Abstract
Colorectal cancer (CRC) is a major cause of morbidity and mortality in the United States. Tumor-stromal metabolic crosstalk in the tumor microenvironment promotes CRC development and progression, but exactly how stromal cells, in particular cancer-associated fibroblasts (CAFs), affect the metabolism of tumor cells remains unknown. Here we take a data-driven approach to investigate the metabolic interactions between CRC cells and CAFs, integrating constraint-based modeling and metabolomic profiling. Using metabolomics data, we perform unsteady-state parsimonious flux balance analysis to infer flux distributions for central carbon metabolism in CRC cells treated with or without CAF-conditioned media. We find that CAFs reprogram CRC metabolism through stimulation of glycolysis, the oxidative arm of the pentose phosphate pathway (PPP), and glutaminolysis, as well as inhibition of the tricarboxylic acid cycle. To identify potential therapeutic targets, we simulate enzyme knockouts and find that CAF-treated CRC cells are especially sensitive to inhibitions of hexokinase and glucose-6-phosphate, the rate limiting steps of glycolysis and oxidative PPP. Our work gives mechanistic insights into the metabolic interactions between CRC cells and CAFs and provides a framework for testing hypotheses towards CRC-targeted therapies.
Keywords: Tumor microenvironment, Metabolomics, Mathematical biosciences, Systems biology, Flux balance analysis
1. Introduction
Colorectal cancer (CRC) remains one of the deadliest cancers in the United States, with a 5-year survival rate of less than 15% for patients with stage IV CRC (Zacharakis et al., 2010). Over 140,000 people are diagnosed with CRC each year, leading to approximately 50,000 deaths (Siegel et al., 2020). Interactions between tumor and stromal cells have long been speculated to promote tumor development and progression. Cancer-associated fibroblasts (CAFs), a dominant cellular component of the tumor stroma, play a significant role in cancer pathogenesis by contributing to the cancer cells’ altered metabolism, a hallmark of CRC (Liotta and Kohn, 2001; Littlepage et al., 2005). Various factors secreted by CAFs, including hepatocyte growth factor (HGF) and neuregulin-1, are known to inhibit therapeutic response in cancer (Räsänen and Vaheri, 2010; Straussman et al., 2012). In addition, increased deposition of matrix proteins (e.g., hyaluronan and collagen) by CAFs has been found to affect drug penetration (Flach et al., 2011; Misra et al., 2003; Loeffler et al., 2006).
Increasing evidence supports the idea of reciprocal metabolic reprogramming among CRC cells and CAFs, but questions remain regarding the mechanism of the metabolic dependencies. For example, it is not clear if the influence of CAFs causes CRC cells to redistribute their carbon fluxes through central carbon metabolism, and whether oncogenes such as KRAS contribute to the metabolic reprogramming. Understanding the characteristics of tumor cells and CAFs in their metabolic ecosystem may provide insight needed to develop optimal cancer therapies.
Here, we applied a systems biology approach that combines metabolomics with constraint-based models to understand how CAFs influence CRC cell metabolism. We profiled metabolic alterations in KRASWT and KRASMUT DLD-1 cells (a CRC cell line) either cultured solely in CRC media or cultured initially in CRC media and then switched to CAF- conditioned media (Fig. 1). Metabolite pool size profiling alone, however, does not explain how cells modulate their reaction fluxes to achieve the balance between anabolism and catabolism. To infer flux distributions, we developed unsteady-state parsimonious flux balance analysis (upFBA), a data-driven modeling approach that integrates constraint-based methods and liquid chromatography-mass spectrometry (LC-MS) metabolomics data (Bordbar et al., 2017; Lewis et al., 2010). We constructed a network of central carbon metabolism similar to a previous study (Shan et al., 2018) and performed upFBA to estimate the intracellular reaction fluxes of KRASWT and KRASMUT CRC cells grown in CRC media and CAF-conditioned media.
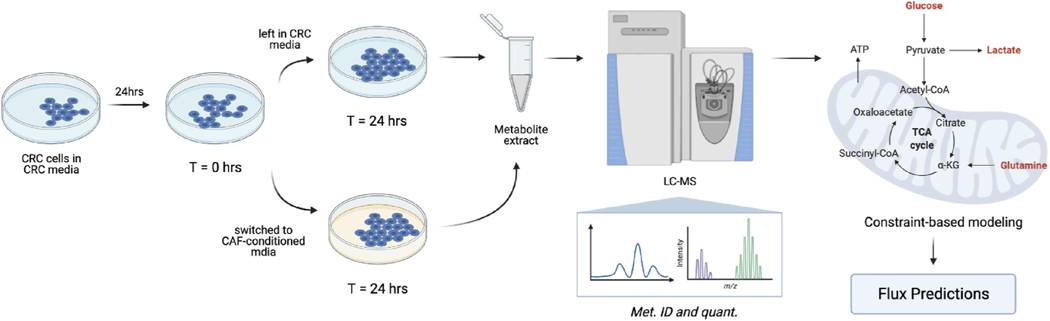
Fig. 1.
Experimental workflow of our study. KRASWT and KRASMUT CRC cells were first cultured in CRC media (DMEM) for 24hrs. At T = 0hrs, they were either left in their normal media, i.e., CRC media, or switched to CAF-conditioned media for another 24hrs. At T = 24hrs, metabolites were extracted from cells and from the media and subjected to LC-MS analysis. For all conditions (KRASWT CRC cells in CRC media, KRASMUT CRC cells in CRC media, KRASWT CRC cells in CAF-conditioned media, and KRASMUT CRC cells in CAF-conditioned media), we measured the fold changes of intracellular metabolites and the secretion/uptake rates of glucose, lactate, and glutamine. These measurements were used to constrain an upFBA model we developed to infer the flux distributions of central carbon metabolism in CRC cells. (See Section 2 for details). This figure was created from BioRender.com.
Our analysis indicates that CAFs play a pivotal role in regulating CRC metabolism through stimulation of glycolysis, the oxidative arm of the pentose phosphate pathway (PPP), and glutaminolysis, as well as inhibition of the tricarboxylic acid (TCA) cycle. Concomitantly, CAFs induce a higher flux through substrate-level phosphorylation and NADH pro- duction. To identify potential therapeutic targets, we subsequently performed unsteady-state flux balance analysis (uFBA) in search of gene deletions that could lead to reduced cancer growth. We found that fewer enzyme knockouts were effective in blocking cancer growth in CAF- conditioned media than in CRC media, which suggests that CAFs generally make CRC cells more resilient to metabolic stress. Yet, inhibition of the hexokinase (HK) or glucose-6-phosphate dehydrogenase (G6PD) reactions, is predicted to lead to reduced cancer cell growth in CAF-conditioned media compared to CRC media as it exploits the metabolic crosstalk mediated by CAFs.
2. Materials and methods
2.1. Cell culture
2.1.1. Culture media
Colorectal cancer media (CRC media) are composed of Advanced DMEM/F12 with 10% FBS, 1% penicillin/streptomycin, 1% Glutamax, and 1% HEPES, 100 ng/ml Noggin (Tonbo, 21–7075-U500), 50 ng/ml EGF (Life Technologies, PGH 0313), 10 μM SB202190 (Sigma, 47 067), 500 nM A-83–01 (Millipore, 616 454–2 MG), 10 mM nicotinamide (Sigma, N0636), 1X B-27 (Sigma Aldrich,17 504 044), 1 mM N-Acetylcysteine (Sigma Aldrich, A7250), and 1X N2 (Sigma Aldrich, 17 502 048). CRC media conditioned by patient-derived cancer associated fibroblasts (CAF-conditioned media) were made by growing CAFs in 10 cm plates up to 70–80% confluency at standard culture conditions (5% CO2, 37 °C). Once the desired confluency was reached, the media were refreshed and allowed to be conditioned by the CAFs for 72 hours. The media were then collected and filtered through a 0.2 μM filter. Conditioned media were aliquoted and stored at −80 °C until used for experiments. CAFs were then collected from the plates and counted with a Bio-Rad T-20 cell counter and trypan blue for normalization purposes. Patient-derived CAFs were isolated from tumor tissue resections of colorectal cancer patients from the USC Norris Comprehensive Cancer Center following Institutional Review Board (IRB) approval and patient consent. The tumor tissue was plated on plastic tissue culture plates to isolate the CAFs and letting the cells grow out over 1–2 passages. These cells were confirmed as CAFs with qPCR and immunofluorescence staining as described previously (Garvey et al., 2020).
2.1.2. Preparation of cells for LC-MS metabolomics
DLD-1 KRASWT and DLD-1 KRASMUT cells were obtained from the Yun lab (Yun et al., 2009) and maintained in McCoy’s 5A media supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum. For LC-MS metabolomics studies, 200,000 cells were seeded in each well of a 6 well plate (3 wells with DLD-1 KRASWT cells and 3 wells with DLD-1 KRASMUT cells) in CRC media. Replicates of this plate were prepared for cell counting purposes and for switching the media after 24 hours. After the DLD-1 KRASWT and KRASMUT cells were grown for 24 hours in CRC media, intracellular and extracellular metabolite extractions were done as described in Section 2.2 below, switched to CAF-conditioned media, or left in CRC media for an additional 24 hours before metabolite extractions (Fig. 1). At each of these time points, plates which were reserved for counting were treated the same way before treated with 0.5% trypsin/EDTA and counted with a TC-20 cell counter. Cell growth rates were calculated using CellPD (Version 1.0) (Juarez et al., 2016), which uses the Levenberg-Marquardt algorithm to perform a least-squares fitting.
2.2. Mass spectrometry-based metabolomics analysis
DLD-1 KRASWT or DLD-1 KRASMUT cells were seeded in 6-well plates at density of 200,000 cells/well. For extraction of intracellular metabolites, cells were washed on ice with 1 ml ice-cold 150 mM ammonium acetate (NH4AcO, pH 7.3). 1 ml of −80 °C cold 80% MeOH was added to the wells, and samples were incubated at −80 °C for 20 minutes. Then cells were scraped off, and supernatants were transferred into microfuge tubes. Samples were pelleted at 4 °C for 5 minutes at 13k rpm. The supernatants were transferred into LoBind Eppendorf microfuge tube; the cell pellets were re-extracted with 200 μl ice-cold 80% MeOH, spun down and the supernatants were combined. Metabolites were dried at room temperature under vacuum and re-suspended in water for LC-MS run. For extraction of extracellular metabolites, 20 μl of cell-free blank and conditioned media samples were collected from wells. Metabolites were extracted by adding 500 μl −80 °C cold 80% MeOH, dried at room temperature under vacuum and re-suspended in water for LC-MS analysis.
For each experiment, biological triplicate samples were randomized and analyzed on a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer coupled to an UltiMate 3000 UHPLC system (Thermo Scientific). The mass spectrometer was run in polarity switching mode (+3.00 kV/−2.25 kV) with an m/z window ranging from 65 to 975. Mobile phase A was 5 mM 2.0 mm NH4AcO, pH 9.9, and mobile phase B was acetonitrile. Metabolites were separated on a Luna 3 μm NH2 100 Å (150 × 2.0 mm) column (Phenomenex). The flowrate was 300 μl/min, and the gradient was from 15% A to 95% A in 18 minutes, followed by an isocratic step for 9 minutes and re-equilibration for 7 minutes.
Metabolites were detected and quantified as area under the curve based on retention time and accurate mass ( ≤ 5 ppm) using the TraceFinder 3.3 (Thermo Scientific) software. Extracellular data were normalized to integrated cell number, which was calculated based on cell counts at the start and end of the time course and an exponential growth equation. Intracellular data were normalized to the cell numbers at the time of extraction. Additionally, both intracellular and extracellular metabolomics data were median-normalized. We calculated the fold changes of metabolite levels by dividing the final abundance of the metabolite (denoted as the T = 24hrs timepoint) by its baseline condition in the CRC media (denoted as the T = 0hrs timepoint). Differential abundance analysis was implemented using the LIMMA Bioconductor package (R version 3.6.2) (Ritchie et al., 2015).
2.3. Constraint-based modeling
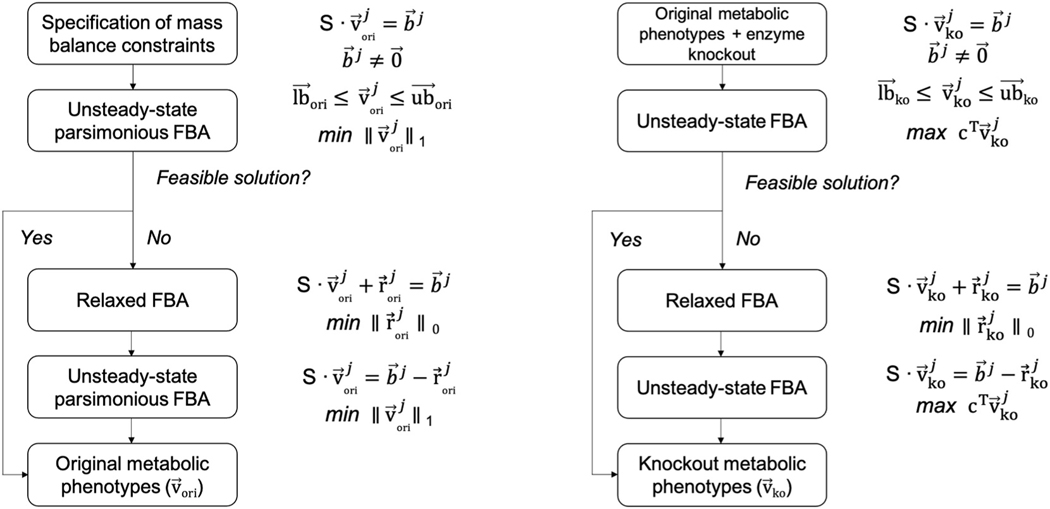
To infer flux distributions, we performed upFBA (Bordbar et al., 2017; Lewis et al., 2010). Compared to existing flux-inference methods, upFBA is more suitable for our study for several reasons. First, upFBA does not require detailed knowledge of enzymatic kinetics. In a typical LC-MS metabolomics experiment, only a subset of metabolites can be experimentally identified and quantified. However, the number of parameters to fit in ODE-based kinetic models are likely to far exceed the number of metabolites for which data are available (Strang, 2016; Roy and Finley, 2017; Wang et al., 2019, 2020b). Second, upFBA can be used to examine media-induced metabolic alterations. As the primary goal of our study is to understand the effects of CAF-conditioned media on CRC metabolism, manipulating the media would defeat the purpose of our study. Hence, 13C metabolic flux analysis (13C-MFA) is not particularly useful here because a 13C-labeled substrate needs to be introduced to the culture medium in a tracing experiment (Long and Antoniewicz, 2019; Antoniewicz, 2018; Ahn and Antoniewicz, 2011). Third, upFBA does not require the steady-state assumption. Both steady-state FBA and 13C-MFA assume that metabolites in the system are at steady state, so these analyses are difficult to interpret when cells undergo a response to perturbations such as a media switch (Dai and Locasale, 2017). Fourth, unlike uFBA (Bordbar et al., 2017), upFBA does not require absolute quantification of metabolites. Instead, it combines knowledge of initial metabolite concentrations and measurements of fold changes to estimate mass balance constraints. Finally, upFBA increases the reliability of the estimated flux distributions by repeatedly sampling the initial metabolite concentrations and generating mass balance constraints from the sampled concentrations and measured fold changes of metabolites.
Our upFBA approach stems from uFBA (Bordbar et al., 2017) but differs in the specification of mass balance constraints as well as objective functions. We specified mass balance constraints by integrating the estimates of initial metabolite concentrations from literature and measurements of relative metabolite changes from our experimental data. Our approach operates in following steps (Fig. 2). First, we sampled initial metabolite concentrations (T = 0hrs) 100 times from the range estimates reported in the literature. Within these 14 metabolites, we chose to constrain seven, namely, glucose 6-phosphate (G6P), fructose 1,6-bisphosphate (FBP), glyceraldehyde 3-phosphate (G3P), phosphoenolpyruvate (PEP), lactate, glutamine, and glutamate (Marín-Hernández et al., 2014; Le Guennec et al., 2012). (See Table S1 for range estimates). Let li and ui denote the lower and upper bounds of the initial concentration of the i-th metabolite for which estimates are available (). The initial concentration of the i-th metabolite in the j-th sampling (), , needs to stay between the lower and upper bounds, i.e., . Thus, the initial concentrations of all metabolites for which range estimates are available in the j-th sampling, , satisfy
| (1) |
Fig. 2.
A constraint-based modeling approach to predicting metabolic phenotypes. The same workflows are carried out repeatedly for each set of initial metabolite concentration that was sampled (j = 1, 2, …100).
Let and denote the normalized abundances of the i-th metabolite at T = 0hrs and T = 24hrs. The fold change of the i-th metabolite is defined as the ratio of to . The mass balance constraint on the i-th metabolite in the j-th sampling, which represents the absolute change in abundance between T = 0hrs and T = 24hrs, can be expressed as
| (2) |
Thus, the mass balance constraints on all metabolites for which range estimates are available in the j-th sampling, , are specified as
| (3) |
Next, we used linear programming to calculate a solution that satisfies the mass balance constraints and flux constraints. To avoid increasing the degrees of freedom of the model, we began by relaxing steady-state conditions for the seven metabolites for which range estimates are available and assuming steady-state conditions for the rest (Bordbar et al., 2017) (Fig. 2). In other words, the mass balance constraints on all metabolites in our model in the j-th sampling, (Fig. 2), can be expressed as
| (4) |
We also applied flux constraints. The uptake/secretion rates of glucose, lactate, and glutamine were experimentally measured, so the fluxes through GLUT, MCT, and ASCT2-mediated transport were all set to fixed values. (See Table S2 for data). The units of uptake/secretion rates were converted from mmol/(cell⋅hr) to mM/hr under the assumption that each DLD-1 cell has an approximate volume of 1000 μM3 (Tzur et al., 2009). Biomass growth rates were calculated based on integrated cell counts measured over three days and also set to fixed values in our models. The upper bounds on all remaining fluxes were set to 500 mM/h. The lower bounds on the fluxes through the remaining reversible and irreversible reactions were set to −500 mM/h and 0 mM/h, respectively. A flux of 500 mM/h was chosen to represent an infinite flux, as such a value is too high to achieve in a real biological system. Lastly, a flux split between glycolysis and the PPP was constrained to be 90/10 based on a previous 13C tracing study performed in cancer cell lines (Zielinski et al., 2017).
Unlike uFBA, which maximizes the biomass objective function, upFBA minimizes the total sum of fluxes through the metabolic network in the j-th sampling, (Fig. 2) (Lewis et al., 2010). Based on the optimality principle, it is reasonable to assume that cells seek to maintain a pre-specified growth rate at the lowest metabolic cost possible (Lewis et al., 2010). In cases where models failed to yield feasible solutions, we performed relaxed FBA (rFBA) which minimizes the number of unmeasured metabolites for which mass balance constraints need to be relaxed (Heirendt et al., 2019) (Fig. 2). As previously suggested (Bordbar et al., 2017), implementing this objective to minimize the sum of the fluxes through the metabolic network produces more accurate results than other common objective functions used in this context. The mass balance constraints for just three metabolites needed to be relaxed in the various upFBA runs (glucose [Glc], fructose 6-phosphate [F6P], and sedoheptulose-7-phosphate [S7P]), and the resulting number of times mass balance constraints were relaxed on these metabolites is shown in Fig. S1 (Supplementary File 1). Following rFBA, we performed another round of upFBA with the relaxed constraints to estimate the reaction fluxes (Fig. 2). Scripts relevant to upFBA, including the reaction network we built for central carbon metabolism, can be found on Github: https://github.com/FinleyLabUSC/CRC-Cell-Constraint-Based-Metabolic-Model.
The single gene deletion analysis operates similarly. We simulated gene deletion by constraining the deleted reaction to zero between T = 0hrs and T = 24hrs. Mathematically, this means that the upper bound and lower bound for the deleted reaction in the j-th sampling were set to 0 mM/h. In contrast, mass balance constraints, and the remaining flux constraints remained the same, except the constraint on the biomass growth rate (Fig. 2). uFBA was then performed to predict the maximal biomass growth rate that could be achieved under the new flux constraints (Fig. 2). Similar to the steps outlined above, we utilized rFBA in cases where models did not produce feasible solutions (Heirendt et al., 2019), following which we performed another round of uFBA to predict the maximal biomass growth rate possible (Fig. 2). Scripts relevant to single gene deletion analysis can be found on GitHub: https://github.com/FinleyLabUSC/CRC-Cell-Constraint-Based-Metabolic-Model.
Both upFBA and uFBA were implemented in MATLAB R2019b using the COBRA Toolbox v3.0 (Heirendt et al., 2019; Vlassis et al., 2014). Linear programming was solved using the GLPK solver (Makhorin, 2000). The resulting fluxes from upFBA and biomass growth rates from uFBA were compared between cells cultured in CRC media and CAF-conditioned media via the Wilcoxon rank sum test (Pearce and Derrick, 2019), which either accepts or rejects the null hypothesis that a reaction has an equal median flux under two media conditions. The adjusted p-values were subsequently computed from the p-values using the Bonferroni correction.
3. Results
3.1. LC-MS profiling reveals media-induced metabolic alterations
To directly assess changes in the levels of CRC cells’ intracellular metabolites, we performed a semi-targeted LC-MS metabolomics analysis of central carbon metabolism in KRASWT and KRASMUT CRC cells cultured in CRC media or CAF-conditioned media. Altogether, 81 metabolites were consistently identified and quantified across all samples. The raw measurements of intracellular metabolites are provided in Supplementary File 2.
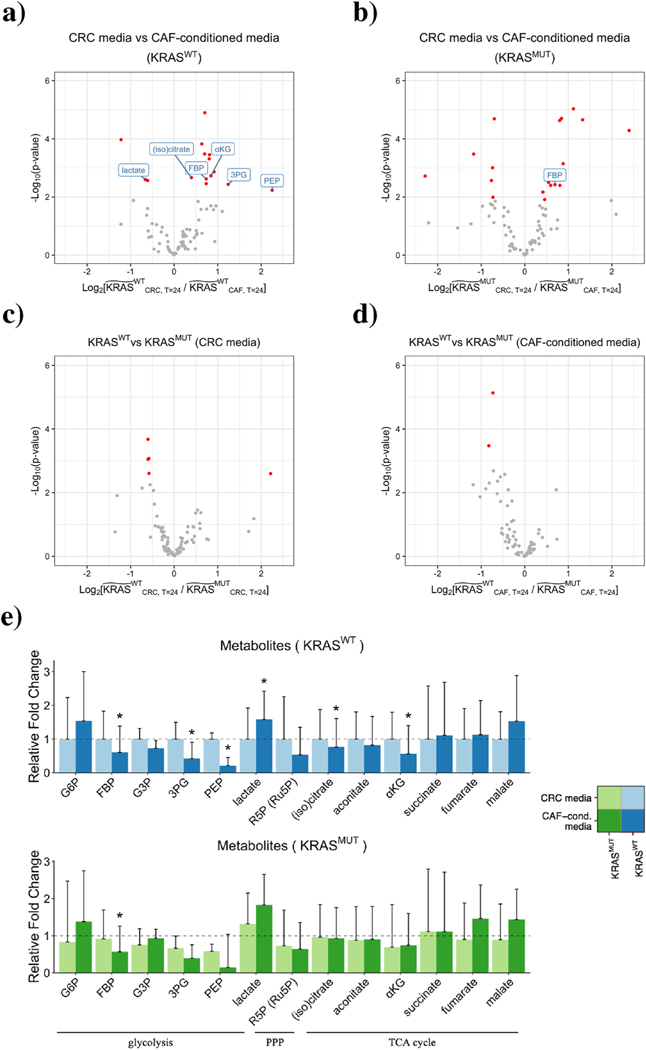
We calculated the fold changes of the metabolite levels and applied moderated t-tests to validate the differential abundance of metabolites (Ritchie et al., 2015). A metabolite was considered significantly different between two groups if its fold change was larger than 1.2 and false discovery rate (FDR) was less than 0.05. A total of 18 metabolites were considered to be significantly different between CRC media and CAF-conditioned media for KRASWT CRC cells; a total of 20 metabolites differed significantly for KRASMUT CRC cells (Fig. 3(a), (b)). In contrast, only 5 metabolites were significantly different between KRASWT and KRASMUT cells in CRC media; only 2 metabolites differed significantly in CAF-conditioned media (Fig. 3(c), (d)). This suggests that CAFs induce more pronounced alterations in CRC metabolism than oncogenic KRAS, as cells of different genotype identities show fewer differences in their metabolic profiles compared to cells grown in different media (Fig. 3(a)–(d)).
Fig. 3.
Distinct metabolic profiles of CRC cells grown under different conditions. a) - d) Comparison of intracellular metabolites between two conditions. The wide tilde in the x-axis label represents the fold change in metabolite abundance between T = 0hrs and T = 24hrs. Metabolites that are significantly different between two groups are highlighted in red (FDR < 0.05, fold change > 1.2). Metabolites unique to glycolysis, PPP, and the TCA cycle are labeled. e) Fold changes of metabolites belonging to glycolysis, PPP, and the TCA cycle in KRASWT (top) and KRASMUT (bottom) CRC cells. (Glc, 6PG, and OAA are not shown due to high coefficients of variation). The heights of the bars represent mean fold changes, and error bars represent standard deviations. A metabolite is considered significantly different between CRC media and CAF-conditioned media if its fold change is larger than 1.2 and FDR is less than 0.05 (asterisks above bars).
Grouping metabolites by pathways (i.e., glycolysis, PPP, and the TCA cycle), we found that no sets of metabolites unique to a particular pathway were consistently regulated in one direction, either up or down (Fig. 3(e)). This, however, does not necessarily indicate that CAFs do not influence CRC metabolism. Metabolic networks are highly intertwined, and for most metabolites, synthesis and breakdown take place simultaneously. The accumulation of a pathway intermediate can be caused by upregulated anabolism and/or downregulated catabolism. Thus, we sought to look at pathway fluxes, rather than individual metabolite levels.
3.2. A data-driven model predicts metabolic fluxes in CRC
To infer intracellular flux distributions, we used a constraint-based model of central carbon metabolism in CRC cells. Our model was adapted from a previously developed constraint-based model (Shan et al., 2018) and included the same 89 metabolites and 73 reactions as described, including the biomass template reaction and the glutaminase (GLNS) reaction, which converts glutamine to glutamate. In addition, we included a reaction to account for the conversion of glutamate to glutamine mediated by glutamine synthetase (GS) (Still and Yuneva, 2017). High GS activity has been observed in glioblastoma, non-small cell lung cancer, luminal breast cancer, as well as pancreatic ductal adenocarcinoma in a glutamine-deprived environment (Cluntun et al.,2017; Tsai et al., 2021). We include the GS reaction since it remains unclear whether CRC cells, irrespective of the genetic identity or tumor microenvironment (TME), acquire glutamine from the environment and/or via GS-mediated de novo synthesis.
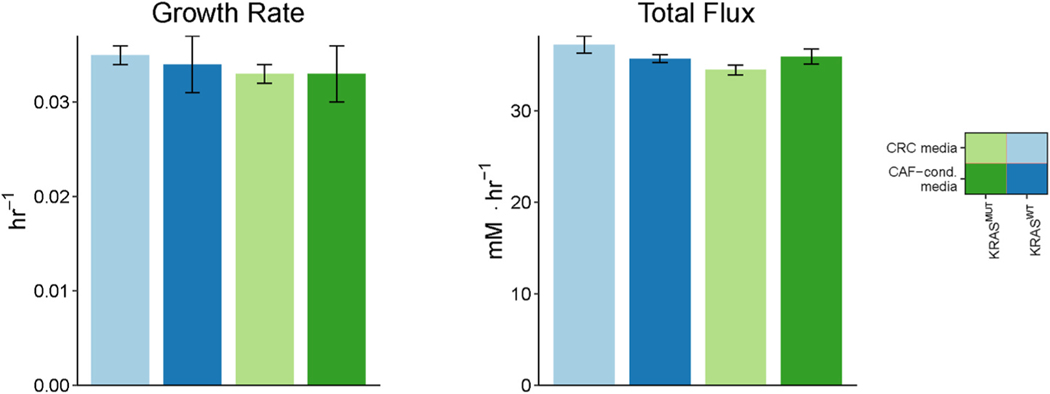
We performed random sampling of mass balance constraints based on knowledge of initial metabolite concentrations from literature and measurements of fold changes of metabolite concentrations between T = 0hrs and T = 24hrs from experimental data. (See Table S1 for initial metabolite concentrations). Biomass growth rates, which we calculated by fitting an exponential growth curve to cell count measurements taken over 3 days using CellPD (Juarez et al., 2016), were used to constrain our models (Fig. 4 and S2; Table S2). As Fig. 4 shows, the growth rate of CRC cells is minimally changed across media and genetic makeups.
Fig. 4.
Experimentally measured growth rates of CRC cells and predicted upFBA-derived minimized sum of fluxes. Growth rates are represented by mean ± standard deviation of three replicates. Data shown are based off of the MultiCellDS name “Live cells” in Fig. S2. The predicted total fluxes are represented by mean ± standard deviation of 100 mass balance constraints.
To compute the exchange fluxes, we also measured the extracellular levels of glucose, lactate, and glutamine. The raw measurements of extracellular metabolites are provided in Supplementary File 3. We observed that the concentration of glucose and glutamine in both CRC media and CAF-conditioned media at T = 24hrs is significantly lower than their blank media counterparts at T = 24hrs, whereas the concentration of lactate is significantly higher. This suggests that CRC cells uptake glucose and glutamine, while they secrete lactate under both media conditions. The experimentally measured extracellular uptake/ secretion rates of glucose, lactate, and glutamine were used to constrain our models. (See Table S2 for data).
For each set of mass balance constraints, we performed upFBA in search of a flux distribution that minimizes the total sum of fluxes through the network (Bordbar et al., 2017; Lewis et al., 2010). Based on the calculated fluxes, we separated reactions into essential reactions and non-essential reactions. Essential reactions are defined as reactions that maintain a non-zero flux under at least one set of mass balance constraints, and non-essential reactions are those that do not. The resulting minimized total sums of fluxes, numbers of essential reactions, and numbers of non-essential reactions are given in Fig. 4 and S3. We did not find significant differences in the total flux through the metabolic reactions or the number of essential reactions across the four conditions (Fig. 4 and S3). Differential flux was validated on all essential reactions using the nonparametric Wilcoxon rank sum test (Pearce and Derrick, 2019). We visualized the distributions of the median fluxes via Voronoi treemaps in Fig. S4. It can be seen that under all conditions, glycolysis accounts for the largest proportion of the total sum of fluxes among the pathways (Fig. S4). Median fluxes, median absolute deviations (MADs), and adjusted p-values are provided in Supplementary File 4. Figs. 5–7 summarize the predicted flux distributions of CRC cells grown in CRC media and CAF-conditioned media. We next discuss in detail these predicted fluxes for glycolysis, PPP, TCA cycle, and glutaminolysis reactions and compare the flux values between the four experimental conditions.
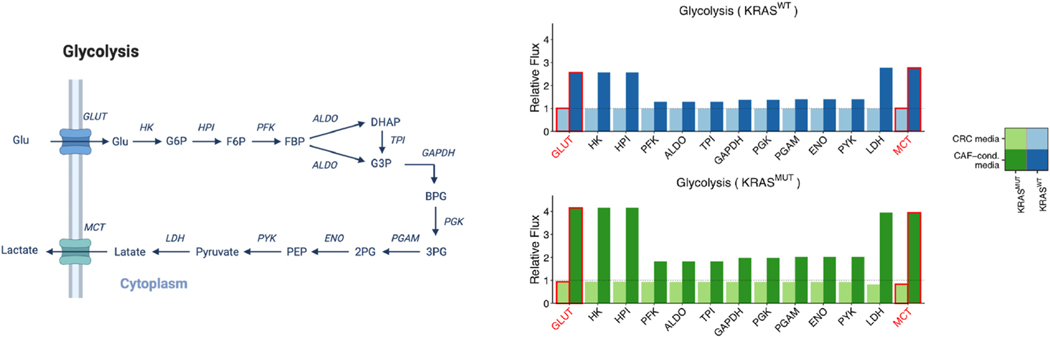
Fig. 5.
Relative fluxes of glycolysis in KRASWT and KRASMUT CRC cells cultured in CRC media compared to CAF-conditioned media. Left, Schematic of the glycolysis pathway for reference. This schematic was created from BioRender.com. Directions of the arrows in the diagram represent directions of the reactions. Right, Predicted fluxes for glycolysis reactions. All reactions have been normalized to the respective flux in KRASWT CRC cells cultured in CRC media. The heights of the bars represent normalized median fluxes, and error bars represent normalized MADs. A flux is considered significantly different between CRC media and CAF-conditioned media if its fold change is larger than 1.2 and adjusted p-value is less than 0.001 (asterisks above bars). (We lower the p-value threshold in order to account for the larger sample size we achieved via simulations than experiments.) We outline in red the fluxes through GLUT and MCT, which are experimentally measured and supplied to the model as fixed constraints.
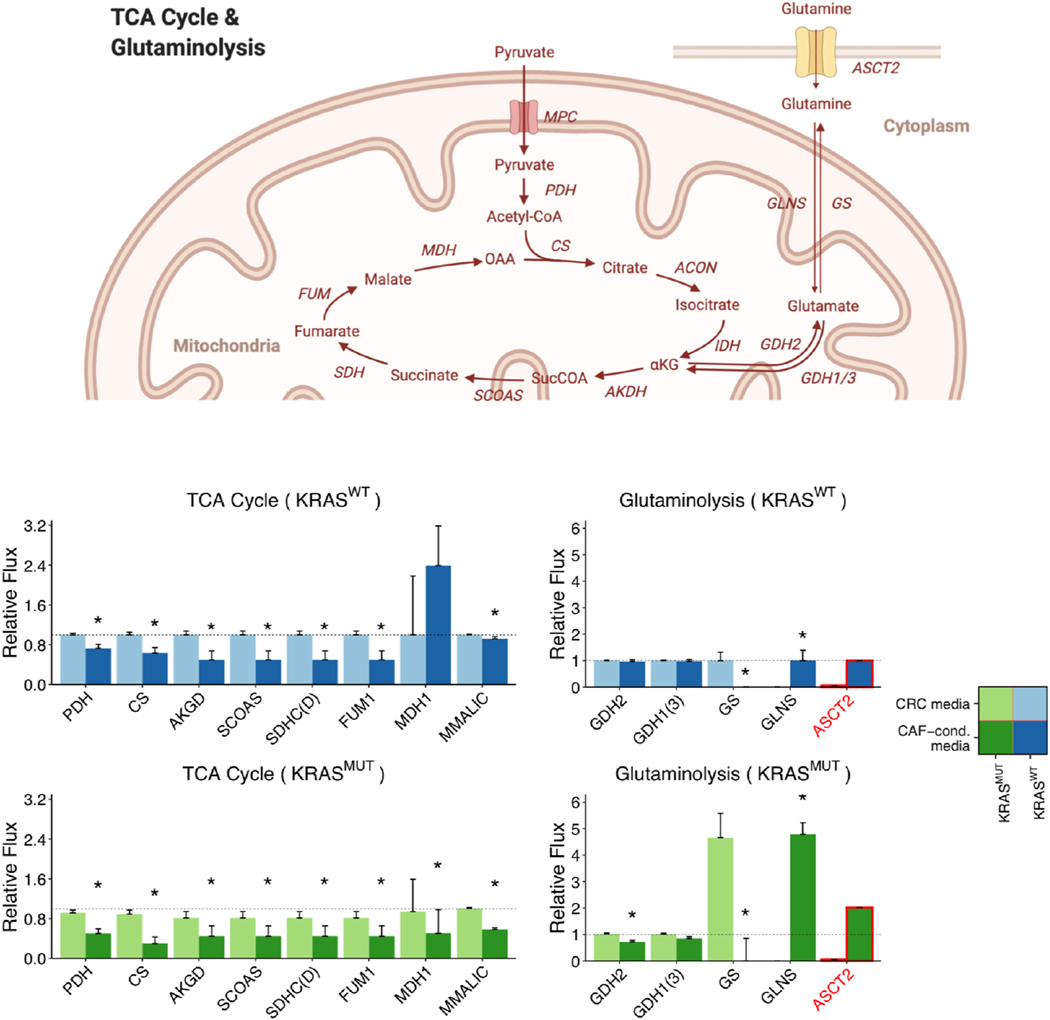
Fig. 7.
Relative fluxes of the TCA cycle and glutaminolysis. KRASWT and KRASMUT CRC cells cultured in CRC media are compared to CAF-conditioned media. Top, Schematic of the TCA cycle and glutaminolysis pathways for reference. This schematic was created from BioRender.com. Directions of the arrows in the diagram represent directions of the reactions. Bottom, Predicted fluxes for TCA cycle and glutaminolysis reactions. All reactions except GLNS and ASCT2 have been normalized to the respective flux in KRASWT CRC cells cultured in CRC media. For visualization, GLNS and ASCT2 have been normalized to the respective flux in KRASWT cells cultured in CAF-conditioned media. The heights of the bars represent normalized median fluxes, and error bars represent normalized MADs. A flux is considered significantly different between CRC media and CAF-conditioned media if its fold change is larger than 1.2 and adjusted p-value is less than 0.001 (asterisks above bars). We outline in red the flux through ASCT2, which is experimentally measured and supplied to the model as a fixed constraint.
3.3. CAFs enhance glycolytic fluxes of CRC cells
Our extracellular metabolomics data indicate that KRASWT and KRASMUT CRC cells cultured in CRC media have comparable glucose uptake and lactate secretion rates, which are mediated by GLUT and MCT, respectively in our model (Fig. 5). (See Table S2 for data). When switched to CAF-conditioned media, KRASWT and KRASMUT cells increase glucose uptake rates by 2.57- and 4.41-fold, respectively, and lactate secretion rates increase by 2.77- and 4.77-fold. This indicates that CAFs play a stimulatory role in glucose uptake and lactate secretion. Similar to CAFs, oncogenic KRAS is reported to upregulate glucose uptake and lactate secretion (Yun et al., 2009; Ying et al., 2012). This effect, however, is only pronounced in CAF-conditioned media, as the glucose uptake and lactate secretion rates differ to a much lesser extent between KRASWT and KRASMUT cultured in the CRC media (Fig. 5).
Regarding intracellular fluxes, upFBA predicts that switching to CAF- conditioned media significantly increases the fluxes through all the glycolytic reactions in both KRASWT and KRASMUT CRC cells (Fig. 5). This suggests that CAFs can upregulate the glycolytic fluxes of CRC cells, and that this interaction occurs by secreted factors and not by contact. Although glucose uptake rates are comparable in KRASWT and KRASMUT cells cultured in CRC media, the flux through the lactate dehydrogenase (LDH) reaction, which dictates lactate production, increases by 1.45-, 2.60-, and 3.50-fold, respectively in KRASMUT cells cultured in CRC media, KRASWT cells grown in CAF-conditioned media, and KRASMUT cells cultured in CAF-conditioned media compared to KRASWT cells grown in CRC media (Fig. 5). This suggests that both oncogenic KRAS and CAFs can upregulate lactate fermentation, although CAFs show a stronger effect.
3.4. CAFs promote NADPH production in CRC cells through oxidative PPP
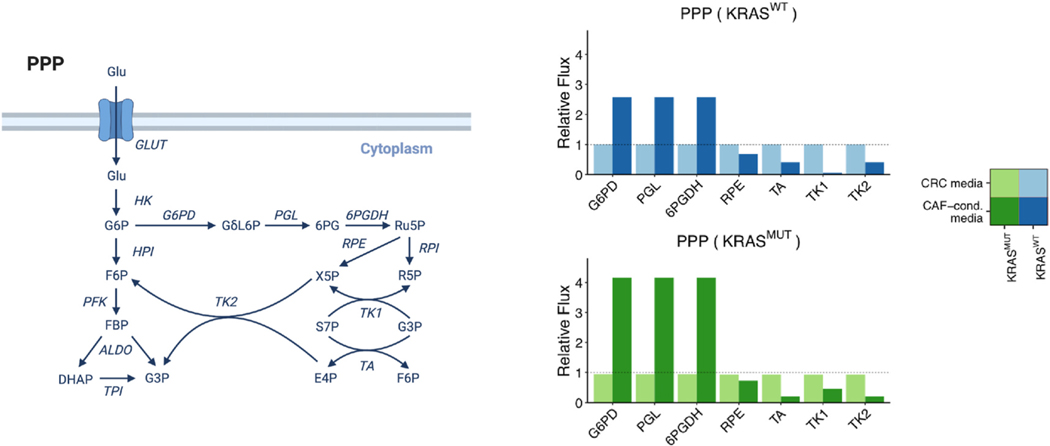
The PPP maintains carbon homeostasis by synthesizing the ribose ring of nucleotides and providing reducing equivalents in the form of NADPH (Frederiks et al., 2008; Stincone et al., 2015). upFBA predicts that KRASWT and KRASMUT CRC cells cultured in CAF-conditioned media have higher fluxes through reactions unique to the oxidative arm of the PPP, i.e., G6PD, lactonase (PGL), as well as 6-phosphogluconate dehydrogenase (6PGDH), than their counterparts cultured in CRC media (Fig. 6). In contrast, the non-oxidative arm of the PPP, comprised of ribose phosphate epimerase (RPE), transaldolase (TA), and trans-ketolases (TK1 and TK2), is barely affected by the growth media in KRASWT cells (Fig. 6). Note that, TK1 proceeds in the direction of S7P → R5P (Fig. 6), which indicates that non-oxidative is also directed to support R5P production in CRC cells. Reduced flux through the TA, TK1, and TK2 reactions in KRASMUT cells in CAF-conditioned media suggests that CRC cells can decouple the oxidative and non-oxidative arms of the PPP to maintain precise control of NAPDH and R5P levels. Interestingly, a similar phenomenon has been observed previously in pancreatic cancer cells (Ying et al., 2012).
Fig. 6.
Relative fluxes of PPP in KRASWT and KRASMUT CRC cells cultured in CRC media compared to CAF-conditioned media. Left, Schematic of the PPP for reference. This schematic was created from BioRender.com. Directions of the arrows in the diagram represent directions of the reactions. Right, Predicted fluxes for PPP reactions. All reactions have been normalized to the respective flux in KRASWT CRC cells cultured in CRC media. The heights of the bars represent normalized median fluxes, and error bars represent normalized MADs. A flux is considered significantly different between CRC media and CAF-conditioned media if its fold change is larger than 1.2 and adjusted p-value is less than 0.001 (asterisks above bars).
3.5. CAFs inhibit the TCA cycle and glutamine anabolism of CRC cells
In eukaryotic cells, pyruvate produced at the end of glycolysis can be transported into the mitochondria to fuel the TCA cycle. We found that switching to CAF-conditioned media inhibits the TCA cycle in CRC cells, as the fluxes through the TCA cycle are in general predicted to be lower for cells in CAF-conditioned media, compared to CRC media (Fig. 7).
In addition to being an important biosynthetic pathway, the TCA cycle also serves to connect glucose and amino acid metabolism. Glutamate, which can be synthesized from glutamine, enters the TCA cycle through its conversion to α-ketoglutarate via glutamate dehydrogenase (GDH) (Nissen et al., 2015). We observed a distinct difference in how CRC cells utilize glutamine in CRC media and CAF-conditioned media, as the measured glutamine uptake rates of KRASWT and KRASMUT CRC cells cultured in CAF-conditioned media are 16 and 39 times higher, respectively, than the glutamine uptake rates of their counter- parts cultured in CRC media (Fig. 7). (See Table S2 for data. Glutamine uptake is mediated by ASCT2 in our model). Furthermore, upFBA predicts that the increased glutamine uptake rates induced by CAFs lead to reprogrammed intracellular glutamine metabolism. CRC cells cultured in CAF-conditioned media divert excessive glutamine via amidohydrolysis mediated by glutaminase (GLNS), whereas CRC cells cultured in CRC media compensate for the reduced glutamine uptake by elevating the flux through the reaction converting glutamate to glutamine, which is mediated by the GS enzyme (Fig. 7).
3.6. CAFs induce reprogramming of NADH, ATP, and pyruvate metabolism in CRC cells
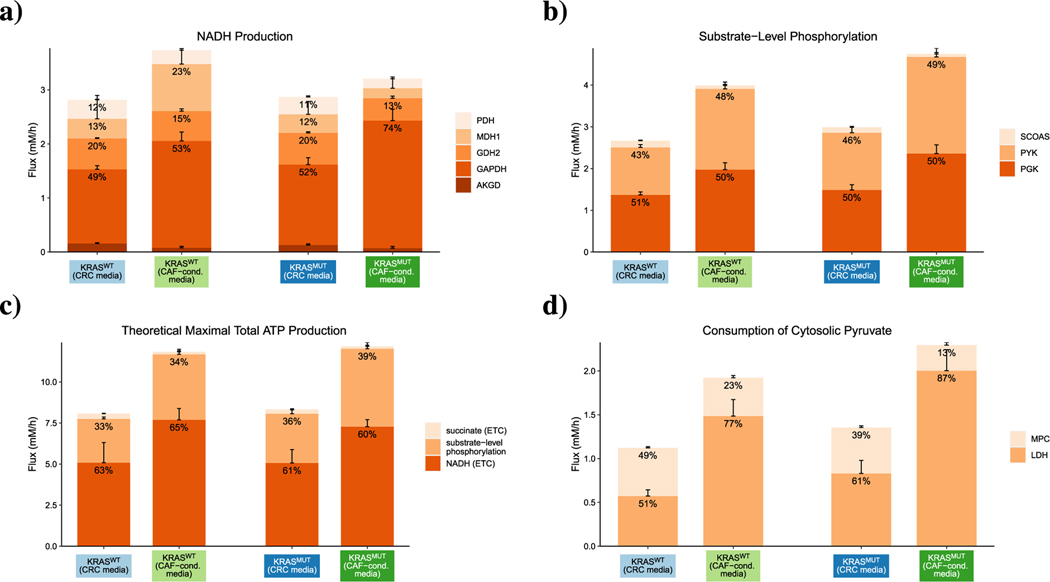
To understand how CAF-induced metabolic alterations affect energy production, we quantified how individual reactions contribute to the production of NADH and ATP as well as the consumption of pyruvate under different conditions. Upon hydrogen transfer to oxygen in oxidative phosphorylation, each NADH maximally produces three ATP molecules, so electron transport from NADH produced in the TCA cycle is widely considered a major source of cellular ATP (Nelson and Lee, 2005). upFBA predicts a stimulatory role of CAFs in NADH production, as the total flux of NADH production is higher in CRC cells cultured in CAF-conditioned media than those cultured in CRC media (Fig. 8(a)). Among the contributing enzymes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is predicted to be the most active, and flux through this reaction further increases in the presence of CAFs (Fig. 8(a)). As is shown in Fig. 8(a), GAPDH not only has a higher absolute flux, but also accounts for a larger fraction of NADH production in CAF-conditioned media.
Fig. 8.
Predicted flux splits of metabolite species. a) NADH production; b) substrate-level phosphorylation; c) theoretical maximum yield of ATP; and d) cytosolic pyruvate consumption in CRC cells. The heights of the bars represent the total sums of absolute flux values. Colors represent enzyme-mediated reactions. Numbers at the top of each stacked bar represent the relative fractions of the total flux devoted to a reaction ( < 8% not shown). Error bars on top of each stacked bar represent the MADs of the fluxes.
Besides oxidative phosphorylation, another source of cellular ATP is substrate-level phosphorylation. Although the relative contributions of the succinyl-CoA synthetase (SCOAS), pyruvate kinase (PYK), and phosphoglycerate kinase (PGK) reactions to overall substrate-level phosphorylation are very similar across all four conditions, the absolute fluxes through these reactions are higher in KRASMUT CRC cells and CAF-conditioned media than in KRASWT CRC cells and CRC media (Fig. 8(b)). Assuming that oxidation of one molecule of NADH or succinate through the electron transport chain (ETC) produces three or two molecules of ATP, respectively (Nelson and Lee, 2005), we then calculated the theoretical total maximum yield of ATP from the given fluxes of reactions that produce ATP and ATP precursors in central carbon metabolism. As is shown in Fig. 8(c), the theoretical total maximum flux of ATP production is higher in CAF-conditioned media than in CRC media. The enhanced theoretical total maximum production of ATP in CAF-conditioned media suggests that CAFs may induce CRC cells to raise the level of ATP production through both substrate-level phosphorylation and oxidative phosphorylation. Note that a higher theoretical total maximum flux of ATP production does not necessarily indicate a higher ATP level, as ATP level is determined by both ATP production and consumption.
Though not a direct energy-carrying molecule, pyruvate serves as a key branching point of central metabolism: it can either be converted to lactate via LDH or transported into the mitochondria via mitochondrial pyruvate carriers (MCPs). upFBA predicts that both oncogenic KRAS and CAFs can enhance lactate fermentation, as the flux through LDH-mediated conversion, both in terms of absolute values and relative fractions, is higher in KRASMUT cells and CAF-conditioned media than in KRASWT cells and CRC media (Fig. 8(d)).
3.7. Gene deletion analysis predicts knockout metabolic phenotypes
Next, we applied flux models to examine how the cancer cells’ growth rate changes in response to enzyme knockouts. To simulate a single-enzyme knockout, we constrained the flux of one reaction in the original metabolic phenotype to zero. Subsequently, we performed uFBA in search of flux distributions that maximize the biomass growth rates under the new constraints. Given that there are 100 sets of mass balance constraints, 74 reactions, 2 genotype identities (KRASWT and KRASMUT), and 2 media conditions (CRC media and CAF-conditioned media), we simulated a total of 100 × 74 × 2 × 2 × 29 600 single-enzyme knockouts.
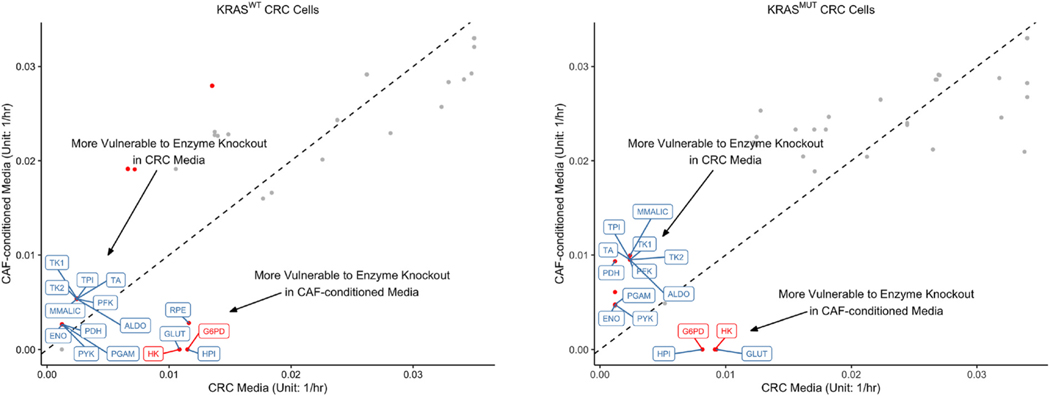
We compared the predictions of the median maximal biomass growth rates upon gene deletion between CRC media and CAF-conditioned media. Similar to the comparison of original metabolic phenotypes, we validated the differences in biomass growth rates via the Wilcoxon rank sum test (Pearce and Derrick, 2019). Median maximal biomass growth rates of single-enzyme-knockout phenotypes, MADs, and adjusted p-values are provided in Supplementary File 5. Among a total of 25 enzyme knockouts leading to different biomass growth rates in KRASWT CRC cells, 16 belong to central carbon metabolism: 9, 5, and 2 are unique to glycolysis, PPP, and the TCA cycle, respectively (Fig. 9). Similarly, out of 19 enzyme knockouts that lead to different biomass growth rates in KRASMUT CRC cells, 15 belong to central carbon metabolism, with 9, 4, and 2 enzyme knockouts from glycolysis, PPP, and the TCA cycle, respectively. In both KRASWT and KRASMUT CRC cells, 11 single-enzyme-knockouts belonging to central carbon metabolism result in reduced biomass growth rates in CRC media. In contrast, only 5 single-enzyme-knockouts result in reduced biomass growth rates in CAF-conditioned media for KRASWT CRC cells, and only 4 single-enzyme-knockouts, for KRASMUT CRC cells (Fig. 9). As CAFs are generally believed to make cancer cells more resilient to enzyme knockouts, it is unsurprising that fewer enzyme knockouts are available for blocking cancer growth in CAF-conditioned media. Nevertheless, it is noteworthy that for both KRASWT and KRASMUT cells, knockouts of HK and G6PD, traditionally considered the rate-limiting steps of glycolysis and PPP, result in lower biomass growth rates in CAF-conditioned media compared to CRC media (Fig. 9). This is likely because CAFs make CRC cells more dependent on glycolysis and PPP, as we have shown in Sections 3.3 and 3.4. Thus, interference with glycolysis and PPP could take a heavier toll on CRC metabolism in presence of CAFs.
Fig. 9.
Predicted maximal biomass growth rates upon gene deletions. uFBA was used to predict the maximal growth rates for KRASWT and KRASMUT CRC cells upon gene deletions. The x- and y-coordinates of each point represent the median maximal biomass growth rates of CRC cells cultured in CRC media and CAF-conditioned media, respectively when the same enzyme is inhibited. The dashed diagonal line represents equal median rates in both media. Gene deletions resulting in significantly different biomass growth rates between CRC media and CAF-conditioned media are represented by red dots (adjusted p-value < 0.001, fold change > 2). Enzymes belonging to glycolysis, PPP, the TCA cycle, and glutaminolysis are labeled.
In contrast to the effects of the medium, few single-enzyme knockouts lead to different biomass growth rates between KRASWT and KRASMUT cells (Fig. S5). In CRC media, the inhibition of RPE is the only enzyme knockout that results in differential cancer growth rates be- tween KRASWT and KRASMUT CRC cells (reduced growth in KRASMUT cells; Fig. S5). In CAF-conditioned media, only three enzyme knockouts, including the knockouts of GAPDH and pyruvate dehydrogenase (PDH), lead to differential cancer growth rates between the two cell lines (Fig. S5). (All lead to reduced growth in KRASWT cells). This is consistent with our findings in Sections 3.1-3.5 and further shows that CAF-conditioned media cause stronger metabolic alterations to CRC cells than oncogenic KRAS.
4. Discussion
4.1. CAFs induce widespread changes in CRC metabolism
In this work, we have studied how CAFs affect CRC metabolism via an integration of LC-MS metabolomics and constraint-based modeling. Our findings of the effects of CAFs on central carbon metabolic pathways can be summarized as follows (Table 1). First, CAFs induce a glycolytic switch in CRC cells. Compared to normal cells, cancer cells have higher rates of glucose uptake and lactate production even in the presence of oxygen and functional mitochondria, a phenomenon known as the Warburg Effect (Liberti and Locasale, 2016; Warburg, 1956). Comparing CRC cells cultured in CRC media and CAF-conditioned media, we have found that CAFs exacerbate the Warburg Effect by enabling cancer cells to uptake glucose and produce lactate at even higher rates. Second, CAFs increase the fluxes through the oxidative arm of the PPP but do not affect the non-oxidative arm of the PPP in CRC cells. The purpose of CAF-induced upregulation of oxidative PPP is perhaps to maintain the redox balance and support the increased demand for nucleotide synthesis in tumor proliferation. Like previous studies, our analysis demonstrates that cells can decouple oxidative and non-oxidative PPP (Ying et al., 2012; Boros et al., 1998). Third, CAFs inhibit the TCA cycle, increase glutamine uptake, and stimulate glutaminolysis in CRC cells. Note that CAF-conditioned media induce widespread metabolic changes in CRC cells despite no appreciable change in CRC cell growth (Fig. 4). Although there are no significant differences in the growth rates between DLD-1 KRASWT and KRASMUT cell lines, there are morphology changes. When the two cell lines are switched to CAF-conditioned media, they appear to have more filopodia-like structures which is indicative of a migratory or invasive state (Mattila and Lappalainen, 2008; Ridley, 2011), compared to the more compact and rounded shape in CRC media (Fig. S6).
Table 1.
Regulation of CRC metabolism by CAFs predicted by upFBA. An upward and downward arrows represent a higher or lower median flux, respectively, through the pathway in CRC cells cultured in CAF-conditioned media compared to CRC cells cultured in CRC media.
| Cell | Treatment | Control | Glycolysis | Oxidative PPP | Non-oxidative PPP | TCA Cycle | Glutaminolysis |
|---|---|---|---|---|---|---|---|
|
| |||||||
| KRASWT | CAF-conditioned | CRC | ↑ | ↑ | - | ↓ | ↑ |
| Media | Media | ||||||
| KRASMUT | CAF-conditioned | CRC | ↑ | ↑ | ↓ | ↓ | ↑ |
| Media | Media | ||||||
At the metabolite level, we have found that CAFs concomitantly switch CRC cells to a phenotype characterized by elevated energy production and lactate fermentation. Lactate was traditionally considered a waste product of glycolysis, but an increasing number of studies found that it can play important roles in promoting metastasis, stimulating angiogenesis, inducing immunosuppression, and maintaining the NAD+/NADH ratio (de la Cruz-López et al., 2019; Dhup et al., 2012; Brand et al., 2016; Luengo et al., 2020). The upregulated conversion of pyruvate to lactate may be driven by the diverse functions of lactate as an oncometabolite (de la Cruz-López et al., 2019; San-Millán and Brooks, 2017). Moreover, we have observed that under both media conditions, CRC cells secrete lactate, despite the high lactate concentration in CAF-conditioned media. The Reverse Warburg Effect, where CAFs secrete lactate to support adjacent cancer cells, has been observed in some cervical and breast cancer cell lines (Fu et al., 2017). Head and neck squamous cell carcinoma (HNSCC), for example, has been found to be inefficient in using lactate as a carbon source (Kumar et al., 2018). Our observations indicate that similar to HNSCC, CRC cells prefer adopting a Warburg Effect over a Reverse Warburg Effect phenotype. While cancer metabolism is typically associated with high energy demand, our analysis overall suggests that cancer cells can modulate their biosynthetic pathways in a more complicated manner in order to adapt to their environments. It remains unclear whether all cancer cell lines adopt a similar metabolic strategy.
Our findings are supported by previous studies on tumor-stromal metabolic crosstalk in other cancer types (Kumar et al., 2018; Knowles et al., 2009). In agreement with our results, CAFs were found to enhance extracellular lactate levels in HNSCC cells via upregulation of glycolysis (Kumar et al., 2018; Knowles et al., 2009). It was further demonstrated that CAF-secreted HGF activates the glycolytic switch in HNSCC cells via paracrine signaling, whereas HNSCC-cell-secreted basic fibroblast growth factor (bFGF) upregulates oxidative phosphorylation and downregulates glycolysis in CAFs (Kumar et al., 2018; Knowles et al., 2009). The metabolic crosstalk between CRC cells and CAFs may have adopted a similar mechanism. In a separate study (Fiaschi and Chiarugi, 2012), it was found that tumor-derived TGFβ1 could increase the level of intracellular reactive oxygen species in CAFs. In return, CAFs secrete HGF, reprogramming the metabolism of cancer cells towards a glycolytic phenotype. It will be interesting to validate the roles of HGF, bFGF, and TGFβ1 in CRC-CAF metabolic interactions and identify additional signaling molecules that contribute to CRC metabolic reprogramming via biochemical assays.
Another noteworthy finding of our study is that CRC cells in CAF-conditioned media consume exogenous glutamine, whereas CRC cells in CRC media synthesize glutamine from glutamate (Fig. 7). It was demonstrated in vivo that CRC cells are addicted to glutamine (Zhao et al., 2019). In agreement with their findings, we find that CRC cells cultured in CAF-conditioned media, which simulate a TME more closely than CRC cells cultured in CRC media, depend more heavily on exogenous glutamine. On the other hand, it was shown in vitro that SW480 and SW620 CRC cells cultured in RPMI-1640 (Invitrogen, 11 875–093) or DMEM (Invitrogen, 11 965–092) media are also dependent on exogenous glutamine (Li et al., 2017). The CRC media used in our study have a higher glucose to glutamine ratio (Glc/Gln = 8.75) compared to the two media mentioned above (RPMI: Glc/Gln = 5.41; DMEM: Glc/Gln = 6.25) (Li et al., 2017), which may have partially contributed to the differences in our observations.
Another reason why our findings differ is that we conducted our study in DLD-1 cells, which were derived from a different patient compared to SW480 and SW620 cells (Ahmed et al., 2013). Cell-line and TME specificity has long been regarded as a cause of metabolic diversity in cancer. SW480 and SW620 cells were found to be more sensitive to glutamine deprivation than HT29 and HCT116 cells (Li et al., 2017). One study showed that glutamine uptake is essential for HNSCC in SCC15 cell lines (Zhang et al., 2020), whereas another demonstrated that glucose, not glutamine, is the dominant energy source of HNSCC in other cell lines (Sandulache et al., 2011). Furthermore, it has been shown pancreatic ductal adenocarcinoma cells, traditionally known for glutamine addiction, could regulate the activity of GS, switching between metabolic states optimal for growth under nutrient-replete and nutrient-deplete conditions (Tsai et al., 2021). Similarly, it remains unclear whether all CRC cells, irrespective of the genetic identity or TME, 1) are addicted to glutamine and 2) can rewire metabolism to adapt to nutrient depletion. Further studies are needed to determine whether our findings can be generalized to other CRC cell lines.
4.2. Gene deletion analysis identifies CAF-dependent targets
In addition to characterizing the metabolic phenotypes of CRC cells grown under different media conditions, we have also made predictions of knockout metabolic phenotypes by performing single gene deletion analysis. Our analysis predicts not only enzyme knockouts that inhibit cancer cells’ growth but also those that affect cancer cells’ growth differently in CAF-conditioned media than in CRC media. We have found that a large number of in silico single-enzyme knockouts result in stronger inhibition of cancer growth in CRC media than in CAF-conditioned media. This suggests that CAFs generally make CRC cells more resilient to metabolic stress. Combining CAF-targeted therapies with these enzyme knockouts may be necessary to achieve optimal therapeutic efficacy.
Meanwhile, we have also found that deletions of HK and G6PD, in particular, result in slower cancer growth in CAF-conditioned media. In the context of metabolic control analysis, this means that HK and G6PD would have higher control coefficients (Moreno-Sánchez et al., 2008). Biologically, this implies that targeting HK and G6PD suppresses tumor growth by exploiting the metabolic dependence of CRC cells on CAFs. Inhibitors of these two enzymes include metformin, a common anti-diabetic drug found to inhibit HK in breast and cervical cancer cell lines (Liu et al., 2017; Marini et al., 2013) and polydatin, a natural molecule found in plants found to inhibit G6PD in HNSCC cell lines (Mele et al., 2018). It will be of great interest to validate the efficacy of metformin and polydatin in CRC cell lines. Clinically, selective blockade of HK and G6PD remains a critical challenge as HK and G6PD are ubiquitously expressed in all mammalian cells. However, emerging targeted therapies such as bispecific antibodies and proteolysis targeting chimeras (PROTACs) may enable us to reevaluate the druggability of these genes as well as their upstream regulators.
4.3. UpFBA sheds light on dynamic metabolic physiology
Method wise, our work demonstrates upFBA as an effective data- driven approach for inferring flux distributions from high-throughput metabolomics data. Metabolite pool size profiling alone may be insufficient for understanding the causes of metabolic alterations due to the intertwined structure of metabolic networks, as we have shown in Section 3.1. While transcriptomics and proteomics data are commonly used to investigate drivers of metabolic/phenotypic differences (Hahne et al., 2010; Wang et al., 2020a; Suh et al., 2015; Nuber et al., 2021), the expression levels of genes encoding the metabolic enzymes do not fully represent their enzymatic activities. Our method serves to complement the tool box available for metabolism research by making direct use of data obtained at the metabolite level. Future work can investigate further the metabolic alterations shown to occur. For example, 13C tracing studies can be applied to determine how glutamine is being utilized. In addition, upFBA can be applied to investigate how metabolism of CRC cells is influenced by CAFs. In this way, we can understand reciprocal metabolic interactions between CRC cells and CAFs. Additionally, here, we focused on KRASMUT cells. However, the same approach can be applied to cells from other genetic backgrounds.
4.4. Limitations and assumptions of this study
We acknowledge some limitations of our study. First, we aim to capture the averaged metabolite dynamics and assume that the rate of change of metabolites is constant between T = 0hrs and T = 24hrs. Similar to uFBA (Bordbar et al., 2017), more frequent sampling will enable us to capture the fast dynamics more accurately by discretizing metabolite time profiles into time intervals for piecewise simulation. Second, we have only compared CRC media and CAF-conditioned media in this work. Media derived from co-cultures of CRC cells and CAFs, however, may diverge from media derived from either cell type alone. It will be interesting to study the metabolism of cancer cells cultured in the media derived from co-cultures of CRC cells and CAFs. In addition, upFBA cannot identify the mechanism(s) underlying flux changes, e.g., whether a change in flux is driven by a change in the substrate concentration or a conformational change in the enzyme. Nevertheless, upFBA shows promise as a bridge connecting metabolomics and fluxomics in systems biology and multi-omics studies.
In addition to these limitations, we acknowledge certain assumptions that can affect the model predictions. Specifically, we consider the reliability of initial metabolite measurements, flux split between glycolysis and PPP, and metabolite steady state. Below, we describe additional model calculations generated when assumptions based on these factors are relaxed.
Reliability of initial metabolite levels.
Although we measured the relative changes of more than 80 metabolites in this study, only 14 of those metabolites are involved in central carbon metabolism. We chose to exclude seven out of these 14 metabolites because the lower and upper bounds of the range estimates found in literature differ by more than one order of magnitude, compared to an average of 3-fold range for the seven metabolites included (Table S3) (Roy and Finley, 2017). To understand the effect of other metabolites on the calculated fluxes, we performed upFBA by including all 14 metabolites, imposing nonzero mass balance constraints for those metabolic species. The predicted median fluxes, MADs, and adjusted p-values are provided in Supplementary File 6 and Figs. S7–S9. More variation was observed in the calculated fluxes through PPP due to the wider range estimates of initial metabolite concentrations (Table S3 and Fig. S8), but qualitatively, our findings remained the same (Table S4). More work is needed to understand the level of uncertainty in range estimates that can be tolerated by upFBA. Such simulations can also investigate the question about how strongly the intracellular metabolite data affects the model predictions, compared to data for extracellular species (i.e., uptake and secretion rates).
Glycolysis and PPP flux split.
In implementing upFBA, we constrained the flux split between glycolysis and PPP to be 90/10 following a previous study (Zielinski et al., 2017). To determine the effect of this constraint, we performed upFBA allowing the glycolysis to PPP flux ratio to vary. Without constraining the flux ratio, upFBA yields a biologically unrealistic solution where there is very little flux entering the oxidative arm of the PPP (Supplementary File 7). Meanwhile, previous 13C tracing studies showed that approximately 16% and 5% of glucose uptake is diverted to PPP in A549 and MCF7 cancer cell lines, respectively (Zielinski et al., 2017). This suggests that incorporating existing biological knowledge into the analysis is essential for the interpretation of the results. Another discrepancy between constraining and not constraining the flux ratio is the variability of fluxes in oxidative PPP. The oxidative PPP fluxes do not differ significantly between the two media conditions, whereas the non-oxidative PPP fluxes in KRASMUT cells are higher in CRC media than in CAF-conditioned media (Fig. S11). The predicted median fluxes, MADs, and adjusted p-values are provided in Supplementary File 7 and Figs. S10–S12.
Metabolite steady state.
To understand the benefits we gained from upFBA, we performed conventional parsimonious FBA (pFBA) on the same metabolic network and assessed the global differences between the calculated fluxes of the upFBA and pFBA models. The predicted fluxes are provided in Supplementary File 8 and Figs. S13–S15. We found considerable differences between the predicted fluxes of non-oxidative PPP, TCA cycle, and glutaminolysis (Figs. 5–7 and S13-S15; Tables 1 and S5-S7). Most notably, upFBA predicts that fluxes through the TCA cycle are either decreased or unchanged in KRASMUT compared to KRASWT cells (Fig. 7 and Table S6), whereas pFBA predicts that the TCA cycle reaction fluxes are either unchanged or increased in KRASMUT cells compared to KRASWT cells (Fig. S15 and Table S7). Compared to pFBA, the findings of upFBA are more consistent with previous studies showing that KRASMUT and BRAFMUT either decrease or have no effect on the expression of TCA cycle enzymes (Ying et al., 2012; Hutton et al., 2016; Haq et al., 2013).
The results of upFBA differ from pFBA because the former considers the measurements of metabolite changes while the latter does not. Between T = 0hrs and T = 24hrs, the intracellular lactate level increases more in KRASMUT than in KRASWT cells (Fig. 3(e)). To explain the increased intracellular fold change of lactate in KRASMUT cells, upFBA infers an elevated flux of pyruvate converted to lactate and decreased flux of pyruvate entering the TCA cycle in KRASMUT cells (Fig. 8(d) and Table S8). Conventional pFBA, however, predicts a simultaneous increase of lactate production (LDH) and mitochondrial transport (MCP), as it bases the optimization solely upon the exchange fluxes without considering the transient changes in intracellular metabolite levels (Fig. S16 and Table S8). Compared to pFBA, upFBA is more data-oriented, with the aim to find a solution that optimizes the objective function and explains the data simultaneously. It enables us to generate insights that may not be deemed obvious based on measurements of metabolite exchange rates alone.
4.5. Conclusions
By combining metabolomics data and constraint-based modeling, we have identified metabolic perturbations that exploit the CAF-induced metabolic changes in CRC cells. Our work not only provides quantitative insights that complement experimental studies to develop therapeutic strategies for reducing CRC cell growth but also drives targeted follow-up experiments that can accelerate biological discovery as well as potential clinical translations.
Supplementary Material
Acknowledgements
This work was supported by the NIH National Cancer Institute grant 1U01CA232137.
Footnotes
Author statement
Junmin Wang: Methodology, Formal analysis, Software, Writing – Original draft, Writing – Review & Editing; Alireza Delfarah: Investigation, Formal analysis, Writing – Review & Editing; Patrick E. Gelbach: Investigation; Emma Fong: Investigation, Formal analysis, Writing – Review & Editing; Paul Macklin: Conceptualization, Funding acquisition, Writing – Review & Editing; Shannon Mumenthaler: Conceptualization, Funding acquisition, Resources, Writing – Review & Editing; Nicholas Graham: Resources, Supervision, Writing – Review & Editing; Stacey D. Finley: Conceptualization, Project administration, Funding acquisition, Resources, Writing – Review & Editing.
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymben.2021.11.006.
References
- Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, Lothe RA, 2013. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2 (9), e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn WS, Antoniewicz MR, 2011. Metabolic flux analysis of cho cells at growth and non-growth phases using isotopic tracers and mass spectrometry. Metab. Eng 13 (5), 598–609. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, 2018. A guide to 13c metabolic flux analysis for the cancer biologist. Exp. Mol. Med 50 (4), 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A, Yurkovich JT, Paglia G, Rolfsson O, Sigurjónsson ÓE, Palsson BO, 2017. Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics. Sci. Rep 7 (1), 46249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros LG, Lee PW, Brandes JL, Cascante M, Muscarella P, Schirmer WJ, Melvin WS, Ellison EC, 1998. Nonoxidative pentose phosphate pathways and their direct role in ribose synthesis in tumors: is cancer a disease of cellular glucose metabolism? Med. Hypotheses 50 (1), 55–59. [DOI] [PubMed] [Google Scholar]
- Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, Kastenberger M, Bogdan C, Schleicher U, Mackensen A, Ullrich E, Fichtner-Feigl S, Kesselring R, Mack M, Ritter U, Schmid M, Blank C, Dettmer K, Oefner PJ, Hoffmann P, Walenta S, Geissler EK, Pouyssegur J, Villunger A, Steven A, Seliger B, Schreml S, Haferkamp S, Kohl E, Karrer S, Berneburg M, Herr W, Mueller-Klieser W, Renner K, Kreutz M, 2016. Ldha-associated lactic acid production blunts tumor immunosurveillance by t and nk cells. Cell Metabol. 24 (5), 657–671. [DOI] [PubMed] [Google Scholar]
- Cluntun AA, Lukey MJ, Cerione RA, Locasale JW, 2017. Glutamine metabolism in cancer: understanding the heterogeneity. Trends Cancer 3 (3), 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Locasale JW, 2017. Understanding metabolism with flux analysis: from theory to application. Metab. Eng 43, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, García-Carrancá A, Manzo-Merino J, 2019. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol 9, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhup S, Dadhich RK, Porporato PE, Sonveaux P, 2012. Multiple biological activities of lactic acid in cancer: influences on tumor growth, angiogenesis and metastasis. Curr. Pharmaceut. Des 18 (10), 1319–1330. [DOI] [PubMed] [Google Scholar]
- Fiaschi T, Chiarugi P, 2012. Oxidative stress, tumor microenvironment, and metabolic reprogramming: a diabolic liaison. Int. J. Cell Biol 762825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach EH, Rebecca VW, Herlyn M, Smalley KSM, Anderson ARA, 2011. Fibroblasts contribute to melanoma tumor growth and drug resistance. Mol. Pharm 8 (6), 2039–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiks WM, Vizan P, Bosch KS, Vreeling-Sindelárová H, Boren J, Cascante M, 2008. Elevated activity of the oxidative and non-oxidative pentose phosphate pathway in (pre)neoplastic lesions in rat liver. Int. J. Exp. Pathol 89 (4), 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Liu S, Yin S, Niu W, Xiong W, Tan M, Li G, Zhou M, 2017. The reverse warburg effect is likely to be an achilles’ heel of cancer that can be exploited for cancer therapy. Oncotarget 8 (34), 57813–57825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey CM, Lau R, Sanchez A, Sun RX, Fong EJ, Doche ME, Chen O, Jusuf A, Lenz H-J, Larson B, Mumenthaler SM, 2020. Anti-egfr therapy induces egf secretion by cancer-associated fibroblasts to confer colorectal cancer chemoresistance. Cancers 12 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne H, Mäder U, Otto A, Bonn F, Steil L, Bremer E, Hecker M, Becher D, 2010. A comprehensive proteomics and transcriptomics analysis of bacillus subtilis salt stress adaptation. J. Bacteriol 192 (3), 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, Frederick DT, Hurley AD, Nellore A, Kung AL, Wargo JA, Song JS, Fisher DE, Arany Z, Widlund HR, 2013. Oncogenic braf regulates oxidative metabolism via pgc1 and mitf. Cancer Cell 23 (3), 302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinken A, Haraldsdóttir HS, Wachowiak J, Keating SM, Vlasov V, Magnusdóttir S, Ng CY, Preciat G, Žagare A, Chan SHJ, Aurich MK, Clancy CM, Modamio J, Sauls JT, Noronha A, Bordbar A, Cousins B, El Assal DC, Valcarcel LV, Apaolaza I, Ghaderi S, Ahookhosh M, Ben Guebila M, Kostromins A, Sompairac N, Le HM, Ma D, Sun Y, Wang L, Yurkovich JT, Oliveira MAP, Vuong PT, El Assal LP, Kuperstein I, Zinovyev A, Hinton HS, Bryant WA, Aragón Artacho FJ, Planes FJ, Stalidzans E, Maass A, Vempala S, Hucka M, Saunders MA, Maranas CD, Lewis NE, Sauter T, Palsson BØ, Thiele I, Fleming RMT, 2019. Creation and analysis of biochemical constraint-based models using the cobra toolbox v.3.0. Nat. Protoc 14 (3), 639–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton JE, Wang X, Zimmerman LJ, Slebos RJC, Trenary IA, Young JD,Li M, Liebler DC, 2016. Oncogenic kras and braf drive metabolic reprogramming in colorectal cancer. Mol. Cell. Proteomics 15 (9), 2924–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez EF, Lau R, Friedman SH, Ghaffarizadeh A, Jonckheere E, Agus DB, Mumenthaler SM, Macklin P, 2016. Quantifying differences in cell line population dynamics using cellpd. BMC Syst. Biol 10 (1), 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles LM, Stabile LP, Egloff AM, Rothstein ME, Thomas SM, Gubish CT, Lerner EC, Seethala RR, Suzuki S, Quesnelle KM, Morgan S, Ferris RL, Grandis JR, Siegfried JM, 2009. Hgf and c-met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clin. Cancer Res 15 (11), 3740–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, New J, Vishwakarma V, Joshi R, Enders J, Lin F, Dasari S, Gutierrez WR, Leef G, Ponnurangam S, Chavan H, Ganaden L, Thornton MM, Dai H, Tawfik O, Straub J, Shnayder Y, Kakarala K, Tsue TT, Girod DA, Van Houten B, Anant S, Krishnamurthy P, Thomas SM, 2018. Cancer- associated fibroblasts drive glycolysis in a targetable signaling loop implicated in head and neck squamous cell carcinoma progression. Cancer Res. 78 (14), 3769–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guennec A, Tea I, Antheaume I, Martineau E, Charrier B, Pathan M, Akoka S, Giraudeau P, 12, 2012. Fast determination of absolute metabolite concentrations by spatially encoded 2d nmr: application to breast cancer cell extracts. Anal. Chem 84 (24), 10831–10837. [DOI] [PubMed] [Google Scholar]
- Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P, Polpitiya AD, Adkins JN, Schramm G, Purvine SO, Lopez-Ferrer D, Weitz KK, Eils R, König R, Smith RD, Palsson BØ, 2010. Omic data from evolved e. coli are consistent with computed optimal growth from genome-scale models. Mol. Syst. Biol 6 (1), 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Song P, Zhu L, Aziz N, Zhou Q, Zhang Y, Xu W, Feng L, Chen D, Wang X, Jin H, 2017. Synthetic lethality of glutaminolysis inhibition, autophagy inactivation and asparagine depletion in colon cancer. Oncotarget 8 (26), 42664–42672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti MV, Locasale JW, 2016. The warburg effect: how does it benefit cancer cells? Trends Biochem. Sci 41 (3), 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC, 2001. The microenvironment of the tumour-host interface. Nature 411 (6835), 375–379. [DOI] [PubMed] [Google Scholar]
- Littlepage LE, Egeblad M, Werb Z, 2005. Coevolution of cancer and stromal cellular responses. Cancer Cell 7 (6), 499–500. [DOI] [PubMed] [Google Scholar]
- Liu Y, Murray-Stewart T, Casero RA Jr, Kagiampakis I, Jin L, Zhang J, Wang H, Che Q, Tong H, Ke J, Jiang F, Wang F, Wan X, 2017. Targeting hexokinase 2 inhibition promotes radiosensitization in hpv16 e7-induced cervical cancer and suppresses tumor growth. Int. J. Oncol 50 (6), 2011–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler M, Krüger JA, Niethammer AG, Reisfeld RA, 2006. Targeting tumor- associated fibroblasts improves cancer chemotherapy by increasing intratumoral drug uptake. J. Clin. Invest 116 (7), 1955–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CP, Antoniewicz MR, 2019. High-resolution 13c metabolic flux analysis. Nat. Protoc 14 (10), 2856–2877. [DOI] [PubMed] [Google Scholar]
- Lueng A., L Z., Gu DY., Sulliva LB., Zagoruly M., D BT., Ferreir R., Naamat A., Al A., Lewi CA., Thoma CJ., Sprange S., Matheso NJ., Vander Heide MG., 2020. Increased demand for nad relative to atp drives aerobic glycolysis. Mol. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhorin A, 2000. Glpk (gnu linear programming kit). Available at. http://www.gnu.rg/software/glpk/glpk.html. [Google Scholar]
- Marín-Hernández A, López-Ramírez SY, Del Mazo-Monsalvo I, Gallardo-Pérez, Rodríguez-Enríquez S, Moreno-Sánchez R, Saavedra E, 2014. Modeling cancer glycolysis under hypoglycemia, and the role played by the differential expression of glycolytic isoforms. FEBS J. 281 (15), 3325–3345. [DOI] [PubMed] [Google Scholar]
- Marini C, Salani B, Massollo M, Amaro A, Esposito AI, Orengo AM, Capitanio S, Emionite L, Riondato M, Bottoni G, Massara C, Boccardo S, Fabbi M, Campi C, Ravera S, Angelini G, Morbelli S, Cilli M, Cordera R, Truini M, Maggi D, Pfeffer U, Sambuceti G, 2013. Direct inhibition of hexokinase activity by metformin at least partially impairs glucose metabolism and tumor growth in experimental breast cancer. Cell Cycle 12 (22), 3490–3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P, 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol 9 (6), 446–454. [DOI] [PubMed] [Google Scholar]
- Mele L, Paino F, Papaccio F, Regad T, Boocock D, Stiuso P, Lombardi A, Liccardo D, Aquino G, Barbieri A, Arra C, Coveney C, La Noce M, Papaccio G, Caraglia M, Tirino V, Desiderio V, 2018. A new inhibitor of glucose-6-phosphate dehydrogenase blocks pentose phosphate pathway and suppresses malignant proliferation and metastasis in vivo. Cell Death Dis. 9 (5), 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Ghatak S, Zoltan-Jones A, Toole BP, 2003. Regulation of multidrug resistance in cancer cells by hyaluronan. J. Biol. Chem 278 (28), 25285–25288. [DOI] [PubMed] [Google Scholar]
- Moreno-Sánchez R, Saavedra E, Rodríguez-Enríquez S, Olín-Sandoval V, 2008.Metabolic control analysis: a tool for designing strategies to manipulate metabolic pathways. J. Biomed. Biotechnol. 597913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson David L., Lee David, 2005. In: Lehninger Principles of Biochemistry, fourth ed. Freeman WH, New York. 2005. [Google Scholar]
- Nissen JD, Pajecka K, Stridh MH, Skytt DM, Waagepetersen HS, 2015. Dysfunctional tca-cycle metabolism in glutamate dehydrogenase deficient astrocytes. Glia 63 (12), 2313–2326. [DOI] [PubMed] [Google Scholar]
- Nuber S, Nam AY, Rajsombath MM, Cirka H, Hronowski X, Wang J, Hodgetts K, Kalinichenko LS, Müller CP, Lambrecht V, Winkler J, Weihofen A, Imberdis T, Dettmer U, Fanning S, Selkoe DJ, 2021. A stearoyl- coenzyme a desaturase inhibitor prevents multiple Parkinson disease phenotypes in α-synuclein mice. Ann. Neurol 89 (1), 74–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce J, Derrick B, 2019. Preliminary testing: the devil of statistics? Reinvention: an International Journal of Undergraduate Research 12. [Google Scholar]
- Räsänen K, Vaheri A, 2010. Activation of fibroblasts in cancer stroma. Exp. Cell Res 316 (17), 2713–2722. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, 2011. Life at the leading edge. Cell 145 (7), 1012–1022. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, 2015. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, Finley SD, 2017. Computational model predicts the effects of targeting cellular metabolism in pancreatic cancer. Front. Physiol 8, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Millán I, Brooks GA, 2017. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the warburg effect. Carcinogenesis 38 (2), 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandulache VC, Ow TJ, Pickering CR, Frederick MJ, Zhou G, Fokt I, Davis- Malesevich M, Priebe W, Myers JN, 2011. Glucose, not glutamine, is the dominant energy source required for proliferation and survival of head and neck squamous carcinoma cells. Cancer 117 (13), 2926–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan M, Dai D, Vudem A, Varner JD, Stroock AD, 2018. Multi-scale computational study of the warburg effect, reverse warburg effect and glutamine addiction in solid tumors. PLOS Comp. Biol 14 (12), 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A, 2020. Cancer statistics, 2020. CA. Cancer J. Clin 70 (1), 7–30. [DOI] [PubMed] [Google Scholar]
- Still ER, Yuneva MO, 2017. Hopefully devoted to q: targeting glutamine addiction in cancer. Br. J. Cancer 116 (11), 1375–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stincone A, Prigione A, Cramer T, Wamelink MMC, Campbell K, Cheung E, Olin- Sandoval V, Grüning N-M, Krüger A, Tauqeer Alam M, Keller MA, Breitenbach M, Brindle KM, Rabinowitz JD, Ralser M, 2015. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Phil. Soc 90 (3), 927–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strang G, 2016. Introduction to Linear Algebra. [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, Cooper ZA, Chapman PB, Solit DB, Ribas A, Lo RS, Flaherty KT, Ogino S, Wargo JA, Golub TR, 2012. Tumour micro-environment elicits innate resistance to raf inhibitors through hgf secretion. Nature 487 (7408), 500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M-J, Tovchigrechko A, Thovarai V, Rolfe MA, Torralba MG, Wang J, Adkins JN, Webb-Robertson B-JM, Osborne W, Cogen FR, Kaplowitz PB, Metz TO, Nelson KE, Madupu R, Pieper R, 2015. Quantitative differences in the urinary proteome of siblings discordant for type 1 diabetes include lysosomal enzymes. J. Proteome Res 14 (8), 3123–3135. [DOI] [PubMed] [Google Scholar]
- Tsai P-Y, Lee M-S, Jadhav U, Naqvi I, Madha S, Adler A, Mistry M, Naumenko S, Lewis CA, Hitchcock DS, Roberts FR, DelNero P, Hank T, Honselmann KC, Morales Oyarvide V, Mino-Kenudson M, Clish CB, Shivdasani RA, Kalaany NY, 2021. Adaptation of pancreatic cancer cells to nutrient deprivation is reversible and requires glutamine synthetase stabilization by mtorc1. Proc. Natl. Acad. Sci. U. S. A 118 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW, 2009. Cell growth and size homeostasis in proliferating animal cells. Science 325 (5937), 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassis N, Pacheco MP, Sauter T, 2014. Fast reconstruction of compact context- specific metabolic network models. PLOS Comp. Biol 10 (1), e1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li M, Zhang C, Cheng H, Gao Y, Deng W, Li T, 2020a. Transcriptome and proteome analyses reveal the regulatory networks and metabolite biosynthesis pathways during the development of tolypocladium guangdongense. Comput. Struct. Biotechnol. J 18, 2081–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Belta C, Isaacson SA, 2020b. How retroactivity affects the behavior of incoherent feedforward loops. iScience 23 (12), 101779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Isaacson SA, Belta C, 2019. Modeling genetic circuit behavior in transiently transfected mammalian cells. ACS Synth. Biol 8 (4), 697–707. [DOI] [PubMed] [Google Scholar]
- Warburg O, 1956. On the origin of cancer cells. Science 123 (3191), 309–314. [DOI] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik J. h., Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA, 2012. Oncogenic kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149 (3), 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JKV, Markowitz S, Zhou S, Diaz Jr, Luis A, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N, 2009. Glucose deprivation contributes to the development of kras pathway mutations in tumor cells. Science 325 (5947), 1555–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharakis M, Xynos ID, Lazaris A, Smaro T, Kosmas C, Dokou A, Felekouras E, Antoniou E, Polyzos A, Sarantonis J, Syrios J, Zografos G, Papalambros A, Tsavaris N, 2010. Predictors of survival in stage iv metastatic colorectal cancer. Anticancer Res. 30 (2), 653–660. [PubMed] [Google Scholar]
- Zhang Z, Liu R, Shuai Y, Huang Y, Jin R, Wang X, Luo J, 2020. Asct2 (slc1a5)- dependent glutamine uptake is involved in the progression of head and neck squamous cell carcinoma. Br. J. Cancer 122 (1), 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhao X, Chen V, Feng Y, Wang L, Croniger C, Conlon RA, Markowitz S, Fearon E, Puchowicz M, Brunengraber H, Hao Y, Wang Z, 2019. Colorectal cancers utilize glutamine as an anaplerotic substrate of the tca cycle in vivo. Sci. Rep 9 (1), 19180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski DC, Jamshidi N, Corbett AJ, Bordbar A, Thomas A, Palsson BO, 2017. Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism. Sci. Rep 7, 41241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.