Abstract
Early detection of pancreatic ductal adenocarcinoma (PDAC) is key to improving patient outcomes; however, PDAC is usually diagnosed late. Therefore, blood-based minimally invasive biomarker assays for limited volume clinical samples are urgently needed. A novel microRNA profiling platform (Abcam Fireplex-Oncology Panel) was used to investigate the feasibility of developing early detection miRNA biomarkers with 20ul plasma from a training set (58 stage II PDAC cases and 30 controls) and two validation sets (34 stage II PDAC cases and 25 controls; 44 stage II PDAC cases and 18 controls). miR-34a-5p (AUC = 0.77, 95% CI 0.66 to 0.87), miR-130a-3p (AUC = 0.74, 95% CI 0.63 to 0.84,), and miR-222–3p (AUC = 0.70, 95% CI 0.58 to 0.81,) were identified as significantly differentially abundant in plasma from stage II PDAC vs. controls. Although none of the miRNAs individually outperformed the currently used serological biomarker for PDAC, CA19–9, combining the miRNAs with CA 19–9 improved AUCs from 0.89 (95% CI 0.81 to 0.95) for CA 19–9 alone to 0.92 (95% CI 0.86 to 0.97), 0.94 (95% CI 0.89 to 0.98), and 0.92 (95% CI 0.87 to 0.97), respectively. Gene Set Enrichment Analyses of transcripts correlated with high and low expression of the three miRNAs in the TCGA PDAC sample set. These miRNA biomarkers, assayed in limited volume plasma together with CA19–9, discriminate stage II PDAC from controls with good sensitivity and specificity. Unbiased profiling of larger cohorts should help develop an informative early detection biomarker assay for diagnostic settings.
Cancer Prevention Relevance Statement:
Development of minimally invasive biomarker assays for detection of pre-malignant disease and early stage pancreatic cancer is key to improving patient survival. This study describes a limited volume plasma miRNA biomarker assay that can detect early stage resectable pancreatic cancer in clinical samples necessary for effective prevention and clinical intervention.
Keywords: microRNA, plasma biomarker, early stage pancreatic cancer, limited volume assay, Fireplex
Introduction:
Pancreatic cancer, presenting as pancreatic ductal adenocarcinoma (PDAC) in more than 90% cases, is the most deadly cancer by organ site with a five year survival rate of only ~9%(1). While only 2.9% of patients with distant PDAC survive 5 years, survival rates increase by over an order of magnitude to 37.4% when the disease is detected early (stages I and II)(1). Recent findings have revealed a significant increase in 5-year survival of patients with stage IA disease from 44.7% in 2003 to 83.7% in 2012. This trend has been suggested to be the result of improved early diagnosis and detection(2). Only patients with early stage, localized PDAC (comprised of stages I and II), are candidates for curative surgical resection, thus detection of these early stage cases is crucial to improving survival rates(3). However, early detection of PDAC remains challenging because most patients do not present with symptoms until after the disease has spread locally or to distant sites. Furthermore, given the location of the pancreas deep in the abdomen, palpating pancreatic tumors is difficult, and imaging may miss tumors depending on their size and location. Together, these facts underscore the need to develop minimally invasive liquid biopsy assays with adequate sensitivity and specificity to detect PDAC early.
The utility of carbohydrate antigen 19–9 (CA 19–9) as a diagnostic biomarker has been extensively studied in PDAC. While serum CA 19–9 levels have a sensitivity and specificity for PDAC of 79–81% and 82–90%, respectively, CA 19–9 is not recommended as a screening marker due to its low positive predictive value (0.5 to 0.9%)(4–6), particularly in asymptomatic populations(5). CA 19–9 is elevated in about 10% to 20% of patients with various benign pancreatobiliary conditions, which result in false positives (7–9). Conversely, 10% of the population lacking 1,4-fucosyl transferase due to germline mutations are unable to produce sialyl Lewis antigen epitopes and hence do not secrete CA 19–9, making the marker unusable for a subset of pancreatic cancer patients due to their false negativity (6,10). Despite these limitations, however, CA19–9 remains the most informative biomarker for a substantial group of patients with pancreatic cancer and no other biomarker has yet surpassed its performance. Among other promising biomarkers described for diagnosis of pancreatic cancer, most have revealed improved specificity and sensitivity in combination with CA19–9, which include cell-free DNA, metabolites, and various RNAs, including microRNAs (miRNAs).
miRNAs are small noncoding RNAs, approximately 22 nucleotides in length, that epigenetically modulate gene expression or translation by predominantly binding to the 3’ UTR of mRNAs resulting in miRNA deadenylation, target cleavage, or translational repression(11–13). It is estimated that miRNAs regulate at least 60% of mRNA transcripts and are conserved across species (14,15). These small RNAs are stable when circulating in various body fluids(16), including blood, and have also been found ensconced in the protective lumen of circulating extracellular vesicles, such as exosomes(17). Additionally, miRNAs regulate critical physiological processes and are frequently deregulated in many diseases, including viral infections, immune-related diseases, neurodegenerative disorders, and cancer (18–20). Taken together, their deregulation in disease and stability in circulation make miRNAs promising circulating biomarkers of disease diagnosis and prognosis.
The utility of circulating miRNAs as biomarkers has been explored in many diseases, including in pancreatic cancer (21). A recent publication has reported compelling evidence about the diagnostic potential of a serum miRNA panel, developed with a neural network analysis, for detection of ovarian cancer with 100% specificity(22). The miRNA neural network outperformed the currently used gold standard biomarker for ovarian cancer, CA125, with a positive predictive value of 91.3% and negative predictive value of 78.6%.
Despite their biological significance due to involvement in critical disease relevant pathways and many publications demonstrating their promising performance as informative biomarkers of disease diagnosis and prognosis, there are multiple challenges to developing and transitioning circulating miRNA biomarker assays to clinical application. These include the relatively large sample volume, in hundreds of microliters, required to perform the currently available and commonly used assays for profiling of miRNAs.
In this study, we report successful application of a novel limited volume assay platform, Abcam Fireplex™ (23), to identify differentially abundant plasma miRNAs between early stage pancreatic cancer cases and controls. This hydrogel particle-based miRNA assay platform was used for 20 microliters of plasma to detect up to 68 miRNAs per sample in a 96 well plate format without requiring extensive prior miRNA isolation and purification steps(23). This assay was recently used to develop a plasma miRNA signature for detection of lung cancer and identify histological subtypes(24). Here we demonstrate successful application of this technology in identifying circulating miRNA biomarkers for detection of early stage resectable PDAC in limited volume plasma with good sensitivity and specificity.
Materials and Methods:
Sample Collection & Patient Characteristics
The study was approved by the Institutional Review Boards at all participating institutions with written informed consent from enrolled patients. This study was conducted in accordance with the U.S. Common Rule regulations. PDAC cases were pathologically confirmed and plasma was collected from these patients at baseline, prior to receiving any treatment, including surgery. Patients with prior history of cancer or concurrent diagnosis with another cancer were excluded from this study. The main cohort included 88 plasma samples from 30 healthy controls and 58 patients with pathologically confirmed stage II PDAC cases from the TexGen repository, a consortium from the Texas Medical Center. Using the TexGen cohort, we identified promising candidate plasma miRNA markers and marker panels. We confirmed and validated the individual candidate markers and marker panel using two independent patient cohorts: one from the University of Pittsburgh (UPMC), which included 25 control plasma samples with benign pancreatic disease and 34 stage II PDAC plasma samples, and the second from the National Cancer Institute Early Detection Research Network (NCI EDRN) pancreatic cancer reference set including plasma samples from18 healthy controls and 44 stage II cases.
Abcam Fireplex™ Platform
We analyzed plasma samples in a 96 well plate using the Abcam Fireplex™ platform with their predesigned oncology panel (Abcam, Cambridge, United Kingdom) in accordance with manufacturer instructions(23). In brief, samples were thawed at room temperature and placed on ice. 20 μl of lysis buffer was placed in each well of a sterile 96 well plate and mixed with 20 μl of plasma. Plates were sealed and incubated for 45 minutes at 60°C while shaking (here and after always 750 RPM). 20 μl of lysed sample was aliquoted into a new plate along with the positive controls provided with the platform. Water served as negative controls. The remaining sample was stored at −20°C. Then 35 μl of particles from the oncology panel were added to a clean filter plate and connected to a vacuum manifold to remove storage buffer. After adding 25 μl each of hybridization buffer and lysed sample, the filter plate was covered and incubated for 60 minutes at 37°C while shaking. The samples were then rinsed twice with 1x Rinse A solution and re-suspended in 1x labeling mix. The plates were incubated for 60 minutes at room temperature while shaking and stored overnight at −20°C. The next day, plates were thawed on ice and rinsed with Rinse B and Rinse A solutions. Then miRNAs were eluted from the probes by adding RNAse free water and incubated for 30 minutes at 55°C while shaking. A clean catch plate was inserted into the vacuum manifold and carefully aligned with the wells so that orientation of the samples could be properly suctioned into the correct wells. The wells were kept hydrated with Rinse A in the empty filter plate and stored at 4°C until needed. In a clean PCR plate, 30 μl of eluent and 20 μl of PCR master mix were added and subjected to PCR in a thermocycler with the following program: 1 cycle at 93°C for 15 seconds; 27 cycles of: 93°C for 5 seconds, 59°C for 15 seconds, 72°C for 60 seconds; 6 cycles of: 93°C for 5 seconds, 63°C for 15 seconds, 72°C for 60 seconds; 1 cycle of 94°C for 4 minutes; 4°C hold. After PCR the Rinse A solution was removed under vacuum and 60 ul of hybridization buffer was added to each well of the filter plate. 20 μl of PCR product was transferred to the filter plate and incubated for 30 minutes at 37°C with shaking while the remaining PCR product was frozen at −20°C. After removing the plate from the filter plate on the shaker, the PCR product in the wells of the plate was rinsed twice with Rinse A, added with 75 μl of reporter mix, covered and incubated at room temperature for 15 minutes while shaking. Then the wells were rinsed twice with Rinse A, mixed with run buffer and read using a BD Accuri cytometer (BD Biosciences, San Jose, CA). The FCS files were uploaded into the Fire Code software (Abcam, Cambridge, United Kingdom) and analyzed by normalizing the MFIs using the three most stable miRNAs across all samples: with miR-17–5p, miR-20a-5p, and miR-93–5p. The customized oncology panel allowed us to measure relative abundances of 68 vendor designed preselected miRNAs (Supplementary Table 1).
Statistical Analysis
Patient characteristics have been summarized using frequency tables and descriptive statistics. The Chi-square test or Fisher’s exact test was used to compare the discrete variable (such as sex and diabetes) differences between cases and controls. For continuous variables such as age, the Wilcoxon test was used. In the miRNA marker screening process, the method of Benjamini and Hochberg (1995) was used to produce the adjusted p-values so that we could control the false discovery rate (FDR) under 10%(25). Logistic regression model was used to combine markers in the panel development. Receiver operation characteristic (ROC) curves were constructed. The area under the curve (AUC) was estimated, and its 95% confidence interval (CI) was estimated using the bootstrapping method. The study used the TexGen cohort for marker discovery. The top three candidate miRNA markers were chosen for the panel development due to their reported clinical importance in published literature and their statistical significance in differentiating cancer cases vs. controls. The performance of the marker panels was assessed using 10-fold cross validation. The performance of the candidate markers and marker panels were further tested using two independent validation cohorts from the University of Pittsburgh (UPMC) and the National Cancer Institute - Early Detection Research Network (NCI-EDRN). All the analyses were performed using the statistical software R 3.3.3 (CRAN, RRID:SCR_003005, https://cran.r-project.org) and Stata release 16 (Stata, RRID:SCR_012763, https://www.stata.com).
Gene Set Enrichment Analysis (GSEA)
TCGA-PAAD miRNA data for gene set enrichment analysis (GSEA) was downloaded from http://gdac.broadinstitute.org/; mRNA data for GSEA was downloaded from the TCGA data portal and normalized using the quantile normalization technique. Both data sets were log2 transformed for subsequent analysis. 178 pancreatic adenocarcinoma cases with the miRNA profiling data were used for further analysis. The level of the 3mir panel (3mir-level) was calculated as the average log2 expression of miR-34a-5p, miR-130a-3p, and miR-222–3p. All samples were divided into 2 groups by median into those with high 3miR-level and those with low 3miR-level. Then the datasets were analyzed using GSEA (26,27). Signal-to-noise and 1000 permutations of the genes were applied in the GSEA analysis with the gene sets obtained from the MSigDB database v6.2 for all hallmark and oncogenic signatures.
Results:
Plasma miRNAs with Significant Differential Abundance between Stage II and Control Samples
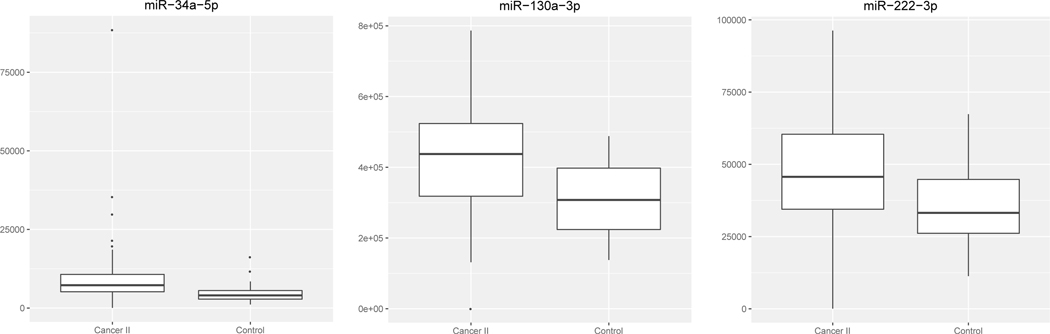
Patient characteristics have been summarized in Table 1. In the training set (TexGen cohort), there were 58 stage II and 30 healthy control samples. Comparisons were performed for each individual miRNA marker present on the Fireplex oncology panel. The distribution for all miRNAs can be found in Supplemental Figure 1. Eleven miRNAs were identified with significant differential abundance between stage II and control patients with FDR <0.1(25). Our top three candidate miRNAs included miR-34a-5p (p < 0.0001), miR-130a-3p (p = 0.0003), and miR-222–3p (0.0019) (Table 2 and Figure 1;). All three of these miRNAs were elevated in cancer v. controls. Neither sex, smoking, nor diabetes were significantly different between cases and controls.
Table 1.
Marginal frequency table by diagnosis for the TexGen, UPMC, and EDRN cohorts.
| TexGen Cohort | UPMC Cohort | EDRN Cohort | Comparison among cohorts | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Level | Control | Cancer | P-value | Control | Cancer | P-Value | Control | Cancer | P-Value | P-value* |
| Diagnosis | Control | 30 (100%) | 0 (0%) | 25 (100%) | 0 (0%) | 18 (100%) | 0 (0%) | 0.2991 | |||
| Stage II PDAC | 0 (0%) | 58 (100%) | 0 (0%) | 34 (100%) | 0 (0%) | 44 (100%) | |||||
| Sex | Female | 29 (72%) | 11 (28%) | 0.260 | 12 (40%) | 18 (60%) | 0.795 | 9 (30%) | 21 (70%) | 1.0 | 0.8447 |
| Male | 28 (60%) | 19 (40%) | 13 (45%) | 16 (55%) | 9 (28%) | 23 (72%) | |||||
| Diabetes | No | 25 (36%) | 45 (64%) | 0.779 | 14 (38%) | 23 (62%) | 0.421 | 14 (33%) | 28 (67%) | 0.190 | 0.05857 |
| Yes | 5 (29%) | 12 (71%) | 11 (50%) | 11 (50%) | 2 (13%) | 13 (87%) | |||||
| Smoking | Current | 5 (100%) | 0 (0%) | 0.126 | 7 (47%) | 8 (53%) | 0.151 | - | - | - | 0.0023 |
| Former | 27 (71%) | 11 (29%) | 10 (59%) | 7 (41%) | - | - | - | ||||
| Never | 25 (57%) | 19 (43%) | 8 (30%) | 19 (70%) | - | - | - | ||||
| Location | Uncinate Process | NA | 0 (0%) | NA | 5 (15%) | NA | 4 (9%) | 0.0120 | |||
| Head | NA | 47 (81%) | NA | 19 (56%) | NA | 27 (61%) | |||||
| Neck | NA | 0 (0%) | NA | 1 (3%) | NA | 0 (0%) | |||||
| Body | NA | 4 (7%) | NA | 2 (6%) | NA | 2 (5%) | |||||
| Tail | NA | 2 (3%) | NA | 4 (12%) | NA | 4 (9%) | |||||
| Overlapping Lesion | NA | 3 (5%) | NA | 3 (9%) | NA | 0 (0%) | |||||
| Not specified | NA | 2 (3%) | NA | 0 (0%) | NA | 7 (16%) | |||||
| Controls | Healthy | 30 (100%) | NA | 0 (0%) | NA | 18 (100%) | NA | ||||
| Chronic pancreatitis (CP) | 0 (0%) | NA | 12 (48%) | NA | 0 (0%) | NA | |||||
| Abnormal image test (benign) | 0 (0%) | NA | 4 (16%) | NA | 0 (0%) | NA | |||||
| Benign stricture; biliary dilation | 0 (0%) | NA | 2 (8%) | NA | 0 (0%) | NA | |||||
| Acute pancreatitis | 0 (0%) | NA | 1 (4%) | NA | 0 (0%) | NA | |||||
| Autoimmune pancreatitis | 0 (0%) | NA | 1 (4%) | NA | 0 (0%) | NA | |||||
| Benign inflammatory changes | 0 (0%) | NA | 1 (4%) | NA | 0 (0%) | NA | |||||
| Common hepatic duct stricture | 0 (0%) | NA | 1 (4%) | NA | 0 (0%) | NA | |||||
| Intraductal papillary mucinous neoplasm (IPMN) | 0 (0%) | NA | 1 (4%) | NA | 0 (0%) | NA | |||||
| CP & IPMN | 0 (0%) | NA | 2 (8%) | NA | 0 (0%) | NA | |||||
| Age: median (Range) | 58.5 (22 – 80) | 64.0 (42 – 84) | 0.025 | 67.0 (43 – 84) | 64.0 (43 – 83) | 0.878 | 64.7 (52 – 85) | 71.2 (50 – 89) | 0.040 | <0.001 | |
Chi-square test was used to compare the diagnosis, sex, diabetes and smoking status marginal frequencies among cohorts. Kruskal-Wallis test was to compare the age difference among cohorts.
Table 2.
AUCs and 95% confidence intervals for the top three significant miRNAs in the TexGen cohort, as well as their performances in the UPMC and EDRN cohorts.
| TexGen Cohort | UPMC Cohort | EDRN Cohort | ||||
|---|---|---|---|---|---|---|
| miRNA | AUC | 95% CI | AUC | 95% CI | AUC | 95% CI |
| mir-34a-5p | 0.77 | (0.66 to 0.87) | 0.65 | (0.50 to 0.79) | 0.66 | (0.51 to 0.80) |
| mir-130a-3p | 0.74 | (0.63 to 0.84) | 0.58 | (0.44 to 0.74) | 0.41 | (0.26 to 0.58) |
| mir-222–3p | 0.70 | (0.59 to 0.82) | 0.60 | (0.46 to 0.74) | 0.54 | (0.38 to 0.70) |
Figure 1.
Distribution of oncology markers showing the top three significant miRNAs between stage II PDAC cases and controls with FDR<0.1 in the TexGen cohort.
Marker Panel Development
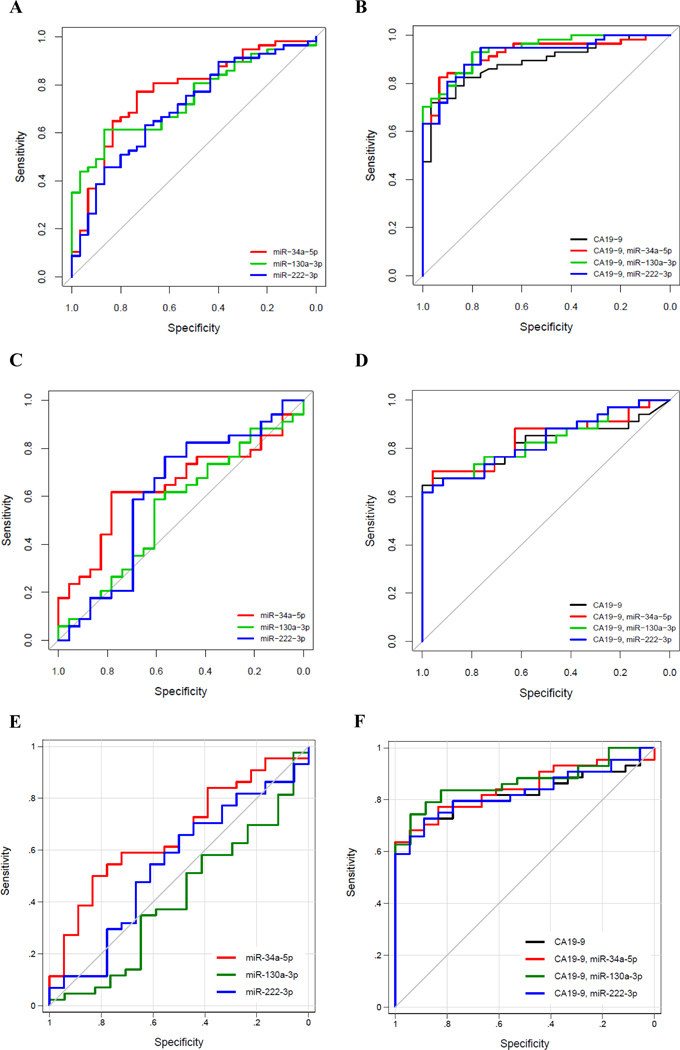
The top three statistically significant differentially abundant miRNAs, discriminating between stage II v. control plasma samples, included: miR-34a-5p, miR-130a-3p, and miR-222–3p, with AUC values of 0.77 (95% CI 0.66 to 0.87), 0.74 (95% CI 0.63 to 0.84), and 0.70 (95% CI 0.59 to 0.82) (Table 2 and Figure 2A), respectively. Consistent with previous studies (28,29), CA19–9 performed well in differentiating stage II cases vs. control cases, with an estimated AUC = 0.89 (95% CI 0.81 to 0.95). Combining CA 19–9 with selected individual miRNA markers, miR-34a-5p, miR-130a-3p, and miR-222–3p, improved the AUC values for each panel to 0.92 (95% CI 0.86 to 0.97), 0.94 (95% CI 0.89 to 0.98), and 0.92 (95% CI 0.87 to 0.97), respectively. (Table 3 and Figure 2B). Among these, the marker panel of CA19–9 with miR-130a-3p improved AUC significantly compared to that using CA19–9 alone (P=0.0390). The performance of the combined CA19–9 and miRNA marker panels remained similar in estimated AUC when using 10-fold cross-validation. Using 10-fold cross validation, CA 19–9 alone had an estimated AUC of 0.88 (95% CI 0.81 to 0.95). When combined with CA 19–9, miR-34a-5p, miR-130a-3p, and miR-222–3p had AUCs of 0.91 (95% CI 0.84 to 0.97), 0.94 (95% CI 0.89 to 0.99), and 0.89 (95% CI 0.82 to 0.96), respectively.
Figure 2.
ROC curves. A. Top 3 miRNA markers in the TexGen cohort comparing stage II vs. control. B. miRNA marker panels in combination with CA 19–9 in TexGen cohort comparing stage II vs. control. C. miRNA markers in the UPMC cohort comparing stage II vs. control. D. miRNA marker panels in combination with CA 19–9 in the UPMC cohort comparing stage II vs. control. E. miRNA markers in the EDRN cohort comparing stages I & II vs. control. F. miRNA marker panels in combination with CA 19–9 in the EDRN cohort comparing stages I & II vs. control.
Table 3.
Maker panel performance.
| Cohort | Model | AUC (95%CI) | *Optimal cutoff [sens, spec] | P-value |
|---|---|---|---|---|
| TexGen | 0.1204 * CA19_9 | 0.89 (0.81, 0.95) | 1.58 [0.82, 0.83] | Reference |
| 0.1186 * CA19_9 + 0.0003 * miR-34a-5p | 0.92 (0.86, 0.97) | 3.32 [0.84, 0.90] | 0.1058 | |
| 0.1229 * CA19_9 + 0.0001 * miR-130a-3p | 0.94 (0.89, 0.98) | 6.22 [0.84, 0.87] | 0.0390 | |
| 0.116 * CA19_9 + 0.0001 * miR-222–3p | 0.92 (0.87, 0.97) | 4.02 [0.88, 0.83] | 0.1285 | |
| UPMC | 0.0164 * CA19_9 | 0.82 (0.69, 0.92) | 1.72 [0.68, 0.96] | Reference |
| 0.0169 * CA19_9 + 0.0001 * miR-34a-5p | 0.84 (0.72, 0.93) | 1.87 [0.71, 0.96] | 0.3146 | |
| 0.0166 * CA19_9 + 0.0001 * miR-130a-3p | 0.83 (0.72, 0.93) | 1.88 [0.68, 0.92] | 0.5282 | |
| 0.017 * CA19_9 + 0.0001 * miR-222–3p | 0.83 (0.72, 0.93) | 1.96 [0.68, 0.92] | 0.5389 | |
| EDRN | .0672*CA19_9 | 0.82 (0.71, 0.91) | 1.80 [0.73, 0.89] | Reference |
| .0679*CA19_9 + 5.4e-05*miR-34a-5p | 0.84 (0.74, 0.93) | 1.80 [0.77, 0.83] | 0.2910 | |
| .0718*CA19_9 + 1.6e-06*miR-130a-3p | 0.87 (0.77, 0.95) | 0.59 [0.84, 0.82] | 0.2373 | |
| .0677*CA19_9 + 4.2e-05*miR-222–3p | 0.83 (0.72, 0.92) | 2.05 [0.73, 0.89] | 0.5631 |
The optimal cutoff was determined to be the point on the ROC curve has the minimum distance to the upper left corner (where sensitivity=1 and specificity=1). By Pathagoras’ theorem this distance is sqrt( (1-sensitivity)2+(1-specificity)2).
Performance of Markers in UPMC and NCI EDRN Validation Sets
The three significant miRNAs from the training set, miR-34a-5p, miR-130a-3p, and miR-222–3p outperformed chance in the UPMC cohort, with significant differences between stage II and control patients (Table 2, Figure 2C). Neither sex, smoking, nor diabetes were significantly different between cases and controls, consisting of patients with benign pancreatic disease, as described in Table 1. When combined with CA19–9, their estimated AUCs were 0.84 (95% CI 0.72 to 0.93), 0.83 (95% CI 0.72 to 0.93), and 0.83 (95% CI 0.72 to 0.93), respectively (Table 3, Figure 2D). However, none of the marker panels had statistically significant improvement in AUC compared to that of CA19–9 alone.
Next, we tested the performance of the three selected miRNA markers and their respective panels with CA19–9 using a blinded validation set of EDRN samples comprising of 44 stage II cases and 18 healthy controls. Marker miR-34a-5p showed significantly higher values for cases than controls (P=0.0470), with an AUC of 0.66 (Table 2, Figure 2E). When combined with CA 19–9, miR-130a-3p (Table 3, Figure 2F) yielded an improved AUC of 0.87 (95%CI 0.77 to 0.95).
Relatively stronger performance of the panels in discriminating stage II from controls in the training set (TexGen cohort) compared with UPMC cohort, could, in part, be a reflection of the fact that the TexGen training set control samples consisted of plasma from disease free healthy individuals while for the UPMC cohort, control samples consisted of plasma from patients with benign pancreatic disease. Improved performance in the training set could also be due to differences in age ranges across cohorts (p < 0.001, Table 1). PDAC risk increases with age, hence this could affect performance in our validation sets, which included control patients of older age. Additionally, given that the sample cohorts were from banked plasma collected at different institutions, which may have had different standard operating protocols (SOP) for sample collection, time to processing and storage, it is possible that varying performance of the candidate miRNAs in detecting resectable disease were partly due to absence of uniform SOPs for bio-banking at respective institutions that affects miRNA detection outcome (30,31). It is imperative that future studies are designed to control for these potential variables by adhering to uniform SOPs for sample collection and storage across institutions. Furthermore, there can be discrepancies in clinical staging, particularly in unresected patients for whom pathological staging is unavailable, where nodal involvement is often underestimated due to current imaging limitations (32–34).
Gene Set Enrichment Analysis (GSEA)
To understand the significance of these three miRNAs in pancreatic cancer, we conducted gene set enrichment analysis (GSEA) of all hallmark genes for 178 pancreatic adenocarcinoma samples from TCGA. Interestingly, samples with high 3miR-levels showed significant enrichment in genes associated with glycolysis (p = 0.0139, Figure 3A). Glycolysis is known to play an important role in cancers, especially in the context of those with Ras mutations (35–37), which are almost ubiquitous in PDAC (38). This finding suggests that high 3miR expression is associated with the genetic pathways facilitating aerobic glycolysis for energy metabolism in early stage PDAC cells. We also conducted GSEA of oncogenic signatures to better understand the role of these three miRNAs in tumorigenesis. Results revealed that high 3miR-expression is associated with gene sets that correlate with the activation of oncogenes, including KRAS and AKT and inactivation of the tumor suppressor gene, RB (Figure 3B). Conversely, samples with low 3miR-expression showed enrichment of genes that are down-regulated in cells with p53 loss of function mutations, also revealed in the hallmark GSEA results. Additionally, KEGG pathway analysis using DIANA-miRPath v3. 0 revealed that these miRNAs are involved in several pancreatic cancer-relevant pathways, such TGF-beta signaling (p = 1.41×10−3), phosphatidylinositol signaling (p = 1.72 ×10−3), FOXO signaling (p = 5.73×10−3), P53 signaling (p = 0.0170), dorso-ventral axis formation (including NOTCH signaling) (p = 0.0186), ERBB signaling (p = 0.0210), and axon guidance (p = 0.0307) pathway, among others (39).
Figure 3.

Gene Set Enrichment Analysis (GSEA) of 178 TCGA PAAD cases with high and low 3miR-levels. A. GSEA of hallmark signatures. B. GSEA of oncogenic signatures.
Discussion:
A panel of miRNA biomarkers that can stratify early stage PDAC patients from control subjects, including those with benign disease, was developed using a novel hydrogel particle-based miRNA profiling platform (Abcam Fireplex) amenable to screening with limited volume body fluid samples. Development of a minimally invasive early detection circulating miRNA biomarker assay using limited volume plasma samples addresses the major challenge of large volume requirement for miRNA profiling in currently available conventional methods. This “proof of concept” study with the vendor designed, prefabricated assay platform for miRNA biomarker assay demonstrates the feasibility of the technology being translated to clinical practice in diagnostic settings to improve chances of curative clinical intervention and survival for patients with early stage PDAC. Although the current assay development was restricted to 68 miRNAs on the Abcam Fireplex Oncology Panel, individual miRNAs (miR-34a-5p, miR-130a-3p, and miR-222–3p) together with the currently used biomarker for pancreatic cancer, CA 19–9, improved performance of the latter in discriminating early stage PDAC patients from controls.. These miRNAs in combination with CA 19–9 yielded AUCs of 0.92, 0.94, and 0.92, respectively, in our training cohort, with miR-34a-5p and miR-222–3p also performing well in both validation cohorts.
All three candidate miRNAs have been previously reported to play roles in disorders of the pancreas and cancer. Importantly, miR-34a has been demonstrated to be a tumor suppressor miRNA in PDAC(40,41) as well as a candidate serum biomarker for the disease(42). Previous studies have also reported that miR-34a-5p impairs PDAC progression through post-transcriptional regulation of Snail1 and Notch1, inhibiting epithelial-mesenchymal transition (43). In addition, miR-34a-5p deficiency mediated loss of TP53 function in lung adenocarcinoma and head and neck cancer was shown to affect critical tumor promoting pathways including cancer associated adrenergic trans-differentiation of sensory nerves(44). These findings have led to the idea of miR-34a is a potential therapeutic target (45,46). Pre-clinical studies with miR-34a miRNA-mimics in mice with PDAC combined with PLK1 siRNA improved survival and decreased the rate of tumor growth (45). Another study has suggested that Genistein, which upregulates miR-34a promoting apoptosis and inhibiting cell growth in PDAC cell lines, may have therapeutic potential against PDAC (46). The GSEA result showing differential enrichment of p53 hallmark signatures between high and low 3miR expressing pancreatic adenocarcinoma cases corroborated previous finding. The level of miR-130a-3p was reported to be elevated in pancreatic islets from hyperglycemic donors and in islets from Goto-Kakizaki rats with type-2 diabetes (47,48). An estimated 50% to 80% of PDAC patients have diabetes or some form of glucose intolerance (49–51), and diabetes itself is a known risk factor for PDAC development (52–54). It would be interesting to determine if circulating levels of miR-130a-3p could be used to identify which diabetic patients are predisposed to develop PDAC. Interestingly, miR-130a was found to be decreased in high-risk IPMN (55), but has not been assessed as a circulating biomarker in blood for such lesions. Finally, elevated levels of miR-222 in tumor tissues has been associated with poor prognosis for patients with pancreatic cancer (56,57). Cell line studies have suggested that miR-222 may play a role in PDAC by targeting cyclin-dependent kinase inhibitor CDNK1C/P57, promoting cell proliferation and viability (58). Overall, GSEA results indicate that the three differentially abundant miRNAs in plasma from early stage PDAC patients, as candidate biomarkers of the disease, play roles in oncogenic signaling mediating downregulation of tumor suppressor genes, activation of glycolysis and cellular growth.
Although the above findings reflect mechanistic involvement of the three miRNAs in deregulating cancer relevant pathways, it is plausible that the three miRNAs do not comprehensively capture the complex and heterogeneously evolving aberrant proteomics landscape of early stage cancer genomes. Additionally, miRNA expression is regulated by diverse cellular and developmental contexts (59) and multiple miRNAs regulate overlapping target genes and their pathways (60). This suggests that highly sensitive and specific early detection PDAC miRNA biomarker signatures will include miRNAs involved in diverse pathways frequently deregulated in the disease. Development of such miRNA signatures would require unbiased global profiling of miRNAs in larger sample cohorts processed under controlled standard operating procedures. It will also be important that the data be analyzed with adequately powered statistical tools, such as neural network analysis. This tool recently helped generate a 7 miRNA panel from next generation sequencing data for detecting epithelial ovarian cancer with high sensitivity and specificity (22). This panel outperformed the gold standard biomarker, CA-125, commonly used for diagnosing ovarian cancer (22). While such additional larger scale discovery and validation studies are warranted for developing a more robust early detection miRNA biomarker signature for PDAC, the current findings demonstrate that limited volume plasma samples, available in clinical diagnostic settings, can be effectively used to assay the differentially abundant miRNAs associated with early stage resectable pancreatic cancer. Reliability of the assay platform, used in this study, was suggested by a recent study reporting that on average, ~70% of miRNAs detected by Fireplex match in directionality and significance to those detected by RNASeq (61). In the future, it will be important to include stage I cases as well as prospective pre-diagnostic samples besides those with benign disease and benign pancreatic neoplasms to assess performance of the miRNA biomarkers described in this study.
In summary, we have identified candidate circulating miRNA biomarkers from limited volume of plasma that can discriminate individuals with early stage resectable pancreatic cancer from cancer free controls, including those with benign pancreatic diseases. Having vetted this technology by using the commercially available Oncology Panel from Fireplex, we plan to use in the future a custom panel with miRNAs, deregulated in pancreatic cancer and/or involved in known pancreatic cancer related pathways, such as those suggested by the GSEA findings in this study. Focusing on differentially abundant circulating miRNAs involved in deregulated biological pathways would be a logical approach to developing minimally invasive early detection biomarker assay for patients not expressing CA19–9 besides improving sensitivity and specificity of CA19–9 as a combined panel, similar to the proteomic markers representing a migratory signature in pancreatic cancer reported by us earlier (62). Further development of the assay platform for more robust plasma miRNA biomarker panel discriminating early stage pancreatic cancer with high sensitivity and specificity is expected to have significant impact on improving clinical care and disease outcomes for patients with this recalcitrant malignancy.
Supplementary Material
Acknowledgments
Funding: This study was supported by funding from the University of Texas MD Anderson Cancer Center Pancreatic Cancer Moonshot Program; the Cancer Prevention Research Institute of Texas (RP180392 to AMK and SS); the National Cancer Institute at the National Institutes of Health (1 U01 CA214263 to SS and AMK, P50CA221707, U01CA196403, and U01CA200468 to AM, and U01 CA200466 to RB). RLD is supported by Dr. John J. Kopchick Fellowship from the University Of Texas MD Anderson UT Health Graduate School of Biomedical Sciences and the National Institutes of Health Translational Genomics and Precision Medicine in Cancer T32 Training Program (5T32CA217789-03) from the National Cancer Institute. This work was also supported in part by The Cancer Prevention Institute of Texas (CPRIT) RP170005, NIH P30 shared resource grant CA125123, and NIEHS grants 1P30ES030285 and 1P42ES0327725.
Financial Support:
Subrata Sen and Ann Killary: NCI 1 U01 CA214263, Cancer Prevention Research Institute of Texas (CPRIT) RP180392
Rachel L. Dittmar: Dr. John J. Kopchick Fellowship from the University Of Texas MD Anderson UT Health Graduate School of Biomedical Sciences, NCI 5T32CA217789-03
Cristian Coarfa: CPRIT RP170005, NIH P30 CA125123, NIEHS 1P30ES030285 and 1P42ES0327725
Randall Brand: NCI U01 CA200466
Anirban Maitra: NCI P50CA221707, U01CA196403, and U01CA200468
Footnotes
The authors declare no potential conflicts of interest.
References:
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70(1):7–30 doi 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Blackford AL, Canto MI, Klein AP, Hruban RH, Goggins M. Recent trends in the incidence and survival of Stage 1A Pancreatic Cancer: A Surveillance, Epidemiology, and End Results analysis. J Natl Cancer Inst 2020. doi 10.1093/jnci/djaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clancy TE. Surgery for Pancreatic Cancer. Hematol Oncol Clin North Am 2015;29(4):701–16 doi 10.1016/j.hoc.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19–9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol 2012;3(2):105–19 doi 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JE, Lee KT, Lee JK, Paik SW, Rhee JC, Choi KW. Clinical usefulness of carbohydrate antigen 19–9 as a screening test for pancreatic cancer in an asymptomatic population. J Gastroenterol Hepatol 2004;19(2):182–6. [DOI] [PubMed] [Google Scholar]
- 6.Tsen A, Barbara M, Rosenkranz L. Dilemma of elevated CA 19–9 in biliary pathology. Pancreatology 2018;18(8):862–7 doi 10.1016/j.pan.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Safi F, Roscher R, Bittner R, Schenkluhn B, Dopfer HP, Beger HG. High sensitivity and specificity of CA 19–9 for pancreatic carcinoma in comparison to chronic pancreatitis. Serological and immunohistochemical findings. Pancreas 1987;2(4):398–403 doi 10.1097/00006676-198707000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Duraker N, Hot S, Polat Y, Höbek A, Gençler N, Urhan N. CEA, CA 19–9, and CA 125 in the differential diagnosis of benign and malignant pancreatic diseases with or without jaundice. J Surg Oncol 2007;95(2):142–7 doi 10.1002/jso.20604. [DOI] [PubMed] [Google Scholar]
- 9.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7(4):248–9 doi 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vestergaard EM, Hein HO, Meyer H, Grunnet N, Jørgensen J, Wolf H, et al. Reference values and biological variation for tumor marker CA 19–9 in serum for different Lewis and secretor genotypes and evaluation of secretor and Lewis genotyping in a Caucasian population. Clin Chem 1999;45(1):54–61. [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136(2):215–33 doi 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, et al. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 2014;56(1):104–15 doi 10.1016/j.molcel.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011;12(2):99–110 doi 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 14.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19(1):92–105 doi 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res 2008;36(Database issue):D154–8 doi 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105(30):10513–8 doi 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9(6):654–9 doi 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 18.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006;11(4):441–50 doi 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics 2012;10(5):246–53 doi 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kai K, Dittmar RL, Sen S. Secretory microRNAs as biomarkers of cancer. Semin Cell Dev Biol 2018;78:22–36 doi 10.1016/j.semcdb.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2(9):807–13 doi 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elias KM, Fendler W, Stawiski K, Fiascone SJ, Vitonis AF, Berkowitz RS, et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. Elife 2017;6 doi 10.7554/eLife.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tackett MR, Diwan I. Using FirePlex. Methods Mol Biol 2017;1654:209–19 doi 10.1007/978-1-4939-7231-9_14. [DOI] [PubMed] [Google Scholar]
- 24.Leng Q, Wang Y, Jiang F. A Direct Plasma miRNA Assay for Early Detection and Histological Classification of Lung Cancer. Transl Oncol 2018;11(4):883–9 doi 10.1016/j.tranon.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995. [Google Scholar]
- 26.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34(3):267–73 doi 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haab BB, Huang Y, Balasenthil S, Partyka K, Tang H, Anderson M, et al. Definitive Characterization of CA 19–9 in Resectable Pancreatic Cancer Using a Reference Set of Serum and Plasma Specimens. PLoS One 2015;10(10):e0139049 doi 10.1371/journal.pone.0139049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Brien DP, Sandanayake NS, Jenkinson C, Gentry-Maharaj A, Apostolidou S, Fourkala EO, et al. Serum CA19–9 is significantly upregulated up to 2 years before diagnosis with pancreatic cancer: implications for early disease detection. Clin Cancer Res 2015;21(3):622–31 doi 10.1158/1078-0432.CCR-14-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grasedieck S, Sorrentino A, Langer C, Buske C, Döhner H, Mertens D, et al. Circulating microRNAs in hematological diseases: principles, challenges, and perspectives. Blood 2013;121(25):4977–84 doi 10.1182/blood-2013-01-480079. [DOI] [PubMed] [Google Scholar]
- 31.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013;59(1):S1–6 doi 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Elbanna KY, Jang HJ, Kim TK. Imaging diagnosis and staging of pancreatic ductal adenocarcinoma: a comprehensive review. Insights Imaging 2020;11(1):58 doi 10.1186/s13244-020-00861-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swords DS, Firpo MA, Johnson KM, Boucher KM, Scaife CL, Mulvihill SJ. Implications of inaccurate clinical nodal staging in pancreatic adenocarcinoma. Surgery 2017;162(1):104–11 doi 10.1016/j.surg.2016.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qayyum A, Tamm EP, Kamel IR, Allen PJ, Arif-Tiwari H, Chernyak V, et al. ACR Appropriateness Criteria. J Am Coll Radiol 2017;14(11S):S560–S9 doi 10.1016/j.jacr.2017.08.050. [DOI] [PubMed] [Google Scholar]
- 35.Racker E, Resnick RJ, Feldman R. Glycolysis and methylaminoisobutyrate uptake in rat-1 cells transfected with ras or myc oncogenes. Proc Natl Acad Sci U S A 1985;82(11):3535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 2012;149(3):656–70 doi 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science 2009;325(5947):1555–9 doi 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491(7424):399–405 doi 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43(W1):W460–6 doi 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007;26(5):745–52 doi 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 2008;7(16):2591–600 doi 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 42.Alemar B, Izetti P, Gregório C, Macedo GS, Castro MA, Osvaldt AB, et al. miRNA-21 and miRNA-34a Are Potential Minimally Invasive Biomarkers for the Diagnosis of Pancreatic Ductal Adenocarcinoma. Pancreas 2016;45(1):84–92 doi 10.1097/MPA.0000000000000383. [DOI] [PubMed] [Google Scholar]
- 43.Tang Y, Cheng YS. miR-34a inhibits pancreatic cancer progression through Snail1-mediated epithelial-mesenchymal transition and the Notch signaling pathway. Sci Rep 2017;7:38232 doi 10.1038/srep38232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amit M, Takahashi H, Dragomir MP, Lindemann A, Gleber-Netto FO, Pickering CR, et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 2020;578(7795):449–54 doi 10.1038/s41586-020-1996-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibori H, Eliyahu S, Krivitsky A, Ben-Shushan D, Epshtein Y, Tiram G, et al. Amphiphilic nanocarrier-induced modulation of PLK1 and miR-34a leads to improved therapeutic response in pancreatic cancer. Nat Commun 2018;9(1):16 doi 10.1038/s41467-017-02283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia J, Duan Q, Ahmad A, Bao B, Banerjee S, Shi Y, et al. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets 2012;13(14):1750–6. [DOI] [PubMed] [Google Scholar]
- 47.Ofori JK, Salunkhe VA, Bagge A, Vishnu N, Nagao M, Mulder H, et al. Elevated miR-130a/miR130b/miR-152 expression reduces intracellular ATP levels in the pancreatic beta cell. Sci Rep 2017;7:44986 doi 10.1038/srep44986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esguerra JL, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One 2011;6(4):e18613 doi 10.1371/journal.pone.0018613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pannala R, Leirness JB, Bamlet WR, Basu A, Petersen GM, Chari ST. Prevalence and clinical profile of pancreatic cancer-associated diabetes mellitus. Gastroenterology 2008;134(4):981–7 doi 10.1053/j.gastro.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Permert J, Ihse I, Jorfeldt L, von Schenck H, Arnqvist HJ, Larsson J. Pancreatic cancer is associated with impaired glucose metabolism. Eur J Surg 1993;159(2):101–7. [PubMed] [Google Scholar]
- 51.Wakasugi H, Funakoshi A, Iguchi H. Clinical observations of pancreatic diabetes caused by pancreatic carcinoma, and survival period. Int J Clin Oncol 2001;6(1):50–4. [DOI] [PubMed] [Google Scholar]
- 52.Batabyal P, Vander Hoorn S, Christophi C, Nikfarjam M. Association of diabetes mellitus and pancreatic adenocarcinoma: a meta-analysis of 88 studies. Ann Surg Oncol 2014;21(7):2453–62 doi 10.1245/s10434-014-3625-6. [DOI] [PubMed] [Google Scholar]
- 53.Haugvik SP, Hedenström P, Korsæth E, Valente R, Hayes A, Siuka D, et al. Diabetes, smoking, alcohol use, and family history of cancer as risk factors for pancreatic neuroendocrine tumors: a systematic review and meta-analysis. Neuroendocrinology 2015;101(2):133–42 doi 10.1159/000375164. [DOI] [PubMed] [Google Scholar]
- 54.Stevens RJ, Roddam AW, Beral V. Pancreatic cancer in type 1 and young-onset diabetes: systematic review and meta-analysis. Br J Cancer 2007;96(3):507–9 doi 10.1038/sj.bjc.6603571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Permuth-Wey J, Chen YA, Fisher K, McCarthy S, Qu X, Lloyd MC, et al. A genome-wide investigation of microRNA expression identifies biologically-meaningful microRNAs that distinguish between high-risk and low-risk intraductal papillary mucinous neoplasms of the pancreas. PLoS One 2015;10(1):e0116869 doi 10.1371/journal.pone.0116869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer 2010;126(1):73–80 doi 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 57.Lee C, He H, Jiang Y, Di Y, Yang F, Li J, et al. Elevated expression of tumor miR-222 in pancreatic cancer is associated with Ki67 and poor prognosis. Med Oncol 2013;30(4):700 doi 10.1007/s12032-013-0700-y. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Y, Wang Y, Yang Y, Liu J, Song Y, Cao Y, et al. MicroRNA-222 Controls Human Pancreatic Cancer Cell Line Capan-2 Proliferation by P57 Targeting. J Cancer 2015;6(12):1230–5 doi 10.7150/jca.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Towler BP, Jones CI, Newbury SF. Mechanisms of regulation of mature miRNAs. Biochem Soc Trans 2015;43(6):1208–14 doi 10.1042/BST20150157. [DOI] [PubMed] [Google Scholar]
- 60.Sohel MMH. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci 2020;248:117473 doi 10.1016/j.lfs.2020.117473. [DOI] [PubMed] [Google Scholar]
- 61.Yeri A, Courtright A, Danielson K, Hutchins E, Alsop E, Carlson E, et al. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genomics 2018;19(1):331 doi 10.1186/s12864-018-4726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balasenthil S, Huang Y, Liu S, Marsh T, Chen J, Stass SA, et al. A Plasma Biomarker Panel to Identify Surgically Resectable Early-Stage Pancreatic Cancer. J Natl Cancer Inst 2017;109(8) doi 10.1093/jnci/djw341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.