Abstract
Pasteurella multocida is composed of three subspecies that are often differentiated by fermentation of sorbitol and dulcitol. We studied 35 dulcitol-negative P. multocida isolates from infected dog and cat bite wounds, 16 of which yielded weak and/or conflicting fermentation reactions in Andrades sorbitol, thus making it difficult to distinguish between the two dulcitol-negative subspecies of P. multocida, i.e., P. multocida subsp. multocida and P. multocida subsp. septica. All isolates and two control strains were further analyzed using a PCR fingerprinting technique with a single primer (M13 core) and assessed for α-glucosidase (α-Glu) activity. Although the PCR fingerprint patterns and α-Glu activity did not correlate well with the sorbitol fermentation reactions, they did correlate well with each other. All strains identified as P. multocida subsp. septica were positive for α-Glu activity and exhibited the group I PCR fingerprint profile. All strains categorized as P. multocida subsp. multocida displayed either the group II or group III PCR fingerprint profile; 9 of 11 of these isolates were α-Glu negative. These data suggest that both PCR fingerprinting and α-Glu activity provide reliable means for differentiating P. multocida subsp. multocida from P. multocida subsp. septica, particularly in strains that produce weak and/or discrepant sorbitol fermentation reactions.
Pasteurella species have been isolated from various animals, either as saprophytes in the nasopharynx or gastrointestinal tract or as primary pathogens (reviewed in reference 17). Human disease is generally associated with some form of animal contact, most commonly a dog bite or cat bite or scratch (13, 14, 27, 30). Taxonomic relationships and nomenclature for this genus have undergone considerable change throughout the years. In 1985, DNA hybridization studies performed by Mutters et al. (22) revealed three homology groups of Pasteurella multocida that differed sufficiently enough from each other that they qualified as different species of Pasteurella. However, because the recommended therapy for human infection with Pasteurella is generally the same regardless of the species involved (reviewed in reference 17), the three groups of P. multocida were assigned different subspecies names for epidemiological purposes. P. multocida subsp. multocida includes the dulcitol-negative, sorbitol-positive isolates; P. multocida subsp. septica includes the dulcitol-negative, sorbitol-negative isolates; and P. multocida subsp. gallicida includes the dulcitol-positive isolates.
Other authors (3, 14) have suggested different ecological niches, as well as potential differences in pathogenicity, for the various Pasteurella species. Pasteurella multocida subsp. multocida and P. multocida subsp. septica are more frequently recovered from “more serious cases of infection” (14), including bacteremia. Whereas P. multocida subsp. multocida can be isolated from both dog- and cat-associated injuries, P. multocida subsp. septica is more frequently isolated from cases with cat contact and may have a greater affinity for the central nervous system (3). Therefore, there may be both epidemiological and clinical importance to the correct identification of these subspecies.
In our ongoing studies of organisms isolated from animal bite wound infections in humans, we have cultured numerous Pasteurella isolates, including P. multocida. In this study, we attempted to differentiate 35 dulcitol-negative P. multocida clinical isolates to the subspecies level using sorbitol fermentation as the differentiating test. We found that a significant number of our clinical isolates gave variable results when assayed for sorbitol and dulcitol fermentation. This is problematic in that sorbitol and dulcitol fermentation are key biochemical tests for the differentiation of P. multocida subspecies as originally described by Mutters et al. (22). To our knowledge, variation in the ability of a single P. multocida isolate to ferment sorbitol and dulcitol has not been addressed elsewhere in the literature.
Since the studies of Mutters et al. (22), there have been many studies on the biochemical characterization of P. multocida subspecies. However, most (if not all) of these studies rely heavily on the use of sorbitol fermentation to differentiate P. multocida subsp. multocida from P. multocida subsp. septica. In the studies of Bisgaard and colleagues (4, 21), sorbitol fermentation is the only biochemical characteristic which clearly and consistently differentiates P. multocida subsp. multocida from P. multocida subsp. septica. Likewise, the studies of Blackall and colleagues (5, 12), show that sorbitol fermentation is also the only biochemical test that consistently differentiates P. multocida subsp. multocida from P. multocida subsp. septica. In other work by that group (6, 7), the authors differentiated P. multocida subsp. multocida from P. multocida subsp. gallicida. Based on citation of their previous work for the differentiation of these organisms, we conclude that sorbitol, as well as dulcitol, fermentation reactions were likely used as key differentiating reactions for these subspecies.
A few exceptions to the use of sorbitol fermentation to differentiate the two dulcitol-negative subspecies of P. multocida have been reported in the literature. Seven sorbitol-negative strains of P. multocida have been described which have not been classified as either P. multocida subsp. multocida or P. multocida subsp. septica due to differences in their trehalose and/or xylose fermentation reactions relative to the P. multocida subsp. septica type strain (12). In their discussion, the authors note that these could be either sorbitol-negative variants of P. multocida subsp. multocida or trehalose-negative P. multocida subsp. septica strains. In another study (25), eight sorbitol-negative strains have apparently been identified as P. multocida subsp. multocida as deduced from the difference between the number of P. multocida subsp. multocida strains and the number of sorbitol-positive strains of P. multocida isolated from dead turkeys cited in the tables. However, because the authors cite the original work of Mutters et al. (22) for methods on biotype and subspecies identification, i.e., methods which rely on sorbitol fermentation for the differentiation of P. multocida subsp. multocida from P. multocida subsp. septica, it is not clear on what basis these eight sorbitol-negative strains were identified as P. multocida subsp. multocida. Thus, based on our review of the literature, it appears that sorbitol fermentation is still used as a key test for the phenotypic differentiation of P. multocida subsp. multocida from P. multocida subsp. septica.
In an effort to ascertain if our sorbitol-variable isolates could be classified as P. multocida subsp. multocida or P. multocida subsp. septica, we used a PCR fingerprinting technique. In addition, we screened our isolates for preformed enzyme activity using the API-ZYM panel to determine if another biochemical marker that would clearly differentiate these isolates could be found. Our results suggest that both PCR fingerprinting and α-glucosidase (α-Glu) activity provide reliable means for differentiating P. multocida subsp. multocida from P. multocida subsp. septica, particularly in strains that produce weak and/or discrepant sorbitol fermentation reactions.
MATERIALS AND METHODS
Strains.
Thirty-five clinical isolates of P. multocida cultured from different sources were obtained from the R. M. Alden Research Laboratory culture collection. Thirty-one of these strains were isolated from infected cat bite wounds in humans; four were isolated from dog bite wounds in humans. All strains had been previously identified as P. multocida using the API-20E test system (BioMerieux, Hazelwood, Mo.). Moeller's ornithine decarboxylase broth (Carr Scarborough Microbiologicals, Decatur, Ga.) and urease Wee-Tabs (Key Scientific, Round Rock, Tex.) were used to confirm ornithine decarboxylase and urease reactions, respectively. Indole reactions were confirmed using the spot indole test (para-dimethylaminocinnamaldehyde). All isolates were positive for catalase, oxidase, indole production, and ornithine decarboxylase; were negative for urease; reduced nitrate to nitrite; and fermented glucose, sucrose, xylose, and mannitol. None of the isolates fermented arabinose, inositol, mellibiose, rhamnose, or amygdalin. The isolates were subcultured from stock cultures (frozen at −70°C in 20% skim milk) onto tryptic soy agar plates supplemented with 5% sheep blood (Hardy Diagnostics, Santa Maria, Calif.) twice before further biochemical or PCR fingerprinting analyses. Control strains included P. multocida subsp. multocida ATCC 12947 (dog isolate) and P. multocida subsp. septica ATCC 51688 (human wound isolate).
Biochemical tests.
The isolates were tested for sorbitol and dulcitol fermentation in Andrades media obtained from various sources (Carr Scarborough Microbiologicals; Remel, Lenexa, Kans.; and Hardy Diagnostics) and in prereduced anaerobically sterilized (PRAS) medium (Anaerobe Systems, Morgan Hill, Calif.) that had been open to the ambient air for inoculation of the cultures. Cultures were incubated at 37°C for up to 14 days. α-Glu activity was determined using the API-ZYM test system (BioMerieux) and Wee-Tabs (Key Scientific) as per the manufacturers' instructions.
PCR fingerprinting analysis.
Several colonies of each isolate were suspended in 1 ml of sterile distilled water to a turbidity approximately equal to a number 3 McFarland standard. Two hundred microliters of the cell suspension was removed, pelleted, resuspended in 100 to 200 μl of Insta-Gene Matrix (Bio-Rad, Hercules, Calif.), and then incubated at 50°C for 15 to 30 min. Cell solutions were vortexed and then heated for 8 to 10 min at 100°C. Cell lysate supernatants containing the DNA extract were centrifuged to remove cellular debris and stored at −20°C until use.
Each cell lysate supernatant was subjected to PCR amplification in 50-μl volumes containing 25 μl of cell lysate supernatant; PCR buffer with 1.5 mM MgCl2 (final concentration) (Perkin-Elmer Cetus, Norwalk, Conn.); 200 μM dATP, dCTP, dGTP, and dTTP (Pharmacia LKB Biotechnology, Piscataway, N.J.); 25 pmol of the single primer, M13 core (5′-GAGGGTGGCGGTTCT-3′) (13); and 2.5 U of Taq DNA polymerase (Perkin-Elmer). Amplification reactions were performed as follows: 40 s at 93°C, 1 min at 50°C, and 40 s at 72°C for 35 cycles, followed by a final extension cycle of 6 min at 72°C. Reaction tubes were held at 4°C prior to analysis. Samples were concentrated to approximately 20 to 25 μl each (Speed-Vac; Savant, Halbrook, N.Y.) prior to electrophoretic separation in 1.2% agarose gels (0.5 by 25 by 20 cm) for 5 h at 3V/cm. Amplified products were detected by staining with ethidium bromide (2 μg/ml). Data were evaluated as described previously (10). Each isolate was analyzed a minimum of three times, twice from the same extract and once from a separate extract, to ensure reproducibility of the results.
RESULTS
Sorbitol fermentation reactions provided ambiguous results for differentiating dulcitol-negative P. multocida subspecies.
Sorbitol and dulcitol fermentation reactions were performed to differentiate P. multocida subsp. multocida and P. multocida subsp. septica. The control strains reacted as expected in both the Andrades and PRAS fermentation reactions; i.e., P. multocida subsp. multocida ATCC 12947 was positive for sorbitol fermentation and negative for dulcitol fermentation, whereas P. multocida subsp. septica ATCC 51688 was negative for both sorbitol and dulcitol fermentation. All isolates tested negative for dulcitol fermentation when tested using the PRAS medium; i.e., the pH was ≥6.0. In contrast, dulcitol fermentation results were variable using the Andrades medium. Three of the isolates (9%) tested both clearly positive (rose to deep rose; pH, <5.8) and negative (no color change; pH, ≥6.5) for dulcitol fermentation on repeat testing with this medium. Fourteen of the 35 clinical isolates (40%) gave at least one weak (pale pink to very pale pink; pH, 6.0 to 6.4) dulcitol fermentation reaction when tested with the Andrades medium, although repeat testing of the weak fermenters generally yielded a clear negative reaction. Thus, when data from the PRAS and Andrades media were examined together to establish a consensus, all 35 isolates were determined to be dulcitol nonfermenters.
Based on the API-20E sorbitol fermentation results, 18 isolates (51%) were sorbitol fermenters and 14 (40%) were nonfermenters (Table 1). The remaining three isolates (9%) gave discrepant results, i.e., positive and negative results on repeat testing using the API-20E system. Using the PRAS sorbitol fermentation medium, 10 isolates (29%) were sorbitol fermenters (pH, ≤5.5), 12 (34%) were nonfermenters (pH, ≥6.0), and 13 isolates (37%) tested weakly positive (pH, 5.6 to 5.9) in at least one test assay. With the Andrades medium, 16 isolates (46%) tested positive for sorbitol fermentation and 4 isolates (11%) were nonfermenters. Eleven isolates (31%) gave at least one weak sorbitol fermentation reaction. Seven isolates (20%) yielded at least one clear positive and negative sorbitol fermentation reaction on repeat testing. When the data from the API-20E, PRAS medium, and Andrades medium were examined together to establish a consensus, 13 isolates (37%) were determined as nonfermenters and 19 (54%) were sorbitol fermenters. The sorbitol fermentation reactions for three of the isolates (9%) remained uncertain.
TABLE 1.
Comparison of sorbitol fermentation results obtained using different test media
| Sorbitol fermentation result | No. of isolates as determined by:
|
||
|---|---|---|---|
| APF-20E | PRAS | Andrades | |
| Positive | 18 | 10 | 16 |
| Negative | 14 | 12 | 4 |
| Weak | 0 | 13 | 11a |
| Discrepant reaction on repeat testing | 3 | NDb | 7a |
Overlapping groups.
ND, not done.
PCR fingerprinting patterns did not correlate with the sorbitol fermentation reactions.
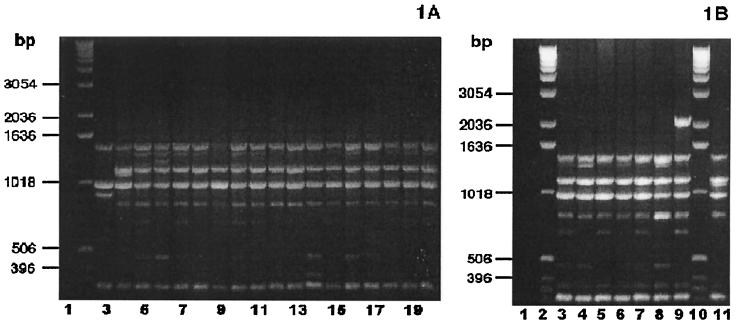
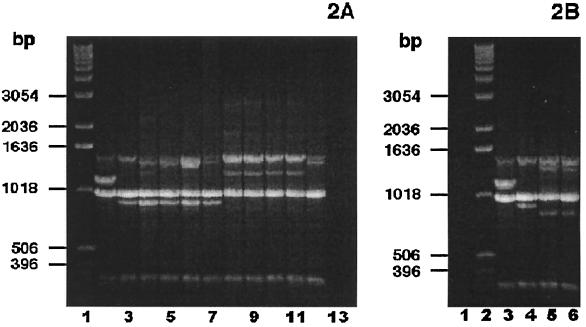
PCR fingerprinting analysis with the single primer, M13 core, was used to group the clinical isolates. Based on the presence or absence of four major bands (band A at ≈1,420 bp, band B at ≈1,130 bp, band C at ≈975 bp, and band D at ≈885 bp), the clinical isolates separated into three groups. Group I isolates (24 of 35) and the control strain P. multocida subsp. septica ATCC 51688 shared bands A, B, and C (Fig. 1 [23 strains are shown]). Group II isolates (4 of 35) and the control strain P. multocida subsp. multocida ATCC 12947 shared bands A, C, and D (Fig. 2A). Group III isolates (7 of 35) were characterized by bands A and C (Fig. 2).
FIG. 1.
P. multocida subsp. septica PCR fingerprint profiles. (A) Thirteen sorbitol-negative, α-Glu-positive and three sorbitol-uncertain, α-Glu-positive P. multocida bite wound isolates, examined by PCR fingerprint analyses using the single primer, M13 core, exhibited the group I PCR fingerprint profile characteristic of P. multocida subsp. septica. (Sorbitol fermentation was based on a consensus of API-20E, PRAS, and Andrades sorbitol fermentation reactions.) Lane 1, negative control (no DNA template); lane 2, DNA ladder; lane 3, P. multocida subsp. multocida ATCC 12947; lane 4, P. multocida subsp. septica ATCC 51688; lanes 5 to 20, bite wound isolates. (B) Seven sorbitol-positive, α-Glu-positive P. multocida bite wound isolates, examined by PCR fingerprint analyses using the single primer, M13 core, showed the group I PCR fingerprint profile characteristic of the P. multocida subsp. septica control strain. Lane 1, negative control (no DNA template); lanes 2 and 10, DNA ladder; lanes 3 to 9, bite wound isolates; lane 11, P. multocida subsp. septica ATCC 51688.
FIG. 2.
P. multocida subsp. multocida PCR fingerprint analyses. (A) Nine sorbitol-positive, α-Glu-negative P. multocida subsp. multocida bite wound isolates, examined by PCR fingerprint analyses using the single primer, M13 core, displayed either the group II or group III PCR fingerprint profile. (Sorbitol fermentation was based on a consensus of API-20E, PRAS, and Andrades sorbitol fermentation reactions.) Lane 1, DNA ladder; lane 2, P. multocida subsp. septica ATCC 51688; lane 3, P. multocida subsp. multocida ATCC 12947; lanes 4 to 7, bite wound isolates demonstrating the group II PCR fingerprint pattern; lanes 8 to 12, bite wound isolates showing the group III PCR fingerprint profile; lane 13, negative control (no DNA template). (B) Two sorbitol-positive, α-Glu-positive P. multocida bite wound isolates, examined by PCR fingerprinting analyses using the single primer, M13 core, exhibited the group III PCR fingerprint profile characteristic of other P. multocida subsp. multocida clinical isolates. Lane 1, negative control (no DNA template); lane 2, DNA ladder; lane 3, P. multocida subsp. septica ATCC 51688; lane 4, P. multocida subsp. multocida ATCC 12947; lanes 5 and 6, bite wound isolates.
The PCR fingerprint patterns did not appear to correlate well with the sorbitol fermentation reactions (Table 2). All of the sorbitol nonfermenters shared bands A, B, and C, i.e., the group I pattern characteristic of the P. multocida subsp. septica control strain. However, eight sorbitol fermenters and the three uncertain fermenters also fell within PCR fingerprint group I. Eleven additional sorbitol fermenters had PCR fingerprint patterns characteristic of either group II or group III.
TABLE 2.
Sorbitol fermentation reactions show poor correlation with PCR fingerprint analyses and α-Glu activity
| Sorbitol fermentation resulta | No. of isolates in PCR fingerprint group:
|
No. of isolates with α-Glu activity:
|
|||
|---|---|---|---|---|---|
| I | II | III | Positive | Negative | |
| Negative | 13 | 0 | 0 | 13 | 0 |
| Positive | 8 | 4 | 7 | 10 | 9 |
| Uncertain | 3 | 0 | 0 | 3 | 0 |
Based on consensus of API-20E, PRAS, and Andrades sorbitol fermentation reactions.
α-Glu activity did not correlate with the sorbitol fermentation reactions but did correlate with the PCR fingerprint patterns.
API-ZYM results from the control and clinical isolates demonstrated that the isolates could be differentiated based on α-Glu activity. P. multocida subsp. multocida ATCC 12947 showed negative α-Glu activity, whereas P. multocida subsp. septica ATCC 51688 was positive for α-Glu activity. Twenty-six of the clinical isolates (74%) showed positive α-Glu activity; nine of the isolates (26%) were negative for this enzyme activity. Because the Wee-Tabs double test tablets (α-Glu and phenylalanine deaminase) often produced negative or weakly positive results for isolates that tested positive using the API-ZYM test system (data not shown), we used the Wee-Tabs single test tablet to test for α-Glu activity (nitrophenol substrate test system). Subsequent testing of 20 select clinical isolates (16 strains that showed a weak or discrepant reaction with the Wee-Tabs double test tablets and four randomly selected isolates) and the two reference strains using the Wee-Tabs single test tablets for α-Glu activity confirmed the API-ZYM results.
As shown in Table 2, the α-Glu activity did not always correlate with the sorbitol fermentation reactions. Although all sorbitol nonfermenters were positive for α-Glu, 10 of the 19 positive sorbitol fermenters and the 3 isolates whose sorbitol fermentation reactions were unclear were also positive for this enzyme. In contrast, the α-Glu activity did correlate well with the PCR fingerprint profiles (Table 3). All of the α-Glu-negative isolates expressed either the group II or group III PCR fingerprint pattern. Twenty-four of the 26 α-Glu-positive isolates exhibited the group I PCR fingerprint pattern; two α-Glu-positive isolates had the group III fingerprint pattern.
TABLE 3.
α-Glu activity shows good correlation with the PCR fingerprint profiles
| α-Glucosidase activity | No. of isolates in PCR fingerprint group:
|
||
|---|---|---|---|
| I | II | III | |
| Positive | 24 | 0 | 2 |
| Negative | 0 | 4 | 5 |
DISCUSSION
The results of this study suggest that sorbitol fermentation may not provide the ideal marker for the differentiation of the two dulcitol-negative P. multocida subspecies, P. multocida subsp. multocida and P. multocida subsp. septica. We have found that a significant number of the P. multocida isolates examined in this study gave weak and/or discrepant results for sorbitol fermentation, even when sorbitol fermentation tubes from different manufacturers were used. Furthermore, when the results of sorbitol fermentation using all three of the test media (API-20E, PRAS, and Andrades) were considered, 3 of the 35 strains still failed to give a clear sorbitol fermentation reaction. Unfortunately, this made it difficult to categorize these organisms, as the sorbitol and dulcitol fermentation reactions have been the basis for differentiating P. multocida subsp. multocida from P. multocida subsp. septica (22).
To assist us in distinguishing between these two subspecies, we used a single-primer (M13 core) PCR fingerprinting technique. In other studies that we and others have performed, this and similar techniques have been successfully employed to differentiate species, including Porphyromonas, Bacteroides fragilis group, Leptospira, Staphylococcus, and Streptococcus (9, 10, 24, 31, 32). In this study, the dulcitol-negative P. multocida isolates clearly separated into three PCR fingerprint groups based on the presence or absence of four primary bands. The group I fingerprint profile was characteristic of the control strain P. multocida subsp. septica ATCC 51688; the group II fingerprint profile was characteristic of the control strain P. multocida subsp. multocida ATCC 12497. Although the group III fingerprint pattern was different from that of each of the control strains used in this study, we regarded the group III isolates as a subgroup of P. multocida subsp. multocida because the majority of these isolates were positive for sorbitol fermentation and negative for α-Glu activity in accordance with the control strain. We also noted that these three fingerprint patterns were substantially different from the fingerprint patterns generated by the American Type Culture Collection type strains of P. canis, P. dagmatis, P. stomatis, P. haemolytica, P. testudinis, and P. pneumotropica (S. Hunt Gerardo, unpublished observations).
Other investigators (33) have used the M13 core primer to distinguish strain-to-strain variation among P. multocida subsp. multocida pig respiratory isolates. The same four primary bands detected in our studies were also found by these investigators. However, none of their fingerprint profiles matched the combination of primary bands expressed by the control strains or clinical isolates analyzed in our study. One possible explanation for these differences may be the differences in the sources of these Pasteurella isolates, i.e., pig respiratory isolates versus isolates from infected dog and cat bite wounds in humans. Another likely contributing factor is the difference in magnesium ion concentrations used in the PCR amplification reaction mixtures. The magnesium ion concentration is known to affect primer annealing, the strand dissociation temperature of the template and the PCR product, product specificity and yield, and the polymerase activity and fidelity (1, 19). It is notable that we used 1.5 mM MgCl2, whereas Zucker et al. (33) used a significantly higher concentration (4.5 mM Mg2+) in their reaction mixture. Thus, differences in assay conditions provide a reasonable explanation for the differences in our results. As both of our assay systems provided reproducible results in our respective laboratories, it is possible that the higher magnesium ion concentration is useful for detecting strain-to-strain variation, whereas the lower concentration is more useful for analyzing the subspecies (species) differences. As Zucker et al. (33) restricted their analysis to P. multocida subsp. multocida, it would have been quite interesting to see what PCR fingerprint profile(s) was produced by P. multocida subsp. septica isolates under the assay conditions used in their laboratory.
Additional PCR methods have been used for the diagnosis and identification of P. multocida clinical isolates (reviewed in reference 15). However, these methods have been specifically designed to detect either strain-to-strain variation among P. multocida isolates or toxin-producing strains of P. multocida, rather than for the subspecies identification of these organisms. For the detection and identification of P. multocida in mixed cultures or clinical specimens by specific PCR, two approaches have been developed. One approach uses the psI gene, which codes for the P6-like protein of P. multocida (20). The other approach uses a sequence unique to P. multocida that was originally detected by subtractive hybridization (29). Although each of these PCR assays is capable of distinguishing P. multocida from other Pasteurella species and closely related genera, neither of them appears to distinguish among the subspecies of P. multocida. Two additional specific PCR approaches have proven useful for distinguishing P. multocida type B, the causative agent of hemorrhagic septicemia (8, 28). These assays are specific for serogroup B of P. multocida and have not been reported to differentiate P. multocida subsp. multocida and P. multocida subsp. septica. Similarly, several PCR assays have been developed for the detection of toxigenic strains of P. multocida (reviewed in reference 15), but these do not differentiate P. multocida subsp. multocida from P. multocida subsp. septica.
There have also been a number of other molecular assays used in the diagnosis and identification of P. multocida (reviewed in reference 15). Specifically, restriction endonuclease analysis, ribotyping, pulsed-field gel electrophoresis, and PCR fingerprinting have all been used for the differentiation of P. multocida isolates. Although each of these methods is useful for detecting strain-to-strain variation among P. multocida isolates, we have found no mention of their utility in specifically differentiating P. multocida subsp. multocida from P. multocida subsp. septica. In fact, Snipes et al. (26) demonstrated that P. multocida subsp. multocida and P. multocida subsp. septica overlap in serotype, restriction endonuclease analysis type, and ribotype expression; i.e., both subspecies could be found within a given serotype, restriction endonuclease analysis type, or ribotype.
In addition to these molecular approaches for differentiating P. multocida isolates, a variety of biochemical reactions have also proven to be useful for their characterization. In particular, lactose, maltose, trehalose, and xylose fermentation reactions have been used to further characterize P. multocida species into biovars. Based on these biochemical characteristics, a number of variants within both P. multocida subsp. multocida and P. multocida subsp. septica have been noted (6, 7, 12). Interestingly, trehalose and xylose fermentation reactions can be variable for both P. multocida subsp. multocida and P. multocida subsp. septica (4, 6, 7, 12, 21). Similarly, although most isolates are maltose negative, maltose-positive strains of both P. multocida subsp. multocida and P. multocida subsp. septica have been identified (23). Although there is no question that these variations have proven to be useful in epidemiological studies of P. multocida infections, these variants have, nevertheless, exhibited the expected sorbitol fermentation reaction for their respective subspecies, with the possible exception of the strains described above (12, 25). Therefore, these observations appear to confirm Mutters' original statement that “Variations in raffinose, lactose, maltose, trehalose, and d-xylose fermentation (are) of no taxonomic consequence” (22).
One of the interesting observations of our study is the difficulty in accurately assessing sorbitol fermentation among our P. multocida isolates. As mentioned above, this poses a significant problem in distinguishing the two dulcitol-negative P. multocida subspecies, as this is the critical biochemical test for their differentiation. In addition to being negative for dulcitol fermentation, all of our isolates were positive for xylose fermentation and negative for arabinose fermentation, further ruling out classification as P. multocida subsp. gallicida. It is also noteworthy that we identified 10 seemingly discrepant isolates (Fig. 1B and 2B), all of which were positive for sorbitol fermentation and α-Glu activity, i.e., 8 with the P. multocida subsp. septica PCR fingerprint pattern (sorbitol fermentation discrepancy) and 2 with the P. multocida subsp. multocida PCR fingerprint pattern (α-Glu discrepancy). In every case that we have seen in the literature, sorbitol-positive, dulcitol-negative isolates have been identified as P. multocida subsp. multocida, irrespective of their xylose, maltose, and/or trehalose fermentation reactions. Therefore, our proposal that eight sorbitol-positive strains might actually be P. multocida subsp. septica, based on their positive α-Glu activity and their PCR fingerprint pattern, is novel.
We acknowledge that the PCR fingerprinting data alone may not provide sufficient evidence to support our hypothesis that this method can be used to differentiate between the two dulcitol-negative P. multocida subspecies. However, the differentiation of isolates based on α-Glu activity and the strong correlation of that activity with the PCR fingerprinting profiles does strengthen our hypothesis. It is noteworthy that Holst et al. (14) reported that 39 of 95 (41%) of their P. multocida subsp. multocida isolates (all sorbitol positive) were positive for α-Glu activity, in contrast to 100% (21 of 21) of the P. multocida subsp. septica isolates (sorbitol negative). Their results suggest that our eight “discrepant” sorbitol-positive, α-Glu-positive strains are P. multocida subsp. multocida rather than P. multocida subsp. septica as we have suggested. However, we find it highly unlikely that strains of different species, e.g., P. multocida subsp. multocida and P. multocida subsp. septica (22), would present with the same polymorphic genomic fingerprint. If anything, when using the M13 core primer, one finds different, distinct PCR fingerprint patterns within a species, as was seen in this and other studies (1, 11, 16, 18, 31). Furthermore, in the study in which Mutters et al. (22) separated the three P. multocida subspecies based on DNA-DNA homology, only 4 dulcitol-negative, sorbitol-negative isolates (P. multocida subsp. septica) were examined, in contrast to 11 dulcitol-negative, sorbitol-positive isolates (P. multocida subsp. multocida). Our data suggest that examination of a larger number of isolates might have revealed that sorbitol fermentation was not a consistent marker for the differentiation of isolates into these two subspecies. Therefore, we believe that the PCR fingerprinting technique provides a more reliable means for differentiating between these two subspecies than the fermentation reactions.
In conclusion, we found that PCR fingerprinting analyses and α-Glu activity gave more consistent results than did sorbitol fermentation reactions for differentiating P. multocida dulcitol-negative isolates. Furthermore, the PCR fingerprinting profiles and α-Glu activity correlated much better with each other than did either one of these results with the sorbitol fermentation reactions. Thus, we propose that PCR fingerprint analysis (using the M13 core primer) and α-Glu activity more accurately reflect the differences in the two subspecies, P. multocida subsp. multocida and P. multocida subsp. septica, than does sorbitol fermentation.
REFERENCES
- 1.Alexander C J, Citron D M, Hunt Gerardo S, Claros M C, Talan D, Goldstein E J C. Characterization of saccharolytic Bacteroides and Prevotella isolates from infected dog and cat bite wounds in humans. J Clin Microbiol. 1997;35:406–411. doi: 10.1128/jcm.35.2.406-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas R M, Bej A K. Polymerase chain reaction. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 418–435. [Google Scholar]
- 3.Biberstein E L, Spenser S J, Kass P H, Hirsh D C. Distribution of indole-producing urease-negative pasteurellas in animals. J Vet Diagn Investig. 1990;3:319–323. doi: 10.1177/104063879100300408. [DOI] [PubMed] [Google Scholar]
- 4.Bisgaard M, Houghton S B, Mutters R, Stenzel A. Reclassification of German, British and Dutch isolates of so-called Pasteurella multocida obtained from pneumonic calf lungs. Vet Microbiol. 1991;26:115–124. doi: 10.1016/0378-1135(91)90048-k. [DOI] [PubMed] [Google Scholar]
- 5.Blackall P J, Fegan N, Pahoff J L, Storie G J, McIntosh G B, Cameron R D A, O'Boyle D, Frost A J, Bará M R, Marr G, Holder J. The molecular epidemiology of four outbreaks of porcine pasteurellosis. Vet Microbiol. 2000;72:111–120. doi: 10.1016/s0378-1135(99)00192-3. [DOI] [PubMed] [Google Scholar]
- 6.Blackall P J, Pahoff J L, Bowles R. Phenotypic characterisation of Pasteurella multocida isolates from Australian pigs. Vet Microbiol. 1997;57:355–360. doi: 10.1016/s0378-1135(97)00111-9. [DOI] [PubMed] [Google Scholar]
- 7.Blackall P J, Pahoff J L, Marks D, Fegan N, Morris C J. Characteristics of Pasteurella multocida isolated from fowl cholera outbreaks on turkey farms. Aust Vet J. 1995;72:135–138. doi: 10.1111/j.1751-0813.1995.tb15033.x. [DOI] [PubMed] [Google Scholar]
- 8.Brickell S K, Thomas L M, Long K A, Panaccio M, Widders P R. Development of a PCR test based on a gene region associated with the pathogenicity of Pasteurella multocida serotype B:2, the causal agent of haemorrhagic septicaemia in Asia. Vet Microbiol. 1998;59:295–307. doi: 10.1016/s0378-1135(97)00199-5. [DOI] [PubMed] [Google Scholar]
- 9.Citron D M, Hunt Gerardo S, Claros M C, Abrahamian F, Talan D, Goldstein E J C. Frequency of isolation of Porphyromonas species from infected dog and cat bite wounds in humans and their characterization by biochemical tests and arbitrarily primed-polymerase chain reaction fingerprinting. Clin Infect Dis. 1996;23(Suppl. 1):S78–S82. doi: 10.1093/clinids/23.supplement_1.s78. [DOI] [PubMed] [Google Scholar]
- 10.Claros M C, Citron D M, Hunt Gerardo S, Goldstein E J C, Schönian G, Montag T, Hampel B, Rodloff A C. Characterization of indole-negative Bacteroides fragilis group species with use of polymerase chain reaction fingerprinting and resistance profiles. Clin Infect Dis. 1996;23(Suppl. 1):S66–S72. doi: 10.1093/clinids/23.supplement_1.s66. [DOI] [PubMed] [Google Scholar]
- 11.Claros M C, Schumacher U, Jacob M, Hunt Gerardo S, Kleinkauf N, Goldstein E J C, Finegold S M, Rodloff A C. Characterization of Bilophila wadsworthia isolates using PCR fingerprinting. Anaerobe. 1999;5:589–593. [Google Scholar]
- 12.Fegan N, Blackall P J, Pahoff J L. Phenotypic characterisation of Pasteurella multocida isolates from Australian poultry. Vet Microbiol. 1995;47:281–286. doi: 10.1016/0378-1135(95)00119-0. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein E J C, Citron D M, Wield B, Sutter V L, Miller T A, Finegold S M. Bacteriology of human and animal bite wounds. J Clin Microbiol. 1978;8:667–672. doi: 10.1128/jcm.8.6.667-672.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holst E, Rollof J, Larsson L, Neilsen J P. Characterization and distribution of Pasteurella species recovered from infected humans. J Clin Microbiol. 1992;30:2984–2987. doi: 10.1128/jcm.30.11.2984-2987.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt M L, Adler B, Townsend K M. The molecular biology of Pasteurella multocida. Vet Microbiol. 2000;72:3–25. doi: 10.1016/s0378-1135(99)00183-2. [DOI] [PubMed] [Google Scholar]
- 16.Hunt Gerardo S, Citron D M, Goldstein E J C. PCR fingerprint analysis for differentiation of Streptococcus pneumoniae reinfection versus relapse. Diagn Microbiol Infect Dis. 2000;36:275–278. doi: 10.1016/s0732-8893(00)00114-0. [DOI] [PubMed] [Google Scholar]
- 17.Hunt Gerardo S, Goldstein E J C. Pasteurella multocida and other Pasteurella species. In: Yu V L, Merigan T C, Barriere S L, editors. Antimicrobial therapy and vaccines. Baltimore, Md: Williams and Wilkins; 1999. pp. 326–335. [Google Scholar]
- 18.Hunt Gerardo S, Marina M, Citron D M, Claros M C, Hudspeth M K, Goldstein E J C. Bilophila wadsworthia clinical isolates compared by polymerase chain reaction fingerprinting. Clin Infect Dis. 1997;25(Suppl. 2):S291–S294. doi: 10.1086/516229. [DOI] [PubMed] [Google Scholar]
- 19.Innis M A, Gelfand D H. Optimization of PCRs. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols—a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 3–12. [Google Scholar]
- 20.Kasten R W, Carpenter T E, Snipes K P, Hirsh D C. Detection of Pasteurella multocida-specific DNA in turkey flocks by use of the polymerase chain reaction. Avian Dis. 1997;41:676–682. [PubMed] [Google Scholar]
- 21.Korbel R, Gerlach H, Bisgaard M, Hafez H M. Further investigations on Pasteurella multocida infections in feral birds. J Vet Med B. 1992;39:10–18. doi: 10.1111/j.1439-0450.1992.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 22.Mutters R, Ihm P, Pohl S, Frederiksen W, Mannheim W. Reclassification of the genus Pasteurella Trevisan 1887 on the basis of deoxyribonucleic acid homology, with proposals for the new species Pasteurella dagmatis, Pasteurella canis, Pasteurella stomatis, Pasteurella anatis, and Pasteurella langaa. Int J Syst Bacteriol. 1985;35:309–322. [Google Scholar]
- 23.Petersen K D, Christensen J P, Bisgaard M. Phenotypic and genotypic diversity of organisms previously classified as maltose positive Pasteurella multocida. Zentbl Bakteriol. 1998;288:1–12. doi: 10.1016/s0934-8840(98)80091-1. [DOI] [PubMed] [Google Scholar]
- 24.Ralph D, McClelland M, Welsh J, Baranton G, Perolat P. Leptospira species categorized by arbitrarily restriction polymorphisms in PCR-amplified rRNA genes. J Bacteriol. 1993;175:973–981. doi: 10.1128/jb.175.4.973-981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snipes K P, Hirsh D C, Kasten R W, Carpenter T E, Hird D W, McCapes R H. Homogeneity of characteristics of Pasteurella multocida isolated from turkeys and wildlife in California, 1985–88. Avian Dis. 1990;34:315–320. [PubMed] [Google Scholar]
- 26.Snipes K P, Hirsh D C, Kasten R W, Hansen L M, Hird D W, Carpenter T E, McCapes R H. Use of an rRNA probe and restriction endonuclease analysis to fingerprint Pasteurella multocida isolated from turkeys and wildlife. J Clin Microbiol. 1989;27:1847–1853. doi: 10.1128/jcm.27.8.1847-1853.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talan D A, Citron D M, Abrahamian F A, Moran G J, Goldstein E J C the Emergency Medicine Animal Bite Infection Study Group. Bacteriology and management of dog and cat bite wound infections presenting to emergency departments. N Engl J Med. 1999;340:85–92. doi: 10.1056/NEJM199901143400202. [DOI] [PubMed] [Google Scholar]
- 28.Townsend K M, Dawkins H J, Papadimitriou J M. REP-PCR analysis of Pasteurella multocida isolates that cause haemorrhagic septicaemia. Res Vet Sci. 1997;63:151–155. doi: 10.1016/s0034-5288(97)90009-6. [DOI] [PubMed] [Google Scholar]
- 29.Townsend K M, Frost A J, Lee C W, Papadimitriou J M, Dawkins H J S. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36:1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber D J, Hansen A R. Infections resulting from animal bites. Infect Dis Clin N Am. 1991;5:663–680. [PubMed] [Google Scholar]
- 31.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welsh J, McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991;19:861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zucker B, Krüger M, Horsch F. Differentiation of Pasteurella multocida subspecies multocida isolates from the respiratory system of pigs by using polymerase chain reaction fingerprinting technique. J Vet Med B. 1996;43:585–591. doi: 10.1111/j.1439-0450.1996.tb00357.x. [DOI] [PubMed] [Google Scholar]