Abstract
The emergence of disease caused by penicillin-resistant and multidrug-resistant pneumococci has become a global concern, necessitating the identification of the epidemiological spread of such strains. The Pneumococcal Molecular Epidemiology Network was established in 1997 under the auspices of the International Union of Microbiological Societies with the aim of characterizing, standardizing, naming, and classifying antimicrobial agent-resistant pneumococcal clones. Here we describe the nomenclature for 16 pneumococcal clones that have contributed to the increase in antimicrobial resistance worldwide. Guidelines for the recognition of these clones using molecular typing procedures (pulsed-field gel electrophoresis, BOX-PCR, and multilocus sequence typing) are presented, as are the penicillin-binding profiles and macrolide resistance determinants for the 16 clones. This network can serve as a prototype for the collaboration of scientists in identifying clones of important human pathogens and as a model for the development of other networks.
The emergence of penicillin-resistant and multidrug-resistant pneumococcal strains has become a global concern. Since the first description of pneumococci with decreased susceptibilities to penicillin in Australia (18) in the late 1960s, penicillin-resistant strains have been found in various parts of the world with increasing frequency. Resistance to non-β-lactam antibiotics such as chloramphenicol, tetracycline, erythromycin, clindamycin, rifampin, and trimethoprim-sulfamethoxazole also has been reported (2, 22) and often is associated with decreased susceptibility to penicillin (1). Penicillin-resistant pneumococci have been reported with particularly high frequencies in Spain (14, 15), South Africa (22), and Hungary (31) since the mid-1970s, in France (29), Iceland (52), and the United States (48) since the middle to late 1980s and early 1990s, and in the Far East during the 1990s (21, 53).
Although the first penicillin-resistant pneumococcus was identified in an adult (18), the first multidrug-resistant strains were found in children (20), and penicillin-resistant strains have been shown to be more common in children wherever penicillin resistance has been studied. Penicillin-resistant (MICs of ≥2 μg/ml) and multidrug-resistant pneumococci are known to be restricted worldwide to a few serogroups, namely, 23, 6, 19, and 9, and serotype 14, which are particularly associated with carriage and disease in children (10, 22).
The era of the global spread of penicillin-resistant pneumococci has seen the rapid development of DNA typing techniques for use in research and epidemiological investigations. Molecular studies have shown that the penicillin-resistant and multidrug-resistant pneumococcal populations are highly dynamic and that resistance is a combination of the spread of resistant clones, the acquisition and loss of resistance genes within those clonal lineages, and the spread of resistance genes to new lineages. Several techniques, including pulsed-field gel electrophoresis (PFGE) (28), BOX-PCR (24, 60), and, more recently, multilocus sequence typing (MLST) (13), have been used to determine the genetic relatedness of pneumococcal isolates. A search of the literature shows that extensive molecular typing studies from various regions of the world have identified a number of clones of antimicrobial-resistant pneumococci that have achieved significant geographic spread within individual countries and, in some cases, across international boundaries.
The Pneumococcal Molecular Epidemiology Network (PMEN) was established in 1997 under the auspices of the International Union of Microbiological Societies with the aim of standardizing the nomenclature and classification of resistant pneumococcal clones worldwide. Specific objectives of the network include the following: (i) international collaboration and training support between laboratories and institutions; (ii) increased knowledge of resistance and spread of pneumococci worldwide; (iii) standardization of nomenclature of national and international clones; (iv) identification of clones using PFGE, BOX-PCR, and MLST; (v) availability of reference isolates of each clone from the American Type Culture Collection (ATCC); and (vi) access to information through a website.
The nomenclature of 16 national and international antibiotic-resistant pneumococcal clones tested using PFGE, BOX-PCR, and MLST is described. Penicillin-binding protein (PBP) profiles and macrolide resistance determinants are also described.
MATERIALS AND METHODS
Bacterial strains.
Representative isolates for each of the 16 S. pneumoniae clones were serotyped by the Quellung reaction, and MICs of penicillin, cefotaxime, erythromycin, clindamycin, chloramphenicol, tetracycline, and trimethoprim-sulfamethoxazole were determined according to National Committee for Clinical Laboratory Standards criteria (39). ATCC accession numbers for the 16 clones are given in Table 1.
TABLE 1.
Properties of 16 pneumococcal clonesa
| Clone | Reference strain (ATCC accession no.) | Serotype | MIC (μg/ml) of:
|
Reference(s) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PEN | CTX | ERY | CLI | CHL | TET | SXT | ||||
| Spain23F-1 | SP264 (ATCC 700669) | 23F | 2 | 0.5 | 0.06 | 0.06 | 16 | 32 | 4/76 | 7, 32, 37 |
| Spain6B-2 | GM17 (ATCC 700670) | 6B | 2 | 1 | 0.12 | 0.12 | 16 | 32 | 4/76 | 38 |
| Spain9V-3 | TL7/1993 (ATCC 700671) | 9V | 2 | 1 | 0.12 | 0.12 | 2 | ≤0.5 | 8/152 | 7, 29 |
| Tennessee23F-4 | CS111 (ATCC 51916) | 23F | 0.12 | 32 | 32 | 0.12 | 2 | ≤0.5 | 4/76 | 33, 49 |
| Spain14-5 | MS22 (ATCC 700902) | 14 | 2 | 1 | 0.06 | 0.12 | 16 | 32 | 2/38 | 8 |
| Hungary19A-6 | HUN663 (ATCC 700673) | 19A | 2 | 0.5 | >32 | >32 | 16 | 16 | 2/38 | 31, 38 |
| South Africa19A-7 | 17619 (ATCC 700674) | 19A | 0.5 | 0.5 | 0.25 | 0.25 | 2 | ≤0.5 | 4/152 | 51 |
| South Africa6B-8 | 50803 (ATCC 700675) | 6B | 0.5 | 0.25 | 0.12 | 0.12 | 2 | ≤0.5 | 1/19 | 51 |
| England14-9 | PN93/872/B (ATCC 700676) | 14 | 0.03 | 0.03 | 32 | 0.12 | 2 | ≤0.5 | 0.12/4.2 | 17 |
| CSR14-10 | 87-029055 (ATCC 700677) | 14 | 8 | 1 | >32 | >32 | 16 | 32 | 0.25/4.8 | 16, 19 |
| CSR19A-11 | 91-006571 (ATCC 700678) | 19A | 4 | 1 | >32 | >32 | 32 | 32 | 4/76 | 16, 19 |
| Finland6B-12 | 43362 Fi10 (ATCC 700903) | 6B | 1 | >32 | >32 | 4 | 32 | 8/152 | 48 | |
| South Africa19A-13 | 51702 (ATCC 700904) | 19A | 8 | 2 | >8 | >8 | 32 | 8 | 8/152 | 51 |
| Taiwan19F-14 | TW31 (ATCC 700905) | 19F | 2 | 1 | 4 | 0.12 | 2 | 16 | 4/76 | 47 |
| Taiwan23F-15 | TW17 (ATCC 700906) | 23F | 1–2 | 1 | >32 | >32 | 2 | 16 | 0.5/9.5 | 47 |
| Poland23F-16 | 178 | 23F | 8 | 8 | >8 | 0.25 | 16 | 16 | 4/76 | 41 |
PEN, penicillin; CTX, cefotaxime; ERY, erythromycin; CLI, clindamycin; CHL, chloramphenicol; TET, tetracycline; SXT, trimethoprim-sulfamethoxazole; CSR, Czech Republic.
DNA fingerprinting. (i) BOX-PCR.
Chromosomal DNA was isolated by the method described by Ausubel et al. (3). DNA amplifications were performed with 50-μl volumes containing 4 mM MgCl2, 2 μM primer AR1 (CTACGGCAAGGCGACGCTGACG) (60), 150 μM each of the four deoxynucleoside triphosphates (Roche Diagnostics, Mannheim, Germany), 20 ng of genomic DNA, 1 U of Taq DNA polymerase (Advanced Biotechnologies, Epsom, United Kingdom), and 5 μl of 10× reaction buffer. The reaction mixture was overlaid with mineral oil and subjected to initial denaturation at 95°C for 5 min in a Perkin-Elmer Cetus thermal cycler. This step was followed by 30 cycles of 1 min at 90°C, 1 min at 52°C, and 2 min at 72° and a final extension at 72°C for 5 min. PCR products were run on a 2% ethidium bromide-containing agarose gel in Tris-acetate-EDTA buffer for 2 h at 120 V.
(ii) PFGE.
Genomic DNA was prepared in situ in agarose blocks as described previously (28, 33) and was digested with SmaI (Life Technologies, Gaithersburg, Md.). The fragments were resolved by PFGE in 1% agarose (SeaKem GTG agarose; BioWhittaker Molecular Applications, Rockland, Maine) in 0.5× Tris-borate-EDTA buffer at 14°C and 6 V/cm in a CHEF-DR III system (Bio-Rad Laboratories, Hercules, Calif.). The parameters for block 1 were an initial pulse time of 1 s increased to 30 s over 17 h; those for block 2 were 5 s increased to 9 s over 6 h.
(iii) MLST.
MLST was carried out as described previously (13). Briefly, internal fragments of the aroE, gdh, gki, recP, spi, xpt, and ddl genes were amplified by PCR from chromosomal DNA using the primer pairs described by Enright and Spratt (13). The amplified fragments were directly sequenced in each direction using the primers that were used for the initial amplification. The sequences at each of the seven loci were then compared with the sequences of all of the known alleles at those loci. Sequences that were identical to the sequence of a known allele were assigned the same allele number, whereas those that differed from the sequence of any known allele, even at a single nucleotide site, were assigned new allele numbers. The assignment of alleles at each locus was carried out using the software available at the pneumococcal MLST website (http://www.mlst.net). The alleles at each of the seven loci define the allelic profile of each isolate as well as their sequence type. Allelic profiles are shown as a series of seven integers which correspond to the alleles at each of the loci, in the order aroE, gdh, gki, recP, spi, xpt, and ddl. The relatedness between the isolates was represented as a dendrogram, constructed by the unweighted pair-group method with arithmetic averages, from the matrix of pairwise differences in the allelic profiles. The allelic profiles of the reference isolates of each clone are deposited in the database at the pneumococcal MLST website.
PBP analysis.
The genes encoding PBP1A, PBP2B, and PBP2X were amplified using primers and conditions previously described (7). Purified DNA fragments were analyzed using the enzymes HinfI and MseI-DdeI for the PBP1A and PBP2X genes and HinfI and StyI for the PBP2B gene. The samples were electrophoresed at 120 V on a 2% agarose gel for 2 h and stained with ethidium bromide.
Macrolide resistance determinants.
A multiplex PCR using previously published primers (55) was used to detect the mefA and ermB genes in S. pneumoniae strains. Amplification reactions were performed with 50-μl volumes containing 1× reaction buffer; 5 mM MgCl2; 200 μM each dATP, dCTP, dGTP, and dTTP; 1 μM each primer; 1 U of Taq DNA polymerase; and 20 ng of DNA. Amplification was performed with a Perkin-Elmer Cetus DNA thermal cycler programmed for 1 cycle of denaturation at 95°C for 2 min; 30 cycles of denaturation at 95°C for 1 min, primer annealing at 56°C for 2 min, and extension at 72°C for 2 min; and 1 cycle of extension at 72°C for 10 min. Amplification products were run on 2% agarose gels and detected by staining with ethidium bromide.
RESULTS
Nomenclature of clones.
The following criteria were established for inclusion of clones in the PMEN: (i) a clone must have a wide geographic distribution throughout a country or have spread internationally; (ii) a clone should be well established within a country over a number of years; (iii) a clone should be resistant to one or more antibiotics that are in wide clinical use; (iv) data on the clone need to be published or in press before ratification by the network; and (v) a representative isolate of the clone must be made available for confirmation by molecular typing that it differs from previously accepted clones, and permission must be granted to deposit it into the ATCC.
The system use for the naming of clones is as follows: countryfirst identified serotype-sequential numbering in network-subsequent described serotype. For example, in Spain23F-1, Spain is the country in which the clone was first identified (based on publication), 23F is the serotype of the clone first identified, and 1 is the clone number.
In large countries, such as the United States and Russia, the naming of clones could be considered by province or state if the clone is predominantly found in one region of the country, for example, Tennessee23F-4.
Several publications have shown that members of a single clone may express different capsular polysaccharides. In the case of capsular switching, the nomenclature is, e.g., as follows: in Spain23F-1-19F, 19F indicates a serotype 19F variant of the Spain23F-1 clone.
DNA typing.
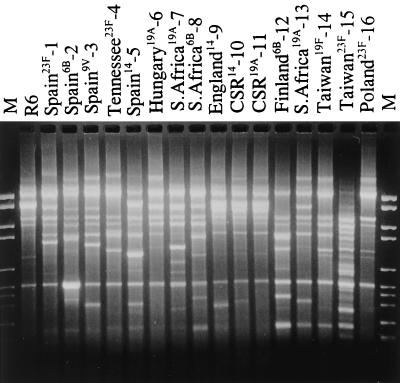
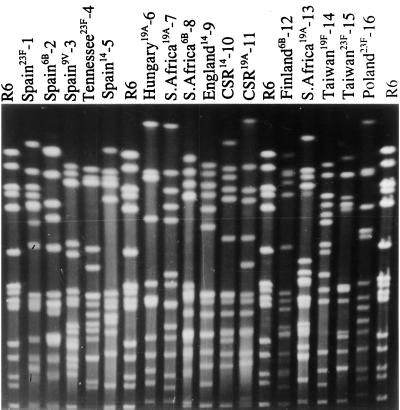
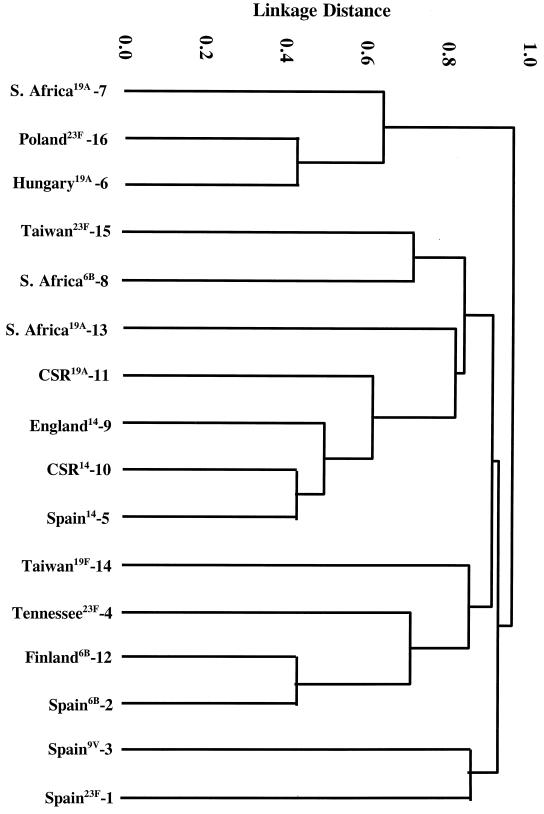
Representative isolates of the 16 clones are shown in Table 1; references indicate the first published reports of the clones. Molecular typing of isolates of each of the 16 clones by BOX-PCR (Fig. 1), PFGE (Fig. 2), and MLST (Table 2; Fig. 3) determined that their genotypes were sufficiently different to distinguish them as distinct clones.
FIG. 1.
BOX-PCR fingerprint patterns of representative isolates of the 16 clones described in Table 1. CSR, Czech Republic. Lanes M, marker VI (Roche Diagnostics).
FIG. 2.
PFGE fingerprint patterns of SmaI restriction digests of representative isolates of the 16 clones described in Table 1.
TABLE 2.
MLST allelic profiles, PBP profiles, and macrolide resistance determinants for 16 clonesa
| Clone | MLST allele number for:
|
Fingerprint pattern for:
|
Presence of erm or mef | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | PBP1A | PBP2B | PBP2X | ||
| Spain23F-1 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 1 | 1 | 1 | − |
| Spain6B-2 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 1 | 2 | 2 | − |
| Spain9V-3 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 1 | 1 | 1 | − |
| Tennessee23F-4 | 1 | 8 | 6 | 2 | 6 | 4 | 6 | 2 | 3 | 1 | mef |
| Spain14-5 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 3 | 4 | 1 | − |
| Hungary19A-6 | 7 | 13 | 42 | 6 | 10 | 6 | 56 | 4 | 3 | 3 | erm |
| South Africa19A-7 | 2 | 13 | 8 | 25 | 25 | 6 | 8 | 5 | 5 | 4 | − |
| South Africa6B-8 | 7 | 22 | 1 | 2 | 5 | 1 | 14 | 6 | 3 | 5 | − |
| England14-9 | 1 | 5 | 4 | 5 | 5 | 1 | 8 | 7 | 3 | 6 | mef |
| CSR14-10 | 1 | 5 | 4 | 1 | 5 | 3 | 3 | 8 | 6 | 7 | erm |
| CSR19A-11 | 7 | 5 | 4 | 26 | 10 | 3 | 55 | 8 | 6 | 7 | erm |
| Finland6B-12 | 5 | 6 | 43 | 2 | 6 | 1 | 28 | 9 | 7 | 8 | erm |
| South Africa19A-13 | 1 | 8 | 9 | 5 | 11 | 1 | 12 | 10 | 8 | 9 | erm |
| Taiwan19F-14 | 15 | 16 | 19 | 15 | 6 | 20 | 26 | 11 | 9 | 10 | mef |
| Taiwan23F-15 | 15 | 29 | 4 | 21 | 30 | 1 | 14 | 11 | 10 | 11 | erm |
| Poland23F-16 | 7 | 13 | 8 | 1 | 10 | 6 | 36 | 12 | 11 | 12 | − |
aroE, shikimate dehydrogenase; gdh, glucose-6-phosphate dehydrogenase; gki, glucose kinase; recP, transketolase; spi, signal peptidase I; xpt, xanthine phosphoribosyltransferase; ddl, d-alanine-d-alanine ligase; erm, ermB gene; mef, mefA gene; −, neither gene was present.
FIG. 3.
Dendrogram constructed from MLST data showing the genetic relatedness of the 16 pneumococcal clones described in Table 1.
Resistance determinants.
PBP fingerprinting results for the PBP1A, PBP2B, and PBP2X genes are shown in Table 2. Twelve patterns were identified for the PBP1A and PBP2X genes, and 11 were identified for the PBP2B gene. The Spain23F-1 and Spain9V-3 clones shared identical PBP profiles, as did the two clones from the Czech Republic (CSR14-10 and CSR19A-11).
The macrolide resistance determinants are shown in Table 2. We identified the ermB gene in 6 of the 16 strains and the mefA gene in 3. The Poland23F-16 clone, for which the erythromycin MIC was >8 μg/ml, harbored neither the ermB nor the mefA gene for macrolide resistance.
DISCUSSION
The global increase in highly penicillin-resistant and multidrug-resistant pneumococci appears, in large part, to result from the spread of individual highly resistant pneumococcal clones. This phenomenon has been particularly highlighted by the worldwide spread of the serotype 23F pneumococcal clone (Spain23F-1), first identified in Spain in the early 1980s (37) and resistant to penicillin, chloramphenicol, and tetracycline (and sometimes erythromycin). This successful clone has now been found in the United States (26, 37), South Africa (23), the United Kingdom (7), South America (5, 6, 12, 43), several other countries in Europe (29, 30, 42, 46, 48, 50, 58, 61), and the Far East (34, 47, 59). Serotype 19F (7), 14 (4), 19A (9), 9N (40), and 3 (40) and serogroup 6 (54) variants of this clone have also been reported in various regions of the world. Of these serotype variants of the Spain23F-1 clone, those expressing the serotype 19F capsule are the most commonly encountered. A recent study conducted in the United States showed that close to 40% of all highly penicillin-resistant pneumococci belong to this Spain23F-1 clone (10).
The penicillin-resistant serotype 9V clone (Spain9V-3), first identified in Spain (7) and France (29), has become widely disseminated throughout the world and has been identified in Italy (30), Sweden (35), Germany (42), the United States (10), the United Kingdom (7), South America (6, 12), and the Far East (34, 47). Serotype 14 (11, 36) and 9A (46) and serogroup 19 (10, 43) variants of this clone have been reported in numerous countries. The serotype 14 variants of the Spain9V-3 clone are the most commonly encountered.
The overwhelming majority of S. pneumoniae isolates for which MICs were about 1.0 μg/ml or higher (107 of 119 isolates, or 89.9%, collected during surveillance from 1996 through 1998) and colonizing the nasopharynges of children attending day care centers in Lisbon, Portugal, belonged to serotype variants of the Spain23F-1 and Spain9V-3 clones (44, 45).
The multidrug-resistant serotype 6B clone (Spain6B-2), recovered in Spain for over a decade, has become the dominant pneumococcus isolated from children in Iceland (52), where it accounts for approximately 75% of multidrug-resistant pneumococci (25). It has also been identified in other countries in Europe (29, 42) and elsewhere, including the United Kingdom (52), the United States (62), and Taiwan (47). Interestingly, Vilhelmsson et al. (63) recently described the emergence of antibiotic-resistant variants of this 6B clone in Iceland which have lost one or more of the antibiotic resistance phenotypes and/or resistance genes, singly or in combination.
Studies from other countries that have a high incidence of antibiotic-resistant pneumococci have identified additional distinctive clones that have presumably emerged within these countries. An example is the serotype 23F pneumococci resistant to extended-spectrum cephalosporins and originally isolated in Tennessee in the United States (33). This clone (Tennessee23F-4), which probably evolved locally and is unrelated to the Spain23F-1 clone, is characterized by a mutation at position 550 in the pbp2X gene which confers high-level cephalosporin resistance. Pneumococci belonging to a multidrug-resistant serotype 14 clone (Spain14-5) have been identified among isolates recovered from six Spanish hospitals between 1990 and 1994 (8). A clone of multidrug-resistant serotype 19A strains first identified in Hungary (Hungary19A-6) (31) has been described in the Czech Republic and Slovakia (16). A second, highly penicillin-resistant clone of serotype 19A (CSR19A-10) and a serotype 14 clone (CSR14-11) have also been described in Slovakia and the Czech Republic (16). An analysis of a wide geographic diversity of resistant pneumococcal isolates in South Africa has identified two clones of serogroup 19A, one intermediately resistant to penicillin (South Africa19A-7) and the other multiply resistant (South Africa19A-13) (51). In addition, a unique penicillin-resistant serotype 6B clone (South Africa6B-8) has emerged locally in South Africa (51). The widespread use of erythromycin may have led to the emergence of a serotype 14 clone (England14-9) resistant only to erythromycin in various parts of the United Kingdom (17). In Finland, pneumococcal strains resistant to multiple antibiotics have been characterized as belonging to a serogroup 6B clone (Finland6B-12) (48). Shi et al. (47) have recently used MLST to describe two antibiotic-resistant clones that have emerged in Taiwan, a serotype 19F clone (Taiwan19F-14) and a serotype 23F clone (Taiwan23F-15). The former clone has been recovered from several countries in the Far East and elsewhere (J. Zhou and B. G. Spratt, unpublished results). Penicillin resistance among pneumococcal isolates has rapidly emerged in Poland during the last decade. Although the pandemic Spain23F-1 and France9V-3 clones are present in that country, a novel serotype 23F clone (Poland23F-16) has clearly contributed to the increase in the prevalence of penicillin-resistant pneumococci nationwide (41).
A search of the literature indicates that the initial characterization of clones within a population of resistant pneumococci can be conveniently carried out using several molecular typing methods, including BOX-PCR (60), repetitive extragenic palindromic-PCR (62), multilocus enzyme electrophoresis (37), and PFGE (52). Most studies have analyzed penicillin-resistant isolates, and each of the above techniques can successfully identify clusters of closely related isolates within a population. However, determination of whether clones are novel or related to previously described clones is more problematic. Some of the clones are relatively old (e.g., 20 years), and substantial variations may have accumulated in the PFGE or BOX profiles of the descendents of the ancestral member of a clone. It can therefore be difficult to decide whether a resistant strain is sufficiently similar by PFGE or BOX-PCR analysis to be considered a member of a known clone or should be considered a distinct and previously unrecognized clone. MLST provides a less ambiguous and highly portable approach to global epidemiological studies by defining the genotypes of antibiotic-resistant isolates based on the alleles at each of seven housekeeping loci (13, 47, 64). MLST studies have shown that isolates of each of the Spain23F-1, Spain6B-2, Spain9V-3, and Spain14-5 clones typically have identical allelic profiles or differ from the typical profile at only one of the seven loci (64). MLST therefore assigns an isolate as a member of one of the resistant clones if it has the allelic profile of the reference isolate of that resistant clone or differs from it at a single locus. Isolates whose allelic profiles differ at three or more of the seven loci used in MLST can be regarded as distinct clones (13, 47, 64). Clearly, PFGE, BOX-PCR, and MLST results do correlate well, although the DNA fragment patterns used in PFGE and BOX-PCR are believed to diversify more rapidly than the allelic profiles used in MLST. Thus, PFGE provides increased levels of discrimination which may be useful for local epidemiological studies, whereas MLST provides a more rigorous way of assigning isolates to one of the major clones.
It should be remembered that similarity of genotype does not necessarily imply that two resistant isolates are members of the same clone. Penicillin resistance has emerged predominantly in a small number of serotypes, and although isolates of the same serotype are often only distantly related in terms of overall genotype (13), most isolates of a few serotypes appear to have very similar genotypes (13, 57). For example, almost all isolates of serotype 9V appear to be similar in genotype, and two resistant clones of this serotype that emerged independently will inevitably appear to be closely related (64). Rigorous assignment of penicillin-resistant isolates to the same clone should therefore also involve an analysis of their PBP1A, PBP2B, and PBP2X genes. PBP analysis of the 16 clones showed that the Spain23F-1 and Spain9V-3 clones harbored identical PBP1A, PBP2B, and PBP2X genes (1-1-1), as previously described by Coffey et al. (7). The presence of identical PBP genes (8-6-7) in the chromosomes of the two unrelated clones from the Czech Republic and Slovakia (CSR14-10 and CSR19A-11) suggests the horizontal transfer of these PBP genes.
Nine of the 16 clones were resistant to erythromycin, and 6 clones carried the ermB resistance determinant, which codes for target modification by 23S rRNA methylase (27). Three isolates showed the M phenotype (macrolide resistant but clindamycin and streptogramin B susceptible) and carried the mefA gene, which codes for an efflux mechanism (56).
In conclusion, the PMEN includes the molecular characterization and nomenclature of 16 important national and international antibiotic-resistant clones that have contributed to the rapid increase in resistance in pneumococci worldwide. The biological basis of the successful spread of these particular clones remains to be established.
ACKNOWLEDGMENT
The establishment and continued work of the network have been made possible by financial support from the ICSU.
REFERENCES
- 1.Allen K D. Penicillin-resistant pneumococci. J Hosp Infect. 1991;17:3–13. doi: 10.1016/0195-6701(91)90072-g. [DOI] [PubMed] [Google Scholar]
- 2.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1992;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 61–63. [Google Scholar]
- 4.Barnes D M, Wittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 5.Camou T, Hortal M, Tomasz A. The apparent importation of penicillin-resistant capsular type 14 Spanish/French clone of Streptococcus pneumoniae into Uruguay in the early 1990s. Microb Drug Resist. 1998;4:219–224. doi: 10.1089/mdr.1998.4.219. [DOI] [PubMed] [Google Scholar]
- 6.Castañeda E, Tomasz A, Vela M C T. Penicillin-resistant Streptococcus pneumoniae in Colombia: presence of international epidemic clones. Microb Drug Resist. 1998;4:233–239. doi: 10.1089/mdr.1998.4.233. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 8.Coffey T J, Berrón S, Daniels M, Garcia-Leoni E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 9.Coffey T J, Enright M C, Daniels M, Wilkinson P, Berrón S, Fenoll A, Spratt B G. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Resist. 1998;4:51–55. doi: 10.1089/mdr.1998.4.51. [DOI] [PubMed] [Google Scholar]
- 10.Corso A, Severina E P, Petruk V F, Mauriz Y R, Tomasz A. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb Drug Resist. 1998;4:325–337. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 11.Doit C, Denamur E, Picard B, Geslin P, Elion J, Bingen E. Mechanisms of the spread of penicillin resistance in Streptococcus pneumoniae strains causing meningitis in children in France. J Infect Dis. 1996;174:520–528. doi: 10.1093/infdis/174.3.520. [DOI] [PubMed] [Google Scholar]
- 12.Echániz-Aviles M, Carnalla-Barajas N, Velàzquez-Meza M E, Soto-Noguerón A, Espinoza-de los Monteros L E, Solórzano-Santos F. Capsular types of Streptococcus pneumoniae causing disease in children from Mexico City. Pediatr Infect Dis J. 1995;14:907–909. [PubMed] [Google Scholar]
- 13.Enright M C, Spratt B G. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 14.Fenoll A, Bourgon M, Munoz R, Vicioso D, Casal J. Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates causing systemic infections in Spain, 1979–1989. Rev Infect Dis. 1991;13:56–60. doi: 10.1093/clinids/13.1.56. [DOI] [PubMed] [Google Scholar]
- 15.Fenoll A, Jado I, Vicioso D, Pérez A, Casal J. Evolution of Streptococcus pneumoniae serotypes and antibiotic resistance in Spain: update (1990–1996) J Clin Microbiol. 1998;36:3447–3454. doi: 10.1128/jcm.36.12.3447-3454.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 17.Hall L M, Whiley R A, Duke B, George R C, Efstratiou A. Genetic relatedness within and between serotypes of Streptococcus pneumoniae from the United Kingdom: analysis of multilocus enzyme electrophoresis, pulsed-field gel electrophoresis, and antimicrobial resistance patterns. J Clin Microbiol. 1996;34:853–859. doi: 10.1128/jcm.34.4.853-859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansman D, Bullen M M. A resistant pneumococcus. Lancet. 1967;ii:264–265. doi: 10.1016/s0140-6736(75)91547-0. [DOI] [PubMed] [Google Scholar]
- 19.Jabes D, Nachman S, Tomasz A. Penicillin-binding protein families: evidence for the clonal nature of penicillin-resistance in clinical isolates of pneumococci. J Infect Dis. 1989;159:16–25. doi: 10.1093/infdis/159.1.16. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs M R, Koornhof H J, Robins-Browne R M, Stevenson C M, Vermaak Z A, Freiman I, Miller G B, Witcomb M A, Isaacson M, Ward J I, Austrian R. Emergence of multiply resistant pneumococci. N Engl J Med. 1978;299:735–740. doi: 10.1056/NEJM197810052991402. [DOI] [PubMed] [Google Scholar]
- 21.Kam K M, Luey K Y, Fung S M, Yiu P P, Harden T J, Cheung M M. Emergence of multiple-antibiotic-resistant Streptococcus pneumoniae in Hong Kong. Antimicrob Agents Chemother. 1995;39:2667–2670. doi: 10.1128/aac.39.12.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klugman K P. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990;3:171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klugman K P, Coffey T J, Smith A, Wasas A, Meyers M, Spratt B G. Cluster of erythromycin-resistant variant of the Spanish multiply resistant 23F clone of Streptococcus pneumoniae in South Africa. Eur J Clin Microbiol Infect Dis. 1994;13:171–174. doi: 10.1007/BF01982193. [DOI] [PubMed] [Google Scholar]
- 24.Koeuth T, Versalovic J, Lupski J R. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res. 1995;5:408–418. doi: 10.1101/gr.5.4.408. [DOI] [PubMed] [Google Scholar]
- 25.Kristinsson K G. Epidemiology of penicillin-resistant pneumococci in Iceland. Microb Drug Resist. 1995;1:121–125. doi: 10.1089/mdr.1995.1.121. [DOI] [PubMed] [Google Scholar]
- 26.Laible G, Spratt B G, Hakenbeck R. Interspecies recombinational events during the evolution of altered PBP2X genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1991;5:1993–2202. doi: 10.1111/j.1365-2958.1991.tb00821.x. [DOI] [PubMed] [Google Scholar]
- 27.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefèvre J C, Faucon G, Sicard A M, Gasc A M. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1993;31:2724–2728. doi: 10.1128/jcm.31.10.2724-2728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lefèvre J C, Bertrand M A, Faucon G. Molecular analysis by pulsed-field gel electrophoresis of penicillin-resistant Streptococcus pneumoniae from Toulouse, France. Eur J Clin Microbiol Infect Dis. 1995;14:491–497. doi: 10.1007/BF02113426. [DOI] [PubMed] [Google Scholar]
- 30.Marchese A, Ramirez M, Schito G C, Tomasz A. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J Clin Microbiol. 1998;36:2944–2949. doi: 10.1128/jcm.36.10.2944-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marton A, Gulyas M, Munoz R, Tomasz A. Extremely high incidence of antibiotic resistance in clinical isolates of Streptococcus pneumoniae in Hungary. J Infect Dis. 1991;163:542–548. doi: 10.1093/infdis/163.3.542. [DOI] [PubMed] [Google Scholar]
- 32.McDougal L K, Facklam R, Reeves M, Hunter S, Swenson J M, Hill B C, Tenover F C. Analysis of multiply antimicrobial-resistant isolates of Streptococcus pneumoniae from the United States. Antimicrob Agents Chemother. 1992;36:2176–2184. doi: 10.1128/aac.36.10.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDougal L K, Rasheed J K, Biddle J W, Tenover F C. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother. 1995;39:2282–2288. doi: 10.1128/aac.39.10.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGee L, Klugman K P, Friedland D, Lee H J. Spread of the Spanish multi-resistant serotype 23F clone of Streptococcus pneumoniae to Seoul, Korea. Microb Drug Resist. 1997;3:253–257. doi: 10.1089/mdr.1997.3.253. [DOI] [PubMed] [Google Scholar]
- 35.Melander E, Ekdahl K, Hansson H B, Kamme C, Laurell M, Nilsson P, Persson K, Söderström M, Mölstad S. Introduction and clonal spread of penicillin- and trimethoprim-sulphamethoxazole-resistant Streptococcus pneumoniae, serotype 9V, in southern Sweden. Microb Drug Resist. 1998;4:71–78. doi: 10.1089/mdr.1998.4.71. [DOI] [PubMed] [Google Scholar]
- 36.Moissenet D, Valcin M, Marchand V, Garabédian E-N, Geslin P, Garbarg-Chenon A, Vu-Thien H. Molecular epidemiology of Streptococcus pneumoniae with decreased susceptibility to penicillin in a Paris children's hospital. J Clin Microbiol. 1997;35:298–301. doi: 10.1128/jcm.35.1.298-301.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 38.Munoz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorensen U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterisation by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 39.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Publication M7–A5. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 40.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multi-drug resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 41.Overweg K, Hermans P W M, Trzcinski K, Sluijter M, de Groot R, Hryniewicz W. Multidrug-resistant Streptococcus pneumoniae in Poland: identification of emerging clones. J Clin Microbiol. 1999;37:1739–1745. doi: 10.1128/jcm.37.6.1739-1745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichmann P, Varon E, Gunther E, Reinert R R, Luttiken R, Marton A, Geslin P, Wagner J, Hakenbeck R. Penicillin-resistant Streptococcus pneumoniae in Germany: genetic relationship to clones from other European countries. J Med Microbiol. 1995;43:377–385. doi: 10.1099/00222615-43-5-377. [DOI] [PubMed] [Google Scholar]
- 43.Rossi A, Corso A, Pace J, Regueira M, Tomasz A. Penicillin-resistant Streptococcus pneumoniae in Argentina: frequent occurrence of an internationally spread serotype 14. Microb Drug Resist. 1998;4:225–231. doi: 10.1089/mdr.1998.4.225. [DOI] [PubMed] [Google Scholar]
- 44.Sá-Leão R, Tomasz A, Santos Sanches I, Brito-Avô A, Vilhelmsson S E, Kristinsson K G, de Lencastre H. Carriage of internationally-spread epidemic clones of Streptococcus pneumoniae with unusual drug resistance patterns in children attending day care centers in Lisbon, Portugal. J Infect Dis. 2000;182:1153–1160. doi: 10.1086/315813. [DOI] [PubMed] [Google Scholar]
- 45.Sá-Leão R, Tomasz A, Santos Sanches I, Nunes S, Alves C R, Brito-Avô A, Saldanha J, Kristinsson K G, de Lencastre H. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J Clin Microbiol. 2000;38:4137–4144. doi: 10.1128/jcm.38.11.4137-4144.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Setchanova L, Tomasz A. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates from Bulgaria. J Clin Microbiol. 1999;37:638–648. doi: 10.1128/jcm.37.3.638-648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Z-Y, Enright M C, Wilkinson P, Griffiths D, Spratt B G. Identification of the three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequencing typing. J Clin Microbiol. 1998;36:3514–3519. doi: 10.1128/jcm.36.12.3514-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sibold C, Wang J, Henrichsen J, Hakenbeck R. Genetic relationships of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect Immun. 1992;60:4119–4126. doi: 10.1128/iai.60.10.4119-4126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sloas M M, Barrett F F, Chesney P J, English B K, Hill B C, Tenover F C, Leggiadro R J. Cephalosporin treatment failure in penicillin- and cephalosporin-resistant Streptococcus pneumoniae meningitis. Pediatr Infect Dis. 1992;11:662–666. [PubMed] [Google Scholar]
- 50.Sluijter M, Faden H, de Groot R, Lemmens N, Goessens W H F, van Belkum A, Hermans P W M. Molecular characterization of pneumococcal nasopharynx isolates collected from children during their first 2 years of life. J Clin Microbiol. 1998;36:2248–2253. doi: 10.1128/jcm.36.8.2248-2253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 52.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 53.Song J-H, Lee N Y, Ichiyama S, Yoshida R, Hirakata Y, Fu W, Chongthaleong A, Aswapokee N, Chiu C-H, Lalitha M K, Thomas K, Perera J, Yee T T, Jamal F, Warsa U C, Vinh B X, Jacobs M R, Appelbaum P C, Pai C H the ANSORP Study Group. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Clin Infect Dis. 1999;28:1206–1211. doi: 10.1086/514783. [DOI] [PubMed] [Google Scholar]
- 54.Song J-H, Yang J-W, Jin J H, Kim S W, Kim C K, Lee H, Peck K R, Kim S, Lee N Y, Jacobs M R, Appelbaum P C the Asian Network for Surveillance of Resistant Pathogens (ANSORP) Study Group. Molecular characterization of multidrug-resistant Streptococcus pneumoniae isolates in Korea. J Clin Microbiol. 2000;38:1641–1644. doi: 10.1128/jcm.38.4.1641-1644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takala A K, Vuopio-Varkila J, Tarkka E, Leinonen M, Musser J M. Subtyping of common pediatric pneumococcal serotypes from invasive disease and pharyngeal carriage in Finland. J Infect Dis. 1996;173:128–135. doi: 10.1093/infdis/173.1.128. [DOI] [PubMed] [Google Scholar]
- 58.Tarasi A, Sterk-Kuzmanovic N, Sieradzki K, Schoenwald S, Austrian R, Tomasz A. Penicillin-resistant and multidrug-resistant Streptococcus pneumoniae in a paediatric hospital in Zagreb, Croatia. Microb Drug Resist. 1995;1:169–176. doi: 10.1089/mdr.1995.1.169. [DOI] [PubMed] [Google Scholar]
- 59.Tarasi A, Chong Y, Lee K, Tomasz A. Spread of the serotype 23F multidrug-resistant Streptococcus pneumoniae clone to South Korea. Microb Drug Resist. 1997;3:105–109. doi: 10.1089/mdr.1997.3.105. [DOI] [PubMed] [Google Scholar]
- 60.Van Belkum A, Sluijter M, de Groot R, Verbrugh H, Hermans P W M. Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1996;34:1176–1179. doi: 10.1128/jcm.34.5.1176-1179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaz Pato M V, de Carvalho C B, Tomasz A the Multicenter Study Group. Antibiotic susceptibility of Streptococcus pneumoniae isolates in Portugal. A multicenter study between 1989 and 1993. Microb Drug Resist. 1995;1:59–69. doi: 10.1089/mdr.1995.1.59. [DOI] [PubMed] [Google Scholar]
- 62.Versalovic J, Kapur V, Mason E O, Shah U, Koeuth T, Lupski J R, Musser J M. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]
- 63.Vilhelmsson S E, Tomasz A, Kristinsson K G. Molecular evolution in a multidrug-resistant lineage of Streptococcus pneumoniae: emergence of strains belonging to the serotype 6B Icelandic clone that lost antibiotic resistance traits. J Clin Microbiol. 2000;38:1375–1381. doi: 10.1128/jcm.38.4.1375-1381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou J, Enright M C, Spratt B G. Identification of the major Spanish clones of penicillin-resistant pneumococci via the Internet using multilocus sequence typing. J Clin Microbiol. 2000;38:977–986. doi: 10.1128/jcm.38.3.977-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]