Abstract
We have developed a PCR-based assay which allows the detection of staphylococci at the genus level by targeting the tuf gene, which encodes the elongation factor Tu. Degenerate PCR primers derived from consensus regions of several tuf genes were used to amplify a target region of 884 bp from 11 representative staphylococcal species. Subsequently, the entire nucleotide sequence of these amplicons was determined. The analysis of a multiple alignment of these sequences revealed regions conserved among staphylococci but distinct from those of other gram-positive bacteria genetically related to staphylococci. PCR primers complementary to these regions could amplify specifically and efficiently a DNA fragment of 370 bp for all of 27 different staphylococcal species tested. There was no amplification with genomic DNA prepared from 53 nonstaphylococcal species tested to verify the specificity of the assay (20 gram positive and 33 gram negative). Furthermore, this assay amplified efficiently all 27 American Type Culture Collection (ATCC) staphylococcal reference strains as well as 307 clinical isolates of staphylococci from the Québec City region. Analysis of the multiple sequence alignment for the 884-bp fragment for the 11 staphylococcal species as well as comparison of the sequences for the 370-bp amplicon from five unrelated ATCC and clinical strains for each of the species S. aureus, S. epidermidis, S. haemolyticus, S. hominis, and S. saprophyticus demonstrated sufficient interspecies polymorphism to generate genus- and species-specific capture probes. This sequence information allowed the development of Staphylococcus-specific and species-specific (targeting S. aureus, S. epidermidis, S. haemolyticus, S. hominis, or S. saprophyticus) capture probes hybridizing to the 370-bp amplicon. In conclusion, this PCR assay is suitable for detection of staphylococci at both genus and species levels.
Staphylococci are one of the most common nosocomial pathogens throughout the world. The incidence of staphylococcal infections has increased in recent years, mainly due to the spreading of multidrug-resistant staphylococcal strains as well as to the growing incidence of infections in immunocompromised patients and in medical-device-related infections (11). Among staphylococci, S. aureus, S. epidermidis, and S. saprophyticus have the greatest pathogenic potential and diversity. S. aureus is responsible for diseases caused by exotoxin production (toxic shock and staphylococcal scalded-skin syndromes) and by direct invasion and systemic dissemination (bacteremia, urinary tract infections [UTI], and septic shock syndrome) (5, 32). For its part, S. epidermidis is widely recognized as an etiologic agent of bacteremia, prosthetic and natural valvular endocarditis, osteomyelitis, UTI, and peritonitis caused by ambulatory dialysis, with a frequent association with the colonization of intravascular catheters and orthopedic devices (5, 32). S. saprophyticus is the second most frequently encountered organism after coliform bacilli (Escherichia coli) in acute UTI (13, 28). S. saprophyticus is often isolated from sexually active young females presenting symptoms of acute UTI (33). Other staphylococcal species (e.g., S. haemolyticus, S. hominis, and S. lugdunensis) are usually found as contaminants of blood cultures but could also be associated with a variety of infections (7, 11, 20).
Several manual and automated methods for the identification of staphylococci are commercially available. These methods include the API identification systems (bioMérieux Vitek) (1, 21) and automated systems such as the MicroScan system (Dade Behring Inc., Deerfield, Ill.) (14). Conventional methods based on biochemical tests to identify staphylococci are often lengthy (at least 30 h) because they require microbial growth. Furthermore, commercially available panels, which are based on functional differences in metabolic pathways, do not allow a reliable distinction between different coagulase-negative staphylococci (CNS). A number of PCR-based methods for the species-specific detection of S. aureus, S. epidermidis, and S. saprophyticus have been reported (4, 8, 26, 27, 31, 35). Regarding PCR assays for detection of staphylococci at the genus level, the 16S rRNA or the HSP60 genes have been used as targets (9, 10). The tuf gene, which encodes the elongation factor Tu (EF-Tu), is involved in peptide chain formation and is an essential constituent of the bacterial genome (12). This fact makes it a target of choice for diagnostic purposes. PCR-based assays in which the tuf gene served as the target sequence have been developed for the genus Enterococcus (17), Mycoplasma fermentans (2), and Mycoplasma pneumoniae (23). We report here the development of a Staphylococcus-specific PCR assay targeting the tuf gene which can detect 27 staphylococcal species with excellent sensitivity and specificity. This PCR assay was coupled with post-PCR hybridization of capture probes allowing Staphylococcus-specific and species-specific detection of S. aureus, S. epidermidis, S. haemolyticus, S. hominis, and S. saprophyticus.
(This study was partially presented at the 99th General Meeting of the American Society for Microbiology, Chicago, Ill., 30 May to 3 June 1999.)
MATERIALS AND METHODS
Bacterial strains.
Twenty-seven staphylococcal strains obtained from the American Type Culture Collection (ATCC) (Manassas, Va.) were used in this study (Table 1). An additional 307 clinical isolates representing 11 different species of staphylococci (S. aureus [n = 36], S. auricularis [n = 3], S. capitis [n = 20], S. cohnii [n = 6], S. epidermidis [n = 19], S. haemolyticus [n = 22], S. hominis [n = 74], S. lugdunensis [n = 18], S. saprophyticus [n = 7], S. simulans [n = 4], S. warneri [n = 33], and Staphylococcus spp. [n = 65]) obtained from the Centre Hospitalier Universitaire de Québec (Pavillon Centre Hospitalier de l'Université Laval, Sainte-Foy, Québec, Canada) were also used in this study.
TABLE 1.
Bacterial strains used in this study
| Species | |
|---|---|
| Staphylococcal (n = 27) | |
| Staphylococcus arlettae ATCC 43957 | |
| Staphylococcus aureus subsp. anaerobius ATCC 35844 | |
| Staphylococcus aureus subsp. aureus ATCC 43300 | |
| Staphylococcus auricularis ATCC 33753 | |
| Staphylococcus capitis subsp. capitis ATCC 27840 | |
| Staphylococcus caprae ATCC 35538 | |
| Staphylococcus carnosus ATCC 51365 | |
| Staphylococcus chromogenes ATCC 43764 | |
| Staphylococcus cohnii subsp. urealyticum DSM 20260 | |
| Staphylococcus delphini ATCC 49171 | |
| Staphylococcus epidermidis ATCC 14990 | |
| Staphylococcus equorum ATCC 43958 | |
| Staphylococcus felis ATCC 49168 | |
| Staphylococcus gallinarum ATCC 35539 | |
| Staphylococcus haemolyticus ATCC 29970 | |
| Staphylococcus hominis ATCC 27844 | |
| Staphylococcus hyicus ATCC 11249 | |
| Staphylococcus intermedius ATCC 29663 | |
| Staphylococcus kloosis ATCC 43959 | |
| Staphylococcus lentus ATCC 29070 | |
| Staphylococcus lugdunensis ATCC 43809 | |
| Staphylococcus saprophyticus ATCC 15305 | |
| Staphylococcus schleiferi subsp. coagulans ATCC 49545 | |
| Staphylococcus sciuri subsp. sciuri ATCC 29060 | |
| Staphylococcus simulans ATCC 27848 | |
| Staphylococcus warneri ATCC 27836 | |
| Staphylococcus xylosus ATCC 29971 | |
| Other gram-positive bacteria (n = 20) | |
| Bacillus subtilis ATCC 27370 | |
| Enterococcus avium ATCC 14025 | |
| Enterococcus durans ATCC 19432 | |
| Enterococcus faecalis ATCC 19433 | |
| Enterococcus faecium ATCC 19434 | |
| Enterococcus flavescens ATCC 49573 | |
| Enterococcus gallinarum ATCC 49573 | |
| Lactobacillus acidophilus ATCC 4356 | |
| Lactococcus lactis ATCC 11454 | |
| Listeria innocua ATCC 33090 | |
| Listeria ivanovii ATCC 19119 | |
| Listeria monocytogenes ATCC 15313 | |
| Macrococcus caseolyticus ATCC 13548 |
| Streptococcus agalactiae ATCC 13813 |
| Streptococcus anginosus ATCC 33397 |
| Streptococcus bovis ATCC 33317 |
| Streptococcus mutans ATCC 25175 |
| Streptococcus pneumoniae ATCC 6303 |
| Streptococcus pyogenes ATCC 19615 |
| Streptococcus salivarius ATCC 7073 |
| Gram-negative bacteria (n = 33) |
| Acinetobacter baumannii ATCC 19606 |
| Bacteroides distasonis ATCC 8503 |
| Bacteroides fragilis ATCC 25285 |
| Bordetella pertussis ATCC 9797 |
| Burkholderia cepacia ATCC 25416 |
| Citrobacter freundii ATCC 8090 |
| Enterobacter aerogenes ATCC 13048 |
| Enterobacter cloacae ATCC 13047 |
| Escherichia coli ATCC 25922 |
| Haemophilus influenzae ATCC 8847 |
| Haemophilus parahaemolyticus ATCC 10014 |
| Haemophilus parainfluenzae ATCC 7901 |
| Hafnia alvei ATCC 13337 |
| Kingella indologenes ATCC 25869 |
| Klebsiella oxytoca ATCC 13182 |
| Klebsiella pneumoniae ATCC 13883 |
| Moraxella catarrhalis ATCC 25240 |
| Morganella morganii ATCC 25830 |
| Neisseria gonorrhoeae ATCC 35201 |
| Neisseria meningitidis ATCC 13077 |
| Proteus mirabilis ATCC 25933 |
| Proteus vulgaris ATCC 13315 |
| Providencia rettgeri ATCC 9250 |
| Providencia stuartii ATCC 29914 |
| Pseudomonas aeruginosa ATCC 27853 |
| Pseudomonas fluorescens ATCC 13525 |
| Salmonella enterica serovar Choleraesuis ATCC 7001 |
| Salmonella enterica serovar Typhimurium ATCC 14028 |
| Serratia marcescens ATCC 8100 |
| Shigella flexneri ATCC 12022 |
| Shigella sonnei ATCC 29930 |
| Stenotrophomonas maltophilia ATCC 13843 |
| Yersinia enterocolitica ATCC 9610 |
The specificity of the PCR-based assay was verified by using a battery of ATCC bacterial strains consisting of 47 gram-positive species (including 27 staphylococcal species) and 33 gram-negative species (Table 1). The 307 clinical isolates of staphylococci from the Centre Hospitalier de l'Université Laval were also tested to further validate the Staphylococcus-specific PCR-based assay. The reference strains as well as the clinical isolates were all identified with the automated MicroScan Autoscan-4 system equipped with the Positive BP Combo Panel Type 6 (Dade-Behring, Mississauga, Ontario, Canada). Bacterial strains were grown from frozen stocks kept at −80°C in brain heart infusion medium containing 10% glycerol and were cultured on sheep blood agar.
PCR primers.
The tuf gene sequences available from public databases were analyzed with Genetics Computer Group (Madison, Wis.) (GCG) programs (version 8.0). Based on multiple sequence alignments, regions of the tuf gene highly conserved among eubacteria were chosen, and PCR primers (Tseq271 and Tseq1138) (Table 2) were derived from these regions using the Oligo primer analysis software (version 5.0; National Biosciences, Plymouth, Minn.). When required, the universal primers contained inosines or degeneracies at one or more variable positions. Oligonucleotide primers were synthesized with a model 394 DNA/RNA Synthesizer (Applied Biosystems, Mississauga, Ontario, Canada).
TABLE 2.
PCR primers used in this study
| Primer | Primer sequenceb | Nucleotide positionsa | Amplicon size (bp) |
|---|---|---|---|
| Universal amplification | 884 | ||
| Tseq271 | 5′-AAY ATG ATI ACI GGI GCI GCI CAR ATG GA-3′ | 271–300 | |
| Tseq1138 | 5′-CCI ACI GTI CKI CCR CCY TCR CG-3′ | 1132–1155 | |
| Staphylococcus specific | 370 | ||
| TStaG422 | 5′-GGC CGT GTT GAA CGT GGT CAA ATC A-3′ | 422–446 | |
| TStag765 | 5′-TIA CCA TTT CAG TAC CTT CTG GTA A-3′ | 765–792 |
The given nucleotide positions are for the E. coli tuf sequence (accession no. J01690).
I, nucleotide analog inosine; K, nucleotides E and T; R, nucleotides A and G; Y, nucleotides C and T.
DNA sequencing.
An 884-bp portion of the tuf gene was sequenced for 11 staphylococcal species: S. aureus, S. auricularis, S. capitis, S. cohnii, S. epidermidis, S. haemolyticus, S. hominis, S. lugdunensis, S. saprophyticus, S. simulans, and S. warneri. Amplification using universal primers (Table 2) was performed from 1 ng of purified genomic DNA as previously described (17). PCR products having the predicted sizes were recovered from an agarose gel stained 15 min with 0.02% methylene blue (Laboratoire MAT, Beauport, Québec, Canada) followed by washing in sterile distilled water for 15 min twice (6). The 884-bp PCR products were recovered from the agarose gel using the QIAquick gel extraction kit (Qiagen Inc., Mississauga, Ontario, Canada). Both strands of the amplicons were sequenced directly with the PRISM Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit using an Applied Biosystems 373A sequencer (Applied Biosystems, Foster City, Calif.). In order to exclude the possibility of sequencing errors attributable to misincorporations by Taq DNA polymerase, each strand was sequenced twice using the PCR products obtained from two independent PCRs.
PCR amplification.
For all bacterial species, amplification was performed from purified genomic DNA or from a bacterial suspension whose turbidity was adjusted to that of a 0.5 McFarland standard (17), except that a 0.2 μM concentration of each of the Staphylococcus-specific primers was used (Table 2). An internal control was integrated into the PCR-based assays to verify the efficiency of the amplifications and to ensure that significant PCR inhibition was absent (17). The Superlinker phagemid pSL1180 (Amersham Pharmacia Biotech, Inc., Baie d'Urfé, Quebec, Canada) linearized by digestion with EcoRI (New England Biolabs, Ltd., Missisauga, Ontario, Canada) and PCR primers derived from the multiple cloning site of the plasmid were used to provide an internal control. As previously described (17), the concentrations of the internal control primers were adjusted to ensure that there was no detrimental effect on the Staphylococcus-specific amplification. We found that concentrations of 0.1 and 0.04 μM were optimal for 30- and 40-cycle PCRs, respectively.
The specificity of the conventional PCR assay was verified by using purified genomic DNA (0.1 ng per reaction) from a battery of ATCC reference strains representing a wide variety of gram-positive (47 species from 8 genera) and gram-negative (33 species from 22 genera) bacterial species (Table 1). A total of 307 clinical strains of staphylococci were also tested to further validate the ubiquity (i.e., the ability to detect all staphylococcal strains) of the Staphylococcus-specific PCR assay. A portion of the amplified PCR mixtures was first visualized on agarose gel under UV light.
Strict precautions to prevent carryover of amplified DNA were used (22). Pre- and post-PCR manipulations were conducted in separate areas. Aerosol-resistant pipette tips were used to handle all reagents and samples. Control reactions to which no DNA was added were routinely performed to verify the absence of DNA carryover.
Phylogenetic analysis.
Multiple sequence alignments were performed using PILEUP from the GCG package (version 10.0) and checked with the editor SeqLab to edit sequences if necessary and to note which regions were to be excluded for phylogenetic analysis. Bootstrap subsets (500 sets) and phylogenetic trees were generated with the neighbor-joining algorithm from David Swofford's PAUP (phylogenetic analysis using parsimony) software (version 4.0b4; Sinauer Associates) and with tree bisection branch swapping. Evolutionary distance values were calculated by Kimura's two-parameter method. The maximum-parsimony analysis was also performed as a heuristic search with tree bisection branch swapping. All trees were resampled with 100 bootstrap replications to test the robustness of the data.
Design of capture probes.
Analysis of the multiple-sequence alignment for the 884-bp portion of the tuf gene for 11 staphylococcal species allowed us to identify regions suitable for the design of species-specific (for S. aureus, S. epidermidis, S. haemolyticus, S. hominis, or S. saprophyticus) or Staphylococcus-specific capture probes. Discriminating mismatches were in the middle of the probes whenever possible. Furthermore, we have also compared sequences for the 370-bp amplicon from five unrelated ATCC and clinical strains for each of the species S. aureus, S. epidermidis, S. haemolyticus, S. hominis, and S. saprophyticus to confirm that chosen regions were well conserved within each target species. All capture probes were designed to have similar melting temperatures to allow uniform hybridization conditions. Oligonucleotides were synthesized with a model 394 DNA/RNA Synthesizer (Applied Biosystems). Capture probe sequences used in this study are listed in Table 3.
TABLE 3.
Hybridization internal probes used in this study
| Probe and specificity | Sequence (5′ to 3′) |
|---|---|
| Staphylococcus-specific | ATT AGA CTA CGC TGA AGC TG |
| Staphylococcus species specific | |
| S. aureus | CGT ATT ATC AAA AGA CGA AG |
| S. epidermidis | CAA AGC TGA AGT ATA CGT AT |
| S. haemolyticus | ATT GGT ATC CAT GAC ACT TC |
| S. hominis | ATT GGT ATC AAG AAA CTT C |
| S. saprophyticus | ATG CAA GAA GAA TCA AGC AA |
PCR amplification and capture probe hybridization.
A 20-μl PCR mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% Triton X-100, 2.5 mM MgCl2, a 0.2 μM concentration of each Staphylococcus-specific primers (TStaG422 and TStaG765) (Table 2), 0.1× PCR digoxigenin (DIG) labeling mix (Roche Diagnostics, Laval, Quebec, Canada), bovine serum albumin (3.3 μg/μl; Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada), and 0.5 U of Taq DNA polymerase (Promega Corp., Madison, Wis.) coupled with the TaqStart Antibody (Clontech Laboratories Inc., Palo Alto, Calif.). For all bacterial species, amplification was performed from purified genomic DNA (0.1 ng per reaction) (17). The 40-cycle PCR amplification and the agarose gel analysis of the amplified products were performed as previously described (26).
Each capture probe was solubilized in 30 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) (Sigma-Aldrich) at a concentration of 0.5 μM. Subsequently, 50 μl of the probe solution was added into each well of a 96-well plate (Microlite 2 flat-bottom microtiter plates; DYNEX Technologies, Inc., Chantilly, Va.) and incubated in the dark overnight to bind noncovalently the oligonucleotide probes. The cationic detergent EDC facilitates the immobilization of the oligonucleotides onto polystyrene surfaces (30). The capture probe solution was discharged, and then each well was filled with 50 μl of saturation buffer (10 mM Tris-HCI [pH 7.5], 150 mM NaCl, 0.05% Tween 20) containing 1% bovine serum albumin for 1 h at 37°C. All washing and incubation steps at room temperature were performed using an automatic plate washer (Wellwash Ascent washer; Labsystem Oy, Helsinki, Finland). Subsequently, the plate was rinsed twice with the saturation buffer and then twice with a washing buffer (100 mM maleic acid, 150 mM NaCl, 0.3% Tween 20 [pH 7.5]).
PCR amplicons labeled with DIG-11-dUTP were denatured by boiling for 4 min. One microliter of the amplicons was added to 49 μl of hybridization solution (1.5 M NaCl, 10 mM EDTA) and then incubated at 45°C for 30 min. Subsequently, each well was washed twice with 200 μl of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% sodium dodecyl sulfate at room temperature and then washed twice with 200 μl of 0.1× SSC plus 0.1% SDS at 45°C four times. Blocking solution (50 μl per well; Roche Diagnostics) was then added, and the plates were incubated for 5 min. The blocking solution containing freshly added anti-DIG alkaline phosphatase (1:1,000; Roche Diagnostics) was subsequently used (50 μl/well), and the plate was incubated for 15 min. The plate was subsequently rinsed twice with 200 μl of washing buffer for 5 min. After a 2-min incubation in 200 μl of detection buffer (100 mM Tris, 100 mM NaCl [pH 9.5]), 20 μl of CSPD ready-to-use solution (Roche Diagnostics) was added into each well and the plate was incubated at room temperature for 5 min followed by 10 min at 37°C. The chemiluminescence signals of each well were read using a microtiter plate luminometer (MLX Dynex Technologies) and recorded in relative light units. The relative light unit ratio of tested sample to the corresponding blank control was then calculated. A ratio greater than 2 was interpreted as a positive hybridization signal. All reactions were performed in duplicate because of occasional well-to-well variations.
Sensitivity tests.
To determine the detection limits (i.e., minimal number of genome copies that can be detected) of the Staphylococcus-specific 30- and 40-cycle PCR assays, serial twofold dilutions of purified genomic DNA from 11 staphylococcal species (S. aureus, S. auricularis, S. capitis, S. cohnii, S. epidermidis, S. haemolyticus, S. hominis, S. lugdunensis, S. saprophyticus, S. simulans, and S. warneri) were tested. To determine the detection limits with capture probes, two ATCC strains of each of the five selected species (S. aureus, S. epidermidis, S. haemolyticus, S. hominis, and S. saprophyticus) were used. The sensitivities of the assays in microtiter plates were compared with those obtained by gel electrophoresis.
Nucleotide sequence accession numbers.
GenBank accession numbers for the partial sequences of the staphylococcal tuf gene (excluding the sequences of the two universal amplification primers) are as follows: AF298796 for S. aureus, AF298797 for S. auricularis, AF298798 for S. capitis, AF298799 for S. cohnii, AF298800 for S. epidermidis, AF298801 for S. haemolyticus, AF298802 for S. hominis, AF298803 for S. lugdunensis, AF298804 for S. saprophyticus, AF298805 for S. simulans, and AF298806 for S. warneri.
RESULTS
Sequencing of a portion of the tuf gene from 11 staphylococcal species.
By using the universal primers Tseq271 and Tseq1138, we were able to amplify the 884-bp portion of tuf for 11 clinically relevant and representative staphylococcal species: S. aureus, S. auricularis, S. capitis, S. cohnii, S. epidermidis, S. haemolyticus, S. hominis, S. lugdunensis, S. saprophyticus, S. simulans, and S. warneri. After purification from agarose gels, the 884-bp PCR product was sequenced for each of the 11 staphylococcal species. In order to facilitate the selection of Staphylococcus-specific primers, we conducted a multiple-sequence analysis using bacterial tuf sequences available in public databases (17). By using this approach, we were able to identify regions conserved in the 11 staphylococcal species but variable in other bacterial species. The Staphylococcus-specific PCR primers were derived from these discriminant regions. A similarity comparison of the tuf sequences for the 11 staphylococcal species showed that the sequence similarities for the 884-bp fragment in these species range from 89 to 97% (Table 4).
TABLE 4.
DNA sequence identities of the staphylococcal tuf genesa
| tuf gene | % Identity with tuf gene from:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. aureus | S. auricularis | S. capitis | S. cohnii | S. epidermidis | S. haemolyticus | S. hominis | S. lugdunensis | S. saprophyticus | S. simulans | S. warneri | |
| S. aureus | 89 | 95 | 90 | 94 | 95 | 94 | 93 | 91 | 89 | 95 | |
| S. auricularis | 87 | 91 | 91 | 92 | 88 | 90 | 88 | 91 | 90 | 90 | |
| S. capitis | 93 | 89 | 92 | 97 | 96 | 95 | 93 | 92 | 92 | 96 | |
| S. cohnii | 86 | 91 | 90 | 92 | 90 | 90 | 90 | 96 | 89 | 90 | |
| S. epidermidis | 93 | 90 | 96 | 90 | 95 | 94 | 93 | 92 | 92 | 94 | |
| S. haemolyticus | 94 | 87 | 95 | 87 | 94 | 97 | 94 | 91 | 90 | 95 | |
| S. hominis | 92 | 89 | 94 | 88 | 94 | 96 | 94 | 90 | 91 | 94 | |
| S. lugdunensis | 90 | 86 | 93 | 87 | 92 | 94 | 95 | 90 | 90 | 93 | |
| S. saprophyticus | 86 | 90 | 89 | 93 | 89 | 87 | 87 | 86 | 91 | 91 | |
| S. simulans | 86 | 88 | 90 | 88 | 90 | 87 | 88 | 87 | 90 | 91 | |
| S. warneri | 94 | 88 | 97 | 88 | 94 | 94 | 92 | 90 | 88 | 89 | |
The numbers in the upper-right triangle are the percentage of identity for the 812- to 815-bp sequences from the 884-bp fragment, and those in the lower-left triangle represent the percentage of identity for the 320-bp sequences flanked by the two Staphylococcus-specific PCR primers. The identity scores were obtained by use of the PILEUP program of the GCG software package.
The selected Staphylococcus-specific PCR primers revealed no more than one mismatch within the tuf sequence from the 11 staphylococcal species. Importantly, at least three mismatches were found in the corresponding regions of the other nonstaphylococcal bacterial species. Since the mismatches with tuf sequences from other bacteria were clustered at the 3′ end of the primers, a position critical for discriminatory PCR amplification, the amplification of bacterial species other than staphylococci could be efficiently prevented.
Amplifications with the Staphylococcus-specific PCR assay.
The specificity of the assay was assessed by performing 30- and 40-cycle PCR amplifications with the panel of species listed in Table 1. The PCR assay was able to detect all of the 27 staphylococcal species tested in both 30- and 40-cycle regimens. For 30-cycle PCR, all bacterial species tested other than staphylococci were negative. For 40-cycle PCR, DNA from Enterococcus faecalis and Macrococcus caseolyticus gave a weakly positive amplification signal (approximately 1,000 to 10,000 times less efficiently than the amplification of staphylococcal DNA). All other species tested remained negative. The internal control was always efficiently amplified when no target DNA was present, thereby showing the absence of significant PCR inhibition. On the other hand, the internal control was not amplified when target DNA was present in a sample. This is explained by the limiting concentration of the internal control primers in order to favor amplification of targeted staphylococcal DNA. Tests performed with a collection of 307 clinical isolates showed a uniform amplification signal for all strains with both the 30- and the 40-cycle PCR assays.
For PCR with 30 cycles of amplification, we found a detection limit of ∼50 genome copies (deduced from the genome size of S. aureus), for all staphylococcal species tested. For PCR assays with 40 cycles of amplification, the detection limit was lowered to a range of 5 to 10 genome copies. The detection limits for the Staphylococcus-specific 40-cycle PCR assay coupled with posthybridization with internal probes was also 5 to 10 genome copies. Therefore, amplicon detection by ethidium bromide-stained agarose gels or by capture probe hybridization yielded similar sensitivities.
Capture probe hybridization to the Staphylococcus-specific amplicons.
A Staphylococcus-specific capture probe as well as S. aureus-, S. epidermidis-, S. haemolyticus-, S. hominis-, and S. saprophyticus-specific capture probes were tested. The Staphylococcus-specific probe hybridized efficiently to amplicons generated from all 11 staphylococcal genomic DNAs tested and did not cross-react with amplicons produced by amplification of E. faecalis and M. caseolyticus, which were both weakly amplified by the Staphylococcus-specific PCR assay.
DISCUSSION
The most frequently encountered staphylococcal species in humans include S. aureus, S. auricularis, S. capitis, S. cohnii, S. epidermidis, S. haemolyticus, S. hominis, S. lugdunensis, S. saprophyticus, S. simulans, and S. warneri (18). Conventional identification of some CNS species is difficult because no phenotypic criteria are available to unequivocally distinguish them (19–21, 27). Furthermore, such tests require 24 to 48 h to provide results after pure cultures are obtained. Even with automated systems, an incubation of up to 24 h is required for CNS species identification, and some additional tests may be required (14, 21, 27).
The development of sensitive DNA-based assays for the identification of staphylococci isolated from clinical specimens may improve the rapidity and the accuracy of the diagnosis of staphylococcal infections. In the present study, we have developed a Staphylococcus-specific PCR-based assay targeting the tuf gene. Based on testing with DNA from a variety of bacterial species, this PCR assay was shown to be specific and sensitive for all 27 staphylococcal species tested. There was some weak cross-amplification (1,000 to 10,000 times less efficiently than staphylococcal DNA) with DNA from E. faecalis and M. caseolyticus observed in the 40-cycle regimen.
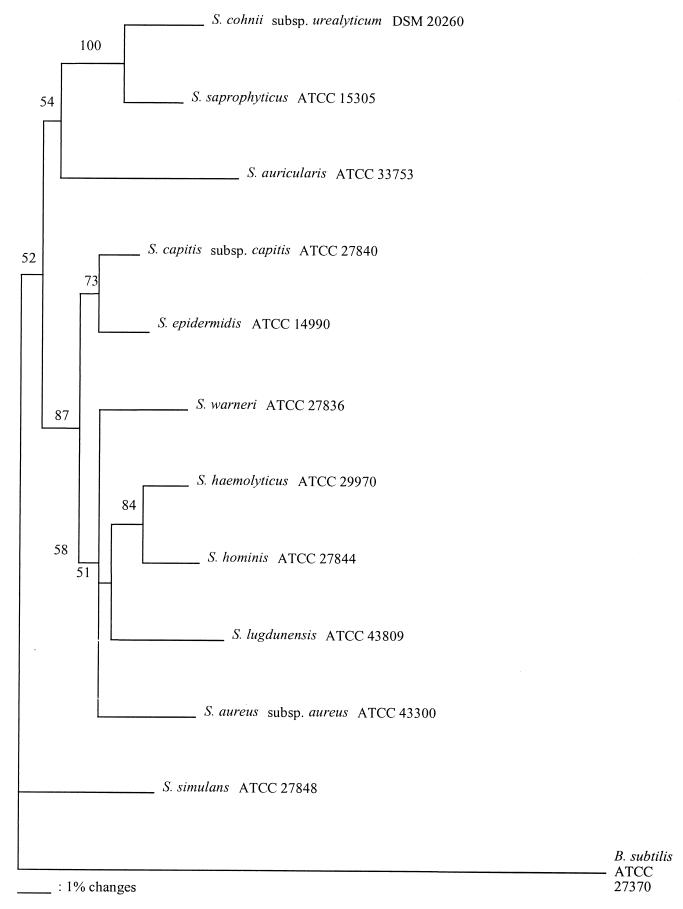
We compared the phylogenetic relationships of staphylococcal species based on the 16S rRNA gene sequence analysis (34) with our data obtained for the 812- to 815-bp tuf partial gene sequence determined from the 884-bp amplification products. We found the same eight cluster groups with both phylogenetic analyses (i.e., S. simulans, S. auricularis, S. saprophyticus, S. lugdunensis, S. haemolyticus, S. warneri, S. epidermidis, and S. aureus) (Table 4 and Fig. 1). Of all species included in our study, S. simulans is the most distant staphylococcal species based on tuf partial gene sequence analysis (Fig. 1). This result is in agreement with phylogenetic studies performed with 16S rRNA (34). Although tuf is well conserved in staphylococci, it has some variable regions which may be exploited for species-specific identification. An example of this variability is demonstrated by the presence of an insertion of three nucleotides unique to four staphylococcal species (S. auricularis, S. cohnii, S. saprophyticus, and S. simulans). The inserted amino acid is glutamic acid (GAA) for S. cohnii, S. saprophyticus, and S. simulans, while it is an aspartic acid (GAC) for S. auricularis. However, this observation does not entirely explain why these four species demonstrate a divergence from the other seven species, as shown in Fig. 1. We generated neighbor-joining trees with and without the trinucleotide insertion and found no significant difference in the phylogenetic relationships among the 11 staphylococcal species (Fig. 1). Based on the neighbor-joining tree created from the 812- to 815-bp tuf partial sequences, including the trinucleotide insertion analysis with the PILEUP program, the most closely related staphylococcal species pairs were (i) S. haemolyticus and S. hominis and (ii) S. capitis and S. epidermidis, with 97% identity for both pairs. On the other hand, the most divergent staphylococcal species pairs were (i) S. auricularis and S. haemolyticus and (ii) S. auricularis and S. lugdunensis, with 88% identities for both pairs (Table 4 and Fig. 1). These observations are in agreement with 16S rRNA gene sequence analysis (34).
FIG. 1.
Unrooted neighbor-joining tree based on the 812- to 815-bp tuf partial sequences including the GAA or GAC insertion, showing the phylogenetic relationships among the 11 staphylococcal species selected for this study. The value on each branch is the percent occurrence of the branching order in 500 bootstrapped trees. The scale bar represents one nucleotide substitution per 100 nucleotides.
We analyzed the sequence polymorphism in the 320-nucleotide sequence encompassed by the Staphylococcus-specific primers in order to design capture probes specific for the selected 11 clinically relevant staphylococcal species. We found that the DNA sequence identities between these 11 species ranged from 86 to 97% (Table 4). There were more nucleotide sequence variations in the tuf sequences than in the corresponding 16S rRNA gene sequences for these 11 staphylococcal species. The internal probes targeting species-specific tuf sequences developed in the present study were all shown to be sensitive (i.e., detected 5 to 10 genome copies) and specific.
Other PCR-based assays for the specific detection of clinically relevant species of staphylococci have been previously developed. These assays target (i) the intergenic spacer between 16S and 23 rRNA genes (15, 29), (ii) the HSP60 gene (22), or (iii) the transfer RNA intergenic spacer (24). The tuf gene, which acts in translation to bring aminoacylated tRNA molecules onto the ribosome, has previously been used for diagnostic purposes. tuf-based PCR detection systems for M. fermentans (2) and M. pneumoniae (23) have been developed. We have also developed previously a PCR-based assay for rapid detection of enterococci (17).
The tuf-based PCR assay developed in this study for staphylococcal species identification will be combined in multiplex with other PCR assays targeting clinically relevant antibiotic resistance genes (e.g., mecA). These multiplex PCR assays could be adapted for direct detection from positive blood cultures or from normally sterile clinical specimens such as blood or urine by using rapid and simple sample preparation protocols similar to those previously developed by our group (3, 16, 25). However, these assays would be of limited value for methicillin-resistant S. aureus screening of nasal or wound swabs, because a mixture of S. aureus and CNS is commonly found in these specimens. When applied for direct detection from clinical samples, these PCR assays have the potential to allow a faster establishment of effective antibiotic therapy and a reduction of empirical treatments with broad-spectrum antibiotics which are expensive and toxic.
In conclusion, we have developed a tuf-based PCR assay coupled with a Staphylococcus-specific probe and five species-specific probes targeting S. aureus, S. epidermidis, S. haemolyticus, S. hominis, and S. saprophyticus. This assay was shown to be specific and highly sensitive. The tuf sequence polymorphisms observed in staphylococci offer a good potential for the development of assays based on biochip technologies or nucleotide sequencing for identification to the species level.
ACKNOWLEDGMENTS
This research project was supported by Infectio Diagnostic (i.D.i.) Inc. (Sainte-Foy, Québec, Canada) and by grant PA-15586 from the Medical Research Council of Canada. Francis Martineau has a scholarship from the Fonds de la Recherche en Santé du Québec. Marc Ouellette is a Canadian Institutes of Health Research Scientist.
We thank Louise Côté, who is the director of the Microbiology Laboratory of the CHUL, for free access to the laboratory and for providing the staphylococcal clinical isolates. We also thank Ann Huletsky and Maurice Boissinot for critical comments regarding the manuscript.
REFERENCES
- 1.Anderson D Y, Vredeveld G N, Brake S R, Buchanan T F, Lewis J F. Evaluation of API 20E strips for identification of coagulase-negative staphylococci from the urinary tract. Am J Med Technol. 1983;12:879–881. [PubMed] [Google Scholar]
- 2.Berg S, Luneberg E, Frosch M. Development of an amplification and hybridization assay for the specific and sensitive detection of Mycoplasma fermentans DNA. Mol Cell Probes. 1996;10:10–14. doi: 10.1006/mcpr.1996.0002. [DOI] [PubMed] [Google Scholar]
- 3.Bergeron M, Ke D, Ménard C, Picard F, Gagnon M, Bernier M, Ouellette M, Roy P, Marcoux S, Fraser W. Rapid detection of group B streptococci in pregnant women at delivery. N Engl J Med. 2000;343:175–179. doi: 10.1056/NEJM200007203430303. [DOI] [PubMed] [Google Scholar]
- 4.Brakstad O G, Maeland J A, Tveten Y. Multiplex polymerase chain reaction for detection of genes for Staphylococcus aureus thermonuclease and methicillin resistance and correlation with oxacillin resistance. APMIS. 1993;101:681–688. doi: 10.1111/j.1699-0463.1993.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 5.Brumfitt W, Hamilton-Miller J. Methicillin-resistant Staphylococcus aureus. N Engl J Med. 1989;320:1188–1196. doi: 10.1056/NEJM198905043201806. [DOI] [PubMed] [Google Scholar]
- 6.Flores N, Valle F, Bolivar F, Merino E. Recovery of DNA from agarose gels stained with methylene blue. BioTechniques. 1992;13:203–205. [PubMed] [Google Scholar]
- 7.Froggatt J W, Johnston J L, Galetto D W. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1989;33:460–466. doi: 10.1128/aac.33.4.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaszewska-Mastalarz A, Zakrzewska-Czerwinska J, Mordarski M. Rapid detection of Staphylococcus saprophyticus using primer specific PCR. Acta Biol Hung. 1997;48:319–322. [PubMed] [Google Scholar]
- 9.Goh S H, Potter S, Wood J O, Hemmingsen S M, Reynolds R P, Chow A W. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J Clin Microbiol. 1996;34:818–823. doi: 10.1128/jcm.34.4.818-823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh S H, Santucci Z, Kloos W E, Faltyn M, George C G, Driedger D, Hemmingsen S M. Identification of Staphylococcus species and subspecies by the chaperonin 60 gene identification method and reverse checkerboard hybridization. J Clin Microbiol. 1997;35:3116–3121. doi: 10.1128/jcm.35.12.3116-3121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grosserode M H, Wenzel R P. The continuing importance of staphylococci as major hospital pathogens. J Hosp Infect. 1991;19(Suppl. B):3–17. doi: 10.1016/0195-6701(91)90197-g. [DOI] [PubMed] [Google Scholar]
- 12.Grunberg-Manago M. Regulation of the expression of aminoacyl-tRNA synthetases and translation factors. In: Neidhardt F C, Curtiss III R, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2 ed. Vol. 2. Washington, D.C.: ASM Press; 1996. pp. 1432–1457. [Google Scholar]
- 13.Gupta K, Scholes D, Stamm W E. Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA. 1999;281:736–738. doi: 10.1001/jama.281.8.736. [DOI] [PubMed] [Google Scholar]
- 14.Hussain Z, Stoakes L, Stevens D L, Schieven B C, Lannigan R, Jones C. Comparison of the MicroScan system with the API Staph-Ident system for species identification of coagulase-negative staphylococci. J Clin Microbiol. 1986;23:126–128. doi: 10.1128/jcm.23.1.126-128.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke D, Ménard C, Picard F, Boissinot M, Ouellette M, Roy P, Bergeron M. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin Chem. 2000;46:324–331. [PubMed] [Google Scholar]
- 17.Ke D, Picard F J, Martineau F, Ménard C, Roy P H, Ouellette M, Bergeron M G. Development of a PCR assay for rapid detection of enterococci. J Clin Microbiol. 1999;37:3497–3503. doi: 10.1128/jcm.37.11.3497-3503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloos W E. Natural populations of the genus Staphylococcus. Annu Rev Microbiol. 1980;34:559–592. doi: 10.1146/annurev.mi.34.100180.003015. [DOI] [PubMed] [Google Scholar]
- 19.Kloos W E, Bannerman T L. Staphylococcus and Micrococcus. In: Murray P, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington. D.C.: American Society for Microbiology; 1995. pp. 282–298. [Google Scholar]
- 20.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kloos W E, Wolfhohl J F. Identification of Staphylococcus species with the API Staph-Ident system. J Clin Microbiol. 1982;16:509–516. doi: 10.1128/jcm.16.3.509-516.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok A Y C, Su S-C, Reynolds R P, Bay S J, Av-Gay Y, Dovichi N J, Chow A W. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int J Syst Bacteriol. 1999;49:1181–1192. doi: 10.1099/00207713-49-3-1181. [DOI] [PubMed] [Google Scholar]
- 23.Luneberg E, Jensen J S, Frosch M. Detection of Mycoplasma pneumoniae by polymerase chain reaction and nonreactive hybridization in microtiter plates. J Clin Microbiol. 1993;31:1088–1094. doi: 10.1128/jcm.31.5.1088-1094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maes N, De Gheldre Y, De Ryck R, Vaneechoutte M, Meugnier H, Etienne J, Struelens M J. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J Clin Microbiol. 1997;35:2477–2481. doi: 10.1128/jcm.35.10.2477-2481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martineau F, Picard F, Ménard C, Roy P, Ouellette M, Bergeron M. Development of a rapid PCR assay specific for Staphylococcus saprophyticus and application to direct detection from urine samples. J Clin Microbiol. 2000;38:3280–3284. doi: 10.1128/jcm.38.9.3280-3284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol. 1998;36:618–623. doi: 10.1128/jcm.36.3.618-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martineau F, Picard F J, Roy P H, Ouellette M, Bergeron M G. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–2893. doi: 10.1128/jcm.34.12.2888-2893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McTaggart L A, Rigby R C, Elliott T S. The pathogenesis of urinary tract infections associated with Escherichia coli, Staphylococcus saprophyticus and Staphylococcus epidermidis. J Med Microbiol. 1990;32:135–141. doi: 10.1099/00222615-32-2-135. [DOI] [PubMed] [Google Scholar]
- 29.Mendoza M, Meugnier H, Bes M, Etienne J, Freney J. Identification of Staphylococcus species by 16S–23S intergenic spacer PCR analysis. Int J Syst Bacteriol. 1998;48:1049–1055. doi: 10.1099/00207713-48-3-1049. [DOI] [PubMed] [Google Scholar]
- 30.Nikiforov T T, Rogers Y-H. The use of 96-well polystyrene plates for DNA hybridization-based assays: an evaluation of different approaches to oligonucleotide immobilization. Anal Biochem. 1995;227:201–209. doi: 10.1006/abio.1995.1271. [DOI] [PubMed] [Google Scholar]
- 31.Saruta K, Hoshina S, Machida K. Genetic identification of Staphylococcus aureus by polymerase chain reaction using single-base-pair mismatch in 16S ribosomal RNA gene. Microbiol Immunol. 1995;39:839–844. doi: 10.1111/j.1348-0421.1995.tb03280.x. [DOI] [PubMed] [Google Scholar]
- 32.Sheagren J N. Staphylococcus aureus: the persistent pathogen. N Engl J Med. 1984;310:1368–1442. doi: 10.1056/NEJM198405243102107. [DOI] [PubMed] [Google Scholar]
- 33.Svanborg C, Godaly G. Bacterial virulence in urinary tract infections. Infect Dis Clin N Am. 1997;11:513–529. doi: 10.1016/s0891-5520(05)70371-8. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi T, Satoh I, Kikuchi N. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int J Syst Bacteriol. 1999;49:725–728. doi: 10.1099/00207713-49-2-725. [DOI] [PubMed] [Google Scholar]
- 35.Zakrzewska-Czerwinska J, Gaszewska-Mastalarz A, Pulverer G, Mordarski M. Identification of Staphylococcus epidermidis using a 16S rRNA-directed oligonucleotide probe. FEMS Microbiol Lett. 1992;79:51–58. doi: 10.1111/j.1574-6968.1992.tb14018.x. [DOI] [PubMed] [Google Scholar]