Abstract
Background
Sarcopenia is the age‐related loss of muscle mass, strength, and function. Epigenetic processes such as DNA methylation, which integrate both genetic and environmental exposures, have been suggested to contribute to the development of sarcopenia. This study aimed to determine whether differences in the muscle methylome are associated with sarcopenia and its component measures: grip strength, appendicular lean mass index (ALMi), and gait speed.
Methods
Using the Infinium Human MethylationEPIC BeadChip, we measured DNA methylation in vastus lateralis muscle biopsies of 83 male participants (12 with sarcopenia) with a mean (standard deviation) age of 75.7 (3.6) years from the Hertfordshire Sarcopenia Study (HSS) and Hertfordshire Sarcopenia Study extension (HSSe) and examined associations with sarcopenia and its components. Pathway, histone mark, and transcription factor enrichment of the differentially methylated CpGs (dmCpGs) were determined, and sodium bisulfite pyrosequencing was used to validate the sarcopenia‐associated dmCpGs. Human primary myoblasts (n = 6) isolated from vastus lateralis muscle biopsies from male individuals from HSSe were treated with the EZH2 inhibitor GSK343 to assess how perturbations in epigenetic processes may impact myoblast differentiation and fusion, measured by PAX7 and MYHC immunocytochemistry, and mitochondrial bioenergetics determined using the Seahorse XF96.
Results
Sarcopenia was associated with differential methylation at 176 dmCpGs (false discovery rate ≤ 0.05) and 141 differentially methylated regions (Stouffer ≤ 0.05). The sarcopenia‐associated dmCpGs were enriched in genes associated with myotube fusion (P = 1.40E‐03), oxidative phosphorylation (P = 2.78E‐02), and voltage‐gated calcium channels (P = 1.59E‐04). ALMi was associated with 71 dmCpGs, grip strength with 49 dmCpGs, and gait speed with 23 dmCpGs (false discovery rate ≤ 0.05). There was significant overlap between the dmCpGs associated with sarcopenia and ALMi (P = 3.4E‐35), sarcopenia and gait speed (P = 4.78E‐03), and sarcopenia and grip strength (P = 7.55E‐06). There was also an over‐representation of the sarcopenia, ALMi, grip strength, and gait speed‐associated dmCpGs with sites of H3K27 trimethylation (all P ≤ 0.05) and amongst EZH2 target genes (all P ≤ 0.05). Furthermore, treatment of human primary myoblasts with the EZH2 inhibitor GSK343 inhibitor led to an increase in PAX7 expression (P ≤ 0.05), decreased myotube fusion (P = 0.043), and an increase in ATP production (P = 0.008), with alterations in the DNA methylation of genes involved in oxidative phosphorylation and myogenesis.
Conclusions
These findings show that differences in the muscle methylome are associated with sarcopenia and individual measures of muscle mass, strength, and function in older individuals. This suggests that changes in the epigenetic regulation of genes may contribute to impaired muscle function in later life.
Keywords: DNA methylation, Sarcopenia, EZH2, Myoblasts
Introduction
Sarcopenia, defined as the loss of muscle mass, strength, and function with advancing age, is associated with a number of adverse physical and metabolic changes that contribute to morbidity, impaired quality of life, increased health care costs, and mortality. Although several operational definitions of sarcopenia are used worldwide, 1 , 2 there is general consensus that defining sarcopenia relies on low muscle function (either weak muscle strength or impaired physical performance, i.e. slower gait speed) in combination with low whole‐body or appendicular lean mass. 1 , 3 , 4 , 5 Prevalence estimates have been reported to be between 1–29% amongst community‐dwelling older adults, 14–33% for those in long‐term care, and 10% for those in acute hospital. 3 , 6
A decline in muscle mass, strength, and function is a fundamental consequence of ageing; however, there is significant variability between individuals in the rate of loss in old age. Some of the variability can be explained by fixed genetic factors, 7 , 8 but much of the remaining variation is unexplained. There is growing evidence that suggests epigenetic processes play a prominent role in the development of many complex diseases. 9 Processes such as DNA methylation induce heritable changes in gene expression without a change in nucleotide sequence. 10 As inter‐individual DNA methylation is specified by the interaction of both genotypic and environmental influences, 11 changes in DNA methylation within skeletal muscle may provide novel insights into the variability in muscle ageing, as well as provide powerful biomarkers when compared with either genotype or lifestyle factors alone. To date, a number of human studies have compared DNA methylation in muscle tissue from young versus old individuals 12 , 13 , 14 , 15 , 16 and reported differential methylation of genes involved in axon guidance, 12 , 13 cytoskeletal function, 13 , 16 cell adhesion, 12 , 13 muscle contraction, 15 calcium signalling, 15 and mTOR signalling. 12 DNA methylation in muscle tissue in individuals with sarcopenia compared with healthy aged‐matched controls has not previously been reported. A comparison of the muscle transcriptome in 119 older men with sarcopenia versus age‐matched controls from Singapore, Hertfordshire UK, and Jamaica demonstrated that the major transcriptional signature of sarcopenia was mitochondrial bioenergetic dysfunction in skeletal muscle, with down‐regulation of oxidative phosphorylation genes. 17 However, whether these transcriptional changes are mediated through changes in DNA methylation is not known. Here, we sought to identify DNA methylation changes in muscle associated with sarcopenia, and its components, namely, grip strength, appendicular lean mass index (ALMi), and gait speed.
Methods
Study design
All participants were recruited from the Hertfordshire Cohort Study (HCS), 18 a retrospective cohort study based in the UK designed to investigate life course influences on muscle function in community‐dwelling older people. DNA was analysed from 40 male participants from the first phase of the study, the Hertfordshire Sarcopenia Study (HSS), 19 , 20 and from the male participants of the second extension phase of the study (n = 43), herein termed HSSe. 21 The 40 males from the HSS and the 43 males from the HSSe were the only samples with sufficient DNA for both genome‐wide methylation analysis and pyrosequencing. All participants gave written informed consent, and the study was approved by the Hertfordshire Research Ethics Committee (07/Q0204/68). Sarcopenia was defined according to the European Working Group on Sarcopenia in Older People (EWGSOP) 3 criteria, with the following thresholds: ALMi (ALM/height2) ≤ 7.23 kg/m2 for men and ≤5.67 kg/m2 for women; grip strength < 30 kg for men and <20 kg for women; and walking speed ≤ 0.8 m/s. Participants were classed as healthy controls (normal ALMi, gait speed, and grip strength) or as having sarcopenia (low ALMi and low gait speed and/or low grip strength).
Procedures
Body composition (appendicular lean mass) was assessed by dual‐energy X‐ray absorptiometry (DXA) (Hologic Discovery, software version 12.5). Isometric grip strength (kilograms) was measured three times in each hand using a Jamar handheld hydraulic dynamometer (Promedics, UK), and the highest value of six measures used. 22 Customary walking speed was measured over a 3 m course. Percutaneous muscle biopsies of the vastus lateralis were conducted after an overnight fast under local anaesthetic using a Weil–Blakesley conchotome. 23
Infinium Human MethylationEPIC BeadChip array
Genomic DNA was extracted from muscle from HSS participants using the QIAamp DNA mini kit (Qiagen) and from HSSe participants using the high salt method. 24 A total of 750 ng of genomic DNA was treated with sodium bisulfite using Zymo EZ DNA Methylation‐Gold kit (ZymoResearch, USA) and hybridized to the Infinium Human MethylationEPIC BeadChip array (Illumina, Inc., USA) at the Centre for Molecular Medicine and Therapeutics (http://www.cmmt.ubc.ca).
Infinium Human MethylationEPIC BeadChip array data processing
EPIC array data were processed using the Bioconductor package minfi 25 in R (version 3.4.2). See details described in Supporting Information, Data S1. After pre‐processing and QC, 77 samples for the muscle tissue analysis remained, and 10 for the myoblast analysis.
Infinium Human MethylationEPIC BeadChip array data analysis
Robust regression models were run using limma (v3.38.3). 26 Models were adjusted for age, and surrogate variables, to account differences in cellular heterogeneity. The analysis was controlled for multiple testing with the Benjamini–Hochberg adjustment for false discovery rate (FDR), using an FDR < 0.05. See full details in Data S1.
Gene ontology, histone, and transcription factor enrichment analysis
Protein–protein interaction (PPI) networks were carried out using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) and visualized in Cytoscape. Large networks were segmented using the MCODE algorithm, 27 and gene ontology (GO) enrichment determined using BiNGO. 28 Enrichment of differentially methylated CpGs (dmCpGs) amongst regions of histone modifications and transcription factor binding sites was assessed using the ChIP‐seq peak data from ENCODE in male human skeletal muscle tissue (https://www.encodeproject.org).
Muscle epigenetic age estimator
Epigenetic age acceleration was calculated as the residuals of regressing the epigenetic age estimated by the muscle epigenetic age estimator (MEAT) 14 over chronological age.
RNA sequencing and analysis
RNA was extracted from frozen muscle tissue using the mirVana™ miRNA Isolation Kit (Ambion, Life Technologies), and RNA sequencing carried out as described in Migliavacca et al. 17
Pyrosequencing methylation analysis
Quantitative DNA methylation analysis was carried out by pyrosequencing, described in Data S1. Primer sequences are shown in Supporting Information, Table S1.
Isolation of myoblast cells from muscle biopsies
Details of myoblast isolation and culture are described in Data S1. Briefly, biopsies were minced (three with sarcopenia and three healthy aged‐matched controls), digested in 0.5 mg/mL collagenase (Sigma), before pre‐plating to remove fibroblasts, and sorting using CD56 MicroBeads (Miltenyi Biotech). 29 Isolation of myogenic cells was confirmed by immunocytochemistry, which showed ≥96% of cells as CD56 positive after sorting. Cells were grown in either proliferation medium [Dulbecco's modified Eagle's medium (DMEM) containing 20% foetal bovine serum (FBS), 10% horse serum (HS), 1% chick embryo extract, and 1% penicillin/streptomycin (P/S)] or differentiation media (DMEM containing 2% HS and 1% P/S). Human primary myoblasts were treated with GSK343 (MedChemExpress), an inhibitor of EZH2, 30 at 20 nM, 200 nM, and 2 μM after the initiation of differentiation over a 10 day period.
Immunocytochemistry
Cells were stained for myosin heavy chain (MYHC), PAX7, and 4′,6‐diamidino‐2‐phenylindole (DAPI) (1 μg/mL). See details in Data S1. To calculate the fusion index, the number of nuclei within myotubes (containing 2+ nuclei) was counted and the ratio of this number to the total number of nuclei was determined.
Metabolic flux assay
Mitochondrial bioenergetics were measured using the Agilent Seahorse XF96 Mito Stress Test (details in Data S1). After measurement, cells were lysed and a protein assay was carried out for normalization. Each data point represents the mean ± standard deviation of six replicates for each condition.
Statistical analysis
All statistical analyses were carried out in R (version 3.4.2). Demographic characteristics were compared between controls and those with sarcopenia using the Mann–Whitney U tests. The hypergeometric distribution probability test was used to test the significance of the overlap between dmCpGs associated with different measures of muscle mass/function. Fisher's exact test was used to test the enrichment of dmCpGs amongst the different histone modifications, chromatin enhancer states, and genomic regions relative to CpG islands. Linear models were fitted to the pyrosequencing data including age as a covariate. Correlation analysis of the methylation and gene expression data was carried out using Spearman's correlation. Statistical analysis of the GSK343‐treated cell cultures was carried out using the Wilcoxon signed‐rank test for the immunocytochemistry and paired t‐test for the metabolic flux assays, with data from sarcopenia and control myoblasts analysed as one group to maximize statistical power.
Results
Participant characteristics
Of the 83 men who had muscle tissue available, 77 samples passed downstream quality control and were included in further analysis. Participant characteristics are given in Table 1. Men who had sarcopenia (n = 11) were older (P = 0.019) and had a lower body mass index (P < 0.001) compared with those who did not have sarcopenia. As expected, men with sarcopenia also had lower measures of muscle mass [total lean mass, appendicular lean mass (ALM), and ALMi; all P < 0.001] and reduced muscle function [gait speed (P < 0.001)] and grip strength (P = 0.001) compared with those who did not have sarcopenia.
Table 1.
Participant characteristics
| Control (n = 41) | Sarcopenia (n = 11) | Total (n = 77 a ) | ||
|---|---|---|---|---|
| Age (years) | 74.49 ± 3.25 | 77.12 ± 3.74 | 75.66 ± 3.55 | * P = 1.90 × 10−2 |
| Height (cm) | 172.32 ± 5.77 | 171.68 ± 9.81 | 172.22 ± 6.12 | P = 9.22 × 10−1 |
| Weight (kg) | 83.45 ± 12.81 | 76.06 ± 12.14 | 79.10 ± 12.32 | *** P = 1.00 × 10−3 |
| BMI (kg/m 2) | 28.04 ± 3.66 | 25.74 ± 2.86 | 26.10 ± 3.47 | **** P = 1.28 × 10−4 |
| Total lean mass (kg) | 55.33 ± 6.94 | 47.82 ± 5.45 | 51.58 ± 7.29 | **** P = 2.96 × 10−7 |
| ALM (kg) | 24.42 ± 2.98 | 20.03 ± 2.68 | 22.35 ± 3.46 | **** P = 4.06 × 10−10 |
| Fat mass (kg) | 24.15 ± 8.02 | 24.61 ± 9.05 | 23.77 ± 7.81 | P = 5.64 × 10−1 |
| Grip strength (kg) | 38.05 ± 6.86 | 29.00 ± 8.72 | 36.19 ± 7.15 | *** P = 1.00 × 10−3 |
| Gait speed (m/s) | 1.08 ± 018 | 0.82 ± 0.26 | 1.05 ± 021 | **** P = 3.61 × 10−4 |
| ALMi (kg/m2) | 8.21 ± 0.76 | 6.77 ± 0.36 | 7.53 ± 0.98 | **** P = 5.92 × 10−15 |
ALM, appendicular lean mass; ALMi, appendicular lean mass index; BMI, body mass index.
Twenty participants were not classed as healthy controls or as having sarcopenia using the European Working Group on Sarcopenia in Older People definition; however, they were included in the continuous analyses of ALMi, grip strength, and gait speed.
P‐value ≤ 0.05.
P‐value ≤ 0.01.
P‐value ≤ 0.001.
P‐value ≤ 0.0001.
Identification of differentially methylated CpGs in muscle biopsies associated with sarcopenia
There were significant (FDR, P < 0.05) associations between DNA methylation and sarcopenia, with 176 CpGs significantly associated with sarcopenia (Tables 2 and S2). The top two dmCpGs were located in an intergenic region on chromosome 1 (cg17974166, FDR = 2.67 × 10−10, and cg01647314, FDR = 1.06 × 10−7) (Figure 1A + 1B); 53.4% of the dmCpGs showed hypermethylation in those with sarcopenia (Figure 1C), with a significant over‐representation of dmCpGs in the OpenSea (P = 0.0188) and CpG island (P = 1.28 × 10−5) regions (Figure 1D).
Table 2.
Top 25 sarcopenia‐associated differentially methylated CpGs
| Probe | logFC | Average methylation | FDR | Gene |
|---|---|---|---|---|
| cg17974166 | 0.1244 | 0.1246 | 2.67 × 10−10 | |
| cg01647314 | 0.0831 | 0.1640 | 1.06 × 10−7 | |
| cg10941472 | 0.0823 | 0.1102 | 1.49 × 10−6 | |
| cg20843809 | −0.1151 | 0.8705 | 4.22 × 10−6 | |
| cg07236061 | −0.1987 | 0.8646 | 2.28 × 10−4 | LINC01331 |
| cg22843429 | −0.0497 | 0.8661 | 2.28 × 10−4 | EMG1 |
| cg03622584 | 0.0528 | 0.6245 | 3.03 × 10−4 | LOC101927934 |
| cg00384701 | 0.0430 | 0.3116 | 3.28 × 10−4 | |
| cg10977501 | −0.0346 | 0.9088 | 3.81 × 10−4 | MYOM2 |
| cg27191906 | 0.0676 | 0.6362 | 4.51 × 10−4 | OR2J2 |
| cg00172812 | 0.1024 | 0.2311 | 4.51 × 10−4 | |
| cg00102685 | −0.0368 | 0.9359 | 5.33 × 10−4 | |
| cg08344114 | −0.0378 | 0.5612 | 7.77 × 10−4 | |
| cg27473406 | −0.0416 | 0.3952 | 7.77 × 10−4 | |
| cg26945376 | 0.0331 | 0.1893 | 7.77 × 10−4 | CLIP1 |
| cg10956589 | 0.0330 | 0.4653 | 8.66 × 10−4 | |
| cg03532253 | −0.0296 | 0.8865 | 9.09 × 10−4 | SUOX |
| cg25441771 | 0.0316 | 0.0468 | 2.20 × 10−3 | MCCC1 |
| cg16307778 | −0.0501 | 0.8696 | 2.75 × 10−3 | |
| cg15744876 | −0.0428 | 0.5705 | 3.17 × 10−3 | |
| cg04344695 | 0.0434 | 0.4816 | 3.66 × 10−3 | CA12 |
| cg27253454 | −0.0323 | 0.7946 | 3.66 × 10−3 | USP43 |
| cg14659184 | −0.0499 | 0.8841 | 4.01 × 10−3 | LINC00925 |
| cg20266770 | 0.0451 | 0.2541 | 4.45 × 10−3 | |
| cg25963061 | 0.0336 | 0.7137 | 4.59 × 10−3 | UBQLNL |
FC, fold change; FDR, false discovery rate.
Figure 1.
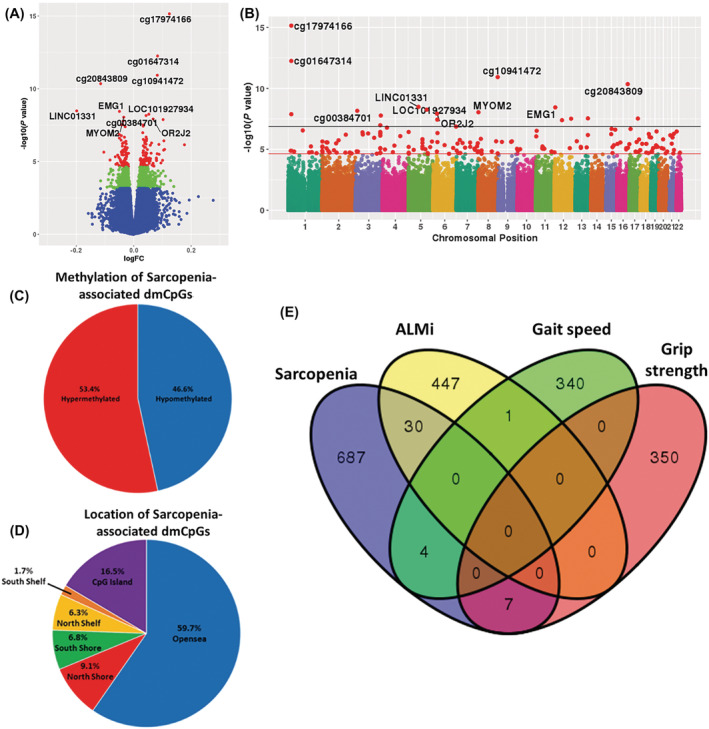
(A) Volcano plot of the differential methylation analysis with respect to sarcopenia, with the significant probes (FDR < 0.05) highlighted in red and probes with an FDR < 0.25 highlighted in green. The top 10 dmCpGs have been annotated. (B) Manhattan plot for the differential methylation analysis with respect to sarcopenia. All significant sarcopenia‐associated dmCpGs are shown as bold red points, with the top 10 dmCpGs annotated. Black line: Bonferroni threshold (P‐value = 1.33 × 10−7); red line: FDR threshold (P‐value = 2.34 × 10−5). (C) Pie chart showing the proportions of dmCpGs showing increased or decreased methylation associated with sarcopenia. (D) Pie chart showing the proportions of the locations of the sarcopenia‐associated dmCpGs. (E) Venn diagram indicating the overlap in the number of CpGs (FDR < 0.2) associated with sarcopenia and the multiple measures of muscle mass.
To understand the contribution of muscle mass, strength, and function to the methylation signature associated with sarcopenia, we analysed DNA methylation with respect to ALMi, grip strength, and gait speed as continuous variables. ALMi was associated with 71 dmCpGs; grip strength with 49 dmCpGs; and gait speed with 23 dmCpGs (Tables S3–S5). The top dmCpG associated with ALMi was cg22350027, located in the body of the phospholipase C‐like protein 2 (PLCL2) gene (FDR = 9.68 × 10−6), while the top dmCpGs associated with grip strength and gait speed were located in the intergenic region of chromosome 17 (FDR = 0.0003) and chromosome 1 (FDR = 0.0017), respectively. There was significant overlap between the dmCpGs associated with sarcopenia and ALMi (P = 3.40 × 10−35), sarcopenia and gait speed (P = 4.78 × 10−3), and sarcopenia and grip strength (P = 7.55 × 10−6) (Table S6, Figure 1E). However, there was no overlap between the dmCpGs associated with ALMi, grip strength, or gait speed.
Sarcopenia is not associated with accelerated epigenetic age
To determine whether the methylation changes associated with sarcopenia represented accelerated muscle ageing, epigenetic age acceleration was calculated. 14 Epigenetic age as determined by MEAT was strongly correlated with chronological age (Supporting Information, Figure S2; Pearson's r = 0.421, P = 0.0001). However, there were no significant associations of accelerated epigenetic ageing with sarcopenia status, ALMi, gait speed, or grip strength (Table S7).
Sarcopenia and the individual measures of muscle mass, strength, and function are associated with multiple differentially methylated regions
Regional analysis identified differentially methylated regions (DMRs) associated with sarcopenia, ALMI, grip strength, and gait speed (Tables S8–S11). Sarcopenia was associated with 141 DMRs (Stouffer < 0.05), with the top DMR located within the promoter region of the Methylcrotonyl‐CoA Carboxylase 1 (MCCC1) gene, consisting of 13 CpGs (Stouffer = 3.62 × 10−12). ALMi was associated with 135 DMRs, with the top DMR located within the zinc finger protein 57 (ZFP57) gene. Grip strength was associated with 28 DMRs, with the top DMR located within the claudin 10 (CLDN10) gene (Stouffer = 8.29 × 10−4). Gait speed was associated with 24 DMRs, with the top DMR located in an intergenic region on chromosome 1 (Stouffer = 7.53 × 10−4). There were three DMRs associated with both sarcopenia and ALMi, but no DMRs that overlapped between sarcopenia and grip strength, or sarcopenia and gait speed.
Correlation between DNA methylation and gene expression in skeletal muscle tissue
Of the 77 samples analysed by EPIC DNA methylation arrays, 34 muscle samples had previously been analysed using total RNAseq. 17 Therefore, to explore the correlation between DNA methylation and gene expression, we examined the sarcopenia‐associated dmCpGs with an FDR < 0.2 annotated to a gene (n = 470), with the RNAseq data, removing transcripts with low expression across all samples. This resulted in 298 transcript–CpG pairs. Correlation analysis of the transcript–CpG pairs revealed 29 with a significant correlation between methylation and expression at an FDR < 0.2 (Table 3). There were multiple dmCpGs within MCCC1, which were associated with transcript levels: three CpGs in the body of the MCCC1 gene (cg08395365, cg22211233, and cg00161968) and one within 200 bp of the MCCC1 TSS (cg00890010) were negatively associated with MCCC1 expression, while cg23476885, located in the body of the MCCC1 gene, was positively associated with MCCC1 expression.
Table 3.
Correlation between DNA methylation and RNA expression (false discovery rate < 0.2)
| Probe | Spearman's ρ | P‐value | FDR | Gene | CpG position |
|---|---|---|---|---|---|
| dmCpGs | |||||
| cg20108671 | −0.6327 | 8.57E‐05 | 0.0260 | SCAPER | 5′UTR |
| cg03854273 | −0.5939 | 2.80E‐04 | 0.0339 | LPCAT3 | Body |
| cg13403462 | −0.5875 | 3.36E‐04 | 0.0339 | NECAB3 | TSS200 |
| cg24859375 | −0.5438 | 1.06E‐03 | 0.0786 | LPCAT3 | 1st exon |
| cg04180086 | 0.5355 | 1.30E‐03 | 0.0786 | IRX4 | Body |
| cg01554606 | −0.5150 | 2.09E‐03 | 0.0981 | MYLK | Body |
| cg08395365 | −0.5001 | 2.92E‐03 | 0.0981 | MCCC1 | Body |
| cg07655627 | −0.4967 | 3.14E‐03 | 0.0981 | NUDT12 | Body |
| cg09166085 | −0.4937 | 3.36E‐03 | 0.0981 | NUDT12 | 3′UTR |
| cg02976617 | −0.4934 | 3.38E‐03 | 0.0981 | NUDT12 | Body |
| cg22211233 | −0.4909 | 3.56E‐03 | 0.0981 | MCCC1 | Body |
| cg23423191 | −0.4860 | 3.95E‐03 | 0.0997 | STK10 | 5′UTR |
| cg00617927 | 0.4756 | 4.90E‐03 | 0.1073 | COL27A1 | TSS1500 |
| cg03073264 | −0.4750 | 4.96E‐03 | 0.1073 | NUDT12 | Body |
| cg08767025 | 0.4652 | 6.03E‐03 | 0.1219 | SOX5 | TSS200 |
| cg22350027 | 0.4597 | 6.72E‐03 | 0.1273 | PLCL2 | Body |
| cg16938504 | −0.4564 | 7.18E‐03 | 0.1279 | RAB40B | TSS1500 |
| cg00890010 | −0.4503 | 8.07E‐03 | 0.1285 | MCCC1 | TSS200 |
| cg13665998 | −0.4484 | 8.36E‐03 | 0.1285 | NUDT12 | 3′UTR |
| cg07666882 | −0.4454 | 8.85E‐03 | 0.1285 | NUDT12 | TSS1500 |
| cg18172877 | 0.4451 | 8.91E‐03 | 0.1285 | IRX4 | 5′UTR |
| cg06105925 | −0.4420 | 9.43E‐03 | 0.1299 | NUDT12 | TSS1500 |
| cg09414187 | 0.4353 | 1.07E‐02 | 0.1406 | CENPV | Body |
| cg20279471 | −0.4292 | 1.19E‐02 | 0.1506 | NUDT12 | Body |
| cg00161968 | −0.4264 | 1.25E‐02 | 0.1519 | MCCC1 | Body |
| cg23476885 | 0.4191 | 1.43E‐02 | 0.1663 | MCCC1 | Body |
| cg22855016 | −0.4108 | 1.65E‐02 | 0.1847 | NFYC | Body |
| cg15020502 | −0.4072 | 1.75E‐02 | 0.1888 | ETS2 | TSS1500 |
| cg00575744 | −0.4053 | 1.81E‐02 | 0.1888 | HLA‐DMB | 1st exon |
| DMRs | |||||
| cg11689786 | −0.5251 | 1.66E‐03 | 0.1623 | MYL6B | 5′UTR |
| cg07655627 | −0.4967 | 3.14E‐03 | 0.1623 | NUDT12 | TSS1500 |
| cg15206834 | −0.4949 | 3.27E‐03 | 0.1623 | NUDT12 | TSS1500 |
| cg09166085 | −0.4937 | 3.36E‐03 | 0.1623 | NUDT12 | TSS200 |
| cg02976617 | −0.4934 | 3.38E‐03 | 0.1623 | NUDT12 | 5′UTR |
| cg03073264 | −0.4750 | 4.96E‐03 | 0.1983 | NUDT12 | 5′UTR |
dmCpGs, differentially methylated CpGs; DMRs, differentially methylated regions; FDR, false discovery rate.
Correlation between the CpGs within the DMRs also showed low congruence between DNA methylation and gene expression. Of the 279 CpGs, located within the DMRs, related to a gene with a normalized readcount > 20 in at least 5 samples, 6 CpGs correlated with gene expression at an FDR < 0.2 (Table 3).
Validation
Sodium bisulfite pyrosequencing was performed to validate cg10941472 (Figure 2A), cg22813735 (Figure 2B), and cg17194069 (Figure 2C), top hits associated with sarcopenia, ALMi, and gait speed, respectively, whose genomic context allowed sequencing primers to be designed. Bland–Altman plots comparing the methylation values between the pyrosequencer and the array for these CpG sites showed that the majority of the data points from the two methods lay within the limits of agreement. Moreover, analysis of the associations between the methylation status of the three CpGs, measured by pyrosequencing, and phenotypic parameters also replicated the array analysis, with cg10941472 significantly associated with sarcopenia status (P = 0.0232), cg22813735 negatively associated with ALMi (P = 0.0105), and cg17194069 negatively associated with gait speed (P = 0.01383).
Figure 2.

Bisulfite pyrosequencing validation of top hits associated with (A) sarcopenia, (B) ALMi, and (C) gait speed. Bland–Altman plots (left‐hand panels) show the differences between the two technologies (pyrosequencing and array), with the majority of the points within the limits of agreement. All three CpGs show significant associations with (A) sarcopenia, (B) ALMi, and (C) gait speed, in the same direction of change as seen on the array.
Sarcopenia‐associated differentially methylated CpGs were enriched in pathways associated with cell adhesion, energy production, and voltage‐gated calcium channels
To gain an understanding of the functional significance of the methylation changes, genes associated with the dmCpGs of FDR < 0.2 were inputted into STRING to generate a PPI network ( Figure S3a). The PPI enrichment P‐value for the network was 0.0121, indicating biological connection between the proteins. Ten significant GO terms were over‐represented in the PPI network (FDR < 0.05; Table S12), with the top term being homophilic cell adhesion (FDR = 0.0014), a process essential for myotube fusion. Subdivision of the network using MCODE identified clusters enriched for GO terms including phosphorus metabolic process (FDR = 1.20 × 10−3), oxidative phosphorylation (FDR = 2.78 × 10−2), and voltage‐gated Ca2+ channel activity (FDR = 1.59 × 10−4) (Table S13, Figure S3b). There was no enrichment of specific pathways amongst the DMRs associated with sarcopenia, but many of these lie within intergenic regions.
Appendicular lean mass index, grip strength, and gait speed‐associated differentially methylated CpGs were enriched in pathways associated with calcium channel activity and cell adhesion
The PPI enrichment P‐value for the ALMi associated dmCpGs was P = 0.00345 ( Figure S4a), although there were no significant GO terms associated with the whole network. Subdivision of the network showed enrichment for pathways involved in calcium channel activity and the sarcoplasmic reticulum (Table S14). For grip strength, the PPI network showed a significant enrichment for cell adhesion (FDR = 0.040), while for gait speed, there was enrichment of cadherin‐associated pathways (FDR ≤ 1.59 × 10−11). There was no enrichment of specific pathways amongst the DMRs associated with ALMi, grip strength, or gait speed.
Sarcopenia‐associated differentially methylated CpGs were enriched in regions overlapping sites of H3K27me3 and EZH2 binding motifs
To gain further insights into the functional significance of the changes in DNA methylation, the dmCpGs were examined for enrichment within regions associated with specific histone modifications or transcription factor binding sites. There was a significant enrichment of sarcopenia‐associated dmCpGs overlapping polycomb repressed regions of the genome (FDR = 4.20 × 10−4) and Histone H3 lysine 27 trimethylation (H3K27me3) modifications (FDR = 4.41 × 10−5), with a significant under‐representation of dmCpGs overlapping regions of Histone H3 lysine 4 monomethylation (H3K4me1) modifications (FDR = 1.20 × 10−3) ( Figure S5).
There was also enrichment for the Enhancer of Zeste Homologue 2 (EZH2) binding motif at the transcription start sites (TSS) and gene bodies of genes with a dmCpG associated with sarcopenia (TSS, FDR = 4.20 × 10−4; Body, FDR = 3.40 × 10−4), ALMi (TSS, FDR = 1.65 × 10−3; Body, FDR = 3.96 × 10−6), and grip strength (TSS, FDR = 2.98 × 10−5; Body, FDR = 5.93 × 10−5) (Table 4). The CCCTC‐binding factor (CTCF) binding motif was enriched at the TSS of genes with a dmCpG associated with grip strength (FDR = 0.0142) and in the gene bodies of genes associated with sarcopenia (FDR = 2.73 × 10−3), ALMi (FDR = 0.0185), gait speed (FDR = 4.88 × 10−3), and grip strength (FDR = 1.23 × 10−5).
Table 4.
ENCODE ChIP‐Seq significance results for overlap with identified differentially methylated CpGs
| No. of genes associated with dmCpGs | Factor | No. of genes enriched | FDR | |
|---|---|---|---|---|
| TSS/5′End (1500 bp upstream, 500 bp downstream) | ||||
| Sarcopenia | 379 | EZH2 | 37 | 4.20 × 10−4 |
| ALMi | 257 | EZH2 | 26 | 1.65 × 10−3 |
| Gait speed | 206 | — | — | — |
| Grip strength | 214 | EZH2 | 27 | 2.98 × 10−5 |
| CTCF | 63 | 1.42 × 10−2 | ||
| Whole gene body | ||||
| Sarcopenia | 379 | EZH2 | 30 | 3.40 × 10−4 |
| CTCF | 147 | 2.73 × 10−3 | ||
| ALMi | 257 | EZH2 | 27 | 3.96 × 10−6 |
| CTCF | 98 | 1.85 × 10−2 | ||
| Gait speed | 206 | CTCF | 85 | 4.88 × 10−3 |
| Grip strength | 214 | CTCF | 99 | 1.23 × 10−5 |
| EZH2 | 21 | 5.93 × 10−5 | ||
ALMi, appendicular lean mass index; dmCpGs, differentially methylated CpGs; FDR, false discovery rate; TSS, transcription start sites.
The EZH2 inhibitor GSK343 increases PAX7 expression, reduces myotube fusion index, and increases oxidative respiration in human primary muscle‐derived myoblasts
As the sarcopenia‐associated dmCpGs were enriched amongst EZH2 target genes, as well as pathways including oxidative phosphorylation and myotube fusion, two pathways critical for myogenic differentiation, we investigated the role that EZH2 plays in the regulation of these pathways in primary human myoblasts in vitro. Human skeletal muscle‐derived myoblasts (n = 6) were treated with the specific EZH2 inhibitor, GSK343, and the expression of MYHC and PAX7 after the induction of differentiation, together with oxidative phosphorylation assessed. GSK343 led to an increase in PAX7 positive cells at D2, D6, and D10 (D2: 200 nM P = 0.046, 2 μM P = 0.028; D6: 20 nM P = 0.046; D10: 20 nM P = 0.028, 200 nM P = 0.028, 2 μM P = 0.028; Figure 3A). Concomitant with the increase in PAX7 staining, treatment of myoblasts with GSK343 resulted in a decrease in fusion index at D6 (Figure 3B and 3C; 200 nM P = 0.043, 2 μM P = 0.043) and at D10 (Figure 3B and 3C; 20 nM P = 0.046, 2 μM P = 0.043) compared with vehicle control‐treated cells. The reduction in fusion index was accompanied by a decrease in myotube area on D6 (Figure 3D; 2 μM P = 0.043), but no change in the intensity of MYHC staining at any time point.
Figure 3.

(A) The effect of pharmacological inhibition of EZH2 with GSK343 on human primary myoblast cells with respect to Pax7+ cell number at day 2, day 6, and day 10 of differentiation. (B) Representative stains of myosin heavy chain staining in the DMSO and GSK343‐treated cells. (C) Quantification of the fusion index after treatment with GSK343 at day 6 and day 10 of differentiation. Fusion index was measured as the percentage of MYHC+ cells with more than two nuclei. (D) Quantification of the area of the myosin heavy chain positive cells at day 6 and day 10 of differentiation. (E) Representative Seahorse trace of the mitochondrial stress test of GSK343‐treated cells after 10 days of differentiation. (F–J) The effect of GSK343 treatment at 200 and 20 nM on cells after 10 days of differentiation on baseline respiration (F), ATP production (G), maximal respiration (H), spare capacity (I), and proton leak (J). Results shown are all n = 6, mean ± SEM. *P < 0.05, **P < 0.01.
The effect of GSK343 treatment on oxidative phosphorylation was assessed by measuring oxygen consumption rate (OCR) assessed at D10 of differentiation using the Seahorse XF96 analyser, as an indicator of oxidative phosphorylation (Figure 3E). Treatment with the GSK343 resulted in an increase in baseline respiration (Figure 3F; 20 nM P = 0.002, 200 nM P = 0.004), ATP production (Figure 3G; 20 nM P = 0.008, 200 nM P = 0.008), maximal respiration (Figure 3H; 20 nM P = 0.003, 200 nM P = 0.013), spare capacity (Figure 3I; 20 nM P = 0.007, 200 nM P = 0.050), and proton leak (Figure 3J; 20 nM P = 0.009, 200 nM P = 0.050).
GSK343 alters DNA methylation at oxidative phosphorylation and myogenesis genes
To investigate whether treatment of the myoblasts with GSK343 affected the methylation of genes involved in oxidative phosphorylation and myogenesis, DNA methylation was assessed in vehicle control (DMSO) and GSK343 (200 nM) treated myoblasts (n = 5), on D10 after the induction of differentiation, when a reduction in myotube fusion and changes in oxidative respiration were observed. Treatment with GSK343 significantly changed the methylation at 112 CpGs out of 2637 probes in the KEGG oxidative phosphorylation geneset (P < 0.05; Table S15), with 78.6% of the CpGs hypomethylated following GSK343 addition. The top dmCpG was cg11189039 located within the NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 (NDUFA6) gene (P = 0.0012), a subunit of Complex 1 in the respiratory chain. Treatment with GSK343 also led to differential methylation at 352 CpGs out of the 9136 probes in the myogenesis geneset (P < 0.05); 78.7% of the CpGs were hypomethylated following EZH2 inhibition. Comparison of the dmCpGs (P < 0.05) changed in response to GSK343 treatment in the cultured myoblasts with those associated with sarcopenia in muscle tissue (FDR < 0.05) revealed that only seven of sarcopenia‐associated dmCpGs were also changed by GSK343 treatment in the cultured myoblasts; in all cases, these CpG sites were hypomethylated by GSK343 treatment.
Discussion
Here, we report widespread changes in the muscle methylome associated with sarcopenia and individual measures of grip strength, lean mass, and gait speed, with significant overlap between the dmCpGs associated with sarcopenia and ALMi, sarcopenia and gait speed, and sarcopenia and grip strength. The dmCpGs associated with sarcopenia, and measures of muscle mass, strength, and function were enriched in genes involved in myotube fusion, oxidative phosphorylation, and voltage‐gated calcium channels. Moreover, examination of the chromatin landscape of the sarcopenia‐associated dmCpGs found that they were localized to EZH2 target genes and regions of H3K27 trimethylation. Furthermore, treatment of human primary myoblasts with a selective EZH2 inhibitor led to changes in oxidative phosphorylation and myotube fusion, together with altered methylation of genes involved in oxidative phosphorylation and myogenesis. Such data suggest that epigenetic changes may play an important role in muscle function and the aetiology of sarcopenia.
Significant changes in DNA methylation at both the single CpG and regional level were found to be associated with sarcopenia, many within intergenic enhancer regions. 31 Of the sarcopenia‐associated dmCpGs that were associated with a gene, cg22843429 was located within the EMG1 gene, an N1‐specific pseudouridine methyltransferase involved in ribosome biogenesis, while cg10977501 was located within MYOM2 (Myomesin 2), a major component of the myofibrillar M‐band of the sarcomere, suggesting that differential methylation at these sites may contribute to the impairments in muscle function during ageing. The most significant DMR associated with sarcopenia was located within the MCCC1 gene, a mitochondrial enzyme, which catalyses the carboxylation of 3‐methylcrotonyl CoA to 3‐methylglutaconyl CoA, a critical step in leucine catabolism. Mutations within this gene result in MCCC1 deficiency, which leads to metabolic acidosis, low plasma glucose and carnitine levels, and loss of muscle tone. 32 In this study, both differential methylation and expression of MCCC1 were observed in sarcopenic muscle, potentially impairing the availability of acetyl CoA and muscle metabolism. Interestingly, there was minimal overlap between the dmCpGs identified in this study (sarcopenia 8.5%, ALMi 5.6%, gait speed 4.3%, grip strength 12.2%) and those reported to differ in muscle tissue from young versus old individuals in the meta‐analysis by Voisin et al. 15 Consistent with this, sarcopenia and its component measures were not associated with accelerated epigenetic muscle ageing, suggesting that sarcopenia is not an accentuated form of muscle ageing but rather involves distinct epigenetic changes.
Comparison of the muscle transcriptome from older individuals with sarcopenia and healthy aged‐matched controls found that the major transcriptional signal associated with sarcopenia was mitochondrial dysfunction. 17 We found that oxidative phosphorylation was also one of the pathways enriched amongst the sarcopenia‐associated dmCpGs, suggesting that altered DNA methylation may mediate or consolidate the transcriptional changes observed in sarcopenic muscle. However, in the 34 samples with both RNAseq and methylation data, only 9.7% of the dmCpGs and 2.2% of the CpGs within the DMRs showed a significant correlation between DNA methylation and gene expression. This low level of correlation between DNA methylation and gene expression suggests that differential methylation of a number of CpGs may be required before there is a change in gene expression.
The sarcopenia‐associated, grip strength‐associated, and gait speed‐associated dmCpGs were also enriched in genes involved in homophilic cell–cell adhesion. Homophilic cell–cell adhesion plays a critical role in myotube fusion. 33 Differential methylation of genes involved in myotube fusion in muscle biopsies from individuals with sarcopenic may impair the satellite cell (SC) repair mechanisms by inhibiting the fusion of the newly formed myotubes with existing myofibres. There was also enrichment amongst the sarcopenia and ALMi‐associated dmCpGs of genes involved in voltage‐gated Ca2+ channels, which are essential for the depolarization‐induced contraction of muscle cells, with impairments in calcium signalling and depolarization being linked to muscle atrophy. 34 In muscle ageing studies, enrichment of homophilic cell adhesion 12 , 13 and calcium signalling 15 have also been reported amongst ageing‐associated dmCpGs, suggesting that although the dmCpGs associated with ageing and sarcopenia differ, there is overlap in the pathways affected.
The sarcopenia‐associated dmCpGs were enriched within EZH2 target genes and sites of H3K27 trimethylation. Hypermethylation of EZH2 target genes have also been reported across a number of tissue types during ageing. 15 , 35 Although in sarcopenia, there was both hypermethylation and hypomethylation of the dmCpGs enriched in EZH2 target genes, suggesting epigenetic dysregulation of the EZH2 pathway in sarcopenia, rather than over‐activity or under‐activity of the EZH2 pathway. To investigate the role of EZH2 in myogenic cells, human primary myoblasts were treated with GSK343, an EZH2 inhibitor. GSK343 treatment led to an increase in ATP production, as well as increases in the maximal and basal rates of respiration, suggesting that inhibition of EZH2 signalling, either directly or indirectly, alters muscle cell metabolism. As differentiation of myoblasts into mature myotubes requires a metabolic switch from a highly glycolytic state to rely on oxidative phosphorylation (OXPHOS), 36 increased oxidative phosphorylation might be expected to lead to an increase in differentiation and fusion index. However, treatment with GSK343 led to a decrease in myotube fusion with a concomitant increase in PAX7 expression. Studies have shown that while EZH2 plays a key role in the homeostasis of the adult muscle SC pool, upon SC differentiation, EZH2 re‐locates from myogenic genes, to repress self‐renewal genes such as PAX7 in differentiated myotubes. 37 Inhibition of EZH2 may result in the de‐repression of self‐renewal genes like PAX7 inhibiting muscle differentiation and myotube fusion in the differentiating myoblasts, despite the increased energetic capacity of the myoblasts.
The functional changes induced by GSK343 were accompanied by changes in the DNA methylation status of genes involved in OXPHOS and myogenesis, suggesting that these pathways are key targets of EZH2 regulation in primary myoblasts. Recent studies have demonstrated that EZH2 can recruit DNA methyltransferases to target genes, with EZH2 knockdown decreasing both H3K27 trimethylation and DNA methylation at these sites. 38 Consistent with these previous findings, GSK343 treatment of the myoblasts altered DNA methylation, with the majority of CpG sites being hypomethylated. Comparison of the sarcopenia‐associated dmCpGs with those altered by GSK343 treatment showed that the methylation of only seven sarcopenia‐associated dmCpGs were affected by GSK343 treatment. The epigenetic landscape however of muscle tissue compared with the cultured myoblasts is likely to differ markedly; GSK343 treatment of myoblasts did however lead to an increase in oxidative respiration, suggesting that it may be possible to reset the epigenetic regulation of oxidative phosphorylation to improve muscle bioenergetics though manipulation of these pathways.
There were limitations to this study. Firstly, we only measured DNA methylation in male participants of the HSS and HSSe, and further studies are required to determine whether similar epigenetic changes occur in females. Secondly, we found that the majority of methylation changes were located within intergenic regions, so further work will be required to determine whether these changes contribute to changes in transcription. Thirdly, although GSK343 is a highly selective inhibitor of EZH2, 30 confirmation that GSK343 treatment inhibited EZH2 activity is required together with the effect of EZH2 on the expression of myogenic and metabolic genes to precisely map the role that EZH2 plays in myotube fusion and mitochondrial bioenergetics. Finally, we tested the role of EZH2 in a myogenic enriched population of cells isolated from muscle biopsies; it would also be highly informative to assess the role of EZH2 in all muscle‐derived cells, which might be more representative of the aged muscle niche, as well as from a larger number of individuals to specifically test whether GSK3343 can alleviate the oxidative phosphorylation impairment observed in sarcopenic muscle.
Conclusions
These findings show widespread changes in the muscle methylome associated with sarcopenia and individual measures of grip strength, lean mass, and gait speed. Furthermore, we show that treatment of human primary myoblasts with an inhibitor of EZH2 led to changes in oxidative phosphorylation and myotube fusion, together with altered DNA methylation of genes involved in oxidative phosphorylation and myogenesis. These findings support the premise that epigenetic processes play a central role in muscle function and suggest that it may be possible to ameliorate impairments in ATP production observed in muscle cells from individuals with sarcopenia though epigenetic manipulation of these pathways.
Conflict of interest
A. Taddei and P. Richardson are full‐time employees of BenevolentAI Ltd. J.D. Holbrook is a full‐time employee of Cambridge Epigenetix. E. Migliavacca and J.N. Feige are full‐time employees of the Société des Produits Nestlé (SPN). K.M. Godfrey and H.P. Patel have received reimbursement for speaking at conferences sponsored by companies selling nutritional products. C. Cooper has received consultancy fees and honoraria from Amgen, Danone, Eli Lilly, GlaxoSmithKline, Medtronic, Merck, Nestlé, Novartis, Pfizer, Roche, Servier, Shire, Takeda, and UCB. E. Antoun, E.S. Garratt, M.A. Burton, S.J. Barton, P. Titcombe, K.M. Godfrey, and K.A. Lillycrop are part of academic research programmes that have received research funding from Abbott Nutrition, Nestec, and Danone. L.D. Westbury, A. Baczynska, H.E. Sydall, E. Dennison, R. Dodds, H.C. Roberts, A.A. Sayer, and S. Shaw declare that they have no conflicts of interest.
Funding
This work was supported by collaborative projects with BenevolentAI Ltd and the Nestlé Institute of Health Sciences (NIHS) and grant funding from the Medical Research Council (MC_U47585827, MC_ST_U2055), Arthritis Research UK, National Osteoporosis Society, International Osteoporosis Foundation, Cohen Trust, NIHR Southampton Biomedical Research Centre, University of Southampton and University Hospital Southampton NHS Foundation Trust, NIHR Musculoskeletal Biomedical Research Unit, and University of Oxford. K.M.G. is supported by the UK Medical Research Council (MC_UU_20/4), the US National Institute On Aging of the National Institutes of Health (award number U24AG047867), the UK Economic and Social Research Council and the Biotechnology and Biological Sciences Research Council (award number ES/M0099X/), the National Institute for Health Research (as an NIHR Senior Investigator (NF‐SI‐055‐0042), and through the NIHR Southampton Biomedical Research Centre, and the European Union's Erasmus + Capacity‐Building ImpENSA Project. H.P.P. is supported by the National Institute for Health Research through the NIHR Southampton Biomedical Research Centre. This report is independent research, and the views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health. The grant funders had no role in the design, collection, analysis, and interpretation of data, writing of the paper, or decision to submit for publication.
Supporting information
Data S1. Supporting information
Figure S1: PCA of the top 50,000 most variable probes on the array to determine whether the samples group based on cohort as the DNA extraction method differed between the 2 cohorts. As there was no clear separation between the two, DNA extraction method was not accounted for in the analysis.
Figure S2: Correlation between epigenetic age as estimated by the muscle epigenetic age estimator MEAT, and chronological age.
Figure S3: (A) Protein–protein interaction (PPI) network generated from the genes associated with a CpG with an FDR < 0.2 with respect to sarcopenia. (B‐C) The PPI network was subdivided into smaller modules using the MCODE algorithm, of which 2 modules are shown.
Figure S4: PPI network of dmCpGs (A) associated with ALMI. Networks were further subdivided using the MCODE algorithm with two modules associated with the ALMI dmCpGs (B + C) shown.
Figure S5: (A) Enrichment of sarcopenia‐associated dmCpGs amongst 15 chromatin states as designated by the Epigenome Roadmap Project in male human skeletal muscle tissue samples. Odds ratio and significance calculated using the Fisher exact test. Heatmap shows the enrichment of the different histone modifications amongst the CpGs in the different chromatin states. (B) Enrichment of sarcopenia‐associated dmCpGs amongst six histone modifications as reported by ENCODE.
Table S1: List of Pyrosequencing primers
Table S2: List of Sarcopenia‐associated dmCpGs
Table S3: List of ALMi‐associated dmCpGs
Table S4: List of gait speed‐associated dmCpGs
Table S5: List of grip strength‐associated dmCpGs
Table S6: Overlap of the differentially methylated CpGs with respect to Sarcopenia, ALMi, Grip strength and Gait speed
Table S7: Association between sarcopenia, muscle mass, strength and function and muscle epigenetic age acceleration
Table S8: List of DMRs associated with sarcopenia
Table S9: List of DMRs associated with ALMi
Table S10: List of DMRs associated with grip strength
Table S11: List of DMRs associated with gait speed
Table S12: Significant GO terms associated with the Sarcopenia PPI network
Table S13: Top 10 GO terms associated with the top 2 sarcopenia‐associated clusters
Table S14: Top 10 GO terms associated with the top 2 clusters associated with the ALMI dmCpGs
Table S15: List of dmCpGs (p < 0.05) following GSK343 treatment after 10 days of differentiation of human primary myoblasts
Acknowledgements
The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. 39
Antoun E., Garratt E. S., Taddei A., Burton M. A., Barton S. J., Titcombe P., Westbury L. D., Baczynska A., Migliavacca E., Feige J. N., Sydall H. E., Dennison E., Dodds R., Roberts H. C., Richardson P., Sayer A. A., Shaw S., Cooper C., Holbrook J. D., Patel H. P., Godfrey K. M., Lillycrop K. A., and the EpiGen Global Research Consortium (2022) Epigenome‐wide association study of sarcopenia: findings from the Hertfordshire Sarcopenia Study (HSS), Journal of Cachexia, Sarcopenia and Muscle, 13, 240–253, 10.1002/jcsm.12876
References
- 1. Studenski SA, Peters KW, Alley DE, Cawthon PM, McLean RR, Harris TB, et al. The FNIH sarcopenia project: rationale, study description, conference recommendations, and final estimates. J Gerontol A Biol Sci Med Sci 2014;69:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen LK, Liu LK, Woo J, Assantachai P, Auyeung TW, Bahyah KS, et al. Sarcopenia in Asia: consensus report of the Asian Working Group for Sarcopenia. J Am Med Dir Assoc 2014;15:95–101. [DOI] [PubMed] [Google Scholar]
- 3. Cruz‐Jentoft AJ, Baeyens JP, Bauer JM, Boirie Y, Cederholm T, Landi F, et al. Sarcopenia: European consensus on definition and diagnosis: report of the European Working Group on Sarcopenia in Older People. Age Ageing 2010;39:412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cruz‐Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyère O, Cederholm T, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2019;48:16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cruz‐Jentoft AJ, Sayer AA. Sarcopenia. Lancet 2019;393:2636–2646. [DOI] [PubMed] [Google Scholar]
- 6. Cruz‐Jentoft AJ, Landi F, Schneider SM, Zúñiga C, Arai H, Boirie Y, et al. Prevalence of and interventions for sarcopenia in ageing adults: a systematic review. Report of the International Sarcopenia Initiative (EWGSOP and IWGS). Age Ageing 2014;43:748–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zillikens MC, Demissie S, Hsu YH, Yerges‐Armstrong LM, Chou WC, Stolk L, et al. Large meta‐analysis of genome‐wide association studies identifies five loci for lean body mass. Nat Commun 2017;8:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matteini AM, Tanaka T, Karasik D, Atzmon G, Chou WC, Eicher JD, et al. GWAS analysis of handgrip and lower body strength in older adults in the CHARGE consortium. Aging Cell 2016;15:792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD, Hanson MA. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr Res 2007;61:5R–10R. [DOI] [PubMed] [Google Scholar]
- 10. Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med 2018;378:1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teh AL, Pan H, Chen L, Ong ML, Dogra S, Wong J, et al. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res 2014;24:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turner DC, Gorski PP, Maasar MF, Seaborne RA, Baumert P, Brown AD, et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle‐derived cells: the role of HOX genes and physical activity. Sci Rep 2020;10:15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zykovich A, Hubbard A, Flynn JM, Tarnopolsky M, Fraga MF, Kerksick C, et al. Genome‐wide DNA methylation changes with age in disease‐free human skeletal muscle. Aging Cell 2014;13:360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voisin S, Harvey NR, Haupt LM, Griffiths LR, Ashton KJ, Coffey VG, et al. An epigenetic clock for human skeletal muscle. J Cachexia Sarcopenia Muscle 2020;11:887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voisin S, Jacques M, Landen S, Harvey NR, Haupt LM, Griffiths LR, et al. Meta‐analysis of genome‐wide DNA methylation and integrative omics of age in human skeletal muscle. J Cachexia Sarcopenia Muscle 2021;12:1064–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Day K, Waite LL, Thalacker‐Mercer A, West A, Bamman MM, Brooks JD, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biol 2013;14:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Migliavacca E, Tay SK, Patel HP, Sonntag T, Civiletto G, McFarlane C, et al. Mitochondrial oxidative capacity and NAD(+) biosynthesis are reduced in human sarcopenia across ethnicities. Nat Commun 2019;10:5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Syddall HE, Aihie Sayer A, Dennison EM, Martin HJ, Barker DJ, Cooper C. Cohort profile: the Hertfordshire cohort study. Int J Epidemiol 2005;34:1234–1242. [DOI] [PubMed] [Google Scholar]
- 19. Patel HP, Syddall HE, Martin HJ, Stewart CE, Cooper C, Sayer AA. Hertfordshire Sarcopenia Study: design and methods. BMC Geriatr 2010;10:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Syddall HE, Simmonds SJ, Carter SA, Robinson SM, Dennison EM, Cooper C, et al. The Hertfordshire Cohort Study: an overview. F1000Res 2019;8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Westbury LD, Dodds RM, Syddall HE, Baczynska AM, Shaw SC, Dennison EM, et al. Associations between objectively measured physical activity, body composition and sarcopenia: findings from the Hertfordshire Sarcopenia Study (HSS). Calcif Tissue Int 2018;103:237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roberts HC, Denison HJ, Martin HJ, Patel HP, Syddall H, Cooper C, et al. A review of the measurement of grip strength in clinical and epidemiological studies: towards a standardised approach. Age Ageing 2011;40:423–429. [DOI] [PubMed] [Google Scholar]
- 23. Patel H, Syddall HE, Martin HJ, Cooper C, Stewart C, Sayer AA. The feasibility and acceptability of muscle biopsy in epidemiological studies: findings from the Hertfordshire Sarcopenia Study (HSS). J Nutr Health Aging 2011;15:10–15. [DOI] [PubMed] [Google Scholar]
- 24. Aljanabi SM, Martinez I. Universal and rapid salt‐extraction of high quality genomic DNA for PCR‐based techniques. Nucleic Acids Res 1997;25:4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aryee MJ, Jaffe AE, Corrada‐Bravo H, Ladd‐Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 2003;4:1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21:3448–3449. [DOI] [PubMed] [Google Scholar]
- 29. Agley CC, Rowlerson AM, Velloso CP, Lazarus NL, Harridge SD. Isolation and quantitative immunocytochemical characterization of primary myogenic cells and fibroblasts from human skeletal muscle. J Vis Exp 2015;e52049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, et al. Identification of potent, selective, cell‐active inhibitors of the histone lysine methyltransferase EZH2. ACS Med Chem Lett 2012;3:1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grunert SC, Stucki M, Morscher RJ, Suormala T, Bürer C, Burda P, et al. 3‐Methylcrotonyl‐CoA carboxylase deficiency: clinical, biochemical, enzymatic and molecular studies in 88 individuals. Orphanet J Rare Dis 2012;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ozawa M. E‐cadherin cytoplasmic domain inhibits cell surface localization of endogenous cadherins and fusion of C2C12 myoblasts. Biol Open 2015;4:1427–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Drey M, Krieger B, Sieber CC, Bauer JM, Hettwer S, Bertsch T, et al. Motoneuron loss is associated with sarcopenia. J Am Med Dir Assoc 2014;15:435–439. [DOI] [PubMed] [Google Scholar]
- 35. Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, et al. Human aging‐associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 2010;20:434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sin J, Andres AM, Taylor DJ, Weston T, Hiraumi Y, Stotland A, et al. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy 2016;12:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, et al. TNF/p38α/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 2010;7:455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ning X, Shi Z, Liu X, Zhang A, Han L, Jiang K, et al. DNMT1 and EZH2 mediated methylation silences the microRNA‐200b/a/429 gene and promotes tumor progression. Cancer Lett 2015;359:198–205. [DOI] [PubMed] [Google Scholar]
- 39. von Haehling S, Ebner N, Anker SD. The Journal of Cachexia, Sarcopenia and Muscle in 2019. J Cachexia Sarcopenia Muscle 2019;10:715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting information
Figure S1: PCA of the top 50,000 most variable probes on the array to determine whether the samples group based on cohort as the DNA extraction method differed between the 2 cohorts. As there was no clear separation between the two, DNA extraction method was not accounted for in the analysis.
Figure S2: Correlation between epigenetic age as estimated by the muscle epigenetic age estimator MEAT, and chronological age.
Figure S3: (A) Protein–protein interaction (PPI) network generated from the genes associated with a CpG with an FDR < 0.2 with respect to sarcopenia. (B‐C) The PPI network was subdivided into smaller modules using the MCODE algorithm, of which 2 modules are shown.
Figure S4: PPI network of dmCpGs (A) associated with ALMI. Networks were further subdivided using the MCODE algorithm with two modules associated with the ALMI dmCpGs (B + C) shown.
Figure S5: (A) Enrichment of sarcopenia‐associated dmCpGs amongst 15 chromatin states as designated by the Epigenome Roadmap Project in male human skeletal muscle tissue samples. Odds ratio and significance calculated using the Fisher exact test. Heatmap shows the enrichment of the different histone modifications amongst the CpGs in the different chromatin states. (B) Enrichment of sarcopenia‐associated dmCpGs amongst six histone modifications as reported by ENCODE.
Table S1: List of Pyrosequencing primers
Table S2: List of Sarcopenia‐associated dmCpGs
Table S3: List of ALMi‐associated dmCpGs
Table S4: List of gait speed‐associated dmCpGs
Table S5: List of grip strength‐associated dmCpGs
Table S6: Overlap of the differentially methylated CpGs with respect to Sarcopenia, ALMi, Grip strength and Gait speed
Table S7: Association between sarcopenia, muscle mass, strength and function and muscle epigenetic age acceleration
Table S8: List of DMRs associated with sarcopenia
Table S9: List of DMRs associated with ALMi
Table S10: List of DMRs associated with grip strength
Table S11: List of DMRs associated with gait speed
Table S12: Significant GO terms associated with the Sarcopenia PPI network
Table S13: Top 10 GO terms associated with the top 2 sarcopenia‐associated clusters
Table S14: Top 10 GO terms associated with the top 2 clusters associated with the ALMI dmCpGs
Table S15: List of dmCpGs (p < 0.05) following GSK343 treatment after 10 days of differentiation of human primary myoblasts


