ID/cover/identity) | Assignment | |
|---|---|---|---|---|---|---|---|---|---|
| R. bromii ATCC 27255 | R. bromii L2-63 | ||||||||
| FMBCY1_000046 | Cyclomaltodextrinase | 631 | – | – | – | GH13_39 | PKD32690.1 (98%/73.72%) | SPE91311.1 (98%/73.63%) | Amy 3 |
| FMBCY1_000250 | Starch-binding protein | 737 | ASec/SPI | CBM26, CBM74 | CDoc | – | PKD28238.1 (100%/68.16%) | SPE92126.1 (99%/69.41%) | ENA1 |
| FMBCY1_000353 | 1,4-α-Glucan branching protein | 697 | – | CBM48 | – | GH13_9 | PKD27046.1 (91%/83.65%) | SPE91075.1 (91%/83.49%) | Amy 13 |
| FMBCY1_000428 | 1,4-α-Glucan branching protein | 658 | – | – | – | GH13_9 | PKD27747.1 (96%/67.72%) | SPE91806.1 (97%/68.23%) | Amy 14 |
| FMBCY1_000504 | Pullulanase, type I | 1236 | Sec/SPI | CBM26 × 2, CBM48 | Doc | GH13_14 | PKD26628.1 (100%/64.47%) | SPE91787.1 (100%/64.71%) | Amy 10 |
| FMBCY1_000688 | α-Amylase | 679 | BSec/SPII | CBM26, CBM74 | – | GH13_28 | PKD28240.1 (84%/70.66%) | SPE92124.1 (45%/69.28%) | Amy 17 |
| FMBCY1_000714 | Starch-binding protein | 521 | Sec/SPI | CBM26 × 2 | Doc | – | PKD32717.1 (99%/53.35%) | SPE91138.1 (99%/55.27%) | NA2 |
| FMBCY1_000715 | α-Amylase | 800 | Sec/SPI | CBM26 | – | GH13_42 | PKD32718.1 (100%/74.01%) | SPE91137.1 (100%/74.13%) | Amy 1 |
| FMBCY1_000734 | Cyclomaltodextrinase | 549 | – | – | – | GH13_20 | PKD28303.1 (99%/74.51%) | SPE92578.1 (99%/74.25%) | Amy 8 |
| FMBCY1_001182 | α-Amylase | 1038 | Sec/SPI | CBM26 | Doc | GH13_28 | PKD29375.1 (99%/59.55%) | SPE92705.1 (99%/58.82%) | Amy 9 |
| FMBCY1_001538 | Pullulanase, type I | 1056 | Sec/SPI | CBM26 | Doc | GH13_14 | PKD30566.1 (99%/64.02%) | SPE92040.1 (99%/63.81%) | Amy 12 |
| FMBCY1_001539 | Pullulanase, type I | 957 | Sec/SPI | CBM48 | LPXTG | GH13_14 | PKD30567.1 (98%/67.37%) | SPE92039.1 (98%/67.80%) | Amy 11 |
| FMBCY1_001797 | Cyclomaltodextrinase | 753 | Sec/SPI | CBM26 | – | GH13_42 | PKD32129.1 (100%/78.78%) | SPE91197.1 (100%/78.12%) | Amy 2 |
| FMBCY1_001956 | α-Amylase | 557 | Sec/SPII | – | – | GH13_36 | PKD27034.1 (99%/63.84%) | SPE91476.1 (99%/63.84%) | Amy 5 |
| FMBCY1_002110 | α-Amylase | 878 | Sec/SPI | CBM26 | Doc | GH13_28 | PKD32488.1 (99%/57.17%) | SPE92332.1 (99%/57.77%) | Amy 16 |
| FMBCY1_002193 | α-Amylase/α-Glucosidase | 511 | – | – | – | GH13_37 | PKD26944.1 (100%/79.06%) | SPE91558.1 (100%/79.45%) | Amy 6 |
| FMBCY1_002258 | α-Amylase | 1648 | Sec/SPI | CBM26 | DCoh | GH13_19 | PKD32588.1 (71%/56.73%) | SPE91379.1 (70%/56.85%) | Amy 4 |
| FMBCY1_002260 | α-Amylase | 1233 | Sec/SPI | CBM26 | DCoh/Doc | GH13_19 | PKD32588.1 (100%/56.97%) | SPE91379.1 (100%/57.09%) | Amy 4 |
| FMBCY1_002303 | Glycogen phosphorylase | 789 | – | – | – | GT35 | PKD26876.1 (99%/87.82%) | SPE91261.1 (99%/87.94%) | NA3 |
| FMBCY1_002304 | 4-α-Glucanotransferase | 495 | – | – | – | GH77 | PKD26877.1 (100%/91.11%) | SPE91262.1 (100%/90.91%) | NA4 |
ASec/SPI: “standard” secretory signal peptides transported by the Sec translocon and cleaved by Signal Peptidase I, BSec/SPII: lipoprotein signal peptides transported by the Sec translocon and cleaved by Signal Peptidase II., CDockerin, DCohesin, ENA: Not assigned
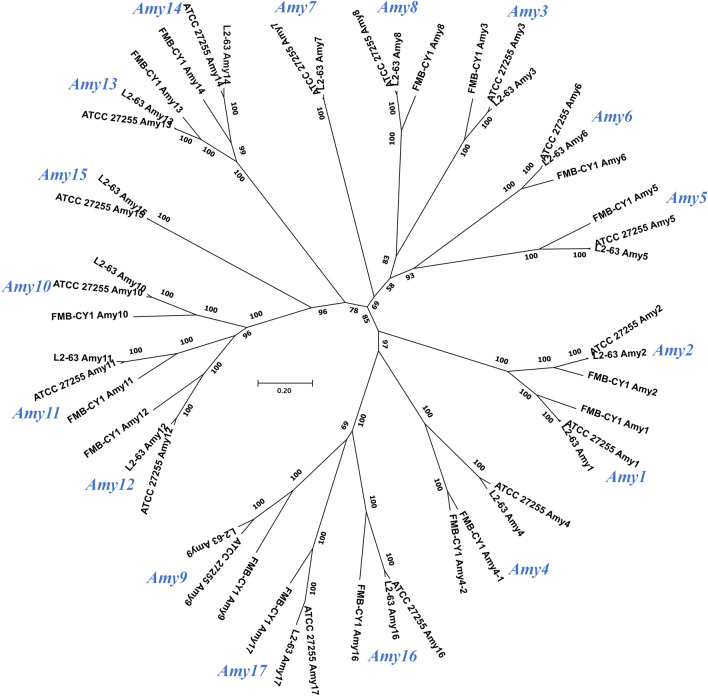
Mukhopadhya et al. (2018) reported that RS-degrading R. bromii L2-63 could form a cell-surface enzyme complex called amylosome by the cohesin and dockerin domains, and 17 amylolytic enzymes (assigned as Amy 1 to 17) were proposed to be involved in the degradation of RS. The enzymes corresponding to the 17 Amys of RS-degrading R. bromii strains ATCC 27255 and L2-63 were also present in the genome of Ruminococcus sp. FMB-CY1, except for Amy 7 and 15 (predicted to be α-amylases and glycogen debranching enzymes, respectively) (Table 1 and Fig. 5). In addition, four unassigned enzymes or proteins were also detected in the genome of FMB-CY1, which were also present in the R. bromii strains ATCC 27255 and L2-63. The sequence coverage of the corresponding Amys of FMB-CY1 to other R. bromii strains was 99%, and the identities ranged from 56 to 91%. The evolutionary trees, including Amys 1 to 17, characterized from the RS-degrading Ruminococcus strain (FMB-CY1, ATCC 27255, and L2-63) revealed that the same protein group of Amys was well divided and close to each other. These results suggest that these Amy enzymes may be functionally analogous in the RS-degrading Ruminococcus strain (Fig. 5). The corresponding Amys of Ruminococcus sp. FMB-CY1 also possessed a dockerin or cohesin domain. Regarding Amy 4, two proteins (FMBCY1_002258 and FMBCY1_002260) were observed, and both α-amylases had signal peptides, CBM26, and cohesin domains. However, the dockerin domain was only found in FMBCY1_002260.
Fig. 5.
Evolutionary trees including Amys 1 to 17 characterized from the RS-degrading Ruminococcus strains. The applied strains are Ruminococcus sp. FMB-CY1, R. bromii ATCC 27255, and R. bromii L2-63. The minimum evolution algorithm with bootstrap replications of 1000 datasets was applied. Scale bar, 0.05 accumulated changes per amino acids
Five scaffolding proteins (Sca 1, 2, 3, 4, and 5) involved in amylosome formation have been previously reported (Mukhopadhya et al., 2018). In Ruminococcus sp. FMB-CY1, five proteins containing a possible cohesin domain were also detected, although the sequence identities were rather low compared to those of R. bromii L2-63 (28% to 44%) (Table S4). Among them, two proteins (FMBCY1_002258 and FMBCY1_002260), corresponding to Amy 4 in R. bromii L2-63, contained α-amylase catalytic domains. Although detailed studies on amylosomes in Ruminococcus sp. FMB-CY1 are required, Ruminococcus sp. FMB-CY1 appears to degrade RS by forming cell-surface amylosomes through the corresponding Amys and scaffolding proteins.
To date, only one species of Ruminococcus (R. bromii) has been reported to degrade RS as a primary degrader in the human gut. In this study, human gut-originated Ruminococcus sp. FMB-CY1, which was able to completely degrade RS, was isolated as a new species of the genus Ruminococcus. The analysis of its RS degradation characteristics and starch-degrading enzymes provides fundamental insights into the degradation of RS by Ruminococcus species, a keystone species in the human gut. In conclusion, the findings presented on this new RS-degrading Ruminococcus species are essential as it is a key species for RS degradation in the human gut, and shed light on the metabolic fate of RS.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2021R1A4A1023437).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of human derivatives were followed. The study of human derivatives was approved by the Institutional Review Board of Kyung Hee University (KHSIRB-17–004).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yeong-Sik Hong and Dong-Hyun Jung contributed equally to this work.
Contributor Information
Yeong-Sik Hong, Email: yeongsik67@naver.com.
Dong-Hyun Jung, Email: dhjung529@gmail.com.
Won-Hyong Chung, Email: whchung@kfri.re.kr.
Young-Do Nam, Email: youngdo98@kfri.re.kr.
Ye-Jin Kim, Email: jinatiger@khu.ac.kr.
Dong-Ho Seo, Email: dhseo@jbnu.ac.kr.
Cheon-Seok Park, Email: cspark@khu.ac.kr.
References
- Armenteros JJA, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. SignalP 50 improves signal peptide predictions using deep neural networks. Nature Biotechnology. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Bello-Perez LA, Flores-Silva PC, Agama-Acevedo E, Tovar J. Starch digestibility: past, present, and future. Journal of the Science of Food and Agriculture. 2020;100:5009–5016. doi: 10.1002/jsfa.8955. [DOI] [PubMed] [Google Scholar]
- Blaak E, Canfora E, Theis S, Frost G, Groen A, Mithieux G, Nauta A, Scott K, Stahl B, van Harsselaar J. Short chain fatty acids in human gut and metabolic health. Beneficial Microbes. 2020;11:411–455. doi: 10.3920/BM2020.0057. [DOI] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. Journal of Biological Chemistry. 2006;281:29321–29329. doi: 10.1074/jbc.M605903200. [DOI] [PubMed] [Google Scholar]
- Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiology and Molecular Biology Reviews. 1998;62:204–229. doi: 10.1128/MMBR.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bren A, Park JO, Towbin BD, Dekel E, Rabinowitz JD, Alon U. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Scientific Reports. 2016;6:1–10. doi: 10.1038/srep24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2008;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature Methods. 2013;10:563. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- DeMartino P, Cockburn DW. Resistant starch: impact on the gut microbiome and health. Current Opinion in Biotechnology. 2020;61:66–71. doi: 10.1016/j.copbio.2019.10.008. [DOI] [PubMed] [Google Scholar]
- Doster E, Lakin SM, Dean CJ, Wolfe C, Young JG, Boucher C, Belk KE, Noyes NR, Morley PS. MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Research. 40: D561-D569 (2019) [DOI] [PMC free article] [PubMed]
- Flint HJ. The impact of nutrition on the human microbiome. Nutrition Reviews. 2012;70:S10–S13. doi: 10.1111/j.1753-4887.2012.00499.x. [DOI] [PubMed] [Google Scholar]
- Fuentes-Zaragoza E, Sánchez-Zapata E, Sendra E, Sayas E, Navarro C, Fernández-López J, Pérez-Alvarez JA. Resistant starch as prebiotic: a review. Starch-Stärke. 2011;63:406–415. doi: 10.1002/star.201000099. [DOI] [Google Scholar]
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Research. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D-H, Seo D-H, Kim G-Y, Nam Y-D, Song E-J, Yoon S, Park C-S. The effect of resistant starch (RS) on the bovine rumen microflora and isolation of RS-degrading bacteria. Applied Microbiology and Biotechnology. 2018;102:4927–4936. doi: 10.1007/s00253-018-8971-z. [DOI] [PubMed] [Google Scholar]
- Jung D-H, Chung W-H, Seo D-H, Kim Y-J, Nam Y-D, Park C-S. Complete genome sequence of Bifidobacterium adolescentis P2P3, a human gut bacterium possessing strong resistant starch-degrading activity. 3 Biotech. 10: 1-9 (2020) [DOI] [PMC free article] [PubMed]
- Jung D-H, Kim G-Y, Kim I-Y, Seo D-H, Nam Y-D, Kang H, Song Y, Park C-S. Bifidobacterium adolescentis P2P3, a human gut bacterium having strong non-gelatinized resistant starch-degrading activity. Journal of Microbiology and Biotechnology. 2019;29:1904–1915. doi: 10.4014/jmb.1909.09010. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35:1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Reau AJ, Meier-Kolthoff JP, Suen G. Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microbial Genomics. 2 (2016) [DOI] [PMC free article] [PubMed]
- Le Leu RK, Brown IL, Hu Y, Bird AR, Jackson M, Esterman A, Young GP. A synbiotic combination of resistant starch and Bifidobacterium lactis facilitates apoptotic deletion of carcinogen-damaged cells in rat colon. The Journal of Nutrition. 2005;135:996–1001. doi: 10.1093/jn/135.5.996. [DOI] [PubMed] [Google Scholar]
- Le Leu RK, Hu Y, Brown IL, Young GP. Effect of high amylose maize starches on colonic fermentation and apoptotic response to DNA-damage in the colon of rats. Nutrition & Metabolism. 2009;6:1–9. doi: 10.1186/1743-7075-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Kim YO, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. International Journal of Systematic and Evolutionary Microbiology. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Research. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuko T, Minami A, Iwasaki N, Majima T, Nishimura S-I, Lee YC. Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Analytical Biochemistry. 2005;339:69–72. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Maziarz MP, Preisendanz S, Juma S, Imrhan V, Prasad C, Vijayagopal P. Resistant starch lowers postprandial glucose and leptin in overweight adults consuming a moderate-to-high-fat diet: a randomized-controlled trial. Nutrition Journal. 2017;16:14. doi: 10.1186/s12937-017-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Mukhopadhya I, Moraïs S, Laverde-Gomez J, Sheridan PO, Walker AW, Kelly W, Klieve AV, Ouwerkerk D, Duncan SH, Louis P. Sporulation capability and amylosome conservation among diverse human colonic and rumen isolates of the keystone starch-degrader Ruminococcus bromii. Environmental Microbiology. 2018;20:324–336. doi: 10.1111/1462-2920.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero N, Conti E, Sánchez B, Walker AW, Margolles A, Duncan SH, Delgado S. Ruminococcoides bili gen. nov., sp. nov., a bile-resistant bacterium from human bile with autolytic behavior. International Journal of Systematic and Evolutionary Microbiology. 71: 004960 (2021) [DOI] [PubMed]
- Raigond P, Ezekiel R, Raigond B. Resistant starch in food: a review. Journal of the Science of Food and Agriculture. 2015;95:1968–1978. doi: 10.1002/jsfa.6966. [DOI] [PubMed] [Google Scholar]
- Sandberg JC, Björck IM, Nilsson AC. Effects of whole grain rye, with and without resistant starch type 2 supplementation, on glucose tolerance, gut hormones, inflammation and appetite regulation in an 11–14.5 hour perspective; a randomized controlled study in healthy subjects. Nutrition Journal. 2017;16:25. doi: 10.1186/s12937-017-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh F, Ali TM, Mustafa G, Hasnain A. Comparative study on effects of citric and lactic acid treatment on morphological, functional, resistant starch fraction and glycemic index of corn and sorghum starches. International Journal of Biological Macromolecules. 2019;135:314–327. doi: 10.1016/j.ijbiomac.2019.05.115. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Research. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH, Louis P, Henrissat B, Juge N, Koropatkin NM. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcus bromii. mBio. 6: e01058–15 (2015) [DOI] [PMC free article] [PubMed]
- Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. The ISME Journal. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Research. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.