Abstract
Lipid nanodiscs (LNDs), comprising a phospholipid bilayer encircled by two molecules of a recombinant membrane scaffold protein, can be targeted to tumors with covalently attached antibodies (Abs) or their fragments. Antibody attachment to click chemistry based PEGylated lipids on LNDs including DOTA allowed PET imaging with the positron emitter 64Cu. Carcinoembryonic antigen (CEA) positive tumors in CEA transgenic mice were chosen as a tumor target. Fab’ fragments, that otherwise are rapidly cleared by the kidney due to their small size, were retained in circulation when conjugated to LNDs. Untargeted PET imaging of 64Cu-DOTA-LNDs revealed low tumor uptake (4–5 %ID/g) in the range expected for the enhanced permeability retention (EPR) effect with high liver uptake (17–21 %ID/g) indicating gut clearance. Fab’-targeted LNDs showed little improvement over untargeted LNDs, but intact IgG targeted LNDs gave high tumor uptake (40 %ID/g) with low liver (8 %ID/g), demonstrating that tumor targeting with antibody conjugated LNDs is feasible.
Keywords: positron emission tomography, carcinoembryonic antigen, lipid nanodiscs, click chemistry
Graphical Abstract
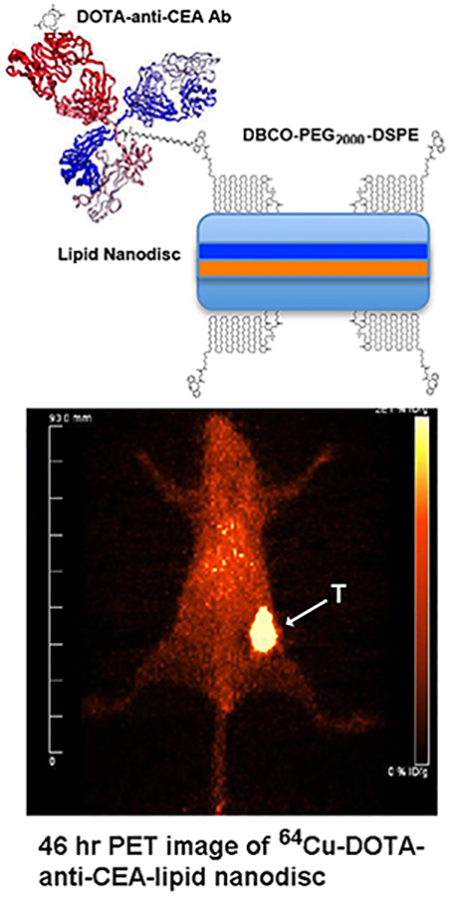
INTRODUCTION
Humanized anti-tumor antibodies have served as versatile platforms for the delivery of imaging1–2 and therapeutic isotopes3, drugs4 and dyes5. However, they have a limited capacity for payloads before their blood clearance and tumor targeting effectiveness are compromised6. As an alternative approach, we7–8 and others9–10 have sought to incorporate antibodies and their fragments into platforms that allow attachment of multiple moieties. Among the possibilities, PEGylated lipid nanoparticles (LNPs) are attractive due to their ability to self assemble into relatively uniform sized nanoparticles (10–300 nm) made from easily derivatized monomeric precursors11. LNPs formulations have been successfully used in many clinical trials12–13, and more recently, shown to have increased tumor accumulation when carrying anti-tumor antibody fragments7–8. In those studies we showed that untargeted LNPs are limited to the enhanced permeability retention (EPR) effect, an effect that has been widely exploited for delivery of untargeted moieties to tumors14. Although, the advent of targeted LNPs has further increased the utility of the LNP platform, it is often limited in scope by the lipids and proteins that it can carry, along with their rapid disassembling under physiological conditions15. In an ongoing search for even more versatile lipid based platforms, we have begun to explore the use of lipid nanodiscs (LNDs).
LNDs comprise a lipid bilayer encircled by two molecules of recombinant membrane scaffold proteins (MSP) that, depending on the size of the MSP, can yield LNDs ranging in diameter from 9–15 nm16. MSPs are truncated versions of apo-A1, a natural component of HDL16–17. Like LNPs, LNDs incorporate a large number of phospholipids that can be derivatized to carry small molecules or proteins18. Unlike LNPs, membrane proteins can be inserted into LNDs to enable structural and biological studies18–20. Given our success with derivatized PEGylated lipids to generate tumor targeted LNPs7–8, we decided to incorporate PEGylated lipids into LNDs as new platforms for targeting tumors with drugs and antibody fragments.
We have selected CEA positive cancer as a model system to take advantage of its high expression in breast21 and colon cancer22 and the availability of a humanized anti-CEA monoclonal antibody (mAb) and their fragments23–24. In order to determine quantitative tumor localization, we derivatized PEGylated lipids and antibody fragments with DOTA to allow in vivo PET imaging. The results demonstrate that antibody targeted LNDs have a 4-fold increased tumor uptake over untargeted LNDs in which uptake is limited by the EPR effect.
RESULTS and DISCUSSION
Generation of DSPE-PEG2000-DBCO LNDs and reaction with azido-monoamide-DOTA and azido-PEG4-Dox
LNDs were synthesized according to Bayburt et al.16. Since the ultimate goal was to target tumors with LNDs, we opted to generate the LNDs with incorporation of the 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-n-[dibenzocyclooctyl(polyethylene glycol)-2000] lipid (DSPE-PEG2000-DBCO) in order that appropriate azido derivatives of DOTA, doxorubicin, anti-CEA mAbs and its F(ab’) fragment could be “clicked” into the LNDs under physiological conditions. The size of the LNDs and resulting number of phospholipids per disc is determined by the size of the recombinant membrane scaffold proteins (MSPs). For this study we chose the commercially available MSP1D1 that generates a LND size of about 10 nm with 160 dimyristyphospholipids (DMPCs) per LND18. As a simple starting point, we chose a molar ratio of 4 DSPE-PEG2000-DBCO per LND to assure conjugation of at least 1 targeting moiety per LND. Since the molecular size of an intact antibody (15 nm) is in the same size range as the LND, we did not want to over-conjugate the LND with antibody. Incorporation of DSPE-PEG2000–DBCO was performed by mixing the lipids in chloroform and drying the lipids into a thin film. Generation of LND was accomplished by sonicating the lipids with PBS until a clear solution was obtained. A scheme outlining the incorporation of DSPE-PEG2000-DBCO into the LND and its reaction with azido-monoamide–DOTA, azido-PEG4-Dox, azido-PEG5-Ab-DOTA, and azido-PEG5-F(ab’)-DOTA are shown in Scheme 1.
Scheme 1.
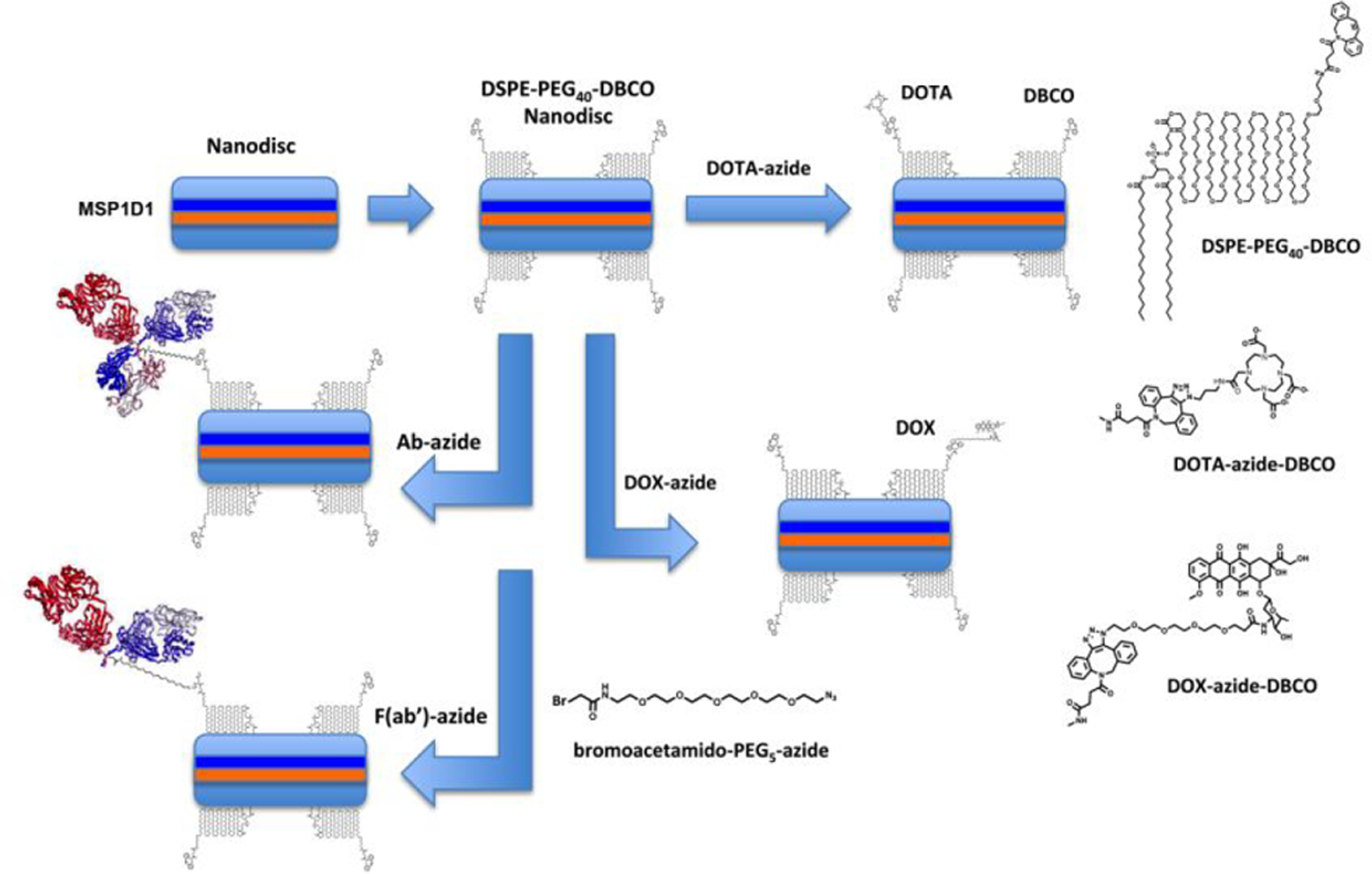
The LND is shown with 2 copies of MSP1D1 and addition of 4 copies of DSPE-PEG40-DBCO (to represent one of the many species of PEG2000). Larger size structures are shown to the right.
The resulting LND preparations were characterized by size exclusion (SEC) HPLC to determine their molecular sizes as read out by their Stokes radii (Fig 1). LNDs comprising DMPC only (Fig 1A) eluted at a later time (24.5 min) than the DSPE-PEG2000-DBCO LND (23.5 min, Fig 1B), demonstrating that the low level of incorporation of DSPE-PEG2000-DBCO had a modest effect on increasing the Stokes radius of the LND. The change in retention time is evidence that DSPE-PEG2000-DBCO has been incorporated into the LND. In addition, the stoichiometry of incorporation of DSPE-PEG2000-DBCO into the LND was determined by 1D 1H NMR as 4.1 (Supplemental Fig S1), in agreement with the molar ratios used to generate LND. Since another goal of this project was to follow the in vivo biodistributions of the LNDs, the DSPE-PEG2000-DBCO lipid was clicked to azido-monoamide-DOTA to allow radiolabeling with the PET radionuclide 64Cu. As shown in Fig 1C, this modification did not significantly affect retention of the LND on SEC HPLC (23.0 vs 23.5 min). Evidence that azido-monoamide-DOTA had reacted with the DBCO moiety of DSPE-PEG2000-DBCO in the LND was provided by monitoring the reaction by 1D 1H NMR in the aromatic region of the spectrum (Supplemental Fig S2). According to this analysis the reaction was almost complete in 3.5 hrs at a ratio of 2.4 azido-monoamide-DOTA per DSPE-PEG2000-DBCO in the LND.
Figure 1. SEC HPLC analyses of LNDs.
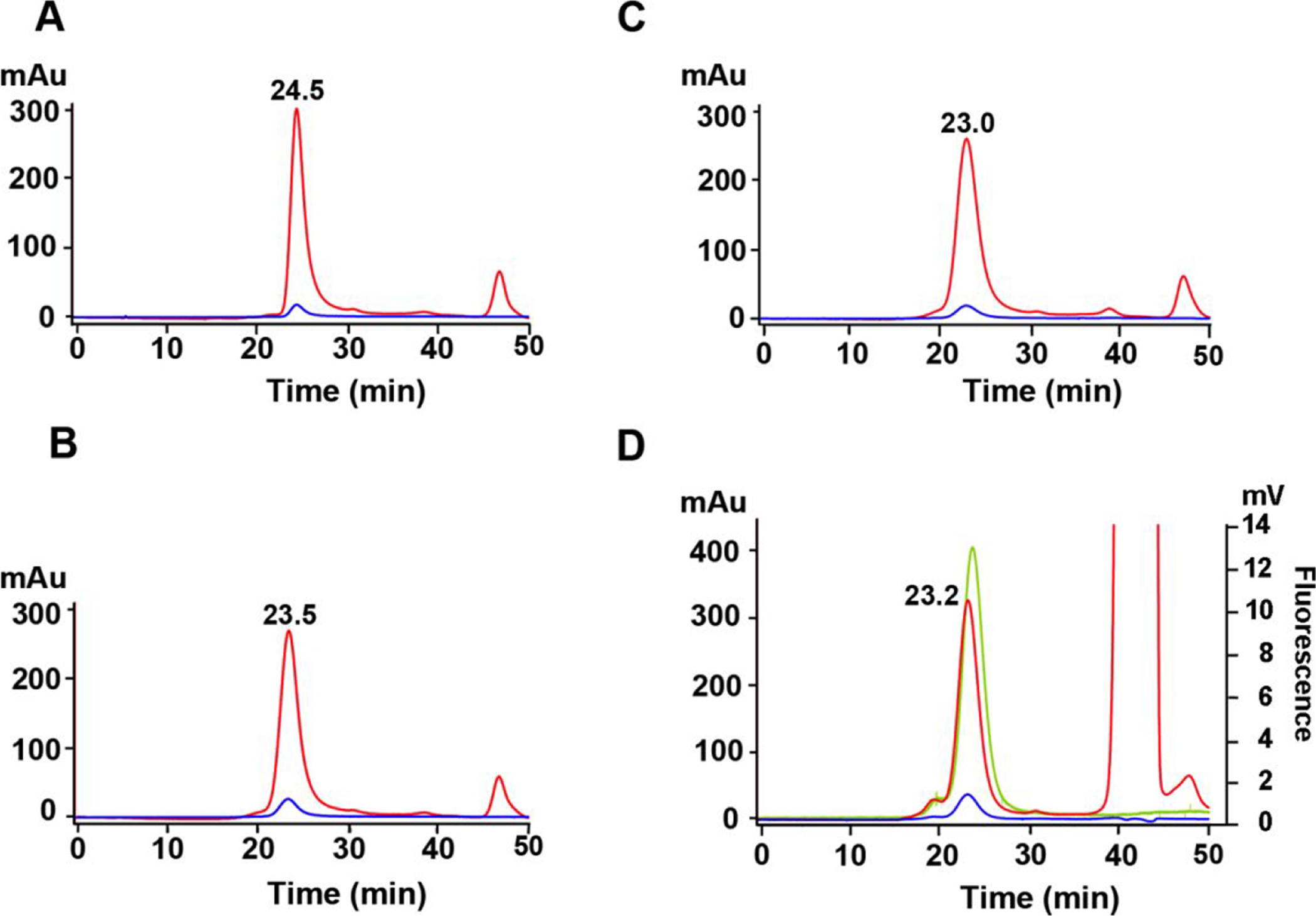
A. DMPC-LND. B. DSPE-PEG2000-DBCO LND. C. Azido-monoamide-DOTA + DSPE-PEG2000-DBCO LND. D. Azido-PEG4-Dox + DSPE-PEG2000-DBCO LND. The large 214 nm peak at 42 min is due to the solvent DMSO. Red= 214 nm. Blue= 240 nm. Green= ex 470, em 595 fluorescence.
In anticipation of future studies in which drugs can be conjugated to the DSPE-PEG2000-DBCO LND, an azido-PEG4-doxorubicin derivative was synthesized (Supplemental Scheme S1) and clicked to DSPE-PEG2000-DBCO. The elution time of this derivative, as monitored by both UV and the intrinsic fluorescence of Dox (Fig 1D), was similar to the other DSPE-PEG2000-DBCO derivatives. However, the fact that the UV and fluorescent peaks are coincident is evidence that azido-PEG4-Dox has clicked to the DSPE-PEG2000-DBCO incorporated in the LND.
Since it was important to prove that both azido-monoamide-DOTA and azido-PEG4-Dox were capable of undergoing Click chemistry, they were reacted with DSPE-PEG2000-DBCO, their products purified by RP-HPLC (Supplemental Fig S3) and their masses confirmed by high resolution mass spectrometry (Supplemental Fig S4). In both cases, the observed masses obtained were consistent with the expected products of Click chemistry. It should be noted that confirmation of masses directly on an LND is made difficult by the fact that the LND is heterogeneous in terms of the number of lipids and the fact that PEG2000 is a heterogeneous polymer as evidenced by the high-resolution mass spectra of the starting material and Clicked products.
Reaction of DSPE-PEG2000-DBCO LNDs with anti-CEA IgG and its azido-F(ab’) fragment
Reduced intact anti-CEA IgG (150 kDa) and its F(ab’) fragment (55 kDa from F(ab’)2 reduced with TCEP) were alkylated on their hinge region sulfhydryl groups with bromoacetamido-PEG5 azide, derivatized with NHS-DOTA and clicked to DSPE-PEG2000-DBCO LND. As a reference point, Fab’ eluted at 29.2 min on SEC HPLC (Fig 2A) and the Clicked product (molar ratio of 4:1) to DSPE-PEG2000-DBCO eluted at 21.0 min, demonstrating that about 90% of the F(ab’) fragment was incorporated into the LND with about 10% of free F(ab’) remaining unconjugated (Fig 2B). In order to demonstrate that the clicked product retained its immunoreactivity, the product was incubated with a 10-fold molar excess of CEA (180 kDa) and chromatographed on SEC HPLC (Fig 2C). Essentially, 100% of the product was shifted to an earlier elution time (17.7 min), demonstrating retention of its immunoreactivity. Similarly, azido-anti-CEA mAb was clicked to DSPE-PEG2000-DBCO and derivatized with NHS-DOTA. Reference anti-CEA M5A on SEC HPLC is shown in Fig 2D (elutes at 25.4 min) and its Clicked product to DSPE-PEG2000-DBCO in Fig 2E (elutes at 23.4 min). The shift in elution time demonstrates successful incorporation into the LNP. Immunoreactivity was fully retained as shown by the addition of CEA (Fig 2F, elutes at 15.6 min).
Figure 2. SEC HPLC analyses of DOTA-anti-CEA-Fab’-LND and DOTA-anti-CEA-LND.
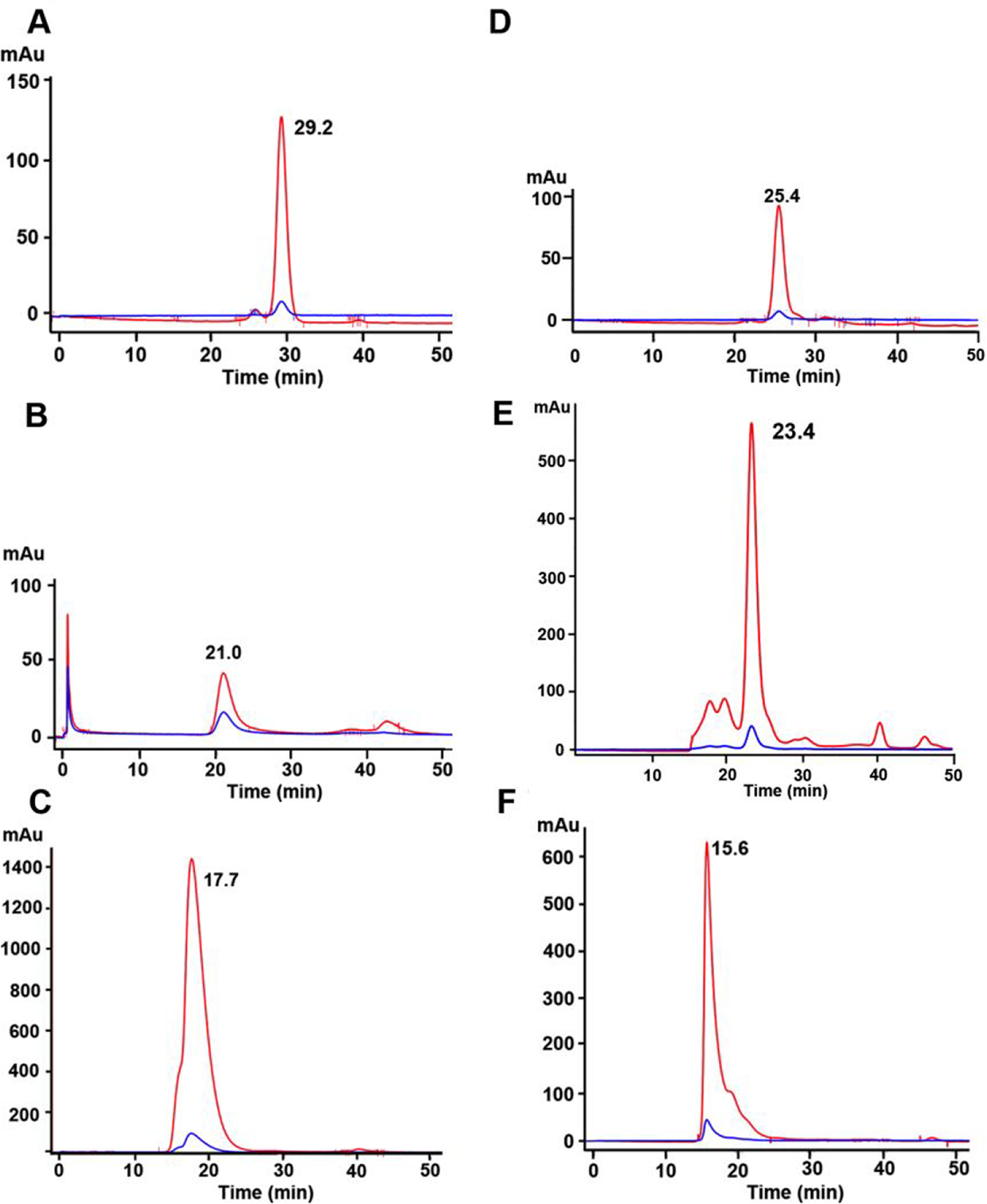
A. DOTA-anti-CEA Fab’-azide. B. DOTA-anti-CEA Fab’-azide clicked to DSPE-PEG2000-DBCO LND. C. Immunoreactivity of DOTA-anti-CEA Fab’-azide clicked to DSPE-PEG2000-DBCO LND after addition of 2-fold excess CEA. D. DOTA-anti-CEA-azide. E. DOTA-anti-CEA-azide clicked to DSPE-PEG2000-DBCO LND. F. Immunoreactivity of DOTA-anti-CEA-azide-DSPE-PEG2000-DBCO LND after addition of two-fold excess of CEA. Red= 214 nm. Blue- 240 nm.
Analysis of LNDs by transmission electron microscopy.
For a more direct measurement of their particle size, each of LNDs was analyzed by TEM (Fig 3). Unmodified DMPC LNDs were visualized as homogeneous discs with a diameter 10–12 nm (Fig 3A). While addition of 4 molar percent DSPE-PEG2000-DBCO had little effect on their apparent size, the particles tended to line up in organized arrays (Fig 3B). Since this interesting phenomenon did not affect their elution on SEC HPLC (Fig 1B), we conclude that the unusual alignment had more to do with the EM fixation process than their solution properties. The intact antibody conjugated LNDs exhibited a fuzzy appearance with increased size (Fig 3C) compared to the Fab’-LNDs (Fig 3D). The fuzzy appearance is likely due to the high flexibility of the IgG molecule attached to a more rigid LND. Notably, the longest dimension on an intact IgG is about 13–14 nm, enough to completely cover one surface of a LND. The increased size of the IgG conjugated LNDs (Fig 3C) is compatible with this possibility. Otherwise, the morphological appearance of the intact IgG and Fab’ conjugated LNDs supports the conclusion that one to two antibodies or fragments were conjugated to each LND.
Figure 3. Transmission EM analysis of LNDs.
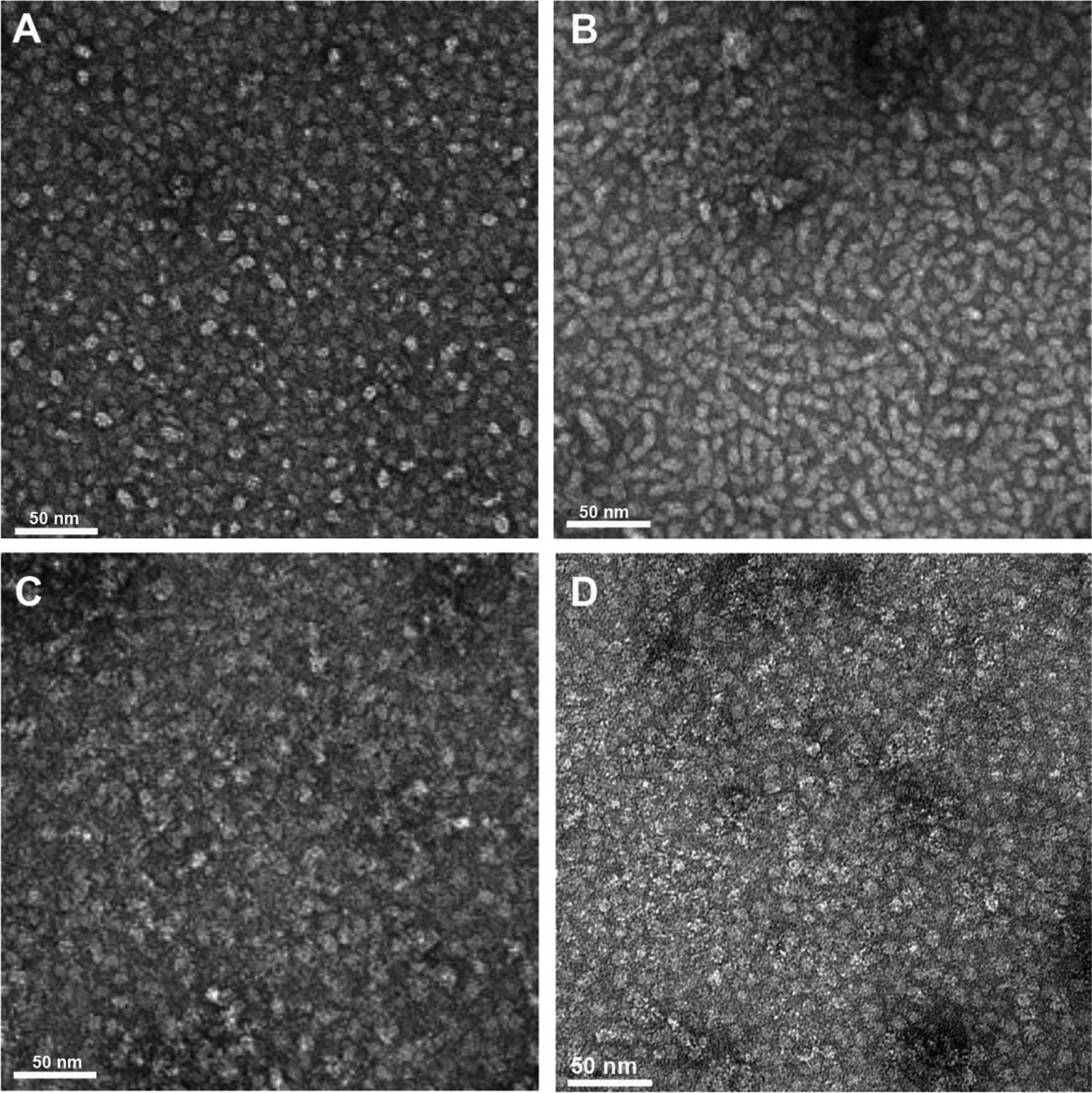
A. DMPC LND (x67,000). B. DMPC LND with 4 mole percent DSPE-PEG2000-DBCO (x67,000). C. Anti-CEA-antibody-azide clicked to DSPE-PEG2000-DBCO LND (x67,000). D. Anti-CEA Fab’-azide clicked to DSPE-PEG2000-DBCO LND (x67,000).
PET imaging
The in vivo behavior of the conjugated LNDs was investigated by conjugation of the promiscuous chelate DOTA either into the lipid moieties directly or into the moieties clicked into DSPE-PEG2000-DBCO LND. The positron emitter 64Cu was chosen for its ready incorporation into DOTA bearing moieties and for its excellent PET imaging characteristics25–26. Since we were interested in targeting the LNDs to tumors, each were radiolabeled and injected into mice bearing CEA positive, orthotopic mammary tumors. We utilized CEA transgenic mice that express CEA in their colon as a model system27 to determine any off target accumulation in normal organs. As a baseline, DSPE-PEG2000-DBCO LNDs were clicked to a commercially available azido-monoamide-DOTA. PET imaging of two mice (Fig 4A–B) with 64Cu-DOTA-labeled/clicked DSPE-PEG2000-DBCO LND exhibited rapid accumulation into the liver and caecum (the initial portion of the large intestine) by 2 hrs after initial distribution into the heart and major veins (a surrogate for blood) at 0 hrs. By 22 hrs the gut uptake has been cleared leaving only the liver on the image. Evidence for gut clearance is evident in mouse 2 where activity is visualized all the way from the descending colon to the anus. In order to calibrate the organ uptake of the PET studies, the mice were euthanized at the terminal time point, the tissues counted and the results expressed as %ID/g to normalize for the organ weight (Fig 4C). The organ uptake and clearance profile is compatible with the fact that the radiolabel is present on the lipid moieties comprising the LND and that lipids are mainly cleared by biliary excretion into the gut. Tumor uptake at the terminal time point ranged from 5–7 %ID/g, a result suggesting untargeted uptake mediate by the EPR effect.
Figure 4. PET imaging of CEA positive tumors in CEA transgenic mice with 64Cu-DOTA-azide clicked to DSPE-PEG2000-DBCO LND.
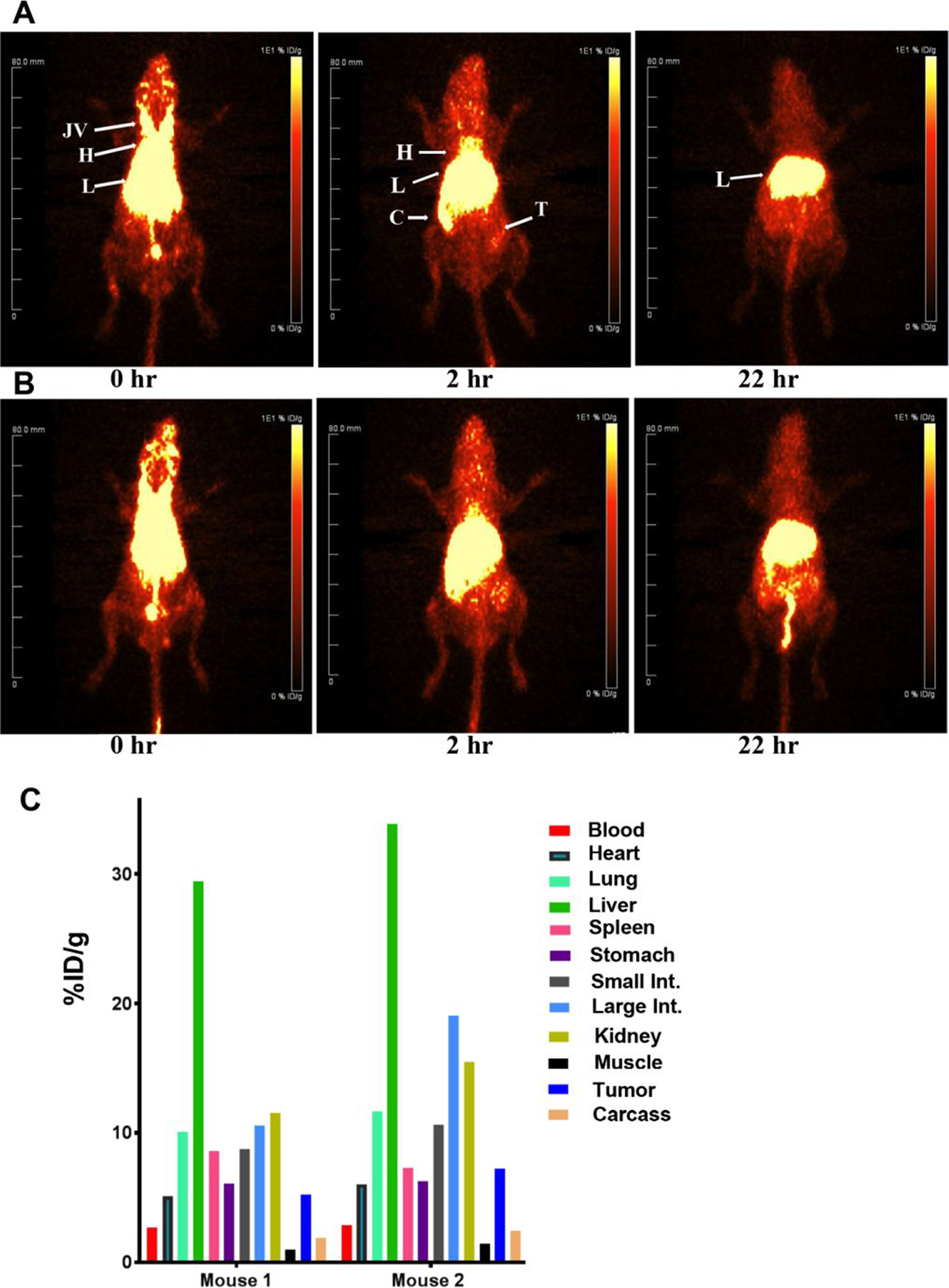
CEA Tg mice bearing CEA transfected E0771 murine mammary cancer cells in their right mammary fat pads were injected with 43 μCi (A) or 46 μCi (B) of DOTA-azide clicked to DSPE-PEG2000-DBCO LNDs and imaged at 0, 2 and 22h. Major organs are indicated: JV= jugular veins, H= heart, L= liver, C= cecum, T= tumor. At the terminal time point, the mice were euthanized and their tissues analyzed for %ID/g (C).
In order to determine if attachment of antibody or antibody fragments to the LNDs would enhance tumor targeting, the above described derivatives of anti-CEA antibody and its Fab’ fragment were clicked to DSPE-PEG2000-DBCO LND. Since we have not previously reported tumor imaging of 64Cu-DOTA-Fab’ anti-CEA with CEA positive tumors in CEA Tg mice, these PET studies were performed first (Supplemental Fig S5). Not unexpectedly, the main route of clearance of this imaging agent is the kidneys, since the molecular size of Fab’ is below the kidney filtration threshold. Quantitation of the uptake into tumor and other tissues reveals slightly higher tumor uptake compared to the untargeted LNDs (8% ID/g vs 4–7 %ID/g). Since the kidney uptake is very high (100% ID/g) and tumor uptake in the range of EPR uptake, this agent is of limited utility for imaging tumors. When the same Fab’ fragment was clicked to DSPE-PEG2000-DBCO LND, radiolabeled with 64Cu and used for PET imaging, high liver rather than kidney uptake was observed due to the increase molecular size (Fig 5A–B). In line with the high liver uptake, the major route of clearance was through the gut at 4hr. Quantitation performed at the terminal time point of 22 hrs, revealed tumor uptake in the range of 4–5 %ID/g (Fig 5C), suggesting that this fragment did not enhance targeting of the LND to tumor. Although the product was clearly immunoreactive in vitro (Fig 2C), it is possible that the Fab’ modified LNDs do not perform well in vivo due to insufficient levels of Fab’ on the LND. In an attempt to test that possibility, the amount of DOTA-Fab’-azide clicked to the DSPE-PEG2000-DBCO LND was tested over the injected dose per mouse from 10–40 μg (0.18–0.72 nmole) per 0.4 nmole of LND (based on an average of 2 DBCO per mole of DSPE-PEG2000-DBCO LND). The results of terminal biodistributions with three levels of DOTA-Fab’-azide clicked to DSPE-PEG2000-DBCO LND demonstrate that tumor targeting is saturated at the 10 μg level with only a slight increase in liver uptake at the highest level (Supplemental Fig S6). Thus, the reason that conjugation of an antibody fragment to the LND did not improve tumor targeting is unclear. Based on the observed results, it is possible liver clearance dominated over tumor uptake. Since it is difficult to block liver uptake, a phenomenon that occurs even with intact antibodies25, there is no obvious solution to this problem.
Figure 5. PET imaging of CEA positive tumors in CEA transgenic mice with 64Cu-DOTA-Fab’-azide clicked to DSPE-PEG2000-DBCO LNDs.
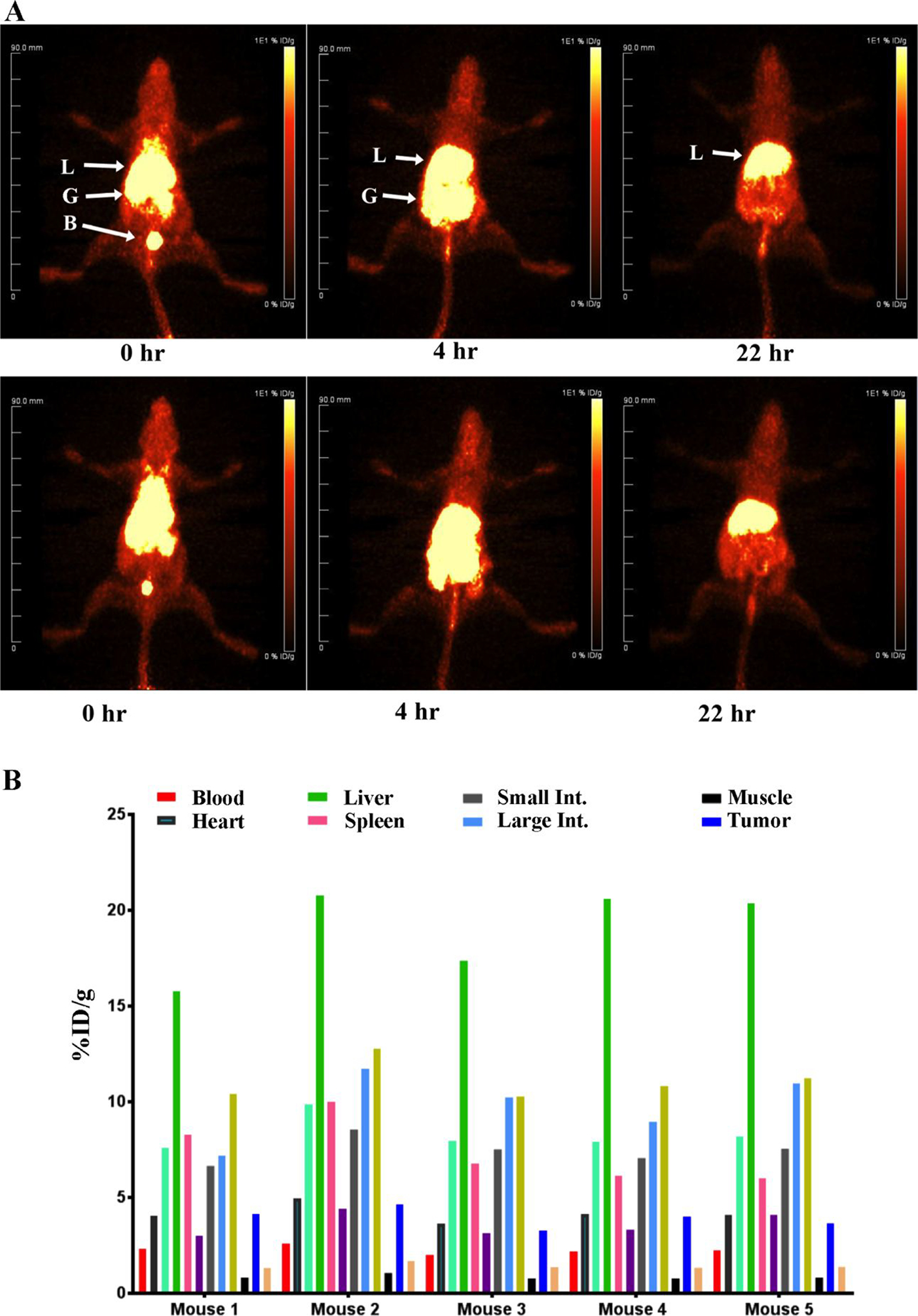
CEA transgenic mice bearing CEA transfected E0771 cells in their right mammary fat pads were injected with 90–100 μCi (A) of 64Cu-DOTA-Fab’-azide clicked to DSPE-PEG2000-DBCO LNDs and imaged at 0, 4 and 22h. Major organs are indicated: L= liver, B= bladder, G= gut. Two out of five imaged mice are shown. At 22 hr the mice were euthanized and tissues analyzed for the listed tissues (B).
In a second attempt to improve in vivo tumor targeting of LNDs, we generated an intact anti-CEA antibody azide derivative at the reduced hinge cysteines and reacted this derivative with NHS-DOTA to allow labeling with 64Cu for PET imaging. Analysis of the product by mass spectrometry revealed incomplete derivatization of the light chain (LC) and complete derivatization of the heavy chain (HC) with up to three PEG5-azides and 3 DOTAs per heavy chain (Supplemental Fig S7). This product was clicked to DSPE-PEG2000-DBCO LND and shown to shift its retention on SEC relative to underivatized LND and to retain the ability to bind CEA (Fig 2). As a point of reference, imaging of 64Cu-DOTA-anti-CEA antibody M5A in CEA bearing CEA Tg mice has been previous reported by us25. In that study, the maximum tumor uptake of 64Cu-DOTA-M5A at 48 hr was about 50 %ID/g. PET imaging of 64Cu-DOTA-M5A-LND revealed a prolonged blood clearance profile with substantial tumor uptake by 22 hrs for this LND (Fig 6A) compared to the other LND derivatives. Given its increased blood retention, this study was continued out to 46 hrs, the usual limit of 64Cu imaging (half-life of 12.7 hrs). At the 46 hr time point, the PET image reveals significant tumor accumulation with reduced blood and liver uptake compared to the earlier time points. The increased blood retention for the Ab-LND vs LND alone is likely due to the ability of the Ab-LND to recirculate via FcRn, a property gained from the Ab and not present in LND alone. The tumor uptake was substantially blocked by addition of a 20-fold excess of cold antibody (Fig 6B). When terminal biodistributions were performed at the 46 hr time point (Fig 6C), tumor uptake was about 40%ID/g, compared to the other LNDs that exhibited tumor uptake in the EPR range (5–7% ID/g). Importantly, the liver uptake (8% ID/g) was about one half to one third of that observed for the Fab’ clicked LND (17–21 %ID/g). Thus, the difference between targeting LND with intact antibody vs its Fab’ fragment, is the dominate liver uptake in the case of the Fab’ fragment. We speculate that some portion of the Fab’ fragment as presented on the LND is strongly recognized by the liver. In contrast, intact antibody as presented on the LND is less affected by liver uptake allowing high tumor uptake.
Figure 6. PET imaging of CEA positive tumors in CEA transgenic mice with 64Cu-DOTA-Ab-azide clicked to DSPE-PEG2000-DBCO LNDs.
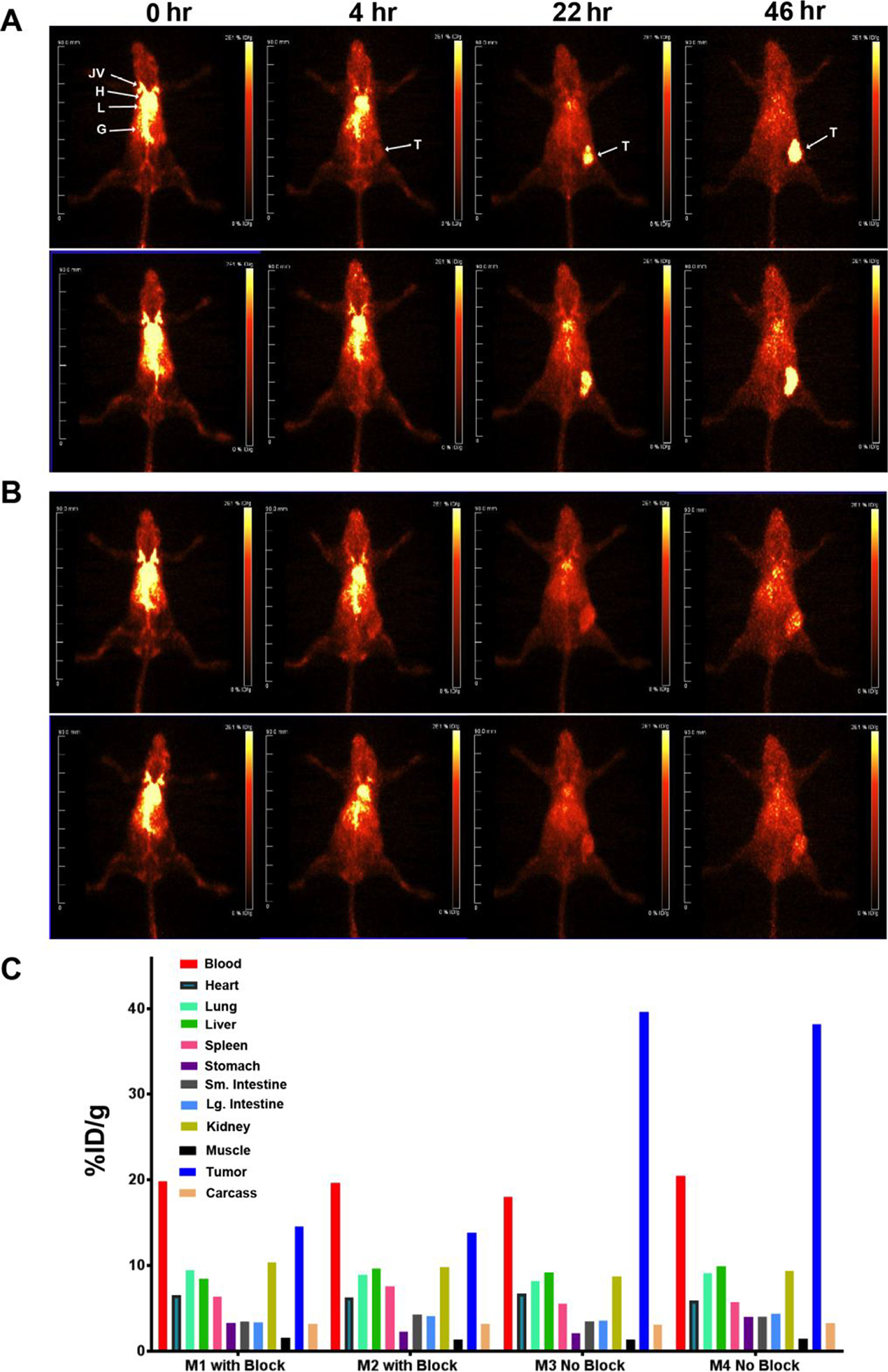
A. Two CEA transgenic mice bearing CEA/E0771 cells in their right mammary fat pads were injected with 40 μCi of 64Cu-DOTA-Ab-azide clicked to DSPE-PEG2000-DBCO LNDs and imaged at 0, 4, 22 and 46h. B. A repeat with a 20-fold excess of blocking Ab. Major organs are indicated: L= liver, B= bladder, G= gut. C. At 46 hr the mice were euthanized and tissues analyzed for the listed tissues.
CONCLUSION
So far there has only been one report of PET imaging in tumor bearing mice with a radiolabeled LND28. In that study the lipid nanodisc was generated by conjugating DOTA to the MSP and radiolabeling with 64Cu. Thus, a major difference in the two studies were their conjugation of DOTA to MSP vs our conjugation of DOTA to a PEGylated DSPE. Minor differences in the composition of the LND were their use of DOPC instead of DMPC plus PEGylated DSPE in our study. However, their low tumor uptake of 5% ID/g reflects an EPR driven accumulation in the tumor, similar to what we observed in untargeted LNDs where DOTA was conjugated to the lipid moieties. The major advance in our study was that the conjugation of an intact anti-tumor antibody to the LND resulted in an eight-fold increase in tumor uptake (40 %ID/g) over the EPR effect (5% ID/g). Importantly, we describe the use of highly efficient Click chemistry to achieve the conjugation of DOTA, drugs, and antibody to LND. By using PET imaging to determine the in vivo biodistributions of the various LNDs, we can rapidly optimize LNDs for tumor targeting and extension to the clinic. Previously, We reported on antibody targeted LNPs in which tumor uptake was 10 %ID/g, a 2-fold increased over untargeted LNPs7. However, the LNP study was based on an antibody to PSMA, rather than to CEA as in this study. Thus, it is possible that antibody targeted LNPs may be further improved depending on the antibody and tumor model. Nonetheless, the 4-fold increase in tumor targeting of Ab-LND over LND alone, is encouraging, and suggests that targeted LNDs may have an advantage over targeted LNPs.
MATERIAL AND METHODS
Materials.
1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[dibenzocyclooctyl(polyethylene glycol)-2000] (ammonium salt; DSPE-PEG2000-DBCO (#880229P) and 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC; #850345) were from Avanti Polar Lipids, Inc, Alabaster, AL. Azido-dPEG4-NHS ester (#10501) and doxorubicin HCl were from Quanta BioDesign, Ltd., Plain City, OH. Bromoacetamido-PEG5-azide (BP-21097) and DSPE-PEG4-NHS ester (BP-22288) were from BroadPharm, Inc., San Diego. SM-2 Biobeads (#1523920) were from BioRad, Hercules, CA. DOTA-NHS (B280) and azido-monoamide-DOTA (B-288) were from Macrocylics, Inc., Plano TX. Sodium cholate (C6445) and tris-(2-carboxyethyl)phosphine (TCEP; C4706). CaptureSelect FcXL resin was from Thermo Fisher Scientific, Waltham, MA. IdeS proteolytic enzyme (FabRICATOR® FragIT MidiSpin kit) was from Genovis AB, Lund, Sweden). Membrane scaffold protein 1D1 (MSP1D1) was produced by Sligar et al.18. Humanized anti-CEA hT84.66-M5A (M5A) was produced by Yazaki et al.25.
Mass spectrometry, electron microscopy and size exclusion HPLC.
Electrospray ionization mass spectrometry (ESI-MS) of small molecules was performed on a Finnigan LTQ or on an Orbitrap Fusion Mass Analyzer (ThermoElectron, San Jose, CA) and molecular weight determination of proteins on a Agilent 6520 QTOF. Electron microscopy was performed on an FEI Tecnai 12 transmission electron microscope equipped with a Gatan Ultrascan 2K CCD camera. LND samples were applied to a glow-discharged 300 mesh Formvar–carbon copper EM grid and stained with 2% uranyl acetate. Size exclusion chromatography (SEC) was performed on a Superdex 200 10/30 column (GE Healthcare Life Sciences) run at 0.5 mL/min in PBS. UV detection was performed at 214 and 280nm on a GE Akta Purifier HPLC. Fluorescence detection was performed on-line on a Jasco Fp-2000 plus set at ex= 470 nm, em= 560 nm.
DSPE-PEG2000-DBCO LND assembly
DSPE-PEG2000-DBCO (0.985 mg, 0.320 μmole) in 330 μL of CHCl3 plus DMPC (8.7 mg, 12.83 μmole) in 4.35 mL of CHCl3 was dried into a thin, uniform film of lipid on the lower walls of a pear shaped 15 mL flask in a Rotavapor (Buchi R-144) for 30 min. The flask was placed in a vacuum dessicator overnight to remove any residual solvent. The dried lipid was dissolved in 320 μL of 100 mM sodium cholate in Milli Q water plus 320 μL of 200 mM phosphate buffer (pH 7.4) with 100 mM NaCl. Lipids were solubilized by sonicating at 60° C for 30 minutes. MSP1D1 (4 mg, 0.08 μmole) in 0.47 mL was added at room temperature on a gentle rocker for 1 hour. After addition of 0.12g of SM-2 Biobeads the sample was incubated at room temperature on a gentle rocker for 2 hours. The solution was sterile filtered and stored in 4° C. Products were characterized by EM and SEC HPLC.
Reaction of DSPE-PEG2000-DBCO with azido-monoamide-DOTA.
DSPE-PEG2000-DBCO (3.00 mg, 0.975 μmole in 0.41 mL PBS) was reacted with azido-monoamide-DOTA (1.16 mg, 1.95 μmole in 0.13 mL of PBS) overnight at 37°C and purified by RP-HPLC on a CN HPLC column (Phenomenex 4.6 × 30 mm) using a linear gradient from 0% B to 100% B over 30 min (A= 0.1% TFA in water, B= 90% MeCN, 10% 0.1% TFA) at a flow rate of 1 mL/min. The purified product (2.68 mg, 75% yield) was analyzed by high resolution mass spectrometry on the Thermo Orbitrap Fusion: (MH)+ obs = 3545.15 for PEG n= 45, (MH)+ exp = 3545.11 for PEG n= 45.
Reaction of DSPE-PEG2000-DBCO with azido-PEG4-Dox.
The synthesis is shown in Supplementary Scheme S1. Azido-dPEG4-NHS ester (26.8 mg, 69.1 μmole) in 400 μL of water, pH adjusted to 6.87 with 1M NaOH was reacted with 400 μL of Doxorubicin HCL (20 mg, 34.5 μmole, in DMSO) under argon at room temperature overnight. The product was purified on a CN HPLC column (Phenomenex 4.6 × 30 mm) using a linear gradient from 0% B to 100% B over 30 min (A= 0.1% TFA in water, B= 90% MeCN, 10% 0.1% TFA) at a flow rate of 1 mL/min. The purified product (20 mg, 60% yield) was analyzed by ESI mass spectrometry (M+Na)+ obs = 839.36, (M+Na)+ exp = 839.37). DSPE-PEG2000-DBCO (2.00 mg, 0.65 μmole in 0.30 mL DMSO) was reacted with azido-PEG4-Dox (0.95 mg, 1.2 μmole in 0.19 mL of DMSO) overnight at 37°C and purified by RP-HPLC on a CN HPLC column (see above for chromatography conditions). The purified product (2.0 mg, 70% yield) was analyzed by high resolution mass spectrometry on the Thermo Orbitrap Fusion: (MH)+ obs = 3549.16 for PEG n= 38, (MH)+ exp = 3566.98 for PEG n= 38, thus the product has the correct mass for M-H2O.
Conjugation of DSPE-PEG2000-DBCO -LND with azido-monoamide-DOTA and azido-dPEG4-Dox.
DSPE-PEG2000-DBCO -LND (16 nmol in 50 μL) was mixed with azido-mono-amide-DOTA (17 nmol in 2 μL) under argon at 37° C for 2 hours. The product was analyzed by SEC HPLC (80% yield by UV). DSPE-PEG2000-DBCO -LND (16 nmol in 50 μL) was mixed with azido-dPEG4-DOX (15 nmol in 29.4 μL of DMSO) under argon at 37° C for 2 hours. The product was analyzed by SEC HPLC (75% yield by UV).
Stoichiometry of DSPE-PEG2000-DBCO incorporation into LND and monitoring of the reaction of azido-monoamide-DOTA with DSPE-PEG2000-DBCO LNDs by 1D NMR.
The spectra were acquired at 37°C on 700 MHz Bruker Ascend instrument equipped with a cryoprobe using a modified zgpr pulse sequence. The modified sequence enabled a long recycle delay (28.5 sec) followed by 1.5 sec presaturation on residual water. The sample was dissolved in D2O-based 50 mM pH 7.4 phosphate buffer. The regions 3.73 to 3.65 ppm were selected for the ethylene protons of PEG2000 and 1.1 to 0.3 ppm for the methyl protons of DMPC, Val, Ile and Leu of MSP1D1. The calculation was based on 160 DMPC/LND with 960 methyl protons and 2 MSP1D1 per LND with 468 methyl protons29. The integration values of these two regions are shown in Supplementary Fig S1. These two regions were chosen because their relative favored T2 relaxation, and less interference from each other of the two targets being studied. Based on the Biological Magnetic Resonance Data Bank, the averaged two Hδ of Pro residues resonate at 3.63 ± 0.4 ppm. Other protons from MSP1D1 are less likely resonate between 3.65–3.73ppm. For the integration of methyl region (1.1 to 0.3ppm), the DSPE methyl group from DSPE-PEG2000-DBCO will resonate in this region. If the molar ratio between DSPE-PEG2000-DBCO and nanodisc is 4:1 (as designed in the experiment), the contribution of DSPE methyl group to this region is only about 1.7% because the total methyl protons of Val, Ile and Leu from two MSP1D1 and 160 DMPC molecules from one nanodisc is 1428. For simplicity, we did not consider the negligible contribution from DSPE methyl group to this region in the molar ratio estimation. Based on the total proton numbers in methyl region, it was deduced the potential integration contribution of total 32 Hδ of Pro to region 3.73–3.65 being 0.044. Considering the total proton number of PEG ethylene group per DSPE-PEG2000-DBCO is 176, the molar ratio between DSPE-PEG2000-DBCO:nanodisc is between 4.12 and 3.94, respectively without and with potential overlaps of 32 Hδ of Pro with PEG2K ethylene groups between 3.73–3.65 ppm.
The reaction of azido-monoamide-DOTA with DSPE-PEG2000-DBCO in the LND (molar ratio of 2.4:1, azide to DBCO) was monitored in the aromatic region of the spectrum (6.5–8.0) over time and compared to the spectrum in the absence of azido-monoamide-DOTA Supplementary Fig S2. The reaction was almost complete at 3.5 hours since the peak intensity did not change after this time point. Peak intensity that were reduced significantly are shown by black arrows, while peaks or new peaks that showed increased intensity are shown in red arrows. The results suggest the click reaction between nanodisc-DBCO and azido-monoamide-DOTA was complete in about 4 hours at 37°C degree.
Generation of anti-CEA Fab’ LND derivatives
M5A (10 mg, 66.7 nmole) was digested with IdeS proteolytic enzyme to generate F(ab’)2 fragments. The digested antibody was loaded onto a 5 mL CaptureSelect FcXL resin column, equilibrated with 1x DPBS and the F(ab’)2 collected as flow through. The M5A-F(ab’)2 was reduced with a 30 fold molar excess of TCEP and 10 mM EDTA for 2 hrs under argon. Excess TCEP was removed by a spin column (Zeba, 7 kDa cutoff, 2 mL). The Fab’ (3.8 mg, 69 nmole) was reacted with a 100 fold molar excess of bromoacetamio-PEG5-azide under argon at 37 °C overnight and dialyzed vs. PBS to remove excess alkylating agent. Fab’-PEG5-azide (1.326 mg, 24 nmole) was reacted with a 10 fold molar excess of DOTA-NHS at RT over night as previously described30, dialysed and concentrated to 0.71 mg/mL. DOTA-Fab’-PEG5-azide (0.214 mg, 3.89 nmole) was clicked to DSPE-PEG2000-DBCO ND (8.0 nmole, 50 μL of 0.985 mg/mL) at 37°C overnight and analyzed by SEC HPLC (75% yield by UV).
Generation of anti-CEA antibody LND.
M5A (10 mg, 66.6 nmole, 400 μL of PBS) was reduced with a 30 molar excess of TCEP at 37° C for 2h under Argon. The TCEP was removed by using a desalting spin column (Zeba, 7 kDa cutoff, 1 mL). The reduced M5A was reacted with a 100 fold molar excess of bromoacetamide-PEG5-azide at RT overnight under Argon. The excess bromoacetamide-PEG5-azide was removed by dialysis (Slide-A-Lyzer, 2 mL, Thermo-Fisher). Conjugation was confirmed by mass spectrometry on an Agilent 6520 QTOF. The analysis revealed one PEG5-azide per light chain and three PEG5-azides per heavy chain with a total of eight per IgG (2 light chains and 2 heavy chains).
M5A- PEG5-azide (2.66 mg, 17 nmole) in 0.45 mL of PBS was reacted with DOTA-NHS (177 nmole, 15.7 μL of 10 mg/mL) in water over night at 37 °C and dialyzed vs PBS to remove excess reagent. DOTA-M5A-PEG5-azide (8 nmole, 219 μL of 2.66 mg/mL) was clicked to DSPE-PEG2000-DBCO ND (8 nmole, 50 μL of 0.985 mg/mL) at 37 °C overnight and analyzed by SEC HPLC (65% yield by UV).
Radiolabeling and immunoreactivity
DOTA-anti-CEA-Fab’-PEG2000-DBCO LND (227 μg, 4.13 nmole of Fab’, 8.0 nmole LND) or DOTA-M5A-PEG2000-DBCO LND (1.20 mg, 8 nmole of M5A, 8.0 nmole of LND) was radiolabeled with 64Cu (5.6 GBq/μmoL; Isotope Production group, Washington University, St Louis, MO) with a target specific activity of ~10 μCi per μg and the radiolabeling yield determined by ITLC after the addition of 10 mm DTPA. The radiolabeling yields for 64Cu-DOTA-PEG2000-DBCO ND, 64Cu-DOTA-Fab’- PEG2000-DBCO ND and 64Cu-DOTA-M5A-PEG2000-DBCO ND were 93%, 70%, and 19%, respectively. In each case the radiolabeled ND was further purified by SEC Superdex to exclude free radionuclide-DTPA from the injected dose. The protein dose per mouse for the 64Cu-DOTA-Fab’-PEG2000-DBCO LND was 20 μg based on the amount of Fab’, while the protein dose per mouse for the DOTA-M5A-PEG2000-DBCO LND was 60 μg based on the amount of M5A.
Animal biodistribution and PET imaging studies in tumor-bearing mice
All animal handling was done in accordance with IACUC protocol 91037 approved by the City of Hope Institutional Animal Care and Use Committee. Female CEA transgenic mice27 were injected in the mammary fat pad with murine breast cancer E0771 cells transfected with CEA. After 10–12 days, the tumor masses were in the range of 0.2–0.6 g. Animals were injected (about 100 μCi/animal) via tail vein with two mice per group. The PET imaging was performed at set time points using a Siemens InVeon microPET system. After the final images were taken, the mice were euthanized, necropsy performed, organs weighed and counted for radioactivity. All data presented are mean values with the standard error (±SEM). The radioactivity has been corrected for background and radioactive decay from the time of injection, allowing organ uptake to be reported as %ID/g.
Supplementary Material
Supplementary Scheme S1. Synthesis of azido-dPEG4-Dox.
Supplementary Fig S1. Estimation of molar ratio of DSPE-PEG2000-DBCO to lipid nanodisc by 1D 1H NMR.
Supplementary Fig S2. Monitoring the reaction between azido-monoamide DOTA and DSPE-PEG2000-DBCO on a lipid nanodisc by 1D 1H NMR.
Supplementary Fig S3. RP-HPLC purification of Clicked products of azido-monoamide-DOTA and azido-PEG4-Dox with DSPE-PEG2000-DBCO.
Supplementary Fig S4. High resolution mass spectrometry of DSPE-PEG2000-DBCO and Clicked products of azido-monoamide-DOTA and azido-PEG4-Dox with DSPE-PEG2000-DBCO.
Supplementary Fig S5. PET imaging of CEA positive tumors in CEA transgenic mice with 64Cu-DOTA-anti-CEA-Fab’.
Supplementary Fig S6. Biodistributions of CEA positive tumors in CEA transgenic mice with increasing amounts of 64Cu-DOTA-anti-CEA-Fab’ LNDs.
Supplementary Fig S7. MS analysis of DOTA-PEG5-azide M5A.
ACKNOWLEDGEMENTS
We wish to thank the City of Hope Electron Microscope Core Facility, whose use of the FEI Tecnai 12 transmission electron microscope was provided by Office of Naval Research N00014-02-1 0958.
Research reported in this publication included work performed in the Small Animal Imaging Core supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Support from the Charles Reames Family (to J.W.) is gratefully acknowledged.
ABBREVIATIONS
- PEG
polyethylene glycol
- LNP
Lipid nanoparticle
- LND
lipid nanodisc
- EPR
enhanced permeability retention
- DSPE-PEG2000
distearoyl phosphatidyl ethanolamine monomethoxy polyethylene glycol
- DSPE-PEG2000-DBCO
distearoyl-sn-glycero-3-phosphoethanolamine-n-[dibenzocyclooctyl(polyethylene glycol)-2000]
- HPLC
high performance liquid chromatography
- SEC
size exclusion chromatography
- NR
non-reducing
- DOTA
1,4,7,10-tetraazacyclododecane-1, 4,7,10-tetraacetic acid
- NHS
N-hydroxysuccinimide
- PET
positron emission tomography
- MIP
maximum intensity projections
- CT
computed tomography
- % ID/g
percent injected dose per gram.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Mestel R, Cancer: Imaging with antibodies. Nature 2017, 543 (7647), 743–746. [DOI] [PubMed] [Google Scholar]
- 2.Scheinberg DA; Strand M; Gansow OA, Tumor imaging with radioactive metal chelates conjugated to monoclonal antibodies. Science 1982, 215 (4539), 1511–3. [DOI] [PubMed] [Google Scholar]
- 3.Larson SM; Carrasquillo JA; Cheung NK; Press OW, Radioimmunotherapy of human tumours. Nat Rev Cancer 2015, 15 (6), 347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zolot RS; Basu S; Million RP, Antibody-drug conjugates. Nat Rev Drug Discov 2013, 12 (4), 259–60. [DOI] [PubMed] [Google Scholar]
- 5.Cilliers C; Nessler I; Christodolu N; Thurber GM, Tracking Antibody Distribution with Near-Infrared Fluorescent Dyes: Impact of Dye Structure and Degree of Labeling on Plasma Clearance. Mol Pharm 2017, 14 (5), 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sievers EL; Senter PD, Antibody-drug conjugates in cancer therapy. Annu Rev Med 2013, 64, 15–29. [DOI] [PubMed] [Google Scholar]
- 7.Wong P; Li L; Chea J; Delgado MK; Crow D; Poku E; Szpikowska B; Bowles N; Channappa D; Colcher D; et al. , PET imaging of (64)Cu-DOTA-scFv-anti-PSMA lipid nanoparticles (LNPs): Enhanced tumor targeting over anti-PSMA scFv or untargeted LNPs. Nucl Med Biol 2017, 47, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong P; Li L; Chea J; Delgado MK; Poku E; Szpikowska B; Bowles N; Minnix M; Colcher D; Wong JYC; et al. , Synthesis, Positron Emission Tomography Imaging, and Therapy of Diabody Targeted Drug Lipid Nanoparticles in a Prostate Cancer Murine Model. Cancer Biother Radiopharm 2017, 32 (7), 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu CM; Kaushal S; Tran Cao HS; Aryal S; Sartor M; Esener S; Bouvet M; Zhang L, Half-antibody functionalized lipid-polymer hybrid nanoparticles for targeted drug delivery to carcinoembryonic antigen presenting pancreatic cancer cells. Mol Pharm 2010, 7 (3), 914–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maruyama K; Takahashi N; Tagawa T; Nagaike K; Iwatsuru M, Immunoliposomes bearing polyethyleneglycol-coupled Fab’ fragment show prolonged circulation time and high extravasation into targeted solid tumors in vivo. FEBS Lett 1997, 413 (1), 177–80. [DOI] [PubMed] [Google Scholar]
- 11.Pozzi D; Colapicchioni V; Caracciolo G; Piovesana S; Capriotti AL; Palchetti S; De Grossi S; Riccioli A; Amenitsch H; Lagana A, Effect of polyethyleneglycol (PEG) chain length on the bio-nano-interactions between PEGylated lipid nanoparticles and biological fluids: from nanostructure to uptake in cancer cells. Nanoscale 2014, 6 (5), 2782–92. [DOI] [PubMed] [Google Scholar]
- 12.Puri A; Loomis K; Smith B; Lee JH; Yavlovich A; Heldman E; Blumenthal R, Lipid-based nanoparticles as pharmaceutical drug carriers: from concepts to clinic. Crit Rev Ther Drug Carrier Syst 2009, 26 (6), 523–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zatsepin TS; Kotelevtsev YV; Koteliansky V, Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside. Int J Nanomedicine 2016, 11, 3077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greish K, Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting. Methods Mol Biol 2010, 624, 25–37. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y; Wang R; Lu X; Lu W; Zhang C; Liang W, Pegylated phospholipids-based self-assembly with water-soluble drugs. Pharm Res 2010, 27 (2), 361–70. [DOI] [PubMed] [Google Scholar]
- 16.Bayburt TH; Sligar SG, Single-molecule height measurements on microsomal cytochrome P450 in nanometer-scale phospholipid bilayer disks. Proc Natl Acad Sci U S A 2002, 99 (10), 6725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingwell BA; Chapman MJ; Kontush A; Miller NE, HDL-targeted therapies: progress, failures and future. Nat Rev Drug Discov 2014, 13 (6), 445–64. [DOI] [PubMed] [Google Scholar]
- 18.Bayburt TH; Sligar SG, Membrane protein assembly into Nanodiscs. FEBS Lett 2010, 584 (9), 1721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efremov RG; Gatsogiannis C; Raunser S, Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM. Methods Enzymol 2017, 594, 1–30. [DOI] [PubMed] [Google Scholar]
- 20.Hagn F; Nasr ML; Wagner G, Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat Protoc 2018, 13 (1), 79–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esteban JM; Felder B; Ahn C; Simpson JF; Battifora H; Shively JE, Prognostic relevance of carcinoembryonic antigen and estrogen receptor status in breast cancer patients. Cancer 1994, 74 (5), 1575–83. [DOI] [PubMed] [Google Scholar]
- 22.Tiernan JP; Ansari I; Hirst NA; Millner PA; Hughes TA; Jayne DG, Intra-operative tumour detection and staging in colorectal cancer surgery. Colorectal Dis 2012, 14 (9), e510–20. [DOI] [PubMed] [Google Scholar]
- 23.Nittka S; Krueger MA; Shively JE; Boll H; Brockmann MA; Doyon F; Pichler BJ; Neumaier M, Radioimmunoimaging of liver metastases with PET using a 64Cu-labeled CEA antibody in transgenic mice. PLoS One 2014, 9 (9), e106921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundaresan G; Yazaki PJ; Shively JE; Finn RD; Larson SM; Raubitschek AA; Williams LE; Chatziioannou AF; Gambhir SS; Wu AM, 124I-labeled engineered anti-CEA minibodies and diabodies allow high-contrast, antigen-specific small-animal PET imaging of xenografts in athymic mice. J Nucl Med 2003, 44 (12), 1962–9. [PMC free article] [PubMed] [Google Scholar]
- 25.Li L; Bading J; Yazaki PJ; Ahuja AH; Crow D; Colcher D; Williams LE; Wong JY; Raubitschek A; Shively JE, A versatile bifunctional chelate for radiolabeling humanized anti-CEA antibody with In-111 and Cu-64 at either thiol or amino groups: PET imaging of CEA-positive tumors with whole antibodies. Bioconjug Chem 2008, 19 (1), 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimer JE; Bading JR; Colcher DM; Conti PS; Frankel PH; Carroll MI; Tong S; Poku E; Miles JK; Shively JE; et al. , Functional imaging of human epidermal growth factor receptor 2-positive metastatic breast cancer using (64)Cu-DOTA-trastuzumab PET. J Nucl Med 2014, 55 (1), 23–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke P; Mann J; Simpson JF; Rickard-Dickson K; Primus FJ, Mice transgenic for human carcinoembryonic antigen as a model for immunotherapy. Cancer Res 1998, 58 (7), 1469–77. [PubMed] [Google Scholar]
- 28.Huda P; Binderup T; Pedersen MC; Midtgaard SR; Elema DR; Kjaer A; Jensen M; Arleth L, PET/CT Based In Vivo Evaluation of 64Cu Labelled Nanodiscs in Tumor Bearing Mice. PLoS One 2015, 10 (7), e0129310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W; Mao A; Wong P; Larsen A; Yazaki PJ; Wong JYC; Shively JE, Characterization of 1,2-Distearoyl-sn-glycero-3-phosphoethanolamine-N-[Methoxy(polyethylene glycerol)-2000] and Its Complex with Doxorubicin Using Nuclear Magnetic Resonance Spectroscopy and Molecular Dynamics. Bioconjug Chem 2017, 28 (6), 1777–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis MR; Kao JY; Anderson AL; Shively JE; Raubitschek A, An improved method for conjugating monoclonal antibodies with N-hydroxysulfosuccinimidyl DOTA. Bioconjug Chem 2001, 12 (2), 320–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Scheme S1. Synthesis of azido-dPEG4-Dox.
Supplementary Fig S1. Estimation of molar ratio of DSPE-PEG2000-DBCO to lipid nanodisc by 1D 1H NMR.
Supplementary Fig S2. Monitoring the reaction between azido-monoamide DOTA and DSPE-PEG2000-DBCO on a lipid nanodisc by 1D 1H NMR.
Supplementary Fig S3. RP-HPLC purification of Clicked products of azido-monoamide-DOTA and azido-PEG4-Dox with DSPE-PEG2000-DBCO.
Supplementary Fig S4. High resolution mass spectrometry of DSPE-PEG2000-DBCO and Clicked products of azido-monoamide-DOTA and azido-PEG4-Dox with DSPE-PEG2000-DBCO.
Supplementary Fig S5. PET imaging of CEA positive tumors in CEA transgenic mice with 64Cu-DOTA-anti-CEA-Fab’.
Supplementary Fig S6. Biodistributions of CEA positive tumors in CEA transgenic mice with increasing amounts of 64Cu-DOTA-anti-CEA-Fab’ LNDs.
Supplementary Fig S7. MS analysis of DOTA-PEG5-azide M5A.


