Abstract
Only a subset of recurrent glioblastoma (rGBM) responds to anti-PD-1 immunotherapy. Previously, we reported enrichment of BRAF/PTPN11 mutations in 30% of rGBM that responded to PD-1 blockade. Given that BRAF and PTPN11 promote MAPK/ERK signaling, we investigated whether activation of this pathway is associated with response to PD-1 inhibitors in rGBM, including patients that do not harbor BRAF/PTPN11 mutations. Here we show that immunohistochemistry for ERK1/2 phosphorylation (p-ERK), a marker of MAPK/ERK pathway activation, is predictive of overall survival following adjuvant PD-1 blockade in two independent rGBM patient cohorts. Single-cell RNA-sequencing and multiplex immunofluorescence analyses revealed that p-ERK was mainly localized in tumor cells and that high-p-ERK GBMs contained tumor-infiltrating myeloid cells and microglia with elevated expression of MHC class II and associated genes. These findings indicate that ERK1/2 activation in rGBM is predictive of response to PD-1 blockade and is associated with a distinct myeloid cell phenotype.
Keywords: PD-1 blockade, MAPK pathway, p-ERK, glioblastoma, biomarker, immunotherapy
Introduction
The prognosis for patients with glioblastoma (GBM) is poor, with a median overall survival (OS) of around 21 months1. Whereas radiation, chemotherapy and tumor-treating fields are established treatments at diagnosis, at recurrence there are no effective therapies. This is attributed partly to the notorious molecular and microenvironmental heterogeneity of GBM2,3, which contributes to erratic and unpredictable responses to therapies. Thereby, the variable response to therapy across patients with GBM often manifests as negative clinical trials in which an elusive subset of patients exhibit response.
Immune checkpoint blockade has seen an unparalleled expansion in cancer therapy leading to long-term remissions, even in cases of advanced metastatic disease. It has been adopted as the standard of care for advanced melanoma, nonsmall cell lung cancer, clear-cell renal cell carcinoma, solid tumors with DNA mismatch repair (MMR) deficiency or microsatellite instability (MSI) and an increasing number of cancers4. In contrast, its success has been limited in GBM5–7, partly due to the diverse and iterative mechanisms of intrinsic and iatrogenic immunosuppression. These include, among other factors, tumor infiltration by immunosuppressive cells, defects in the antigen processing and presentation machinery, sequestration of T cells in the bone marrow and frequent use of immunosuppressive medications such as corticosteroids8–12. Whereas negative trials showed an overall lack of efficacy of immune checkpoint blockade for GBM patients, durable clinical responses have been reported in some patients5,13,14. We recently reported an analysis of patients with recurrent GBM (rGBM) treated with adjuvant PD-1 blockade, where we uncovered molecular features associated with response to this immunotherapy13. Response was based on imaging and pathological criteria; responsive patients exhibited prolongation of survival independent of other therapies and known clinical or molecular prognostic variables. BRAF- and PTPN11 activating mutations, which drive MAPK/ERK pathway signaling15,16, were enriched in rGBM that responded to PD-1 blockade. Somatic mutations in these MAPK pathway genes were encountered in approximately only 30% of patients who responded to PD-1 inhibitors in our GBM cohort, and only 2–3% of GBMs harbored such mutations in the The Cancer Genome Atlas unselected population. Whereas these mutations provide biological clues to the GBM biology associated with response to PD-1 inhibitors, they have limited value as predictive biomarkers given that 70% of responder patients were not identified by these mutations13. To identify tumors that are susceptible to PD-1 blockade beyond those harboring BRAF or PTPN11 mutations, we explored whether MAPK pathway activation is present in rGBM that respond and exhibit prolonged survival following PD-1 blockade. Thus, we analyzed ERK1/2 phosphorylation (p-ERK), the active form of downstream effectors of the MAPK signaling, in specimens from patients with rGBM that were treated with PD-1 blockade, as well as from those who did not undergo immunotherapy. Furthermore, we evaluated the phenotypic and cellular differences of the tumor microenvironment of patients with elevated ERK1/2 activation through multiplex immunofluorescence and single-cell RNA-sequencing (scRNA-seq) of human GBMs.
Results
p-ERK predicts survival following PD-1 blockade in rGBM.
To investigate whether MAPK pathway activation is associated with response to anti-PD-1 therapy, we evaluated p-ERK using immunohistochemistry (IHC) in GBM. To maximize the rigor of this analysis, we titrated the p-ERK antibody using tumor samples, nontumoral human brain and breast cancer microarrays that express this phospho-antigen17 (Extended Data Fig. 1a–c). To determine MAPK activity in each specimen, we performed quantitative image analysis of the density of p-ERK+ cells within tumoral regions as outlined by a neuropathologist who was blinded to the clinical data (Extended Data Fig. 1d). p-ERK staining and quantification were performed in tumor specimens from patients with rGBM that underwent adjuvant PD-1 blockade (n = 29) (samples were naïve to immunotherapy), and from patients with rGBM that underwent surgery at recurrence but did not undergo immunotherapy (n = 33). Baseline clinical characteristics and known prognostic factors were similar between groups (Table 1 and Supplementary Tables 1 and 2).
Table 1.
Demographics and clinical data of GBM patients included in the discovery cohort.
| Patient characteristics | |||
|---|---|---|---|
| Adjuvant PD-1 blockade (n=29) | No immunotherapy (n=33) | Total (n=62) | |
|
| |||
| Demographics | |||
| Age at recurrence or PD-1 therapy initiation, years, mean ± s.d. | 55.7±13.7 | 61.1±12.9 | 58.5±13.5 |
| Gender, n (%) | |||
| Male | 18 (62.07) | 23 (69.67) | 41 (66.13) |
| Female | 11 (37.93) | 10 (30.3) | 21 (33.87) |
| Karnofsky performance status, median (range) | 80 (50–100) | 80 (60–100) | 80 (50–100) |
| MGMT promoter methylation status, n (%) | |||
| Methylated | 12 (41.38) | 6 (18.18) | 18 (29.03) |
| Unmethylated | 17 (58.62) | 12 (36.36) | 29 (46.77) |
| Unknown | 0 (0) | 15 (45.45) | 15 (24.19) |
| IDH status, n (%) | |||
| Wild type | 24 (82.76) | 28 (84.85) | 52 (83.87) |
| Mutant | 5 (17.24) | 5 (15.15) | 10 (16.13) |
s.d: standard deviation
We first investigated whether p-ERK staining is associated with response to PD-1 blockade. For this, patients were classified as responders based on our previous definition of response13, in which they had to fulfill at least one of the two criteria below:
Tissue sampled during surgery after PD-1 immunotherapy showed a robust reactive inflammatory infiltrate and scant to no tumor cells.
Tumor size determined by magnetic resonance imaging (MRI) that was either stable or decreasing over at least 6 months from the initiation of PD-1 blockade.
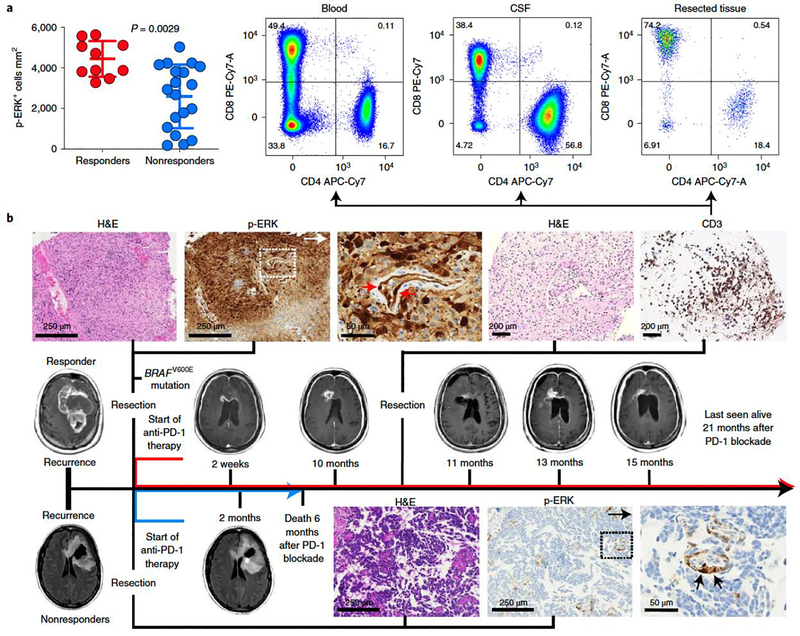
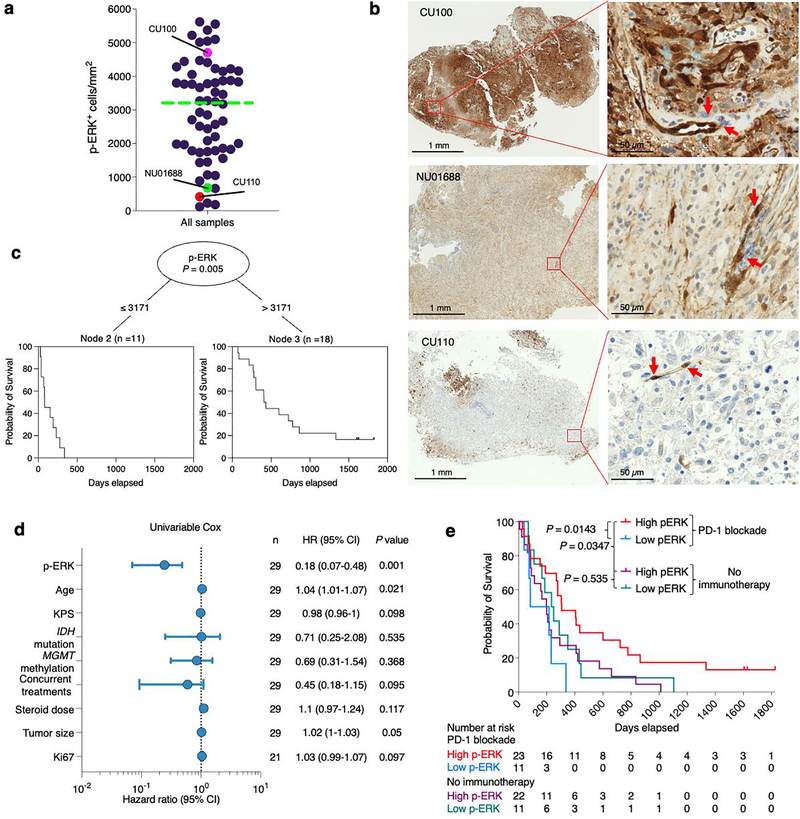
Quantification of p-ERK cell density in tumoral regions revealed that responder patients had increased density of p-ERK+ cells relative to nonresponder patients (P = 0.0029, Mann–Whitney U-test; Fig. 1a). As a representative example, we present a patient with an MGMT-unmethylated, IDH-wild-type rGBM that had a BRAFV600E mutation and who underwent resection followed by PD-1 blockade (Fig. 1b). This tumor showed strong positivity for p-ERK. Although the patient maintained stable disease for 9 months, he developed hydrocephalus and an associated inflammatory profile in his cerebrospinal fluid (CSF). Ten months after surgery and initiation of PD-1 blockade, the growth of a right periventricular enhancing lesion was noticed which, following biopsy, revealed scant tumor cells and an abundant CD3+ T-cell infiltrate consisting of both CD4+ and CD8+ T cells, and a similar lymphocytic pattern in the CSF (Fig. 1b). The patient continued with immunotherapy for a total of 21 months from recurrence before being lost to follow-up. In contrast, we also present a patient with an MGMT-methylated, IDH-mutant rGBM that had minimal p-ERK staining. This patient underwent PD-1 blockade after surgery but exhibited radiographic progression after 2 months and died 4 months later (Fig. 1b).
Figure 1. ERK1/2 activation is a predictive biomarker of radiographic response to anti-PD-1 immunotherapy in patients with recurrent GBM.
a, Dot plot showing the quantification of p-ERK+ cells before PD-1 blockade initiation in responder and nonresponder patients as previously defined13 (n = 29 GBM tumors). P = 0.0029, two-sided Mann–Whitney U-test. Data are presented as mean ± s.d. Each dot represents an independent patient sample. b, Examples of responder and nonresponder patients from results in a showing the corresponding MRI, associated H&E staining and immunostaining for p-ERK in the pretreatment sample. Top: in a responder patient, a biopsy performed in a gadolinium-enhancing lesion 10 months after treatment with anti-PD-1 therapy showed few tumor cells and a profuse CD3+ T-cell infiltrate. Flow cytometry analysis in the CSF and brain tumor showed that many of these T cells were CD4+ and CD8+. This patient experienced stable disease for at least 21 months after immunotherapy initiation. Bottom: a nonresponder patient with the corresponding MRI, H&E and p-ERK immunostaining in the pretreatment sample. Arrows indicate p-ERK+ endothelial cells. The experiment was performed using 29 tumor samples in one standardized run per patient.
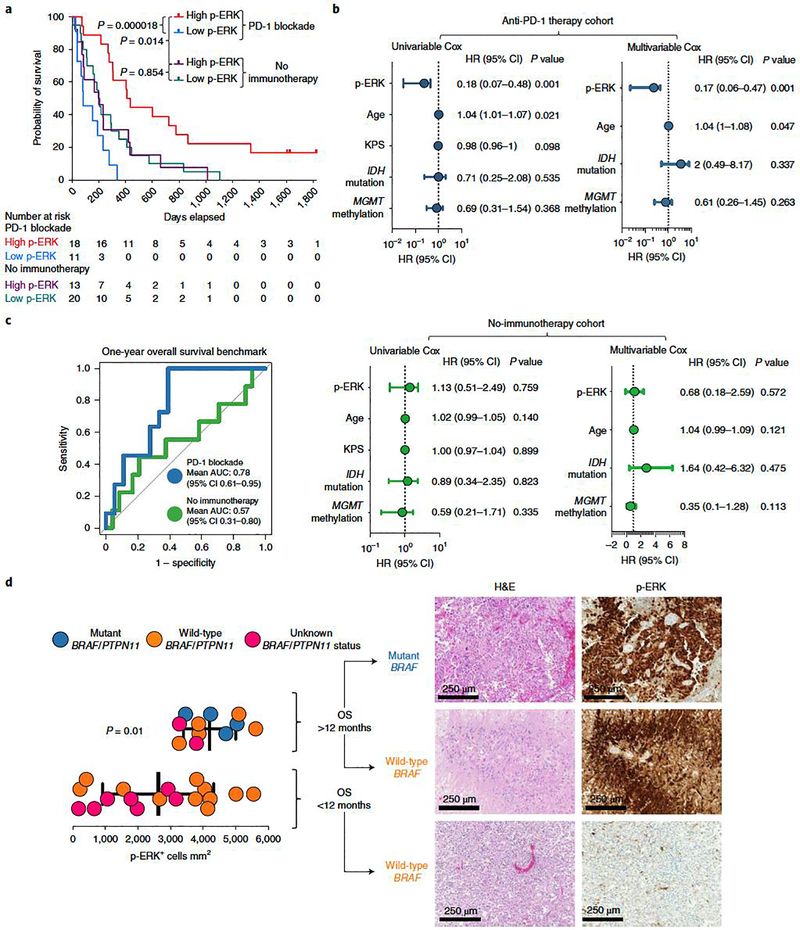
A recognized limitation of MRI in assessment of GBM progression is that brain inflammatory changes derived from immunotherapy may resemble disease progression, a phenomenon designated as pseudoprogression18,19, as exemplified on a biopsy specimen obtained 10 months after PD-1 blockade initiation (Fig. 1b). To investigate the predictive value of p-ERK for GBM susceptibility to PD-1 blockade independent of radiographic response, we performed analyses of OS based on p-ERK cell density. Tumor samples of 29 patients with rGBM treated with PD-1 inhibitors and of 33 patients that did not receive immunotherapy were classified as either high- or low-p-ERK tumors. The cut-point value used to classify high- and low-p-ERK tumors was derived from the median (3,207 p-ERK+ cells mm2) of all values for p-ERK cell density, as determined by quantitative image analysis of all specimens (n = 62) (Extended Data Fig. 2a,b). Based on the date of initiation of PD-1 blockade at recurrence, GBM patients treated with immunotherapy and with high-p-ERK tumors demonstrated a longer OS, with a hazard ratio (HR) of 0.23 compared to GBM patients with low-p-ERK tumors (95% confidence interval (CI) 0.079–0.69, P = 0.000018, log-rank test; Fig. 2a). In this regard, the median OS of patients with GBM and high-p-ERK tumors was 13.89 months (422.5 days) compared to 2.72 months (83 days) for patients whose tumors had low p-ERK cell density. Conditional inference trees for cut-point optimization were used to explore the interaction between p-ERK and anti-PD-1 therapy with respect to OS, employing the cohort that received PD-1 blockade. The optimal cut-point value of p-ERK cell density based on OS led to a value (3,171 cells mm2) of high versus low p-ERK across patients that is close to the median p-ERK value (3,207 cells mm2) used to dichotomize p-ERK (Extended Data Fig. 2c). In contrast, p-ERK was not associated with survival in patients with rGBM that did not receive immunotherapy (Fig. 2a). For this control cohort, survival was calculated from the date of surgery that led to the recurrent tumor specimen used for IHC staining. In this cohort, the high-p-ERK group had a median OS of 6.57 months (200 days) versus 6.65 months (202.5 days) in the low-p-ERK group (HR = 1.06, 95% CI 0.52–2.14, P = 0.854, log-rank test). Patients with high-p-ERK tumors treated with PD-1 blockade also exhibited prolonged survival compared to those with high-p-ERK tumors who did not receive immunotherapy (HR = 0.41, 95% CI 0.17–0.96, P = 0.014, log-rank test; Fig. 2a). As of the final analysis, three patients classified as high p-ERK were still alive and receiving immunotherapy.
Figure 2. ERK1/2 phosphorylation evaluated by semiautomatic IHC quantification shows that is a predictive biomarker following PD-1 blockade in recurrent GBM.
a, Kaplan–Meier curve comparing OS of patients with recurrent GBM defined as having either high- or low-p-ERK tumors, counting from initiation of PD-1 blockade (anti-PD-1 therapy group, n = 29 patients) and from surgery at recurrence (no-immunotherapy group, n = 33 patients); P values, two-sided log-rank test. b, Forest plots representing univariable and multivariable survival analyses using a Cox proportional hazard model evaluating prognostic variables and p-ERK+ cell density on survival in the anti-PD-1 therapy cohort (top) and the no-immunotherapy cohort (bottom), presented as HR (95% CI). P values by two-sided Wald test. c, ROC curve of sensitivity and 1 – specificity displaying mean AUC (95% CI) for the anti-PD-1 therapy and no-immunotherapy cohorts (n values as in a) d, Left: dot plot comparing the quantification of p-ERK+ cell density between tumors of patients with GBM harboring either BRAF/PTPN11 mutations (n = 4 tumors), wild-type BRAF/PTPN11 (n = 5 tumors) or unknown BRAF/PTPN11 status (n = 2 tumors) that had OS >12 months with those that had either wild-type tumors (n = 11) or unknown BRAF/PTPN11 status (n = 7 tumors) and lived <12 months after initiation of immunotherapy. Right: H&E and p-ERK immunostaining of three GBM samples. From top to bottom: a BRAF mutated tumor and a wild-type BRAF tumor from patients that lived >12 months, and a wild-type BRAF tumor from a patient that lived <12 months. P values by two-sided Mann–Whitney U-test. Data are presented as mean ± s.d. Each dot represents an independent patient sample.
To further assess p-ERK staining as a predictive biomarker for PD-1 immunotherapy, we performed univariable and multivariable analyses using the Cox proportional hazard model to evaluate the association of p-ERK cell density with survival. Univariable Cox analysis revealed that p-ERK cell density was associated with increased OS in patients treated with PD-1 blockade (HR = 0.18, 95% CI 0.07–0.48, P = 0.001, Wald test) but not in patients not treated with immunotherapy (HR = 1.13, 95% CI 0.51–2.49, P = 0.759, Wald test; Fig. 2b). We also investigated whether age, Karnofsky performance score (KPS), IDH mutations, MGMT promoter methylation, concurrent treatments, steroid use, tumor size or Ki67 labeling was associated with OS in the cohort treated with PD-1 blockade, and found that age (HR = 1.04, 95% CI 1.01–1.07, P = 0.021) was weakly associated with shorter survival (Extended Data Fig. 2d). Next, we investigated the influence of prognostic factors for patients with GBM, such as age, MGMT promoter methylation status and IDH mutations, in our cohorts as part of a multivariable Cox model. This confirmed that p-ERK was associated with survival independently of these prognostic factors in patients treated with PD-1 inhibitors (HR = 0.17, 95% CI 0.06–0.47, P = 0.001, Wald test; Fig. 2b). The predictive value with respect to OS was also assessed using p-ERK by treatment interaction term (PD-1 blockade versus no immunotherapy). This analysis showed a notable association between p-ERK and OS (HR = 0.18, 95% CI 0.06–0.56, P = 0.003, Wald test). This supports the conclusion that elevated p-ERK is associated with OS prolongation only in the context of PD-1 inhibition.
To investigate the sensitivity and specificity of p-ERK as a predictive biomarker for response to PD-1 blockade, we generated receiver operating characteristic (ROC) curves using 1-year OS as benchmark. In this context, the area under the curve (AUC) for the anti-PD-1 therapy cohort was 0.78 (95% CI 0.61–0.95) and 0.57 (95% CI 0.31–0.80) for the no-immunotherapy cohort (Fig. 2c). Overall, these survival analyses indicate that high p-ERK is associated with a survival benefit only in the context of anti-PD-1 therapy, and therefore is predictive but not prognostic as a biomarker.
Next, we investigated the feasibility of using p-ERK staining for predicting response to PD-1 blockade through scoring by a neuropathologist who was blinded to outcomes and treatments. The pathologist designated the samples as high or low based on the staining pattern in tumor regions. Whereas the pathologist’s scoring showed a trend similar to the quantification of p-ERK (HR = 0.35, 95% CI 0.095–1.3, P = 0.0143, log-rank test; Extended Data Fig. 2e), the association between high-p-ERK tumors with OS was stronger when this biomarker was quantified employing software. This analysis suggests that computer-based quantification of p-ERK might be more reproducible and reliable than scoring by pathologists.
We investigated whether elevated p-ERK cell density and its association with long-term survival following PD-1 blockade can occur irrespective of BRAF/PTPN11-activating mutations. Interestingly, patients that survived >12 months from initiation of PD-1 blockade had elevated p-ERK+ cell density compared to those that survived <12 months (P = 0.01, Mann–Whitney U-test; Fig. 2d). Of those patients that survived >12 months, five had wild-type BRAF and PTPN11, four had activating mutations in these genes and two had unknown BRAF/PTPN11 mutational status. These results show that patients who experienced a longer OS in the context of anti-PD-1 therapy consistently exhibited high levels of p-ERK regardless of the genetic status of the BRAF/PTPN11 genes.
Assessment of p-ERK integrity and antibody specificity.
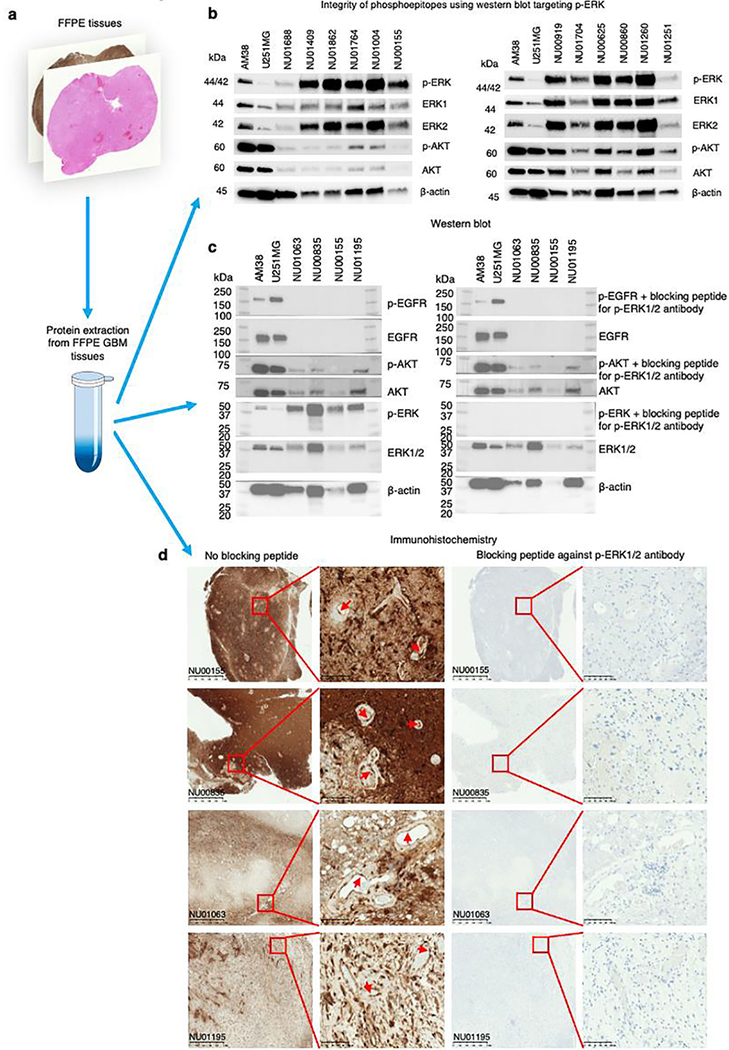
Given that ischemic time can lead to degradation of phospho-epitopes such as p-ERK, and to rule out nonspecific staining as potential confounders of the survival analysis, we investigated the integrity and specificity of p-ERK staining in our samples. We evaluated the preservation of p-ERK in a set of formalin-fixed, paraffin-embedded (FFPE) GBM samples that were used for survival analysis (Extended Data Fig. 3a). Using immunoblot, we detected p-ERK corresponding to its molecular weight in these samples, confirming the integrity of the p-ERK phospho-epitope (Extended Data Fig. 3b). Additionally, we performed a peptide competition assay by immunoblot and IHC and confirmed unequivocal specific staining of p-ERK in FFPE GBM samples, ruling out any kind of cross-reactivity by the p-ERK antibody (Extended Data Fig. 3c,d).
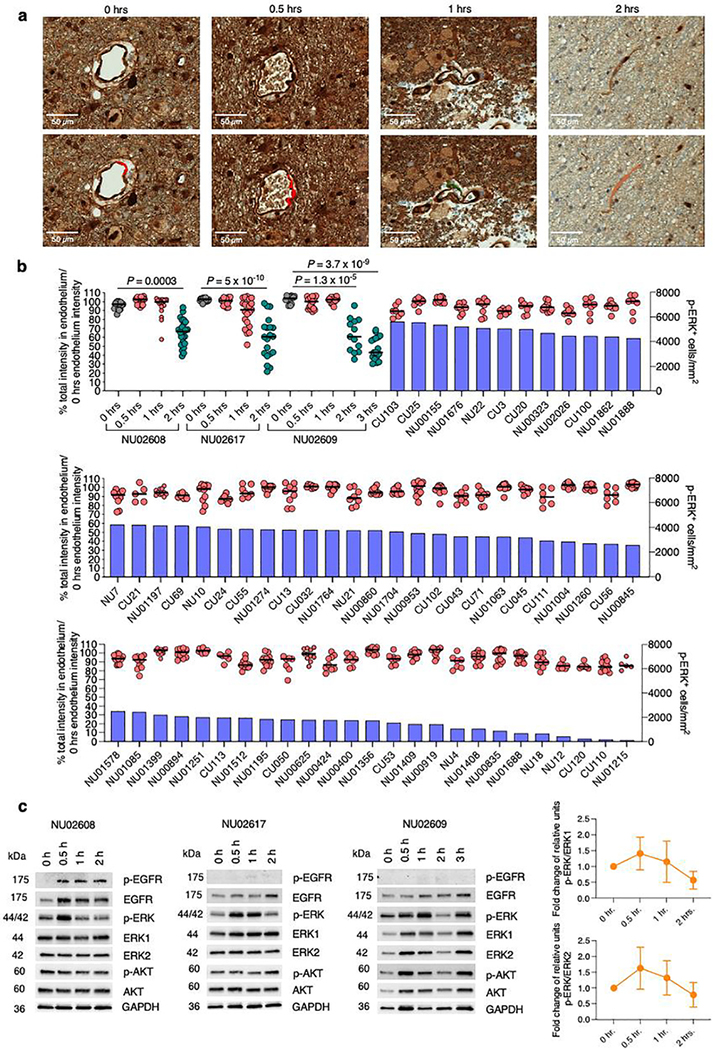
Given that preanalytical variables such as delay in fixation time can affect phosphorylated proteins20, we studied the degradation dynamics of p-ERK in GBM samples subjected to different ischemic times. We obtained fresh GBM tumor specimens and fixed them at different time intervals following surgical resection (0, 0.5., 1 and 2 h). Then, we stained these samples against p-ERK using the same antibody dilution. We analyzed endothelial cells as a common denominator across samples, because p-ERK is strongly expressed by these cells21. We compared the intensity of endothelial p-ERK staining for each tumor sample fixed at different ischemic time points and for tumor samples in the discovery cohort (Extended Data Fig. 4a). We found that the intensity of endothelial p-ERK in GBM samples from our discovery cohort used for the survival analysis remained relatively stable, without an important decrease relative to 0 min of ischemia time and for up to 1 h of ischemic time. Endothelial p-ERK staining exhibited degradation starting at 2 h (Extended Data Fig. 4b). Additionally, we performed immunoblot evaluation of phospho-ERK, phospho-AKT and phospho-EGFR in tumor samples subjected to different ischemic times (Extended Data Fig. 4c). Collectively, these results show that p-ERK staining is specific. Also, the p-ERK epitope was preserved for up to 1 h of ischemic time and was detectable in samples from the discovery cohort.
Time of biopsy influences prediction of response by p-ERK.
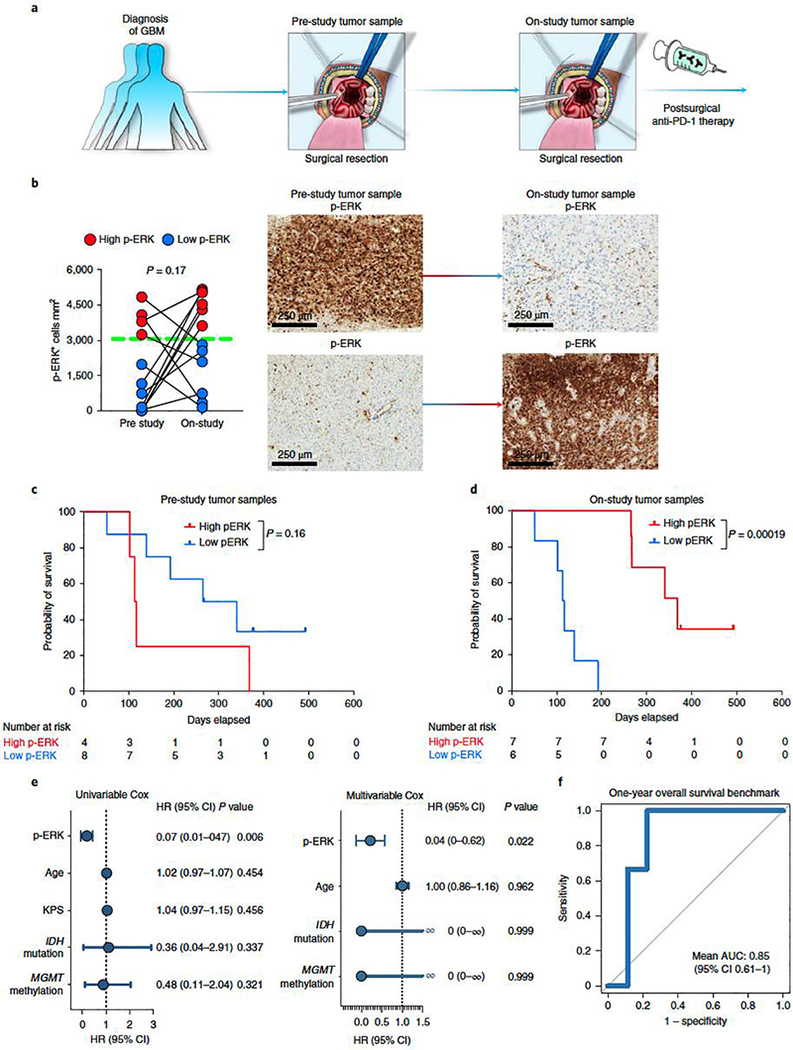
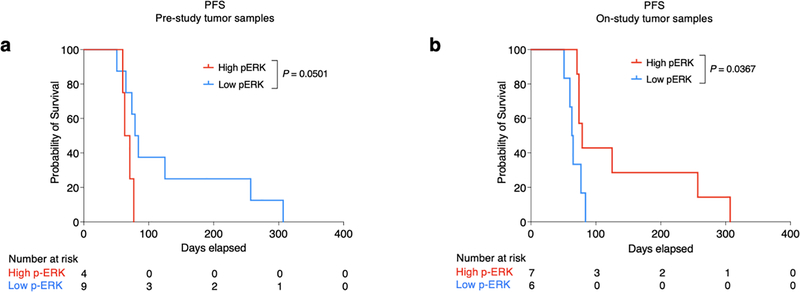
Tumor cell phenotype and the associated microenvironment can change over time, particularly between newly diagnosed GBM and recurrent disease2,22,23. To investigate whether the timing of tissue used for p-ERK determination relative to initiation of PD-1 blockade influences the predictive properties of this biomarker, we used paired samples obtained at different time points before initiation of PD-1 blockade from a clinical trial14, which included an arm that underwent adjuvant PD-1 blockade (Supplementary Table 3). We compared p-ERK from GBM specimens obtained during the surgery that preceded enrollment on this trial (pre-study samples) versus samples from the same patients obtained during surgery at recurrence as part of the trial, weeks before initiation of anti-PD-1 therapy (on-study tumor samples) (Fig. 3a). This cohort also served as an independent validation set where tumor samples and clinical outcomes were collected prospectively. For this validation cohort, the designation of high- versus low-p-ERK gliomas was done using the same quantification method and cut-point value (3,207 cells mm2) employed to partition groups in the discovery cohort. Though we found no systematic overall change in p ERK-high versus -low designation between the two time points (P = 0.17, Wilcoxon rank test), p-ERK cell density fluctuated considerably between pre- and on-study time points for most tumors, including 91.6% of cases that were initially designated as high p-ERK that became low p-ERK, and 62.5% that were initially designated as low that became high p-ERK, in the group of on-study samples (Fig. 3b). Next, we performed survival analysis to evaluate the performance of p-ERK in pre-study and on-study samples to predict OS following anti-PD-1 therapy. Based on the staining of pre-study samples, patients from the low-p-ERK group exhibited marginally longer progression-free survival (PFS) compared to the high-p-ERK group (P = 0.0501, log-rank test; Extended Data Fig. 5a), and no difference in OS (P = 0.16, log-rank test; Fig. 3c). In contrast, when using on-study samples for staining and analysis, the high-p-ERK group exhibited longer PFS (P = 0.0367, log-rank test; Extended Data Fig. 5b) as well as longer OS (P = 0.00019, log-rank test; Fig. 3d) compared to the low-p-ERK patients. The median OS of patients with GBM with high-p-ERK tumors was 12.09 months (368 days) compared to 3.78 months (115 days) for patients whose tumors had low p-ERK activation. These results suggest that tumors acquired shortly before PD-1 blockade initiation are more reliable in predicting the response of patients with rGBM to PD-1 blockade. Additionally, we performed the Cox proportional hazard model to test the association of p-ERK cell density with survival using the on-study tumor samples. In this independent GBM cohort, p-ERK cell density was associated with OS in both univariate (HR = 0.07, 95% CI 0.01–0.47, P = 0.006, Wald test) and multivariable analysis (HR = 0.04, 95% CI 0–0.62, P = 0.022, Wald test; Fig. 3e). We also employed ROC curves to determine the ability of p-ERK cell density to distinguish between patients with rGBM living >1 year in the validation cohort. The mean AUC was 0.85 (95% CI 0.61–1.0), further demonstrating the ability of p-ERK to discern which GBM patients would be appropriate candidates for anti-PD-1 therapy (Fig. 3f).
Figure 3. Validation of pretreatment p-ERK staining correlates with OS in an independent recurrent GBM cohort treated with adjuvant PD-1 blockade.
a, Cartoon showing the time points of surgical tumor acquisition relative to treatment with PD-1 blockade identified as prestudy or on-study tumor samples. b, Left: change in p-ERK cell density in pre-study and on-study tumor samples. n = 12 paired tumor samples. Red and blue dots represent high- and low-p-ERK tumors, respectively; dashed green line represent the cut-point value used to partition high- and low-p-ERK tumors. P value calculated using two-sided Wilcoxon signed-rank test. Each dot represents an independent patient sample. Right: representative IHC micrographs showing the change in p-ERK immunostaining between paired tumor samples. c,d, Kaplan–Meier plots showing OS in high- versus low-p-ERK groups treated with adjuvant PD-1 blockade evaluating pre-study (c) and on-study tumor samples (d). P values by two-sided log-rank test. e, Forest plots for on-study tumor samples representing univariable and multivariable survival analysis using a Cox proportional hazard model presented as HR (95% CI). n = 13 patients with GBM, P values by two-sided Wald test. f, ROC curve of sensitivity and 1 – specificity displaying mean AUC (95% CI) for the validation GBM cohort. n = 13 patients with GBM.
In sum, these results emphasize the predictive power of p-ERK from GBM samples acquired close to initiation of PD-1 blockade. Furthermore, these results validate initial observations that high p-ERK is associated with prolonged survival in patients treated with adjuvant PD-1 blockade.
High-p-ERK GBMs exhibit abundant infiltrating microglia.
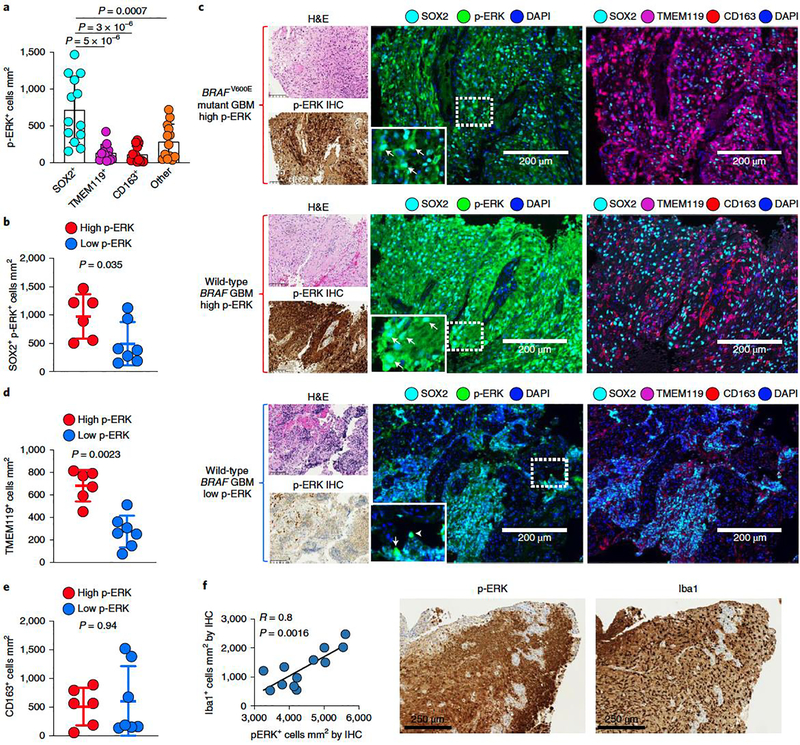
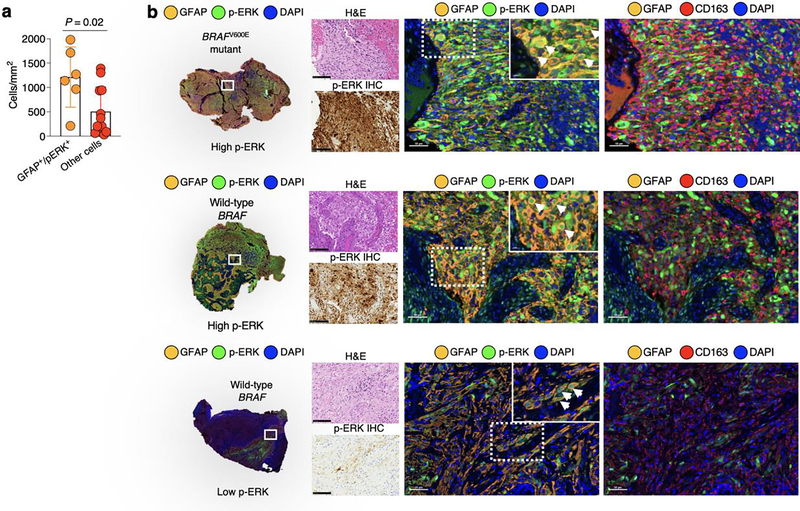
To investigate which cell populations contribute to p-ERK expression in GBM, we performed multiplex immunofluorescence in GBM samples evaluating p-ERK and the predominant cellular populations in the tumor microenvironment, including SOX2 (tumor marker24), TMEM119 (microglial marker22,25,26), CD163 (macrophage marker26,27) and DAPI to label nuclei. By quantifying these cell populations, we found that p-ERK was detected predominantly in SOX2+ cells relative to TMEM119+ cells (P = 5 X 10−6), CD163+ cells (P = 3 X 10−6) and other cells (SOX2−TMEM119−CD163− cells, P = 0.0007, one-way ANOVA; Fig. 4a). Multiplex staining utilizing glial fibrillary acidic protein (GFAP) to label tumor cells also confirmed that most p-ERK staining was predominantly associated with tumor cells/astrocytes (Extended Data Fig. 6a,b). We also found higher numbers of SOX2+ p-ERK+ cells in high- compared to low-p-ERK gliomas (P = 0.035, Mann–Whitney U-test; Fig. 4b). As examples, we present a BRAFV600E mutated GBM and a wild-type BRAF/PTPN11 tumor, both of which had high levels of p-ERK expressed by SOX2+ cells (Fig. 4c). In contrast, in a rGBM classified as having low levels of ERK1/2 activation by IHC, the majority of SOX2+ cells were negative for p-ERK (Fig. 4c). These results show that the majority of p-ERK+ cells in gliomas are tumor cells, with smaller contributions from immune and stromal cells.
Figure 4. Multiplex immunofluorescence of recurrent GBM samples shows p-ERK positivity in SOX2+ cells and associated myeloid cell infiltration.
a, Bar plot showing the contribution to p-ERK expression from SOX2+, TMEM119+, CD163+ and other cells (SOX2−TMEM119−CD163− cells). Differences among cell types were evaluated using one-way ANOVA with post hoc Tukeýs multiple comparisons test (n = 13 tumors). b, Dot plot showing comparison of SOX2+ p-ERK+ cells mm2 between high- and low-p-ERK tumors (n = 13 tumors). c, Representative images of three different tumor samples derived from results in a and b. From top to bottom: a BRAFV600E GBM sample with high-p-ERK staining, a wild-type BRAF/PTPN11 GBM with high-p-ERK staining and a wild-type BRAF/PTPN11 GBM displaying low-p ERK staining. For the three tumor samples: left, H&E and p-ERK IHC images of the same tumor region; middle, multiplex immunofluorescence images showing the markers for SOX2, p-ERK and DAPI; right, multiplex immunofluorescence images showing the markers for SOX2, TMEM119, CD163 and DAPI. Arrowheads indicate SOX2+ p-ERK+ cells. The experiment was done using 13 tumor samples in one standardized run per patient. d, Dot plot showing comparison of TMEM119+ between high- and low-p-ERK tumors (n = 13 tumors). e, Dot plot showing comparison of CD163+ cells mm2 between high- and low-p-ERK tumors (n = 13 tumors). f, Left, scatter plot showing the correlation of p-ERK+ cells mm2 with Iba1+ cells mm2 obtained by software-based quantification of IHC-stained tumor samples. Right, representative images of p-ERK and Iba1 immunostaining of the same tumor region (n = 12 tumors). P values by two-sided Mann–Whitney U-test (b,d,e) or Pearson’s correlation (f). Data are presented as mean ± s.d. (a,b,d,e). Each dot represents an independent patient sample (a,b,d–f).
Considering the abundance of myeloid cells in the GBM microenvironment27,28, and their implication in the response and resistance to immune checkpoint inhibitors in GBM7,29–31, we analyzed whether p-ERK cell density is associated with infiltration of these immune cells in GBM. We found that numbers of TMEM119+ cells were elevated in GBM with high- relative to low-p-ERK tumors (P = 0.0023, Mann–Whitney U-test; Fig. 4d). In contrast, no difference was found in the numbers of glioma-infiltrating CD163+ cells between gliomas with high versus low p-ERK levels (P = 0.94, Mann–Whitney U-test; Fig. 4e). We also stained glioma samples with the macrophage/microglia cell marker Iba1 by IHC and evaluated its association with p-ERK cell density. We found that p-ERK cell density derived from IHC quantification (which was used to predict response to PD-1 blockade) correlated with Iba1+ cell density across specimens (R = 0.8, P = 0.0016, Pearson r correlation; Fig. 4f).
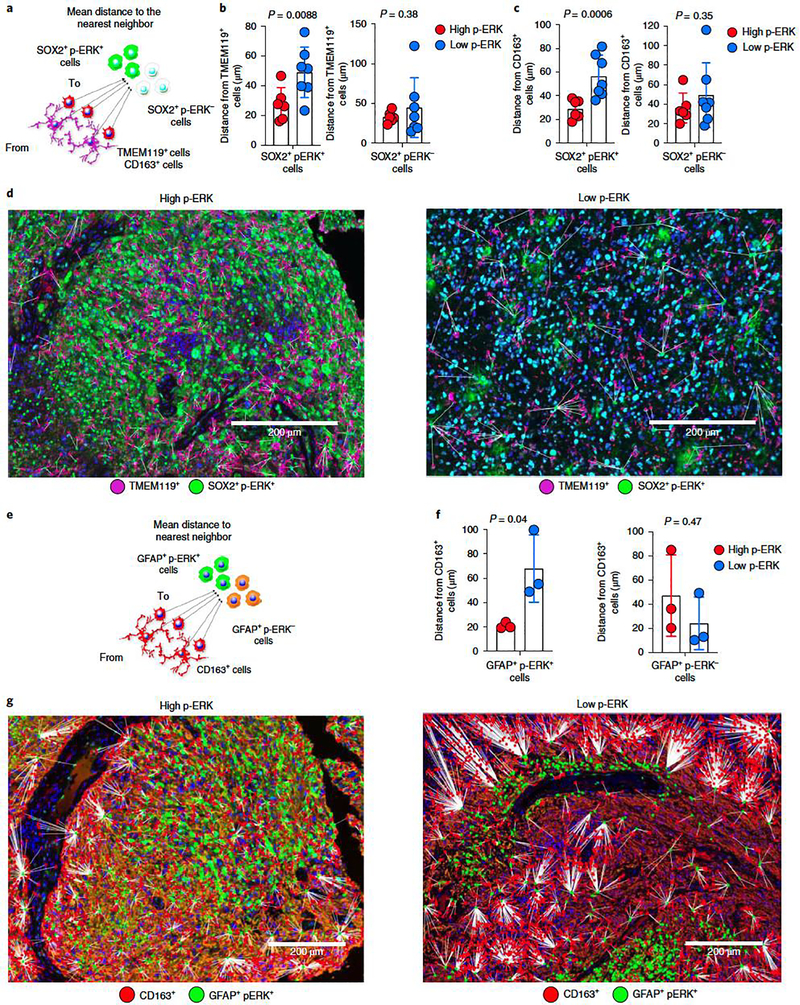
Next, we performed spatial analyses to further investigate the interaction between SOX2+ p-ERK+ cells and glioma-infiltrating myeloid cells. We calculated the distances between TMEM119+ and CD163+ cells to SOX2+ p-ERK+ cells (Fig. 5a). We found that TMEM119+ cells were closer to SOX2+ p-ERK+ cells in high compared to low-p-ERK tumors (P = 0.0088, unpaired t-test; Fig. 5b). Conversely, we did not find differences in distances between TMEM119+ cells and SOX2+ p-ERK− cells between high and low-p-ERK groups (P = 0.38, unpaired t-test; Fig. 5b). Similarly, distances between CD163+ cells and SOX2+ p-ERK+ were shorter in tumors with high p-ERK than in those with low p-ERK (P = 0.0006, unpaired t-test; Fig. 5c), yet no difference was noted in the distance between CD163+ cells and SOX2+ p-ERK− cells between high- and low-p-ERK tumors (P = 0.35; unpaired t-test; Fig. 5c). Representative images for this spatial analysis are presented in Fig. 5d. Similarly, when using GFAP+ to label tumor/astrocytes to measure the distance between tumor cells and myeloid cells (Fig. 5e), we found that the distance from CD163+ cells to GFAP+ p-ERK+ was shorter for tumors with high p-ERK than for those with low p-ERK (P = 0.04, unpaired t-test), whereas no differences in these distances were noted for GFAP+ p-ERK− cells (P = 0.47, unpaired t-test; Fig. 5f). Representative images for this analysis are presented in Fig. 5g. These results suggest that gliomas with an elevated number of p-ERK+ cells are associated with high infiltration of myeloid/ microglial cells. Compared to other solid tumors in which there is separation between immune cells and tumor regions32, the elevated number of p-ERK+ tumor cells near glioma-infiltrating myeloid cells suggests a high degree of mixing between these cell populations.
Figure 5. Spatial analysis of tumor cells expressing p-ERK and their associated myeloid cells.
a, Cartoon representing distances from TMEM119+ and CD163+ cells to SOX2+ p-ERK+/p-ERK− cells. b,c, Bar plots comparing the mean distances from TMEM119+ (b) and CD163+ (c) cells to SOX2+ p-ERK+/p-ERK− cells in high- versus low-p-ERK tumors. Dots represent tumor samples (n = 13 tumors). d, Representative multiplex immunofluorescence images illustrating spatial dimensions between TMEM119+ cells and SOX2+ p-ERK+ in a high- and a low-p-ERK GBM. The experiment was done in 13 tumor samples in one standardized run per patient. e, Cartoon representing distances from CD163+ cells to GFAP+ p-ERK+/p-ERK− cells. f, Bar plots comparing mean distances from CD163+ cells to GFAP+ p-ERK+/p-ERK− cells in high- versus low-p-ERK tumors. Dots represent tumor samples (n = 6 tumors). g, Representative multiplex immunofluorescence images illustrating spatial dimensions between CD163+ cells and GFAP+ p-ERK+ in a high- (left) and a low-p-ERK GBM (right). The experiment was performed on six tumor samples in one standardized run per patient. P values by two-tailed unpaired t-test (b,c,f). Data are presented as mean ± s.d. (b,c,f).
Microglia from high-p-ERK GBMs have elevated MHC class II.
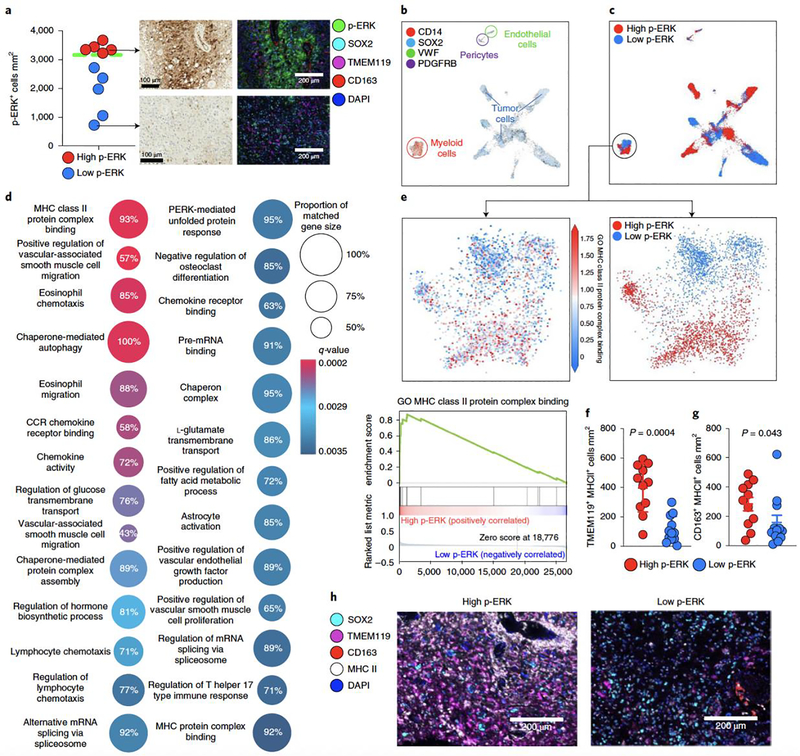
Given the association of myeloid cell infiltration we observed with ERK1/2 activation in tumor cells, we investigated the phenotype of these immune cells using scRNA-seq. We analyzed 28,194 cells from ten GBM specimens24 that were paired with p-ERK IHC staining. p-ERK cell density was quantified using the same methodology as in the discovery and validation GBM cohort, and we used the same cut-point value to designate tumors as high versus low p-ERK resulting in five p-ERK-high tumors and five p-ERK-low tumors (Fig. 6a). Next, we performed unsupervised clustering of all cells without employing cell type markers (Extended Data Fig. 7). Subsequently, we annotated the clusters based on the expression of validated cell markers used for single-cell transcriptomics for tumor cells (SOX2), myeloid cells (CD14), endothelial cells (VWF) and pericytes (PDGFRB)24. Whereas tumor cells clustered into several sample-dependent groups, the myeloid cell compartment, pericytes and endothelium derived from all GBM samples clustered uniformly (Fig. 6b shows cell phenotype, Fig. 6c shows high- versus low-p-ERK designation from IHC in tumor and myeloid cells, and Extended Data Fig. 7 shows the tumor sample uniform manifold approximation and projection (UMAP) plot). We focused our analysis on myeloid cells, given their increased infiltration in tumors with elevated p-ERK and their immunomodulatory roles in the context of immune checkpoint blockade in GBM7,29–31. To investigate transcriptional differences within the myeloid cell population between high- and low-p-ERK tumors, we performed gene set enrichment analysis (GSEA33) with the gene ontology (GO) set collection. Twenty-seven GO pathways were considerably enriched in myeloid cells from high-p-ERK tumors (false discovery rate (FDR), q < 0.05; Fig. 6d and Supplementary Table 4). Interestingly, the top gene set enriched in myeloid cells infiltrating high-p-ERK tumors was the GO term ‘MHC class II protein complex binding’ (normalized enrichment score (NES) = 2.916, q = 0.000258; Fig. 6d,e), among several other GO themes related to lymphocyte chemotaxis, chemokines and antigen presentation (Fig. 6d). In contrast, we did not find significant gene sets enriched in myeloid cells from low-p-ERK tumors. Given the enrichment of the MHC class II protein complex binding GO term in myeloid cells from high-p-ERK GBMs, we investigated whether these myeloid cells also express the MHC class II (MHC II) molecule. We employed multiplex immunofluorescence staining to evaluate SOX2 (tumor cell marker), CD163 (myeloid cell marker), TMEM119 (microglial marker), MHC II and DAPI. This analysis revealed that TMEM119+ cells from high-p-ERK tumors had elevated MHC II protein expression relative to tumors with low p-ERK (P = 0.0004, Mann–Whitney U-test; Fig. 6f). We also observed MHC II expression by CD163+ cells from high-p-ERK tumors relative to those with low p-ERK (P = 0.043, Mann–Whitney U-test; Fig. 6g). Representative images for this analysis are presented in Fig. 6h. Collectively, these results suggest that tumors with elevated activation of ERK1/2 are characterized by a particular microenvironment in which myeloid cells, mostly microglia, express MHC II and a gene signature related to this antigen-presenting molecule.
Figure 6. scRNA-seq in patients with GBM from high- and low-p-ERK groups.
a, Quantification of, and representative, p-ERK IHC images and multiplex immunofluorescence images of GBM samples used in the analysis. Dashed green line represent the cut-point value used to partition high- and low-p-ERK tumors in the discovery and validation cohorts. n = 10 tumor samples. b, UMAP graph showing the expression of cell markers for tumor cells (SOX2), myeloid cells (CD14), endothelial cells (VWF) and pericytes (PDGFRB). The color key indicates expression levels. Each dot represents an individual cell. n = 28,194 cells contained in ten tumor samples. c, UMAP graph showing the overlapping annotations derived from high- and low-p-ERK IHC-stained tumors obtained from software-based quantification in all cells (top) and in the myeloid cell compartment (bottom). d, GO terms analysis. Differentially expressed gene signatures of the myeloid cell population infiltrating high- and low-p-ERK GBM samples. Twenty-seven differentially expressed GO terms are represented in a dot plot, with dot size corresponding to the percentage of genes that matched the GO term. Dot color corresponds to the q-value of enrichment. e, Top: UMAP plot showing expression of the MHC II protein binding complex gene signature in myeloid cells. Bottom: GSEA plot showing enrichment of the GO term MHC II protein binding complex within myeloid cells. n = 3,153 myeloid cells from ten tumor samples. f,g, Beeswarm plots showing the cell density of TMEM119+ MHC II+ cells (f) and CD163+ MHC II+ cells (g) between high- and low-p-ERK tumors (n = 23). P values by two-sided Mann–Whitney U-test. h, Representative multiplex immunofluorescence images illustrating the expression of MHC II by TMEM119+ and CD163+ cells in a high- and a low-p-ERK GBM sample. SOX2, TMEM119, CD163, MHC II and DAPI are included as cell markers. The experiment was performed in 23 tumor samples in one standardized run per patient. Data presented as mean ± s.d. (f,g).
Discussion
This study supports previous observations that only a fraction of patients with GBM treated with PD-1 blockade exhibit radiographic responses and survival benefit. Whereas a recent clinical trial of PD-1 blockade in rGBM failed to show an increase in OS, the study reported positive responses in 7.8% of patients based on radiographic criteria, with prolonged durability of response as typically seen in other solid tumors5. Given that the majority of patients discontinued immunotherapy due to tumoral progression as assessed by radiographic response, which can be related to pseudoprogression5, the percentage of patients with GBM who might benefit from PD-1 blockade could be higher. Moreover, recent studies report encouraging results for the efficacy of neoadjuvant PD-1 blockade14,34. Predictive biomarkers of response to PD-1 inhibitors in other cancers include PD-L1 protein expression, high tumor mutational burden (TMB), MMR deficiency and the status of the tumor immune microenvironment35–37. Studies have reported efficacy of immune checkpoint inhibitors in patients with GBMs harboring germline mutations in the MMR pathway and POLE gene38–40. However, these biomarkers (high-TMB, MSI-high, POLE mutations, tumor-infiltrating lymphocytes and PD-L1 expression) are not informative for the majority of GBMs5–7. MMR-deficient hypermutant tumors that arise as a result of temozolomide treatment do not seem to benefit from immune checkpoint inhibitors41–43. Indeed, recent evidence indicates that mutations derived from chemotherapy-induced MMR deficiency tend to be subclonal and less immunogenic43,44. Thus, to date there are no reliable markers for response to PD-1 blockade for GBM.
We previously reported that rGBMs responsive to PD-1 blockade were enriched in BRAF and PTPN11 mutations. Nevertheless, these mutations were present in only 2–3% of GBM and ~30% of responsive patients13. In this study, we report that patients that respond to PD-1 blockade exhibit activation of ERK1/2, including those that do not have BRAF/PTPN11 mutations. The median OS in rGBM is approximately 5–9 months, with that of resectable tumors closer to 9 months45,46. We observed that 62.5% of patients with GBM in the high-p-ERK group were alive at 1 year after initiation of immunotherapy compared to 0% in the low-p-ERK group. This survival benefit is unlikely to be explained by survivorship bias from resectable patients, because all underwent surgery and it seems to be independent of other prognostic clinical and molecular variables. Furthermore, these results were validated in an independent cohort in which data were collected prospectively as part of a clinical trial. Nevertheless, adoption of p-ERK as a biomarker will require prospective validation.
We identified robust differences in the microenvironment associated with p-ERK/MAPK signaling in GBM, in particular an increased number of tumor-infiltrating myeloid cells and microglia in proximity to p-ERK+ tumor cells in GBM that have elevated p-ERK responsive to PD-1 blockade. Similarly, we showed that development of mouse transgenic gliomas in the absence of CD8+ T cells leads to elevated intratumoral p-ERK1/2, and that this signaling correlates with a robust increase tumor infiltration by Cd11b+ and Iba1+ myeloid cells and a proinflammatory tumor microenvironment47.
While some studies have characterized immunosuppressive glioma-infiltrating myeloid cells30,48, others have described antitumoral activities of myeloid cells in the setting of cancer immunotherapy, including in glioma31,49,50. In a recent preclinical study, transgenic gliomas in CD8a KO mice had prolongation of survival after PD-1 blockade31. Of note, murine gliomas with genetic ablation of CD8a had an increased percentage of proinflammatory CD11b+ MHC II+ cells and peripheral expansion of CD4+ T cells compared to their wild-type CD8a counterparts. This evidence highlights the possible direct contribution of myeloid cells and microglia to response to PD-1 blockade in gliomas31,47.
This investigation also suggests that infiltrating myeloid cells in high-p-ERK tumors might have different antigen presentation capabilities via MHC II. Indeed, scRNA-seq from myeloid cells from tumors with high p-ERK showed enrichment of GO terms related to lymphocyte chemotaxis, chemokines, antigen presentation and predominantly MHC II-related genes, a finding confirmed at the protein level with multiplex immunofluorescence. Some similarities exist between high-p-ERK tumors and a subset of GBMs associated with increased numbers of macrophages and high levels of MHC I and II defined to have mesenchymal (MES)-like states of both cancer cells and macrophages23. Whether p-ERK is associated with a specific GBM cellular state remains to be determined. Nonetheless, this evidence supports the idea that a subgroup of patients with GBM and increased expression of MHC II might benefit from immunotherapy. In this regard, it has been suggested that patients with rGBM and low mutational burden have enrichment of inflammatory gene signatures, including an MHC II gene set and longer survival after immunotherapy51. These findings concur with a study of metastatic melanomas in which increased expression of MHC II-associated genes was associated with response to anti-PD-1 immunotherapy52. Mechanistically, antigen presentation by MHC II to CD4+ T cells has been shown necessary in the induction of an effective antitumoral response during immune checkpoint inhibition53. In particular, peripheral CD4 T cells from GBM patients treated with neoadjuvant PD-1 blockade had elevated expression of CD152 (CTLA4) and CD127 (IL7-Rα) but downregulation of PD-114. These findings, together with our results, suggest an interplay between MHC II-mediated antigen presentation and peripheral activated memory CD4+ T cells that increases after anti-PD-1 therapy in patients with rGBM.
This study also suggests that, in the case of high-p-ERK tumors, there is an abundance of TMEM119+ microglial cells22,25,26. The expression of MHC II in tumor-infiltrating myeloid cells was more robust for TMEM119+ cells compared to CD163+ cells in high-p-ERK tumors. Although microglia have low levels of MHC II expression in homeostatic states54,55, HLA-DR is upregulated in human glioma-infiltrating microglia compared to noncancerous controls26. On the other hand, it has been suggested that impaired MHC II expression by glioma-infiltrating microglia leads to evasion from CD4+ T cells56,57, which could explain the lack of response to PD-1 blockade in some patients with GBM. Whereas previous studies have demonstrated the ability of human and murine microglia to impair tumor growth31,58,59, their exact function in the context of immune checkpoint inhibition remains poorly defined. With the incremental use of single-cell technologies, it will be informative to explore the impact of immune checkpoint blockade on the proportions and functionality of the diverse myeloid cell populations of gliomas22. Considering that tumoral immune composition varies among patients with GBM2,23,27,28, it is possible that a heterogenous clinical response to PD-1 blockade derives from phenotypic differences of tumor-associated myeloid cells that are modulated by interchangeable cellular states described for glioma cells3.
We did not find an association between p-ERK and the proportions of CD3+ T cells (high-p-ERK median, 0.0056%; low-p-ERK median, 0.0058%, P = 0.77, Mann–Whitney U-test). This is not surprising, because we studied immunotherapy-naïve specimens; T-cell sequestration in the bone marrow and scant tumor-infiltrating effector T cells are characteristic of patients with GBM9,28, and T-cell infiltration remains modest in studies employing neoadjuvant PD-1 blockade14. In other solid tumors, neoadjuvant PD-1 blockade induced an expansion of peripheral T-cell clones that were able to infiltrate tumors between 2 and 4 weeks after initiation of immunotherapy60. In our cohort, we noticed an increase in CD4+ and CD8+ T cells in the CSF and brain tissue in a patient with high-p-ERK GBM months after continuous PD-1 blockade (Fig. 1b). Though we acknowledge that this is a description of one case only, it is possible that longer treatment periods with anti-PD-1 antibody may induce more robust T-cell infiltration and activity. Due to the interval (2–3 weeks) between neoadjuvant PD-1 immunotherapy and tissue acquisition7,14, current studies in GBM may not have captured this phenomenon.
One of the limitations of the current study is that it does not establish causality. Therefore, the mechanism underlying p-ERK as a predictive biomarker of PD-1 blockade remains to be determined. Although we found differences in OS between patients with high- and low-p-ERK GBM in the discovery and validation cohorts, we acknowledge that a further limitation is the number of patients analyzed and the retrospective design of the study. Therefore, prospective validation of this biomarker evaluating anti-PD-1 therapy is important. Additionally, given the potential therapeutic benefit of neoadjuvant PD-1 blockade in GBM7,14,34, another unsolved question is whether p-ERK1/2 remains predictive of response in specimens exposed to this immunotherapy.
In summary, these results suggest that p-ERK1/2 is indicative of response to adjuvant PD-1 blockade in patients with rGBM that harbor a unique tumor immune microenvironment composed of myeloid/microglial cells expressing MHC II. This offers an opportunity to apply immunotherapy with a personalized approach for GBM, providing therapeutic benefit for a subset of patients while avoiding futile treatments for others.
Methods
Study design and patient selection.
This is a retrospective cohort clinical study followed by a validation cohort of a prospective clinical trial that evaluates the association of p-ERK as biomarker for response to anti-PD-1 therapy in recurrent GBM patients. As part of the discovery cohort, a control group that did not receive PD-1 blockade was included for survival comparisons with the group that received the immunotherapy. For the discovery cohort, patients were at least 18 years old with diagnosis of recurrent GBM treated with either pembrolizumab or nivolumab or without immunotherapy from two institutions: Northwestern University (n= 40) and Columbia University (n= 22). Institutional review board (IRB) approval was acquired from both institutions. At Northwestern University all participants underwent consent for the Tumor Bank IRB (STU00095863 and STU00206457). At Columbia University, all patients had either informed consent (prospective patients) or waiver of consent (retrospective patients) under the Columbia IRB-AAAJ9652 [Herbert Irving Comprehensive Neurological Cancer Center Database), which enrolls both prospective and retrospective patients. The study was conducted in accordance with the institutional ethical regulations and the Declaration of Helsinki principles. No compensation was provided to participants. Clinicopathological patient data are provided in the Supplementary Table 1, and 2.
Inclusion criteria:
Diagnosis of histologically confirmed recurrent GBM.
Age ≥ 18 years old.
Treatment with PD-1 inhibitor (nivolumab or pembrolizumab) following surgery for resection of recurrent GBM.
Patients must have received first line treatment with temozolomide and radiotherapy.
Exclusion criteria:
Patients that did not have adequate tumor tissue for immunohistochemistry assessment at recurrence.
Patients that did not have clinical data available for analysis.
For the validation cohort, tumor samples of recurrent GBM patients (n = 13) were analyzed from Cloughesy T et al. (NCT02852655)14. We studied the adjuvant arm of this prospective controlled clinical trial. As mentioned in the original publication, patients received pembrolizumab 200 mg by intravenous infusion every 3 weeks after recovery from surgery until either tumor progression or an adverse event requiring study drug discontinuation. Clinical data is accessible in the referenced article and in Supplementary Table 3.
Survival analysis.
The primary endpoint in the discovery and validation cohorts is OS. Secondary endpoints included OS rate at 12 months and PFS. OS is defined as the time of initiation of anti-PD-1 therapy to date of death from any cause for the immunotherapy cohort. For the no immunotherapy cohort, OS is defined as the time of recurrence to the date of death from any cause. OS was censored for GBM patients who were alive at the time of the cutoff date for the discovery and validation cohorts. Clinical and molecular variables influencing survival were explored as part of a Cox proportional hazard model, including age, Karnofsky performance score (KPS), IDH mutational status, MGMT methylation, steroid dose at the time of immunotherapy, size of residual tumor determined by MRI measurement, Ki67, and concurrent treatments. Partition of the groups into high and low p-ERK tumors was determined by using the median of the values derived from the software-based quantification of p-ERK cell density of all the GBM patients in the discovery cohort (immunotherapy group and no immunotherapy cohort). The same value used to define high and low p-ERK tumors in the discovery cohort was applied to the validation cohort to define the same groups. Additionally, to evaluate the ability of p-ERK as a biomarker to select patients that could benefit from treatment with PD-1 blockade in the clinical setting, a neuropathologist scored the tumor samples from 0 to 3, for which specimens were defined as high (2 or 3) and low (0 or 1) p-ERK tumors. For both methods of partitioning into high and low p-ERK groups, software-based quantification of p-ERK cell density and neuropathological assessment of p-ERK-expressing tumor regions, we employed two statistical methods for survival analysis: 1) two-sided log-rank test; 2) Wald test resulting from univariable and multivariable Cox proportional hazard models with the quantification and the score of p-ERK as a variable. Multivariable models looked at the association between p-ERK and survival after adjusting for variables of interest as additive effects. The proportional hazards assumption was checked through the assessment of scaled Schoenfeld residuals. Error bars on forest plots indicate 95% confidence intervals for the hazard ratios.
To calculate the mean AUC values in the discovery and validation cohort, we employed the R package pROC61 in which we evaluated the ability of p-ERK cells/mm2 to predict survival at 12 months after initiation of anti-PD-1 therapy in the discovery and validation cohort. To explore the interaction between p-ERK and immunotherapy with respect to OS, cut-point optimization using conditional inference trees with the R package partykit62,63 (v.1.2–9) was performed. In optimizing the cut-point value for p-ERK, we used the PD-1 blockade treatment arm to avoid confounding due to differences in OS between arms (PD-1 blockade and No immunotherapy).
GBM samples.
Tumor samples from participating institutions used for analysis for the discovery cohort (Northwestern University and Columbia University) were collected by the dedicated Brain Tumor Bank staff and clinical pathology cores that have standard operating procedures (SOPs) in which tissue is fixed and catalogued in a timely fashion. Slides were cut from blocks shortly prior to staining, so no unstained slides were stored for long periods.
Determination of the integrity of p-ERK from FFPE embedded tumors.
Based on the availability of tissue blocks from the Nervous System Tumor Bank, we obtained tissue scrolls from a representative set of 7 low and 5 high p-ERK tumors from the same FFPE GBM tissues used for survival analyses (n=12). We extracted phosphoproteins and total proteins using the QProteome FFPE Tissue Kit following manufacturer’s protocol (Qiagen, cat. 37623). We performed western blot against p-ERK1/2 using the same antibody used to perform IHC in GBM tissues and 2 GBM cell lines representing positive (AM38) and negative (U251MG) controls. To assess the integrity of an additional phosphorylated protein in these tumors, we evaluated the phospho-AKT (Cell Signaling Technology (CST), clone: D9E, dilution 1:1000), ERK1 (Thermo Fisher Scientific, dilution 1:1000), ERK2 (CST, dilution 1:1000), and β-actin (CST, dilution 1:1000).
Peptide competition assay.
The phospho-p44/42 (ERK1/2) antibody (CST) was incubated overnight at 4° C with a specific blocking peptide in a 1:10 dilution that saturated the p-ERK antibody (phospho-p44/42 MAPK (ERK1/2) blocking peptide, catalog #1150, CST). As controls for the blocking peptide, this was also incubated with the antibodies detecting p-EGFR and p-AKT. Western blot was performed to evaluate p-ERK (CST, dilution 1:1000), ERK1/2 (CST, dilution 1:1000), p-EGFR (CST, clone: D7A5, dilution 1: 1000), EGFR (CST, clone: D38B1, dilution 1: 1000), p-AKT (CST, clone: D9E, dilution 1:1000), AKT (CST, clone: C67E7, dilution 1:1000), and β-actin (CST, clone 8H10D10, dilution 1:1000). Additionally, the specificity of the p-ERK1/2 antibody was evaluated by IHC in the same set of samples used to perform the peptide competition assay by western blot.
Western Blot.
Western blotting was performed using conventional protocols. In brief, cells from fresh tumor specimens were lysed with M-PER buffer (Thermo Fisher Scientific) which contained protease and phosphatase inhibitors (Thermo Fisher Scientific). After quantification of proteins acquired from either FFPE GBM tissues or fresh tumor specimens, these were loaded in a 4–20% Mini-PROTEAN TGX Stain-Free protein gel (Biorad) for electrophoresis. Then, blots were blocked for 1 hour at room temperature with 5% milk in 10 mM Tris-HCl pH 7.5, 150 mM NaCl containing 0.1% Tween 20 (TBST) and incubated at room temperature for 1 hour with the corresponding primary antibodies. Blots were washed after incubation with the primary antibodies and then incubated with an anti-rabbit IgG HRP-linked antibody (CST, dilution 1:5000) for 1 hour at room temperature. Finally, antigen-antibody reaction was detected using Clarity Western ECL substrate kit (catalog #1705061, Biorad) following manufacturer’s protocols.
Evaluation of the effect of ischemic time on p-ERK stability.
3 human tumor specimens were obtained during neurosurgery, and immediately divided into similar size portions and subjected to different ischemic times (0 hr., 0.5 hr., 1 hr., and 2 hrs.) before fixation. Next, samples were processed and embedded in paraffin blocks that were cut to get tissue sections. IHC staining was performed in these sections with the same dilution used to stain all GBM samples in the study. Stained slides were scanned to perform image analysis of endothelial cells. Depending on the number of vessels in each tumor, 5 to 24 ROIs each corresponding to one endothelial cell were acquired for tumor samples subjected to different ischemic times and from tumor of the discovery cohort. The intensity of endothelial cells fixed immediately after resection (0 hr. of ischemic time) was used as the reference to compare the intensity of all the tumor samples.
Immunohistochemistry staining.
IHC and H&E staining was performed using standard immunoperoxidase staining on FFPE tissue sections of 5 μm thick from resected recurrent tumors. Sections were stained against mouse anti-phospho-p44/42 (ERK1/2) (CST, dilution 1:500), mouse anti-Iba1 (Abcam, dilution 1:1000) mouse anti-CD3 (DakoCytomation, dilution 1:200). The procedure was performed in a DAKO Autostainer Link 48 slide stainer (Agilent Technologies). Paraffin sections were deparaffinized with xylene in the stainer and then underwent heat-mediated antigen retrieval with sodium citrate buffer. Sections were counterstained with hematoxylin, dehydrated, and mounted with coverslips. The slides were scanned and digitalized with the Hamamatsu K.K. Nanozoomer 2.0 HT and were visualized with the NDP.view2 Viewing software. A board-certified neuropathologist evaluated the staining digitally to ensure the appropriate quality of the tumor tissue.
IHC image analysis.
For the quantification of p-ERK, a neuropathologist outlined the tumoral regions on each sample in a blinded fashion regarding treatments, survival outcomes, and other clinical characteristics. HistoQuest version 6.0 software (TissueGnostics) was employed for the quantification of p-ERK cell density. We employed a nuclear segmentation method, a ring mask with interior radius of −0.45 μm and exterior radius of 0.91 μm, and a cytoplasmic cell mask. For each cell, the nucleus was segmented and a cytoplasmic region surrounding the nucleus was defined. To identify positive nuclei, we adjusted the software parameters to detect hematoxylin alone (color RGB: 30, 45, 84) and DAB chromogen (color RGB: 94, 48, 14) combined with hematoxylin. To consider a cell positive for p-ERK, stained cells had to exceed the mean intensity threshold of 150 in the cytoplasmic compartment after the machine identified a nucleus. For the quantification of Iba1, we employed a nuclear segmentation method with cell mask segmentation without using a ring mask. Then, we taught the machine to recognize the positive signal derived from the DAB chromogen in the color separation parameters (color RGB: 81, 41, 16). Positive Iba1 signal was the result of a mixture between the DAB chromogen and hematoxylin (color RGB: 102, 109, 128). A mean intensity threshold of 130 was set to define a cell positive for Iba1. We analyzed the tumor regions that the neuropathologist delineated. p-ERK and Iba1 cell density was defined as the number of p-ERK+ or Iba1+ cells in a given area (mm2). For the tumor samples that had 2 or more spatially separated tumor regions, these tissues were quantified, and the resulting values were averaged to provide a single value for a tumor sample.
Tumor sample processing for flow cytometry data acquisition and analysis.
GBM tumor samples were collected and kept in cold PBS before processing the samples for FACS analysis. Tumor samples were gently dissociated using 70 μm cell strainers (Fisher Scientific 352350) in cold FACS buffer (PBS + 2 mM EDTA + BSA 2%) and washed two times in FACS buffer (50 mL). Cells were incubated for 5 minutes with human Fc block (BD, 564219) on ice and then incubated with antibodies at 4°C for 30 mins. Cells were washed, and 7AAD (BD, 559925) was added (1:100 dilution) before FACS acquisition.
Blood samples were collected on heparin tubes and mix at a 1:1 ratio with DMEM medium at room temperature to perform lymphocyte (BD, NC9587917) PBMC isolation. PBMC were washed in FACS buffer and stained similarly to the tumor samples. Antibodies used were APC anti-CD45 (Biolegend, dilution 1:200), PE-Cy7 anti-CD8a (eBiosciences, dilution 1:200), APC-eFluor 780 anti-CD4 (eBiosciences, dilution 1:200). Data were acquired on BD Fortessa LSRII and analyzed by first gating live cells, then CD45+ cells, and finally CD4+ cells in the X axis and CD8+ in the Y axis of the scatter plot. Analyses were conducted using FlowJo v. 10.6.2.
Multiplex immunofluorescence.
Sections of 5-μm thickness were obtained from FFPE tumors. Deparaffinization of the slides was done with xylene and then rehydrated in histological grade ethanol and fixed with 3% hydrogen peroxide in methanol before antigen retrieval using pH6 citrate buffer or pH9 EDTA buffer. For the first panel, the primary antibodies used were: p-ERK (CST, clone D13.14.4E, 1:1600), CD163 (Abcam, clone EPR19518, 1:600), MHC II (Abcam, 1:400), TMEM119 (Sigma-Aldrich, 1:250), SOX2 (Abcam, clone EPR3131, 1:5000). Tyramide signal amplification (TSA) visualization was done using the Opal 7-color IHC Kit (NEL821001KT, Akoya Biosciences). Opal 520 (1:100), Opal 540 (1:200), Opal 570 (1:800), Opal 620 (1:150), Opal 690 (1:100) and DAPI were used to pair primary antibodies. For the second panel, the primary antibodies used were the following: p-ERK (CST, clone D13.14.4E, 1:1600), GFAP (Abcam, clone EPR1034Y, 1:300), CD163 (Abcam, clone EPR19518, 1:100). TSA visualization was done using the Opal 7-color IHC Kit (NEL821001KT, Akoya Biosciences). Opal 520 (1:200), Opal 620 (1:250), Opal 690 (1:600), and DAPI were used. Multiplex staining was performed with an antigen retrieval step, protein blocking, epitope labeling, and signal amplification between each cycle. Finally, Spectral DAPI (Akoya Biosciences) was used to counterstain the slides and were mounted with long lasting aqueous-based mounting medium.
Imaging and analysis of multispectral images.
For the first panel, multispectral imaging (MSI) was performed using the Vectra 3 Automated Quantitative Pathology Imaging System from Akoya Biosciences. Whole slide images were acquired after autoadjusting focus and signal intensity. MSI was acquired in the tumor regions delineated by the neuropathologist at 20x of original magnification. Then, we created a spectral library for all fluorophores to subject acquired multispectral images to spectral unmixing to visualize the signal of each marker (SOX2, p-ERK, TMEM119, CD163, MHC II, DAPI) in inForm Tissue Finder software 2.4.9 (Akoya Biosciences). Nuclear-based cell segmentation using DAPI was performed as well as phenotyping of the cell markers.
For the second panel, the stained slides were scanned using Vectra Polaris (Akoya Biosciences). Images were visualized using Phenochart (v1.0.12) to select multiple regions of interest (ROIs) encompassing the tumoral regions delineated by the neuropathologist to maintain consistency with the IHC quantification analysis. The selected ROIs were uploaded to inForm 2.4.9 (Akoya Biosciences) to subject the images to spectral unmixing. Then, after adjusting the parameters to identify nucleus of the analyzed cells, cell segmentation was performed to determine the nuclear and cytoplasmic compartments on each cell. For the two panels, we used a machine-learning algorithm within inForm in which cells were automatically assigned to a specific phenotype (SOX2+, TMEM119+, CD163+, p-ERK+, MHC II+) (CD163+, GFAP+, p-ERK+, CD163−, GFAP−, p-ERK−). The processing and analysis of images from all tumor samples were exported to cell segmentation tables. Exported files from inForm were processed in R using R packages Phenoptr and PhenoptrReports to merge and create consolidated single files for each tumor sample. Consolidated files had double cell phenotypes as outputs that were employed for further quantification and spatial analyses using the Phenoptr R addin.
Cell quantification and spatial analysis of multiplex immunofluorescence images.
The consolidated files obtained using Phenoptr were analyzed to quantify the density of SOX2+ p-ERK+, SOX2+ p-ERK−, TMEM119+, CD163+, TMEM119+ MHC II+, CD163+ MHC II+, GFAP+, GFAP+ p-ERK+ cells, CD163+ p-ERK+ cells, and GFAP− CD163− p-ERK+ cells. For the spatial analysis, mean distances between the nearest neighbors were calculated from myeloid cells (TMEM119+ and CD163+ cells) to SOX2 p-ERK+, SOX2 p-ERK−, GFAP+ p-ERK+, and GFAP+ p-ERK− cells. The spatial map viewer addin within R allowed the visualization of nearest cell neighbor between selected phenotypes in a single field of a high and low p-ERK tumor. Cartoons were created using Adobe Illustrator v.22.1.0.
scRNA-seq processing.
Single-cell RNA-seq data was obtained from 10 GBM cases previously published by Yuan, et al.24 All datasets were first filtered to remove genes coding for mitochondrial and ribosomal proteins. Count matrices for each case were then merged keeping the union of genes with genes with zero total counts being discarded. Raw counts were then normalized to log2(1 + TPK), as described in Yuan, et al.24,. Z-scores for gene expression were calculated based on these normalized counts. For visualization: first Principal Components Analysis (PCA) was applied to reduce the total dimensionality to 5% of the number of genes, then Uniform Manifold Approximation and Projection (UMAP)64 with default parameters to non-linearly reduce that into a two-dimensional embedding. Agreeing with the previous analysis from Yuan, et al.24, four cell types were readily identified using the standard markers of CD14, VWF, PDGFRB, and SOX2 for myeloid, endothelial, pericytes, and tumor cells, respectively.
scRNA-seq analysis.
Differential expression in the myeloid compartment was calculated by computing Welch’s t-statistic between cells from p-ERK-low and p-ERK-high cases. The single-cell subtype score method from Patel, et al.65 was used to calculate signature scores for each of the gene sets. GSEA was performed using the Prerank module of gseapy33, ranking by signal-to-noise of the normalized expression values across cells and using default parameters. Gene sets were obtained from MSigDB v7.1, utilizing the C5: GO gene sets collection66.
Statistical analysis.
GraphPad Prism v6.0c and 8, Python 3.6, R v. 4.0.2, and Microsoft Excel v16.33 were used for statistical analyses. No statistical method was used to predetermine sample size. Patients were not randomized and were excluded from analysis if there was insufficient tumor sample for biomarker assessment. Neither investigators nor patients were blinded to treatments in the discovery and validation cohorts. Numerical data is reported as mean ± sd. For the analysis requiring two-sided nonparametric and parametric calculations, Mann-Whitney U-test and Student t test, respectively, were used for non-paired observations. Wilcoxon signed-ranked test was used for non-parametric calculations comparing matched samples. Pearsońs r was used to correlate raw data values of the indicated variables depicting linear relationships. One-way ANOVA was used for multiple comparisons and P values were adjusted using Tukey or Dunn test for multiple comparisons where it was appropriate. P value threshold of 0.05 was considered statistically significant.
Data availability
Single-cell RNA–sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE103224 and GSE141383. Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
Code is available in https://github.com/RabadanLab/GBMsinglecell.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
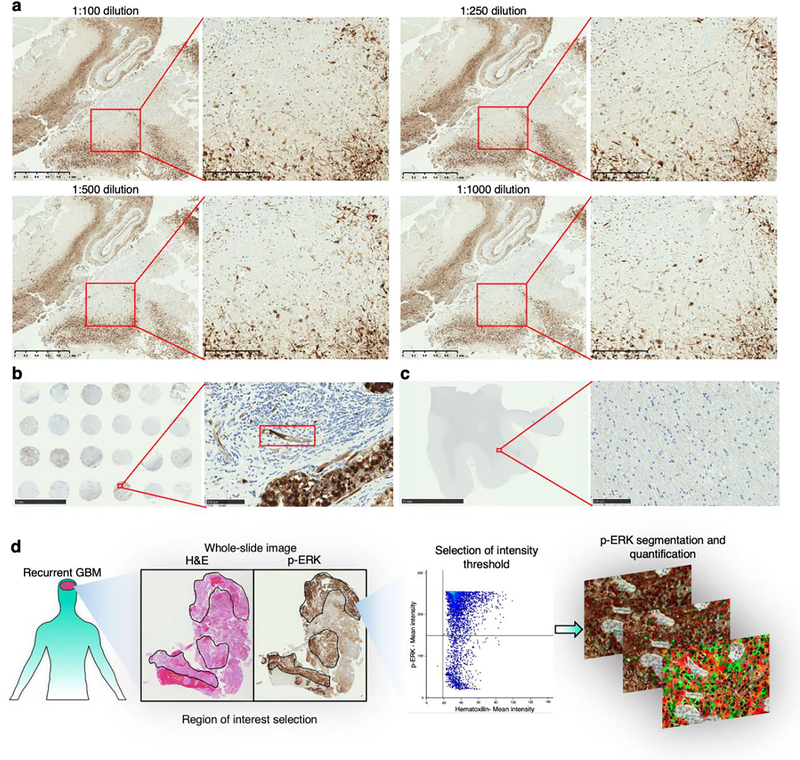
Extended Data Fig. 1 |. Optimization of the staining technique and quantification of p-ERK.
a, Titration of the p-ERK antibody (clone: D13.14.4E) using different dilutions performed in GBM samples. We show the same region of a GBM sample stained with the indicated dilutions of the p-ERK antibody with a low and high magnification image for each dilution. b, (left) Microarray containing breast cancer tissues stained with p-ERK antibody (1:500 dilution) representing a positive control. (right) Magnification of one the breast cancer tissues showing specific staining in the endothelium (red rectangle). c, (left) Nontumoral brain tissue stained with p-ERK antibody (1:500 dilution) representing a negative control. (right) Magnification of the white matter showing p-ERK staining with minimal background. Dilution titration and staining of positive and negative controls were performed as a single experiment in one standardized run. d, Workflow used for the software-based quantification of p-ERK+ cells.
Extended Data Fig. 2 |. Quantification and cut point optimization of p-ERK+ cell density in tumoral regions.
a, Dot plot showing the distribution of p-ERK quantification of all GBM samples treated and nontreated with PD-1 blockade N = 62 tumors). b, From top to bottom, micrographs showing one high p-ERK tumor sample and two low p-ERK tumor samples with positive staining in the endothelial cells (red arrows). In the dot plot, the magenta dot represents CU100 patient, the green dot represents NU01688 patient, and the red dot represents CU110 patient. IHC images are representative of 62 independent GBM samples. c, Conditional inference trees analysis for cut-point optimization in the GBM cohort treated with PD-1 blockade reveals a cut-point value similar to the median of all tumor samples. d, Forest plot representing the univariable analysis using a Cox regression model evaluating the clinical and molecular prognostic factors that might confound the association between survival p-ERK and presented as Hazard ratio (95% CI). N = 29 GBM patients. P value by two-sided Wald test. e, Kaplan-Meier curve comparing OS of recurrent GBM patients scored as either high or low p-ERK by assessment of a neuropathologist counting from initiation of PD-1 blockade (anti-PD-1 therapy group, N = 29 GBM patients) and from surgery at recurrence (no-immunotherapy group, N = 33 GBM patients). p-ERK scores in tumor regions were designated as follows: 0–1 were considered as low, and 2–3 as high; P value by two-sided log-rank test.
Extended Data Fig. 3 |. Preservation of the p-ERK epitope and peptide competition assay neutralizing the p-ERK1/2 antibody tested in FFPE GBM samples.
a, Protein extraction from FFPE GBM tissues for assessment of selected phosphoproteins. b, Western blot targeting p-ERK, ERK1, ERK2, p-AKT, AKT, and β-actin in a subset of GBM samples used for survival analysis. Western blotting was done as a single experiment in 12 independent GBM samples and 2 GBM cell lines. c, Peptide competition assay in which p-ERK1/2 antibody was neutralized with a blocking peptide employing extracted proteins obtained from GBM samples. The peptide competition assay was assessed by western blot. One western blot was incubated with the neutralized p-ERK antibody and the other with the free p-ERK antibody. d, Peptide competition assay employing IHC using the same GBM samples used to perform western blot employing the neutralized and free p-ERK antibody to perform the staining. The experiments were done in 4 independent GBM samples and 2 GBM cell lines as a single experiment.
Extended Data Fig. 4 |. Evaluation of the ischemic time on p-ERK degradation by IHC and western blot, and comparison to the samples used in this study.
a, Representatives images of the analysis conducted to evaluate p-ERK degradation in endothelial cells of GBM samples at different periods of ischemic time. For this, 3 human tumor specimens were obtained during surgery, and immediately divided into similar size portions, which then were subjected to different ischemic times before processing. Specific endothelial cells subjected to analysis are labeled with colors assigned by the software. b, Blue bars represent p-ERK+ cells mm2 in tumor regions, and dots represent p-ERK intensity on individual endothelial cells within the same samples used to evaluate the effect of ischemic time on p-ERK degradation, and tumor samples used for survival analysis (PD-1 immunotherapy cohort and no immunotherapy cohort). Each dot represents one ROI analyzing one endothelial cell. Green dots (N = 24, 20, 13 endothelial cells from NU02608, NU02617, and NU02609, respectively) represent a statistically significant group compared to the group of 0 hrs. of ischemic time represented as gray dots (N = 18, 19, 14 endothelial cells from NU02608, NU02617, and NU02609, respectively). All samples were normalized to the average of values of the three 0 hrs. groups. P values by two-sided Kruskal Wallis test with post hoc Dunn’s multiple comparison test. c, Western blot showing p-ERK and other phosphoproteins in samples subjected to different ischemic times. Densitometry analysis for p-ERK western blot was performed using ERK1 and ERK2 staining. For this densitometry, every patient had density normalized by 0 minutes of ischemic time. N = 3 GBM samples. Error bars represent SEM. Western blot was done as a single experiment in 3 independent GBM samples.
Extended Data Fig. 5 |. Progression-free survival of the validation cohort from the Cloughesy T et al.14 clinical trial.
a, b, Kaplan-Meier showing progression-free survival following PD-1 blockade based on p-ERK high vs low for pre-study (a) and on-study (b) tumor samples. N = 13 GBM patients. P values by two-sided log rank test.
Extended Data Fig. 6 |. Multiplex immunofluorescence staining of recurrent GBM samples employing GFAP marker.
a, Bar plot showing the comparison of GFAP+ p-ERK+ cells and other cells expressing p ERK+. N = 6 tumor samples. P value by two-sided Mann Whitney U test. Data is presented as mean ± s.d. b, Representative images of three different tumor samples derived form results in a. From top to bottom: a BRAFV600E GBM sample having high p-ERK staining, a wild-type BRAF/PTPN11 GBM having high p ERK staining, and a wild-type BRAF/PTPN11 GBM displaying low p-ERK staining. For the three tumor samples: (left) H&E and p-ERK IHC images of the same tumor region. (middle), Multiplex immunofluorescence images showing the markers for GFAP, p-ERK, and DAPI. (right) Multiplex immunofluorescence images showing the markers for GFAP, CD163, and DAPI. Experiment was done using a tumor sample in one standardized run per patient.
Extended Data Fig. 7 |. Single-cell RNA seq of GBM patients with high and low p-ERK IHC staining.
UMAP representation of 28,194 individual cells from 10 GBM patients measured with scRNA-seq (left). UMAP graph showing the representation of 3,153 myeloid cells derived from the 10 GBM patients (right). Each dot represents an individual cell.
Supplementary Material
Acknowledgments
This work was supported by NIH grant nos. 1R01NS110703-01A1 and 5DP5OD021356-05 (both A.M.S.); no. P50CA221747 SPORE for Translational Approaches to Brain Cancer (principal investigator M. S. Lesniak, with support to A.M.S.); developmental funds from the Robert H. Lurie Cancer Center Support Grant (no. P30CA060553, A.M.S.); Vagelos Precision Medicine Award (F.M.I., R.R. and J.Z.), U54CA193313 (R.R.), U54CA209997 (R.R.), R35CA253126 (R.R.); Keep Punching (F.M.I.); The William Rhodes and Louise Tilzer-Rhodes Center for Glioblastoma at New York-Presbyterian Hospital (F.M.I.); NIAID nos. 1R01AI099195 (U.B.) and R01AI134988 (U.B.). V.A.A. is financially supported by the Mexican government through the Mexican National Council for Science and Technology and the Plan of Combined Studies in Medicine of the National Autonomous University of Mexico. A.X.C. is funded by the Medical Scientist Training Program (no. T32GM007367). We thank T. Sudhakar for sample collection at Columbia University; K. McCortney, R. Javier and J. Walshon from the Nervous System Tumor Bank supported by the P50CA221747 SPORE for Translational Approaches to Brain Cancer; and B. Shmaltsuyeva for immunohistochemistry at the Northwestern University Pathology Core Facility funded by Cancer Center Support Grant (no. NCI CA060553). We thank L. Kai for technical support on multiplex staining performed at the Immunotherapy Assessment Core at Northwestern University and in the Flow Cytometry & Cellular Imaging Core Facility, which is supported in part by NIH through MD Andersońs Cancer Center Support Grant no. CA016672, the NCÍs Research Specialist 1 (no. R50 CA23707, J.K.B.) and NIH grant nos. R01-CA120813 and 1R01-CA237418 (both A.B.H.).
Competing interests
F.M.I. is a consultant for Merck, Novocure, Regeneron, Abbvie, Alexion, Guidepoint and Tocagen. He has received clinical or research support (including equipment or material) from Bristol Myers Squibb and Merck. A.M.S., F.M.I. and R.R. are co-authors for the following patent filed by Columbia University: Systems and methods for predicting clinical responses to immunotherapies. A.M.S., V.A.A, F.M.I. and R.R. are co-authors of the following patent filed by Northwestern University: 5. Methods for treating glioblastoma. A.M.S. has received a consulting honorarium from Abbvie. R.R. is a member of the SAB of AimedBio and a founder of Genotwin. R.V.L. has received support from Roche-Genentech for meeting travel to present study results; honoraria for advisory boards for AstraZeneca, Abbvie and Ziopharm; and honoraria for medical editing for EBSCO publishing, Medlink Neurology and American Physician Institute. He has also received honoraria for consultation with Eisai and Abbvie, and honoraria for creating and presenting CME Board review material for the American Physician Institute. R.V.L. received drug support (but no additional support) from BMS for an investigator-initiated trial. A.B.H. is a consultant for Caris Life Sciences and WCG Oncology Advisory Board, receives royalties and milestone payments from Celldex Therapeutics and DNAtrix and receives clinical or research support (including equipment and materials) from Celularity, Codiak Biosciences, Moleculin and Carthera. The remaining authors declare no competing interests.
References
- 1.Stupp R, et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 318, 2306–2316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 32, 42–56.e46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neftel C, et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 178, 835–849.e821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong J, Chehrazi-Raffle A, Reddi S & Salgia R Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer 6, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reardon DA, et al. Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial. JAMA Oncol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nayak L, et al. Randomized phase II and biomarker study of pembrolizumab plus bevacizumab versus pembrolizumab alone for recurrent glioblastoma patients. Clin Cancer Res (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot J, et al. Window-of-opportunity clinical trial of pembrolizumab in patients with recurrent glioblastoma reveals predominance of immune-suppressive macrophages. Neuro Oncol 22, 539–549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrieta VA, et al. The possibility of cancer immune editing in gliomas. A critical review. Oncoimmunology 7, e1445458 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chongsathidkiet P, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med 24, 1459–1468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson CM, Choi J & Lim M Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol 20, 1100–1109 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Arrieta VA, et al. Can patient selection and neoadjuvant administration resuscitate PD-1 inhibitors for glioblastoma? J Neurosurg, 1–6 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Woroniecka K, et al. T-Cell Exhaustion Signatures Vary with Tumor Type and Are Severe in Glioblastoma. Clin Cancer Res 24, 4175–4186 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med 25, 462–469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cloughesy TF, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 25, 477–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhillon AS, Hagan S, Rath O & Kolch W MAP kinase signalling pathways in cancer. Oncogene 26, 3279–3290 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Ruess DA, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 24, 954–960 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Svensson S, et al. ERK phosphorylation is linked to VEGFR2 expression and Ets-2 phosphorylation in breast cancer and is associated with tamoxifen treatment resistance and small tumours with good prognosis. Oncogene 24, 4370–4379 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Roth P, Valavanis A & Weller M Long-term control and partial remission after initial pseudoprogression of glioblastoma by anti-PD-1 treatment with nivolumab. Neuro Oncol 19, 454–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada H, et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol 16, e534–e542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vassilakopoulou M, et al. Preanalytical variables and phosphoepitope expression in FFPE tissue: quantitative epitope assessment after variable cold ischemic time. Lab Invest 95, 334–341 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Nagasawa-Masuda A & Terai K ERK activation in endothelial cells is a novel marker during neovasculogenesis. Genes Cells 21, 1164–1175 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Pombo Antunes AR, et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci 24, 595–610 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Hara T, et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 39, 779–792.e711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan J, et al. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med 10, 57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett ML, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113, E1738–1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sankowski R, et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat Neurosci 22, 2098–2110 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Friebel E, et al. Single-Cell Mapping of Human Brain Cancer Reveals Tumor-Specific Instruction of Tissue-Invading Leukocytes. Cell 181, 1626–1642.e1620 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Klemm F, et al. Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 181, 1643–1660.e1617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saha D, Martuza RL & Rabkin SD Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer Cell 32, 253–267.e255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goswami S, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med 26, 39–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao G, et al. Anti-PD-1 Induces M1 Polarization in the Glioma Microenvironment and Exerts Therapeutic Efficacy in the Absence of CD8 Cytotoxic T Cells. Clin Cancer Res 26, 4699–4712 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keren L, et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387.e1319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schalper KA, et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat Med 25, 470–476 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellmann MD, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 378, 2093–2104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cristescu R, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johanns TM, et al. Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discov 6, 1230–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukas RV, et al. Clinical activity and safety of atezolizumab in patients with recurrent glioblastoma. J Neurooncol 140, 317–328 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Bouffet E, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol 34, 2206–2211 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Ahmad H, Fadul CE, Schiff D & Purow B Checkpoint inhibitor failure in hypermutated and mismatch repair-mutated recurrent high-grade gliomas. Neurooncol Pract 6, 424–427 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51, 202–206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Touat M, et al. Mechanisms and therapeutic implications of hypermutation in gliomas Nature 580, 517–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351, 1463–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wick W, et al. Lomustine and Bevacizumab in Progressive Glioblastoma. N Engl J Med 377, 1954–1963 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Azoulay M, et al. Benefit of re-operation and salvage therapies for recurrent glioblastoma multiforme: results from a single institution. J Neurooncol 132, 419–426 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Robert KJ, et al. CD8+ T-cell-mediated immunoediting influences genomic evolution and immune evasion in murine gliomas. Clin Cancer Res 26, 4390–4401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen P, et al. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer Cell 35, 868–884.e866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutter G, et al. Microglia are effector cells of CD47-SIRPα antiphagocytic axis disruption against glioblastoma. Proc Natl Acad Sci U S A 116, 997–1006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strauss L, et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci Immunol 5(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gromeier M, et al. Very low mutation burden is a feature of inflamed recurrent glioblastomas responsive to cancer immunotherapy. Nat Commun 12, 352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 25, 1916–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alspach E, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574, 696–701 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mrdjen D, et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity 48, 380–395.e386 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Mundt S, et al. Conventional DCs sample and present myelin antigens in the healthy CNS and allow parenchymal T cell entry to initiate neuroinflammation. Sci Immunol 4(2019). [DOI] [PubMed] [Google Scholar]
- 56.Qian J, et al. TLR2 Promotes Glioma Immune Evasion by Downregulating MHC Class II Molecules in Microglia. Cancer Immunol Res 6, 1220–1233 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Schartner JM, et al. Impaired capacity for upregulation of MHC class II in tumor-associated microglia. Glia 51, 279–285 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Sarkar S, et al. Therapeutic activation of macrophages and microglia to suppress brain tumor-initiating cells. Nat Neurosci 17, 46–55 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Sarkar S, et al. Microglia induces Gas1 expression in human brain tumor-initiating cells to reduce tumorigenecity. Sci Rep 8, 15286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang J, et al. Compartmental Analysis of T-cell Clonal Dynamics as a Function of Pathologic Response to Neoadjuvant PD-1 Blockade in Resectable Non-Small Cell Lung Cancer. Clin Cancer Res 26, 1327–1337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.T, H., K, H. & A, Z. Unbiased Recursive Partitioning: A Conditional Inference Framework. Journal of Computational and Graphical Statistics 15(3), 651–674 (2006). [Google Scholar]
- 63.T, H. & A, Z. partykit: A Modular Toolkit for Recursive Partytioning in R. Journal of Machine Learning Research 16, 3905–3909 (2015). [Google Scholar]
- 64.McInnes L, Healy J, Saul N & Großberger L UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software 3(2018). [Google Scholar]
- 65.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA–sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE103224 and GSE141383. Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author on reasonable request.