Sir,
Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacterales (KPC-EB) represent a major threat because of their wide spread in healthcare facilities, difficult-to-treat multi-drug-resistant phenotypes, and association with high mortality rates. Active surveillance by screening hospitalized patients in high-risk wards using rectal swabs represents the practice recommended by international guidelines to counteract the spread of these multi-drug-resistant strains [1,2].
Ceftazidime-avibactam (CZA) has demonstrated both excellent in vitro and in vivo activity against KPC-EB. However, there is increasing evidence of in vivo selection of CZA-resistant strains [3]. The D179Y mutation in the omega loop domain of the KPC enzyme is the most frequently encountered mechanism to date and is associated with relevant issues regarding its phenotypic detection. Indeed, this mutation in KPC-2 and KPC-3 is associated with weak carbapenemase activity and expression of extended-spectrum-β-lactamase phenotype. This results in the inability of the main phenotypic carbapenemase detection methods to identify these KPC mutants. Immunochromatographic assays are also unable to detect these KPC mutants, probably due to changes in the antigenic set-up of the enzyme [4].
Herein, we evaluated the prevalence of rectal colonization by carbapenemase-producing Enterobacterales (EB) and rate of CZA resistance among KPC-EB isolates in a cohort of patients admitted to an intensive care unit (ICU) during a six-month period. Furthermore, we evaluated the performance of five phenotypic methods recommended by EUCAST and CLSI to detect main carbapenemases in EB.
The examined department underwent conversion to COVID-19 ICU after about three months from the beginning of the study. Rectal screening swabs, collected for new admissions and for inpatients on a weekly basis using the Fecal Swab™ system (Copan, Brescia, Italy), were inoculated on Brilliance CRE medium (Oxoid Ltd, Hampshire, UK) by automated direct plating using Wasp® instrument (Copan, Brescia, Italy). Bacterial species identification was performed on overnight colonies using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) analysis (Bruker DALTONIK GmbH, Bremen, Germany) and carbapenemase production in EB isolates was investigated by the genotypic assay Xpert Carba-R on GeneXpert platform (Cepheid, Sunnyvale, CA, USA). KPC-EB isolates were tested for susceptibility to CZA by E-test method (BioMérieux, Marcy l’Ètoile, France) whereas susceptibility to major antimicrobials was performed using a commercial microdilution system (Panel NMDR, Microscan WalkAway 96 Plus, Beckman Coulter, Switzerland). KPC-producing Klebsiella pneumoniae (KPC-Kp) was the predominant carbapenemase-producing EB identified by rectal screening (41 of 42, 97.6%; of which one isolate was Verona Integron-encoded Metallo-β-lactamase (VIM)-co-producer). Overall, 21 patients were colonized by KPC-Kp resistant to CZA (minimum inhibitory concentration (MIC) >256 mg/L), of which 11 (52.4%) were COVID-19 patients.
Among the 21 patients colonized by CZA-resistant KPC-Kp, 19 had no previous documented colonization by CZA-susceptible KPC-Kp and were not exposed to CZA. The mean time to positivity for CZA-resistant KPC-Kp was 12 days after ICU admission (median value seven days, range three to 70 days). Overall, CZA-resistant KPC-Kp isolates in other sites (blood culture N = 3; bronchoalveolar lavage N = 3) after nine to 69 days (mean 24.2 days) from the date of rectal colonization were documented in six patients.
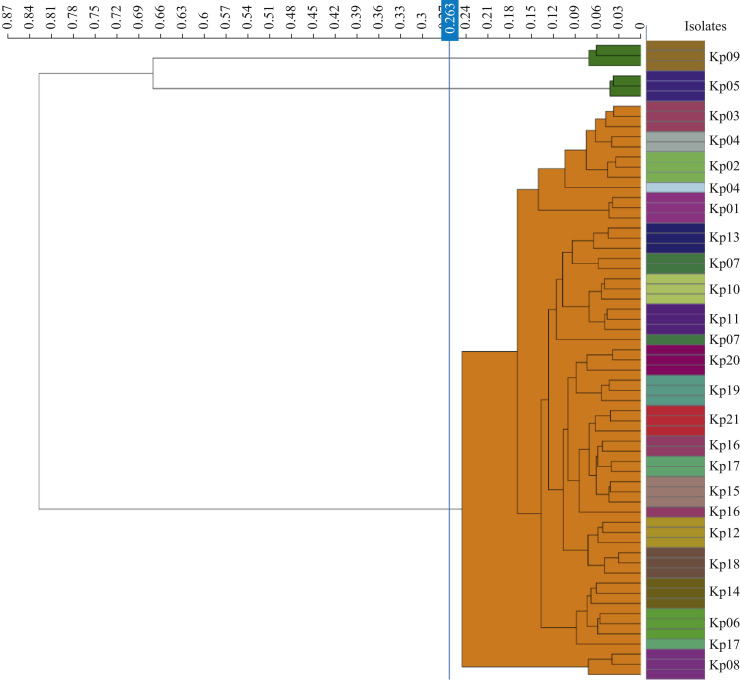
The 21 CZA-resistant KPC-Kp isolates expressed an extended-spectrum-β-lactamase phenotype, showing susceptibility towards carbapenems (meropenem MIC 2 mg/L, imipenem ≤1 mg/L, ertapenem 1 mg/L). All phenotypic carbapenemase detection methods (CarbaNP, Disc Diffusion Synergy test (KPC, MBL and OXA-48 Confirm Kit, Rosco Diagnostica), modified carbapenem inactivation method (mCIM), and lateral flow immunoassay (NG-Test CARBA 5- NG Biotech, France; RESIST-5 O.O.K.N.V-Coris Bioconcept, Belgium) failed to detect KPC carbapenemases conferring CZA resistance. Cluster analysis using Fourier-transform infrared (IR) spectroscopy on IR Biotyper system (Bruker GmbH, Bremen, Germany) was performed as previously described [5]. IR biotyping showed that 19 of the 21 CZA-resistant KPC-Kp isolates belonged to a single outbreak clone (Figure 1 ). Sequencing of the bla KPC gene was performed as previously described [4] and revealed D179Y mutation in the bla KPC-2 gene.
Figure 1.
Fourier-transform infrared spectroscopy biotyping of ceftazidime-avibactam-resistant Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae isolates. The blue line represents the automatic cut-off value (0.263). In the dendogram, isolates belonging to the cluster are shaded orange, singletons are shaded green. The specimens were analysed in triplicate using the IR Byotyper as previously described (Rakovitsky et al., J Clin Microbiol 2020;58:e00098-20).
This study was conducted in accordance with the Declaration of Helsinki. Formal ethical approval was obtained by our Center's institutional review board.
The highlights of this study are the following findings: (1) intra-hospital spread of CZA-resistant KPC-33 producing isolates was relevant; (2) EB harboring KPCD179Y escape screening of surveillance cultures if phenotypic carbapenemase detection methods are used. Considering isolates expressing KPC mutants with impaired carbapenemase activity as alert bacterial strains for which infection control measures should be undertaken is still uncertain. Indeed, these strains do not express a wide multi-drug resistance pattern since they remain susceptible to carbapenems. However, we speculate that the spread of these KPC mutants located on resistance plasmids, might co-habit together with the wild-type KPCs soon, generating extended-drugs-resistant phenotypes. Moreover, it should be also considered that KPCD179Y and other KPC mutants have been associated with co-resistance towards CZA and the recently approv=ed cefiderocol [6].
In conclusion, international guidelines regarding methods to screen carbapenemase-producing EB and to detect carbapenemases require an urgent update, especially after the SARS-CoV-2 pandemic and its impact on incessant hospitals reorganization and antimicrobial prescription [7]. In areas and hospital settings with high circulation of KPC mutants, genotypic carbapenemase detection methods should be preferred and recommended. If they are not available, screening for CZA resistance on isolates grown on selective medium and tested negative to phenotypic carbapenemase detection tests could be a viable alternative. CZA resistance screening could be alternatively performed using traditional antimicrobial susceptibility testing or implementing the use of CZA-based selective medium [8].
Conflict of interest statement
The authors have no conflicts of interest to declare.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Centers for Disease Control and Prevention . 2015. Facility guidance for control of carbapenem-resistant Enterobacteriaceae (CRE) November 2015 update – CRE Toolkit.https://www.cdc.gov/hai/pdfs/cre/cre-guidance-508.pdf Available at: [last accessed December 2021] [Google Scholar]
- 2.Tacconelli E., Cataldo M.A., Dancer S.J., De Angelis G., Falcone M., Frank U., et al. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin Microbiol Infect. 2014;20:1–55. doi: 10.1111/1469-0691.12427. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Wang J., Wang R., Cai Y. Resistance to ceftazidime-avibactam and underlying mechanisms. J Glob Antimicrob Resist. 2020;22:18–27. doi: 10.1016/j.jgar.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Bianco G., Boattini M., Iannaccone M., Bondi A., Ghibaudo D., Zanotto E., et al. Carbapenemase detection testing in the era of ceftazidime/avibactam-resistant KPC-producing Enterobacterales: a 2-year experience. J Glob Antimicrob Resist. 2021;24:411–414. doi: 10.1016/j.jgar.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Rakovitsky N., Frenk S., Kon H., Schwartz D., Temkin E., Solter E., et al. Fourier transform infrared spectroscopy is a new option for outbreak investigation: a retrospective analysis of an extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae outbreak in a neonatal intensive care unit. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00098-20. e00098-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianco G., Boattini M., Comini S., Iannaccone M., Bondi A., Cavallo R., et al. In vitro activity of cefiderocol against ceftazidime-avibactam susceptible and resistant KPC-producing Enterobacterales: cross-resistance and synergistic effects. Eur J Clin Microbiol Infect Dis. 2022;41:63–70. doi: 10.1007/s10096-021-04341-z. [DOI] [PubMed] [Google Scholar]
- 7.Arcari G., Raponi G., Sacco F., Bibbolino G., Di Lella F.M., Alessandri F., et al. Klebsiella pneumoniae infections in COVID-19 patients: a 2-month retrospective analysis in an Italian hospital. Int J Antimicrob Agents. 2021;57:106245. doi: 10.1016/j.ijantimicag.2020.106245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadek M., Poirel L., Tinguely C., Nordmann P. A selective culture medium for screening ceftazidime-avibactam resistance in Enterobacterales and Pseudomonas aeruginosa. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00965-20. e00965-20. Erratum in: J Clin Microbiol 2020;58: PMID: 32580951. [DOI] [PMC free article] [PubMed] [Google Scholar]