Abstract
Background
Age‐related muscle dysfunctions are common disorders resulting in poor quality of life in the elderly. Probiotic supplementation is a potential strategy for preventing age‐related sarcopenia as evidence suggests that probiotics can enhance muscle function via the gut–muscle axis. However, the effects and mechanisms of probiotics in age‐related sarcopenia are currently unknown. In this study, we examined the effects of Lactobacillus casei Shirota (LcS), a probiotic previously reported to improve muscle function in young adult mice.
Methods
We administered LcS (1 × 108 or 1 × 109 CFU/mouse/day) by oral gavage to senescence‐accelerated mouse prone‐8 mice for 12 weeks (16‐ to 28‐week‐old). Sixteen‐week‐old and 28‐week‐old SMAP8 mice were included as non‐aged and aged controls, respectively. Muscle condition was evaluated using dual‐energy X‐ray absorptiometry for muscle mass, holding impulse and grip strength tests for muscle strength, and oxygen consumption rate, gene expressions of mitochondrial biogenesis, and mitochondrial number assays for mitochondria function. Inflammatory cytokines were determined using enzyme‐linked immunosorbent assay. Gas chromatography–mass spectrometry was utilized to measure the short‐chain fatty acid levels. The gut microbiota was analysed based on the data of 16S rRNA gene sequencing of mouse stool.
Results
The LcS supplementation reduced age‐related declines in muscle mass (>94.6%, P < 0.04), strength (>66% in holding impulse and >96.3% in grip strength, P < 0.05), and mitochondrial function (P < 0.05). The concentration of short‐chain fatty acids (acetic, isobutyric, butyric, penic, and hexanoic acid) was recovered by LcS (>65.9% in the mice given high dose of LcS, P < 0.05) in the aged mice, and LcS attenuated age‐related increases in inflammation (P < 0.05) and reactive oxygen species (>89.4%, P < 0.001). The high dose of LcS supplementation was also associated with distinct microbiota composition as indicated by the separation of groups in the beta‐diversity analysis (P = 0.027). LcS supplementation altered predicted bacterial functions based on the gut microbiota. Apoptosis (P = 0.026), p53 signalling (P = 0.017), and non‐homologous end‐joining (P = 0.031) were significantly reduced, whereas DNA repair and recombination proteins (P = 0.043), RNA polymerase (P = 0.008), and aminoacyl‐tRNA biosynthesis (P = 0.003) were increased. Finally, the genera enriched by high‐dose LcS [linear discriminant analysis (LDA) score > 2.0] were positively correlated with healthy muscle and physiological condition (P < 0.05), while the genera enriched in aged control mice (LDA score > 2.0) were negatively associated with healthy muscle and physiological condition (P < 0.05).
Conclusions
Lactobacillus casei Shirota represents an active modulator that regulates the onset and progression of age‐related muscle impairment potentially via the gut–muscle axis.
Keywords: Probiotics, Gut–muscle axis, Age‐related sarcopenia, Gut microbiota, Short‐chain fatty acid
Introduction
The issue of aging and its associated diseases is becoming increasingly important worldwide. Sarcopenia is a disease that frequently occurs in older adults and is characterized by the loss of muscle mass and function. Additionally, it increases the risk of poor quality of life due to its negative outcomes, such as falls, disability, fractures, and mortality. Therefore, maintaining muscle function is an important component of healthy aging. 1
Age‐related sarcopenia can be induced through a cycle of inflammation, oxidative stress, and mitochondrial dysfunction. 2 In the cycle, age‐related inflammation results in mitochondrial damage and further increases in the level of reactive oxygen species (ROS) and pro‐inflammation. 2 Recently, the gut microbiota has been suggested to be involved in inducing the cycle of age‐related sarcopenia due to a novel concept referred to as the gut–muscle axis. 3 The gut–muscle axis describes how the gut microbiota impacts muscle mass and function by influencing inflammation and levels of short‐chain fatty acids (SCFAs). 3 Given that gut microbiota dysbiosis frequently occurs in older individuals, triggering inflammation and anabolic resistance, age‐related alternations in gut microbiota composition may contribute to sarcopenia in older adults via the gut–muscle axis. 3 Indeed, an increasing number of studies provide evidence that muscle function can be modulated by gut microbiota. 4 Lahiri et al. 5 demonstrated the existence of a gut microbiota‐skeletal muscle axis by showing that the skeletal muscle of germ‐free mice exhibited atrophy that could be reversed by applying microbial metabolites. Moreover, a clinical study of post‐transplant patients with haematological malignancies also indicated that the gut microbiota seemed to improve skeletal muscle via the gut–muscle axis. 6 However, a question remained as to how to improve sarcopenia via the gut–muscle axis.
Oral supplementation of probiotics is a potential strategy to promote gut–muscle axis‐induced prevention of sarcopenia, since many studies have demonstrated that probiotics can regulate gut microbiota, produce SCFAs, and induce anti‐ROS and anti‐inflammatory effects. For example, Munukka et al. 7 revealed that the administration of SCFA‐producing probiotics modulated the gut microbiota composition, reduced systemic inflammation, and increased muscle mass in high‐fat fed mice. Additionally, Lactobacillus helveticus has been shown to decrease age‐related oxidative stress and alternations in the gut microbiota composition in a mouse model. 8 A mixed supplement of multiple Lactobacilli probiotics also reduced inflammation and muscle atrophy markers. 9 Moreover, one of the most well‐known probiotics, Lactobacillus casei Shirota (LcS), has significant anti‐ROS and anti‐inflammatory effects in numerous mouse and human studies. 10 , 11 , 12 Recently, LcS was linked to the induction of healthy gut microbiota, SCFA, and muscle. 10 , 13 , 14 Although these studies were not performed in aged subjects, they provide a basis for our hypothesis that supplemental probiotics may help protect against age‐related sarcopenia.
In the present study, we selected LcS as the probiotic to test this hypothesis, because LcS has been used as a probiotic since 1930 with several studies related to human health. 10 , 11 , 12 , 13 , 14 LcS was administered over 12 weeks to pre‐aged and aged senescence‐accelerated mouse‐prone 8 (SAMP8) mice, which are used as an animal model of age‐related muscle impairment. 15 The effect of LcS and its underlying mechanisms were evaluated on the basis of muscle and mitochondrial conditions, inflammatory cytokines, ROS, SCFA, and gut microbiota.
Methods
Animals and Lactobacillus casei Shirota
SAMP8 mice were provided by Prof Ming‐Fu Wang (Providence University, Taiwan) and housed under a 12/12 h light/dark cycle at 22–24°C and 40–60% humidity. A commercially diet (local supplier) and sterile water were provided to mice ad libitum. LcS was purchased from Yakult Co., Ltd., Taiwan, and administered to mice at 1 × 108 and 1 × 109 CFU/200 μL after it was subcultured.
Twenty‐four mice were randomly and equally divided in to four groups, namely, NA (16‐week‐old non‐aged controls), A (aged mice administered saline), S1X (aged mice administered low‐dose 1 × 108 CFU LcS/mouse/day), and S10X (aged mice administered high‐dose 1 × 109 CFU LcS/mouse/day). The dose of LcS was based on the previous study of LcS regarding anti‐inflammation. 16 The sample size was based on previous studies regarding the composition of gut microbiota in mice. 17
The NA group was humanely sacrificed at 16 weeks old, and the other groups were sacrificed after 12 weeks of treatment at 28‐weeks‐old. The in vivo trials were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of Shih Chien University (IACUC‐10509).
Appearance of senescence
The degree of apparent senescence was assessed based on a graded scoring system described previously. 18 Briefly, senescence related to reactivity, passivity, skin and hair, eyes, and spine was classified into 11 categories. Scores in each category ranged from 0 to 4 as rated by four well‐trained investigators. Mice with more serious senescence received higher scores. The criteria for the graded scoring system are listed in Supporting Information, Table S1.
Body composition
Body composition was determined using dual‐energy X‐ray absorptiometry (DEXA) scans collected on a GE Lunar PIXImus2 densiometer (GE, Madison, WI, USA) three days before sacrifice. The data were analysed using software included by the manufacturer.
Muscle strength
Muscle strength was evaluated using the four‐limb hanging and grip strength tests. The procedure for the tests followed the description in a previous study. 19 The results are represented as holding impulse (weight (g) × duration (s) of holding) and grip force (gm f), respectively.
Oxygen consumption rates
Oxygen consumption rates (OCRs) were detected by an XF24 Extracellular Flux analyser (Seahorse Bioscience, North Billerica, MA, USA). Briefly, the gastrocnemius was homogenized using a Polytron homogenizer in ice‐cold mitochondrial assay solution (MAS) buffer (115 mM KCl, 10 mM KH2PO4, 2 mM MgCl2, 3 mM HEPES, and 1 mM EGTA) containing 0.2% fatty acid‐free bovine serum albumin. Nuclei and cell debris were removed by centrifugation at 700g for 10 min followed by centrifugation at 8000g for 10 min to collect the mitochondria. The mitochondria pellets were re‐suspended in MAS, and the isolated mitochondria were plated in XF24 V7 microplates (Seahorse Bioscience) at a concentration of 10 μg of protein per well. After centrifugation at 2200g for 20 min at 4°C, 625 μL of MAS (containing 5 mM glutamate, 5 mM malate, and 5 mM succinate) was added to each well at 37°C. The XF24 plate was transferred to an XF24 Extracellular Flux analyser at 37°C and equilibrated for 10 min. Subsequently, ADP (3.2 μM), oligomycin (4 μM), FCCP (4 μM), and a cocktail of rotenone (4 μM) and antimycin A (2 μM) were injected into the assay plate, sequentially. All reagents in the assay were purchased from Sigma‐Aldrich (Saint Louis, MI, USA).
Gene expression levels
RNA was extracted from tissues using RNeasy Mini kit (Qiagen, Hilden, Germany). RNA was reverse‐transcribed using an iScript cDNA Synthesis Kit (Bio‐Rad, Hercules, CA, USA), following the manufacturer's instructions. We performed quantitative PCR in a MyiQ Single‐Color Real‐Time PCR Detection System (Bio‐Rad). The primers for the genes (Purigo, Taipei, Taiwan) are listed in Table S2. mRNA levels of the genes were normalized to β‐actin.
Mitochondrial DNA copy number
Total DNA was isolated from tissues using DNeasy Blood & Tissue Kits (Qiagen). Quantitative PCR was performed in a MyiQ Single‐Color Real‐Time PCR Detection System (Bio‐Rad). The level of the COXII gene was normalized to an 18S rRNA gene to evaluate mitochondrial quantity. The primers for the genes (Purigo) are listed in Table S2.
Plasma cytokines
The level of inflammation‐related cytokines in serum was determined using the Mouse TNF‐alpha ELISA MAX Standard and ELISA MAX™ Standard Set Mouse IL‐10 (Biolegend, San Diego, CA, USA) according to the manufacturer's instructions. Absorbance was read at 450 nm using an ELISA reader (BioTek, Winooski, VT, USA).
Protein carbonyl content
The concentration of protein carbonyls was determined using a protein carbonyl assay kit (Cayman Chemicals, Ann Arbor, MI, USA) according to the manual. Briefly, 200 mg of gastrocnemius was homogenized in pH 6.7 phosphate buffer, and then debris was removed by centrifugation at 10 000g for 15 min at 4°C. The sample was incubated with dinitrophenyl‐hydrazine for 1 h at RT. Then, the protein was precipitated with trichloroacetic acid and resuspended in guanidine hydrochloride. After centrifugation at 10 000g for 10 min at 4°C, the absorbance of the supernatant was measured at 370 nm using an ELISA reader (BioTek).
Determination of short‐chain fatty acid levels
A 300 ± 1 mg stool sample was mixed with 1.2 mL of 1 M HCL and sonicated for 10 min in an ice water bath. After centrifugation at 2300 g for 20 min at 4°C, the supernatant was collected and extracted with 1 mL ethyl acetate for 3 min and centrifuged at 13 200 g for 15 min. Finally, the supernatant was filtered using a 0.22 μm membrane and injected into GC–MS. GC–MS analysis was performed using an Agilent 7890 gas chromatograph system coupled with an Agilent 5975C mass spectrometer. The system utilized a HP‐FFAP capillary column. A 1 μL aliquot of the analyte was injected in split mode (5:1). Helium was used as the carrier gas, the front inlet purge flow was 3 mL/min, and the gas flow rate through the column was 1 mL/min. The initial temperature was kept at 100°C for 1 min, then raised to 150°C at a rate of 5°C per minute, then raised to 200°C at a rate of 10°C per minute, then kept for 8 min at 240°C at a rate of 40°C per minute. The injection, transfer line, quad, and ion source temperatures were 250°C, 260°C, 150°C, and 230°C, respectively. The energy was −70 eV in electron impact mode. The mass spectrometry data were acquired in full‐scan mode with the m/z range of 20–350 after a solvent delay of 3 min.
16S rRNA gene sequencing and data analysis
The procedure for the analysis followed the description in our previous study. 20 Briefly, the V3–V4 region of the 16S rRNA gene was amplified by PCR with the primers 341 F (5′‐CCTAYGGGRBGCASCAG‐3′) and 806 R (5′‐GGACTACNNGGGTATCTAAT‐3′) to develop the amplicon libraries following the Illumina 16S Metagenomic Sequencing Library Preparation manual protocol. The amplicons were paired‐end sequenced (2 × 250) using an Illumina HiSeq 2000 platform according to the manufacturer's protocol. The paired forward and reverse reads that passed quality control were merged and mapped to the SLIVA database to construct operational taxonomic units at 97% identity through the UPARSE pipeline (drive5, Tiburon, California).
Data analysis was performed using Quantitative Insights Into Microbial Ecology, and chimeric sequences were removed using ChimeraSlayer. Sequences with ≥97% similarity were assigned to the same operational taxonomic unit. Alpha diversity analysis (Shannon and Simpson index) was assessed using Quantitative Insights Into Microbial Ecology. Beta diversity was analysed using principal coordinate analysis, and statistical significance of beta diversity was analysed by PERMANOVA. Linear discriminant analysis (LDA) effect size (LEfSe) was performed online using the Galaxy workflow framework.
Statistical analyses
Data are presented as the mean ± standard error of the mean. Bacterial data were analysed using non‐parametric one‐way analysis of variance and Dunn post hoc, and the other data were analysed using one‐way analysis of variance with a Tukey's honestly significant difference post hoc test. Spearman's correlation coefficient was used to evaluate the relationship between the bacteria and physiological parameters and between the bacteria and level of SCFA. A P value < 0.05 was considered statistically significant.
Results
Lactobacillus casei Shirota delays the appearance of senescence and age‐related muscle mass deposition
The SAMP8 mouse is used as an animal model in many age‐related studies because it begins aging at 4 months old. 18 , 19 In the present study, food intake (Figure 1A) and body weight (Figure 1B) were stable during the experiment and were not different between the groups. The senescence score in the A group was approximately four times higher than the NA group (Figure 1C). Moreover, the muscle and fat properties were nearly 40% lower and 50% higher in the A compared with the NA group, respectively (Figure 1D). Thus, we first confirmed that the appearance of senescence and age‐related loss in muscle mass occurred in 16‐week‐old to 28‐week‐old SAMP8 mice. Since aging presented in the SAMP8 mice, the effect of LcS on the appearance of senescence and age‐related muscle mass loss was investigated in this mouse model. The results showed that LcS supplementation decreased the senescence scores and increased muscle mass in aged SAMP8 mice (Figure 1C,D). LcS also lowered the age‐related increase in fat mass (Figure 1D). The statistical powers of senescence score and fat and muscle properties were 1.00, 0.94, and 0.93, respectively.
Figure 1.
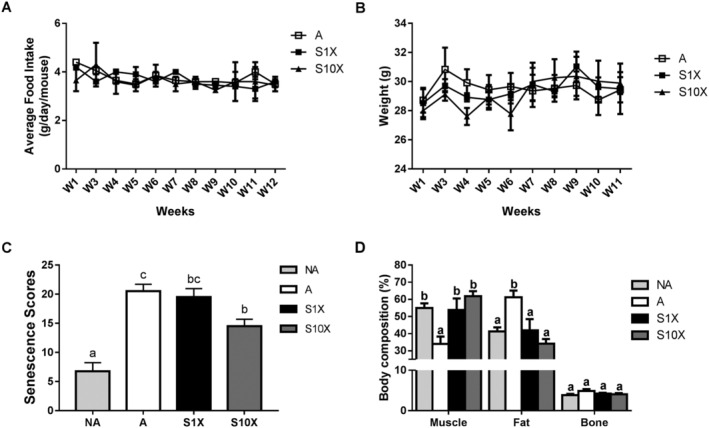
Food intake, body weight, senescence scores, and body composition. Average food intake (A) and weight (B) were measured from Weeks 1 to 12. Senescence scores (C) and body composition (D) were measured in Week 12. Different superscript letters (a, b, & c) differ significantly (P < 0.05) according to one‐way analysis of variance with Tukey's honestly significant difference post hoc test. n = 6.
Age‐related decline in muscle strength was improved by Lactobacillus casei Shirota
In addition to muscle mass loss, weaker muscle strength is also an important feature of age‐related muscle disorders. Therefore, we conducted the holding impulse and grip force assays to evaluate muscle strength. Similar to muscle mass, the lower holding impulse (Figure 2A) and grip force (Figure 2B) in the A group compared with the NA group indicated that aging significantly reduced muscle strength in this study. In the aged mouse groups (A, S1X, and S10X), the holding impulse was highest in the S10X group followed by the S1X and A groups, respectively (Figure 2A). Grip force was also higher in the S1X and S10X groups compared with the A group (Figure 2B). The higher holding impulse and grip force in LcS‐treated SAMP8 mice indicated that LcS maintained muscle strength in the aged mice. The statistical powers of holding impulse and grip force were 1.00 and 0.79, respectively.
Figure 2.
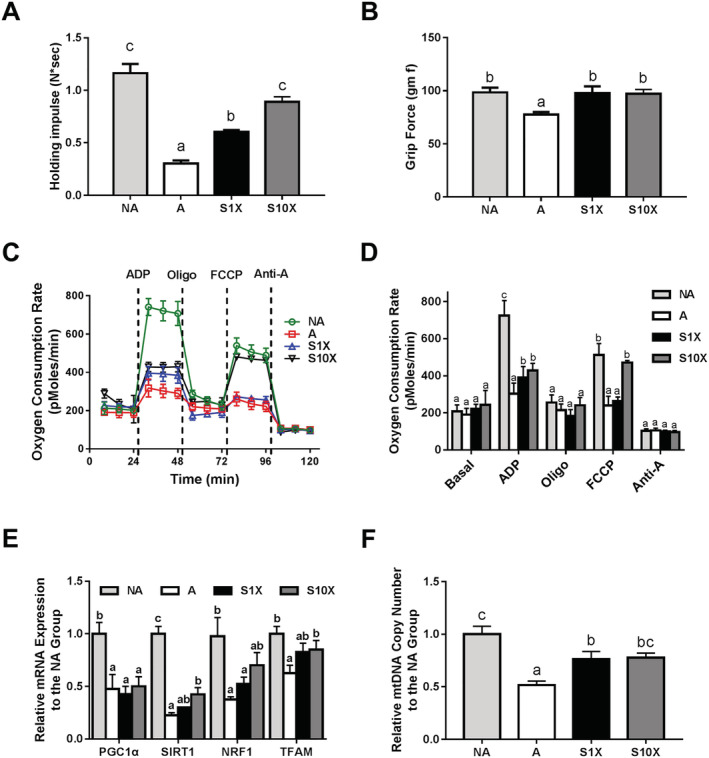
Muscle strength and mitochondria function. (A) holding impulse and (B) grip force to evaluate the muscle strength of SAMP8 mice; (C) graphical description of cellular respiration; (D) average oxygen consumption rate in each step of real‐time respirometer analysis performed with XF24 analyser; (E) expression of mitochondrial biogenesis genes; and (F) mtDNA copy number. Different superscript letters (a, b, & c) differ significantly (P < 0.05) according to one‐way analysis of variance with Tukey's honestly significant difference post hoc test. n = 6.
Lactobacillus casei Shirota attenuated age‐related mitochondrial disorders
Since age‐related muscle loss and weakness could be caused by mitochondrial dysfunction, we further investigated mitochondrial function in the muscle of SAMP8 mice. OCRs were measured using the Seahorse XFe analyser (Figure 2C,D). The A group had lower OCR values than the NA group in the stages after ADP and FCCP injection (0.42 and 0.47, respectively). In these two stages, the OCR values of the S10X group were 1.41 and 1.96 times higher than the A group (Figure 2D). The results show that LcS reduced decreases in phosphorylating respiration induced by ADP and maximal respiration in response to uncoupling by FCCP. Therefore, age‐related mitochondrial dysfunction could be reduced by LcS supplementation. The statistical powers of OCR values of post‐ADP and post‐FCCP were 1.00. Mitochondrial dysfunction and aging are typically associated with dysregulation of mitochondrial biogenesis. Therefore, we measured the expression of PGC1α, SIRT1, NRF1, and TFAM, which are involved in mitochondrial biogenesis. The results showed that these genes were significantly down‐regulated in the A group compared with the NA group (Figure 2E). Although the levels of PGC1α were not different between the A and LcS‐treated groups, the levels of SIRT1, NRF1, and TFAM were significantly up‐regulated in the S10X group compared with the A group. Slightly higher levels of SIRT1, NRF1, and TFAM were also observed in the S1X group. Because the mitochondrial copy number is another marker for assessing mitochondrial disorders, we also determined the mitochondrial copy number by measuring the ratio of mitochondrial and nuclear DNA copy numbers. The mitochondrial copy number of the A group was 51.7% that of the NA group. Moreover, mice given LcS at both the high and low dose had a significantly higher mitochondrial copy number than aged control mice (A group) (Figure 2F). Taken together, LcS prevented age‐related mitochondrial dysfunction in muscle, especially in mice given high‐dose LcS. The statistical powers of the levels of PGC1α, SIRT1, NRF1, and TFAM, and mitochondria copy number were 0.89, 1.00, 0.84, 0.68, and 1.00, respectively.
Age‐related alternations in ROS and inflammation‐related cytokines were attenuated by LcS
Age‐relate sarcopenia can be derived from a cycle of age‐related inflammation, mitochondrial dysfunction, and increases in ROS. Because LcS was able to reduce age‐related muscle decline and mitochondrial dysfunction, we measured the levels of inflammation‐related cytokines and ROS to evaluate whether LcS prevented sarcopenia by disrupting this cycle. The results showed that the proinflammatory cytokine, TNF‐α (Figure 3A), and the anti‐inflammatory cytokine, IL‐10 (Figure 3B), were up‐regulated and down‐regulated, respectively, in the muscle of the A group compared with the NA group. Age‐related changes in TNF‐α and IL‐10 in the muscle were reduced by LcS (Figure 3A,B). The similar pattern of TNF‐α and IL‐10 expression was also observed in the serum samples from the four groups (Figure 3C,D). The statistical powers of the levels of serum TNF‐α and IL‐10 and muscle TNF‐α and IL‐10 were all 1.00. Another component of the sarcopenia‐inducing cycle, ROS, was measured. ROS levels were approximately seven times higher in the A group compared with the NA group (Figure 3E). The levels of ROS in the S1X and S10X groups were about one‐fifth of the A group (Figure 3E). Therefore, LcS significantly decreased the age‐related increases in inflammation and ROS in the aged mice. The statistical power of the level of ROS was 1.00.
Figure 3.

Levels of inflammation‐related cytokines in serum and muscle and reactive oxygen species in muscle. (A, B) The relative expression of TNF‐α (A) and IL‐10 (B) in muscle; (C, D) the serum levels of TNF‐α (C) and IL‐10 (D); and the level of reactive oxygen species in muscle. Different superscript letters (a, b, & c) differ significantly (P < 0.05) according to one‐way analysis of variance with Tukey's honestly significant difference post hoc. n = 6.
Lactobacillus casei Shirota diminished age‐related alternations in short‐chain fatty acids
Given that SCFAs are important mediators of the gut–muscle axis, we investigated the effect of aging and LcS on SCFA levels by measuring acetic, propionic, isobutyric, butyric, isovaleric, penic, and hexanoic acids in the stool. The levels of SCFAs measured in this study significantly decreased during aging as indicated by the lower levels of SCFAs in the A group compared with the NA group, except for propionic and isocaleric acids (Figure 4A–G). Compared with the A group, the S10X group had higher levels of acetic, isobutyric, butyric, penic, and hexanoic acids, and the SCFA levels increased slightly in the S1X group (Figure 4A–G). These results indicate that LcS supplementation could maintain SCFA levels in the aged mice. The statistical powers of the levels of acetic, isobutyric, butyric, penic, and hexanoic acids were 0.15, 0.88, 1.00, 0.90, and 0.83.
Figure 4.

Short‐chain fatty acids in stool. Different superscript letters (a, b, c) differ significantly (p < 0.05) according to one‐way analysis of variance with Tukey's honestly significant difference post‐hoc test. n = 6.
Lactobacillus casei Shirota modified the gut microbiota in aged mice
Since we demonstrated that LcS affected the muscle and SCFA levels in the stool, we could assume that LcS might improve age‐related muscle disorders through the gut–muscle axis. Therefore, we investigated the effect of LcS on gut microbiota. The results were evaluated by at least 54 830 reads per sample. The results of the Shannon (Figure 5A) and Simpson (Figure 5B) analysis revealed that the alpha diversities were not different among the four groups. The microbial communities of all groups were separated, especially between NA and A (P = 0.027), NA and S1X (P = 0.027), NA and S10X (P = 0.026), A and S10X (P = 0.027), and S1X and S10X (P = 0.031) (Figure 5C). In all groups, the most two classified bacteria were bacteroidetes and firmicutes (Figure 5D), and the ratio of firmicus/bacteroidete was not different (Figure 5E). However, the dominant bacterial genus was different between the groups (Figure 5F). These results suggest that LcS could direct the composition of gut microbiota to a preferred condition, but did not prevent age‐related alternations in gut microbiota. Therefore, we further analysed the abundant bacterial genus and predicted bacterial function in the three aged groups (A, S1X, and S10X groups) to avoid the disruption of age. Odoribacter and Oscillibacter were the dominant bacterial genera (top 35) and had significantly higher relative abundance in the A group than S10X group (Figure 5G). Lachnospiraceae_UCG_006 and Erysipelatoclostridium were the dominant bacterial genera with higher relative abundance in the S10X and S1X groups, respectively, than the A group. The results of LEfSe analysis showed that 23, 5, and 11 genera were enriched in the S1X, S10X, and A groups, respectively (Figure 5H). Moreover, the functions of the microbiota were then predicted using PICRUST database. The level 1 KEGG pathways predicted significant differences in cellular processes and genetic information processing between the A and S10X groups (Table 1). The cellular processes pathway was enriched in the A group (P < 0.05), whereas the genetic information processing pathway was enriched in the S10X group (P < 0.05) (Table 1). To identify the functional pathways more specifically, we analysed the pathways in KEGG level 3. Pathways related to apoptosis, p53 signalling, ubiquitin system, DNA repair and recombination proteins, non‐homologous end‐joining, RNA polymerase, and aminoacyl‐tRNA biosynthesis were significantly different among three groups. The pathways of apoptosis, p53 signalling, and non‐homologous end‐joining were higher in the A group compared with the S10X group (Table 1). Significant pathways related to cellular processes and genetic information processing were enriched in the S10X compared with the A group, including DNA repair and recombination proteins, RNA polymerase, and aminoacyl‐tRNA biosynthesis (Table 1). The gut microbiota community and function prediction results indicate that LcS supplementation could regulate the gut microbiota composition, which might produce functional changes in the host.
Figure 5.
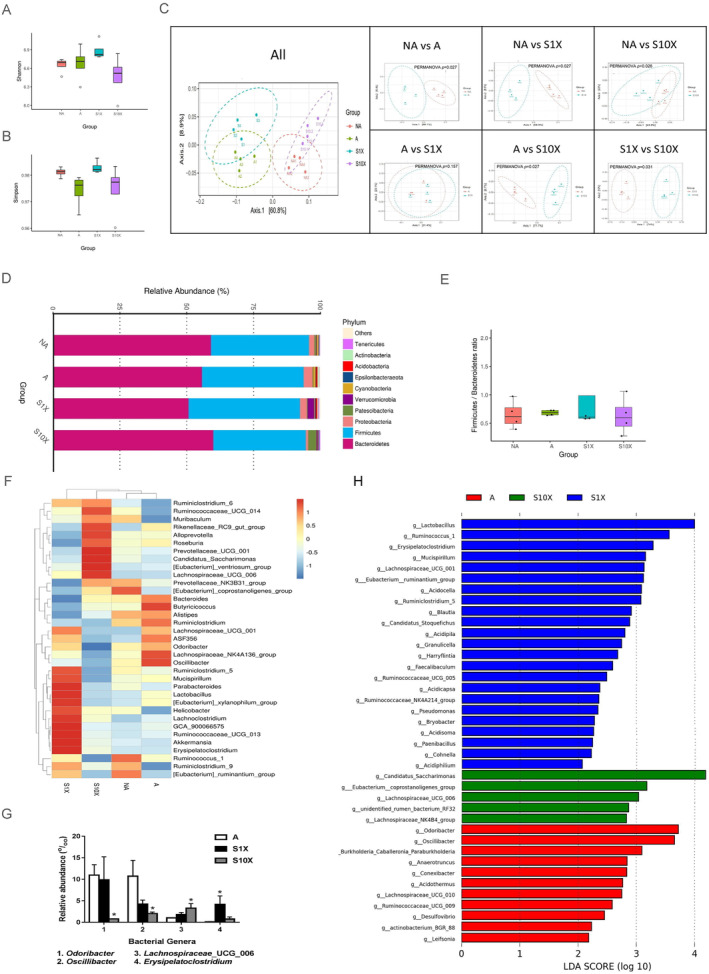
Analysis of gut microbiota. (A) Shannon analysis; (B) Simpson analysis; (C) PCoA analysis; (D) heatmap of phylum; (E) ration of firmicus/bacteroidete; (F) heatmap of genus; (G) relative abundance of dominant bacterial genera with significant difference between the A group and either the S1X or S10X groups (P < 0.05); (H) LEfSe analysis of A, S1X, and S10X groups. n = 4.
Table 1.
The functions predicted with significance in KEGG Levels 1 and 3
| KEGG—level 1 | P | A | S1X | S10X |
|---|---|---|---|---|
| Cellular processes | 0.06 | 3.73E‐02 ± 5.11E‐04 | 3.36E‐02 ± 2.12E‐03 | 3.00E‐02 ± 2.35E‐03* |
| Genetic information processing | 0.036 | 1.93E‐01 ± 1.84E‐03 | 1.95E‐01 ± 2.00E‐03 | 2.01E‐01 ± 2.19E‐03* |
| KEGG—level 3 | ||||
| Apoptosis | 0.026 | 2.67E‐05 ± 1.57E‐06 | 2.86E‐05 ± 7.83E‐06 | 1.64E‐05 ± 1.42E‐06* |
| p53 signalling pathway | 0.017 | 1.06E‐05 ± 2.84E‐06 | 1.25E‐05 ± 1.33E‐06 | 2.98E‐06 ± 1.39E‐06* |
| Ubiquitin system | 0.037 | 4.91E‐05 ± 7.13E‐06 | 5.77E‐05 ± 1.07E‐05 | 2.32E‐05 ± 5.67E‐06 |
| DNA repair and recombination proteins | 0.043 | 2.76E‐02 ± 2.79E‐04 | 2.79E‐02 ± 3.32E‐04 | 2.90E‐02 ± 3.80E‐04* |
| Non‐homologous end‐joining | 0.031 | 7.30E‐05 ± 1.03E‐05 | 1.25E‐04 ± 3.61E‐05 | 3.41E‐05 ± 1.08E‐05* |
| RNA polymerase | 0.008 | 1.51E‐03 ± 1.83E‐05 | 1.56E‐03 ± 1.31E‐05 | 1.63E‐03 ± 2.46E‐05* |
| Aminoacyl‐tRNA biosynthesis | 0.003 | 1.15E‐02 ± 8.97E‐05 | 1.16E‐02 ± 8.48E‐05 | 1.21E‐02 ± 1.20E‐04* |
Significantly different from the A group.
LcS decreased bacterial genera negatively correlated with muscle and mitochondrial function and positively correlated with senescence appearance, ROS, and inflammation
As our results revealed that LcS improved physiological phenomena and regulated the gut microbiota, we further investigated correlations in LcS effects in aged mice using Spearman's correlation. The bacterial genus was analysed only when LDA scores > 2.0 in the A, S1X, and S10X groups. In the A group, the enriched bacterial genera were all negatively correlated with muscle and mitochondrial function, including Odoribacter, Oscillibacter, Burkholderia_Caballeronia_Paraburkholderia, Anaerotruncus, Conexibacter, Acidothermus, Lachnospiraceae_UCG_010, Ruminococcaceae_UCG_009, Desulfovibrio, actinobacterium_BGR_88, and Leifsonia. These bacterial genera were also positively correlated with either serum or muscle proinflammatory cytokine, TNF‐α, and negatively correlated with either serum or muscle anti‐inflammatory cytokine, IL‐10, expect Leifsonia. The level of ROS was positively correlated with Anaerotruncus, Conexibacter, Lachnospiraceae_UCG_010, and Ruminococcaceae_UCG_009. In the S1X group, Ruminococcus_1, Lachnospiraceae_UCG_001, Acidocella, Granulicella, Ruminococcaceae_UCG_005, Ruminococcaceae_ NK4A214 group, Bryobacter, Acidisoma, and Cohnella were linked to unhealthy physiological phenomena, as opposed to Erysipelatoclostridium and Candidatus_Stoquefichus. The enriched bacterial genera, Candidatus_Saccharimonas, Lachnospiraceae_UCG_006, and unidentified_rumen_bacterium_RF32, in S10X mice were positively correlated with muscle ability, mitochondrial function, and IL‐10, and negatively correlated with the appearance of senescence and levels of TNF‐α or ROS (Table 2).
Table 2.
Correlation between the relative abundance of bacterial genera and different parameters
| A | S1X | S10X | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Odoribacter | Oscillibacter | Burkholderia_Caballeronia_Paraburkholderia | Anaerotruncus | Conexibacter | Acidothermus | Lachnospiraceae_UCG_010 | Ruminococcaceae_UCG_009 | Desulfovibrio | actinobacterium_BGR_88 | Leifsonia | Ruminococcus_1 | Erysipelatoclostridium | Lachnospiraceae_UCG_001 | Acidocella | Candidatus_Stoquefichus | Acidipila | Granulicella | Ruminococcaceae_UCG_005 | Ruminococcaceae_NK4A214_group | Bryobacter | Acidisoma | Cohnella | Candidatus_Saccharimonas | Lachnospiraceae_UCG_006 | unidentified_rumen_bacterium_RF32 | |
| Senescence appearance | 0.857** | 0.885** | 0.699* | 0.824** | 0.820** | 0.742** | 0.827** | 0.584* | 0.501 | 0.699* | 0.790** | 0.634* | −0.658* | 0.680* | 0.551 | −0.697* | 0.561 | 0.547 | 0.665* | 0.495 | 0.570 | 0.459 | 0.791** | −0.637* | −0.673* | −0.727** |
| Muscle proportion | −0.483 | −0.601* | −0.495 | −0.712** | −0.610* | −0.609* | −0.522 | −0.709** | −0.716** | −0.392 | −0.350 | −0.462 | 0.545 | −0.315 | −0.221 | 0.466 | −0.442 | −0.521 | −0.392 | −0.427 | −0.371 | −0.416 | −0.283 | 0.469 | 0.552 | 0.535 |
| Holding impulse | −0.729** | −0.816** | −0.727** | −0.740** | −0.827** | −0.784** | −0.770** | −0.521 | −0.663* | −0.647* | −0.657* | −0.606* | 0.504 | −0.578* | −0.523 | 0.533 | −0.602* | −0.650* | −0.612* | −0.537 | −0.572 | −0.527 | −0.593* | 0.655* | 0.725** | 0.689* |
| Grip force | −0.599* | −0.704* | −0.235 | −0.661* | −0.475 | −0.292 | −0.549 | −0.645* | −0.332 | −0.336 | −0.351 | −0.123 | 0.771** | −0.434 | −0.243 | 0.847** | −0.307 | −0.089 | −0.477 | −0.242 | −0.282 | −0.132 | −0.477 | 0.228 | 0.357 | 0.401 |
| PGC‐1α | 0.235 | 0.102 | −0.129 | −0.051 | −0.113 | 0.100 | −0.131 | −0.027 | −0.167 | −0.095 | 0.225 | −0.161 | 0.007 | −0.112 | −0.315 | −0.137 | −0.107 | −0.179 | −0.144 | 0.011 | −0.011 | 0.121 | 0.118 | 0.084 | −0.014 | 0.124 |
| SIRT‐1 | −0.802** | −0.858** | −0.663* | −0.815** | −0.791** | −0.692* | −0.868** | −0.576* | −0.668* | −0.814** | −0.650* | −0.599* | 0.550 | −0.795** | −0.695* | 0.635* | −0.705* | −0.652* | −0.814** | −0.633* | −0.645* | −0.454 | −0.751** | 0.736** | 0.669* | 0.839** |
| NRF‐1 | −0.377 | −0.613* | −0.581* | −0.859** | −0.639* | −0.495 | −0.476 | −0.645* | −0.666* | −0.341 | −0.327 | −0.746** | 0.282 | −0.472 | −0.395 | 0.277 | −0.557 | −0.508 | −0.501 | −0.176 | −0.596* | −0.611* | −0.402 | 0.570 | 0.539 | 0.614* |
| TFAM | −0.245 | −0.462 | −0.310 | −0.821** | −0.399 | −0.196 | −0.406 | −0.804** | −0.491 | −0.193 | −0.080 | −0.510 | 0.441 | −0.399 | −0.239 | 0.413 | −0.410 | −0.260 | −0.427 | 00.000 | −0.446 | −0.387 | −0.280 | 0.399 | 0.392 | 0.453 |
| Mitochondria copy number | −0.594* | −0.762** | −0.594* | −0.821** | −0.756** | −0.623* | −0.631* | −0.698* | −0.418 | −0.563 | −0.682* | −0.476 | 0.678* | −0.462 | −0.389 | 0.715** | −0.339 | −0.406 | −0.490 | −0.353 | −0.303 | −0.190 | −0.661* | 0.510 | 0.552 | 0.602* |
| Serum TNF‐α | 0.657* | 0.755** | 0.776** | 0.723** | 0.817** | 0.833** | 0.744** | 0.610* | 0.582* | 0.653* | 0.733** | 0.601* | −0.594* | 0.448 | 0.424 | −0.564 | 0.424 | 0.663* | 0.518 | 0.554 | 0.389 | 0.358 | 0.545 | −0.601* | −0.685* | −0.636* |
| Serum IL‐10 | −0.648* | −0.739** | −0.531 | −0.721** | −0.591* | −0.460 | −0.731** | −0.675* | −0.467 | −0.425 | −0.418 | −0.469 | 0.680* | −0.501 | −0.404 | 0.639* | −0.457 | −0.373 | −0.612* | −0.368 | −0.513 | −0.410 | −0.448 | 0.417 | 0.473 | 0.510 |
| Muscle TNF‐α | 0.545 | 0.622* | 0.708** | 0.632* | 0.617* | 0.623* | 0.769** | 0.511 | 0.653* | 0.592* | 0.412 | 0.664* | −0.245 | 0.573 | 0.549 | −0.263 | 0.656* | 0.720** | 0.683* | 0.511 | 0.702* | 0.643* | 0.385 | −0.713** | −0.678* | −0.625* |
| Muscle IL‐10 | −0.650* | −0.685* | −0.755** | −0.600* | −0.624* | −0.669* | −0.840** | −0.452 | −0.646* | −0.656* | −0.492 | −0.685* | 0.273 | −0.636* | −0.595* | 0.308 | −0.667* | −0.749** | −0.760** | −0.600* | −0.735** | −0.658* | −0.425 | 0.692* | 0.664* | 0.651* |
| ROS | 0.371 | 0.459 | 0.537 | 0.656* | 0.638* | 0.480 | 0.678* | 0.740** | 0.172 | 0.393 | 0.471 | 0.389 | −0.774** | 0.280 | 0.238 | −0.642* | 0.184 | 0.343 | 0.232 | 0.039 | 0.159 | 0.070 | 0.424 | −0.462 | −0.697* | −0.320 |
| Propionic acid | −0.161 | −0.252 | −0.164 | −0.561 | −0.335 | −0.192 | −0.452 | −0.836** | −0.105 | −0.189 | −0.139 | −0.049 | 0.734** | −0.133 | 0.029 | 0.673* | −0.071 | −0.100 | −0.074 | 0.127 | −0.004 | 0.088 | −0.214 | 0.308 | 0.580* | 0.135 |
| Isobutyric acid | −0.294 | −0.413 | −0.185 | −0.533 | −0.296 | −0.271 | −0.360 | −0.748** | −0.274 | −0.082 | −0.160 | −0.070 | 0.643* | −0.084 | 0.125 | 0.644* | −0.096 | −0.089 | −0.144 | −0.028 | −0.107 | −0.132 | −0.109 | 0.154 | 0.441 | 0.127 |
| Butyric acid | −0.790** | −0.874** | −0.691* | −0.775** | −0.813** | −0.719** | −0.773** | −0.529 | −0.586* | −0.617* | −0.678* | −0.594* | 0.538 | −0.615* | −0.546 | 0.585* | −0.610* | −0.553 | −0.655* | −0.504 | −0.620* | −0.548 | −0.676* | 0.629* | 0.657* | 0.688* |
| Isovaleric acid | −0.154 | −0.308 | −0.061 | −0.505 | −0.182 | −0.032 | −0.381 | −0.861** | −0.123 | −0.057 | 0.011 | 0.091 | 0.741** | −0.063 | 0.086 | 0.708* | −0.014 | 0.046 | −0.175 | 0.035 | −0.007 | 0.102 | −0.062 | 0.105 | 0.308 | 0.056 |
| Pentanoic acid | −0.189 | −0.427 | −0.317 | −0.772** | −0.474 | −0.260 | −0.466 | −0.854** | −0.382 | −0.232 | −0.157 | −0.336 | 0.706* | −0.308 | −0.135 | 0.673* | −0.200 | −0.221 | −0.273 | 0.099 | −0.143 | −0.051 | −0.243 | 0.294 | 0.531 | 0.340 |
| Hexanoic acid | −0.039 | −0.130 | 0.239 | −0.127 | 0.007 | 0.182 | −0.069 | −0.500 | 0.149 | 0.100 | 0.073 | 0.501 | 0.792** | 0.203 | 0.229 | 0.689* | 0.336 | 0.379 | 0.077 | 0.002 | 0.446 | 0.586* | 0.047 | −0.333 | −0.182 | −0.165 |
P < 0.05,
P < 0.01.
The dominant bacteria (top 35) enriched in the A, S1X, or S10X groups showed different correlation status to the predicted functional pathways.
The correlation of the predicted functional pathway and dominant bacteria (top 35) that were enriched in one of the aged mice groups were analysed. The results demonstrated that the dominant bacteria enriched in the A group, Odoribacter and Oscillibacter, presented the similar pattern that was positively correlated with apoptosis and negatively correlated with DNA repair and recombination proteins, RNA polymerase, and aminoacyl‐tRNA biosynthesis. RNA polymerase pathway was positively correlated with Erysipelatoclostridium (enriched in the S1X group) and Lachnospiraceae_UCG_006 (enriched in the S10X group). Lachnospiraceae_UCG_006 was also negatively correlated with the predicted functional pathways including p53 signalling, ubiquitin system, and non‐homologous end‐joining (Table 3).
Table 3.
Correlation between the relative abundance of predicted functional pathways and dominant bacterial genera (top 35) that were enriched in the A, S1X, or S10X groups
| Apoptosis | p53 signalling pathway | Ubiquitin system | DNA repair and recombination proteins | Non‐homologous end‐joining | RNA polymerase | Aminoacyl‐tRNA biosynthesis | ||
|---|---|---|---|---|---|---|---|---|
| A | Odoribacter | 0.860** | 0.399 | 0.517 | −0.692* | 0.427 | −0.748** | −0.790** |
| Oscillibacter | 0.762** | 0.301 | 0.483 | −0.706* | 0.406 | −0.776** | −0.783** | |
| S1X | Erysipelatoclostridium | −0.280 | 0.098 | 0.007 | 0.392 | 0.147 | 0.594* | 0.406 |
| S10X | Lachnospiraceae_UCG_006 | −0.510 | −0.755** | −0.615* | 0.420 | −0.601* | 0.587* | 0.559 |
P < 0.05,
P < 0.01.
Discussion
This is the first study to provide evidence of the effects of probiotics on the onset and progression of age‐related muscle impairment via the gut–muscle axis. In the present study, our results demonstrated that LcS, a probiotic, maintained muscle and mitochondrial functions and regulated the gut microbiota composition in aged SAMP8 mice. LcS also reduced age‐related changes in inflammatory cytokines and SCFAs, which are reported signal mediators in the gut–muscle axis. Moreover, the bacterial genera regulated by LcS were correlated with younger muscle condition and physiological phenomena in aged SAMP8 mice. Given that the concept of the gut–muscle axis is based on the correlation between gut microbiota and muscle function, our results suggest that LcS regulates the onset of age‐related sarcopenia via the gut–muscle axis. This indicates that LcS plays an important role in gut microbiota modification and causes changes in the production of mediators including SCFAs, inflammatory cytokines, and ROS, which subsequently maintains mitochondrial function and ultimately delays the progression of muscle deposition in aged mice.
Probiotics are reported to influence muscle mass and strength. For example, Bindels et al. 9 revealed that oral supplementation with Lactobacillus species reduced sarcopenia in a mouse model of acute leukaemia. In elite athletes, probiotics increased skeletal muscle function. 21 However, there is limited evidence of the benefits of probiotics in the muscles of older subjects. Although our previous study indicated that Lactobacillus paracasei PS23 attenuated age‐related sarcopenia, the correlation between the microbiota and muscle function was not investigated due to a lack of gut microbiota data. 19 In the present study, we not only demonstrated that another probiotic, LcS, improved age‐related sarcopenia, but we also analysed how LcS regulates the gut microbiota to determine whether LcS utilizes the gut–muscle axis to achieve its anti‐sarcopenia effects. Our results show that the LcS‐enriched bacterial genera were positively correlated with better physiological and muscle conditions. Moreover, LcS inhibited several bacterial genera in aged mice, and the abundance of these genera was negatively correlated with better physiological and muscle conditions. Interestingly, some bacterial genera enriched in the aged control group (A group), such as Odoribacter, Oscillibacter, Anaerotruncus, and Ruminococcaceae_UCG_009 have been linked to diminished health in older adults in previous studies. 22 Since the effects of oral LcS supplementation on the muscle and gut microbiota were observed and linked, our results suggest that LcS affects the muscle through the gut–muscle axis.
In addition to our observed correlation between LcS‐regulated muscle function and gut microbiota, our results involving gut–muscle mediators, such as inflammatory cytokines, SCFAs, and ROS, 3 provide further evidence identifying the gut–muscle axis as the mechanism of LcS‐induced anti‐sarcopenia given that LcS supplementation significantly increased the levels of the anti‐inflammatory cytokine, IL‐10, and several SCFAs and decreased the levels of the proinflammatory cytokine, TNF‐α,and ROS. Inflammation mediates communication between the gut and muscle. 23 Aging can induce inflammation by influencing intestinal function and gut microbiota composition, resulting in muscle dysfunction. 3 Moreover, reductions in SCFAs are a general phenomenon in older adults since aging decreases SCFAs‐producing bacteria in the gut. 3 The age‐related decline in SCFAs not only leads to inflammation but also mitochondrial dysfunction, followed by ROS increases and physiological disorders due to reduced SCFAs, particularly butyrate, which is well‐known for its anti‐inflammatory and mitochondrial maintaining effects. 23 Increase of ROS is an inducer of age‐related sarcopenia, because ROS can damage the mitochondria resulting in sarcopenia. 23 Our results also demonstrate that the bacteria genera enriched in aged control mice were negatively correlated with levels of SCFA and IL‐10 and positively correlated with levels of TNF‐α and ROS. We further analysed the correlation of gut microbiota and potential mediators of the gut–muscle axis to investigate how LcS regulated the gut microbiota to initiate the gut–muscle axis. Odoribacter and Oscillibacter—that were the only two genera that were the dominant genera (top 35) and significantly higher in the A group than S10X groups (LDA score > 2 and P < 0.05)—were positively correlated with inflammation, frailty, sarcopenia, and unhealthy physical condition in the mice and human elder. 22 , 24 Lachnospiraceae_UCG_006 was another dominant genus significantly different between the A and S10X groups, which was enriched in the S10X group. Although Lachnospiraceae_UCG_006 was rarely discussed in the study with old subjects, it was reported as a genus contributing to the anti‐inflammation condition in young subjects. 25 Therefore, based on the present study, inflammation should be involved in the gut–muscle axis. Therefore, one possible mechanism of LcS in age‐related sarcopenia is that LcS modulates the gut microbiota, especially Odoribacter, Oscillibacter, and Lachnospiraceae UCG 006, to attenuate the age‐related inflammation so that muscle retains healthy mitochondria and good functions, even in the elderly.
Short‐chain fatty acids generated from fermentation by gut microbiota in the large intestine is known to exert antioxidant properties, immunomodulatory function, and regulate energy metabolism. 26 The observed increases of SCFAs are likely to be involved in the anti‐inflammation states in the groups treated with LcS, but the correlation of dominant bacteria and butyric acid was only partially supported through previous studies. In this study, Lachnospiraceae UCG 006—which was reported as a producer of butyric acid 25 —was also significantly positively correlated with the levels of butyric acid. To our knowledge, the correlation of butyric acid and Odoribacter and Oscillibacter was not reported in the aged subjects, but these bacteria were indicated to produce butyric acid in some studies with non‐aged experimental animals. Thus, how LcS regulated butyric acid production still needs further investigation in subsequent studies.
Reactive oxygen species increase is an inducer of age‐related sarcopenia because ROS can damage the mitochondria, resulting in sarcopenia. 3 In the present study, LcS significantly suppressed the level of ROS. The level of ROS was positively correlated with the bacteria genera enriched in the A group and negatively correlated with those in the S10X group, although only part of the correlation was significant (P < 0.05). Among the dominant and significantly changed bacteria, only Lachnospiraceae UCG 006 was significantly negatively correlated with ROS. The negative association of Lachnospiraceae UCG 006 and ROS was also reported in a previous study. 27 Thus, the LcS‐shaped gut microbiota composition might lower the ROS in the aged SAMP8 mice.
In addition to LcS‐regulated gut microbiota, LcS might achieve the preventing effect on age‐related sarcopenia via reducing oxidative and inflammatory stress. LcS was reported to attenuate the oxidative and inflammatory stress in the 2,2′‐azobis(2‐amidinopropane) dihydrochloride‐treated enterocytes‐like epithelial cells. 11 Matsumoto and colleagues also revealed that LcS could reduce the pro‐inflammatory cytokines in the monocytes isolated from large intestinal lamina propria and RAW264·7 cells in vitro. 28 Thus, LcS might be able to directly decrease the ROS and inflammation in the aged SAMP8 mice.
Taken together, the precise mechanisms of LcS in extenuating age‐related sarcopenia progression were through the gut–muscle axis, where LcS either interacts with intestinal cells or regulates the gut microbiota to suppress the age‐related inflammation, ROS, and apoptosis, to promote SCFA production, thereby preventing mitochondria dysfunction in muscle, and finally slowing the progression of age‐related sarcopenia.
The gut–muscle axis could be characterized by predicted function pathways based on the KEGG pathway related to the dominant bacteria in the mice. In the A group, the enriched functions were apoptosis and p53 signalling pathways, which are related to age‐related sarcopenia. 29 Apoptosis and induction of p53 result in muscle mitochondrial dysfunction and mass loss, because they drive the increases in inflammation and ROS that can damage mitochondria and decrease muscle function due to energy deficiencies in muscle. 6 DNA repair and recombination proteins were the other pathways predicted to be involved in the LcS‐derived gut–muscle axis. This is because DNA repair was decreased during aging due to alteration of gut microbiota, which has been suggested to drive age‐related inflammation and diseases, 30 and the S10X group had the higher level of DNA repair, and recombination proteins pathway and lower level of age‐related sarcopenia than the A group indicated. Since LcS attenuated these two functional pathways, ROS levels, and age‐related systemic inflammation by reducing serum TNF‐α and inducing serum IL‐10, we could assume that LcS prevented age‐related sarcopenia via the gut–muscle axis. Thus, LcS may help maintain muscle function in aged mice by influencing the gut microbiota to reduce apoptosis, p53 signalling, inflammation, and ROS. However, the relationship of the other predicted pathways and age‐related disorders were either opposite or unreported in the previous studies. The ubiquitin system is responsible for the post‐translational modification of proteins. The increase of ubiquitin‐proteasome was positively correlated with the levels of inflammation and apoptosis. 31 However, the ubiquitin system was employed by autophagy that was considered to be an anti‐aging process. 32 Aminoacyl‐tRNA biosynthesis showed pro‐aging and anti‐aging effects in the different studies. 33 Thus, further studies will be needed to understand the linkages of these pathways and sarcopenia.
Although our results indicate that LcS attenuates age‐related sarcopenia through the gut–muscle axis in SAMP8 mice, a question remained as to whether SAMP8 mice were an appropriate model for studying age‐related sarcopenia and the gut–muscle axis. SAMP8 mice are characterized by early onset of age‐related alternations and with decreasing survival rate at as early as 8 months old. 34 , 35 Among all SAMP mouse strains, they show the most senescence deterioration in skeletal muscle function compared with senescence‐resistant (SAMR1) control mice. 34 The studies published by other groups suggest pre‐sarcopenia features, such as insulin resistance 36 and mitochondria dysfunction, 37 in the skeletal muscle of 6‐month‐age SAMP8 mice, and the process of sarcopenia initiated between 5 or 6‐month‐age to 12‐month‐age in SAMP8 mice. 15 , 34 Our previous study also reported significant muscle atrophy in the SAMP8 mice at 7 months old compared with 4 months old. 19 Furthermore, the death of SAMP8 mice begins at about 8 months old 35 ; thus, the study finished when the mice reach 28 weeks old to avoid survivorship bias. Accordingly, administrating LcS to SAMP8 mice from 16 to 28 weeks old is appropriate for investigating how LcS prevented age‐related sarcopenia. However, data are limited regarding the difference in gut microbiota between healthy and unhealthy aged SAMP8 mice. In humans, Claesson et al. 22 analysed the faecal microbiota composition of 178 healthy and unhealthy elderly subjects and suggested that the co‐abundance groups of Alistipes and Oscillibacter increased in unhealthy elderly individuals. They revealed that increased Alistipes and Oscillibacter were accompanied by calf circumference decreases, muscle loss, and proinflammatory cytokine increases. The increase of Oscillibacter co‐abundance groups also reduced the number of genera that produce butyrate. Interestingly, in the present study, Oscillibacter was enriched in the A group compared to the S1X and S10X groups. Moreover, the abundance of Alistipes was higher in the A than S10X group. Since similar gut microbiota differences were observed in healthy and unhealthy aged subjects in human subjects and SAMP8 mice, SAMP8 mice appear to be a suitable animal model for studying sarcopenia and the gut–muscle axis in aged subjects.
In this study, two doses of LcS were applied to investigate the dose‐effect because the dose‐dependent manner was already observed in the LcS studies. Oral administration of LcS at a higher dose had better effects on recovery of hand functions after distal radius fracture in elders. 38 The higher dose of LcS also showed stronger prevention in indomethacin‐induced small intestinal injury. 39 In the present study, the high dose of LcS administration showed better efficiency because more results of the S10X were different from the A groups than the S1X group. Moreover, the S10X had a better condition than the S1X group in the holding impulse, OCR‐FCCP, muscle TNF‐α, acetic acid, butyric acid, and hexanoic acid. The gut microbiota results also suggested that the impact of a high dose of LcS was larger than the lower dose of LcS because the significant difference of beta diversity was observed between the A and S10X groups but not between the A and S1X groups. In addition, the most dominant and significant bacteria taxa, Odoribacter, Oscillibacter, and Lachnospiraceae_UCG_006, and the apoptosis‐related pathways showed more differences between the A and S10X groups than the A and S1X groups. Taken together, the effect of LcS on attenuating age‐related sarcopenia followed a dose‐dependent manner and was better at the higher dose.
Recently, an increasing number of studies have focused on the effects and mechanism of probiotics in the improvement of disease states. Although the design of these studies has been broadly similar, the different probiotics or analysed tissue could result in different outcomes. For example, Tsilingiri et al. 40 reported that Lactobacillus plantarum NCIMB8826 and Lactobacillus rhamnosus GG showed different influences in the tissue with inflammatory bowel disease. They also revealed that the effects of L. plantarum NCIMB8826 were different on health and inflammatory bowel disease tissues. Moreover, there are some common weaknesses in this kind of study. First, the studies usually only contain a proportion of the members in the gut‐organ axis that involves gut microbiota, mediators of the gut–organ axis, and the target organ. Thus, the data could not provide a correlation between the members to draw a whole picture. Second, the link between the animal model and humans might be weak because the probiotics altered dominant bacterial genera in the animal model did not differ between healthy and unhealthy elderly human participants. Hence, there is concern whether the effect and mechanism of probiotics could be reproduced in humans. In the present study, we analysed gut microbiota, SCFA, an inflammatory cytokine, and the function of muscle and mitochondria to demonstrate that the full pathway of LcS‐induced gut–muscle axis was initiating from the regulation of gut microbiota, then translating the signal to muscle using inflammatory cytokine, and finally influencing the function of muscle by maintaining mitochondria. Moreover, LcS significantly reduced the level of Oscillbacter and Alistipes, accompanied by calf circumference decreases, muscle loss, and pro‐inflammatory cytokine increases in the aged humans. Thus, LcS should have a high possibility to attenuate age‐related sarcopenia. Accordingly, this study demonstrated two novel insights in age‐related sarcopenia: (1) the preventing effect of probiotics on age‐related sarcopenia exists and is accomplished via gut–muscle axis and (2) oral LcS supplementation has great potentialities to prevent humans from the threaten of age‐related sarcopenia.
Because the study addressed potential members in gut–muscle axis and revealed a strong correlation between them, our results have provided a reasonable strength of evidence to prove that oral LcS supplementation extenuates age‐related sarcopenia via the gut–muscle axis. However, there is a limitation that the experiments could not directly demonstrate the effect of gut microbiota on age‐related sarcopenia. Although a faecal/microbiota transplantation experiment may help address this limitation, the gut microbiota composition was established by a daily LcS intervention. Furthermore, LcS can directly modulate ROS and inflammation in the intestinal cells. 11 , 28 Therefore, the donor's gut microbiota population might not be easily established in the recipient, and the same effects of LcS supplementation might reappear by faecal/microbiota transplantation. Thus, faecal/microbiota transplantation was not performed in this study. Nevertheless, results of faecal/microbiota transplantation experiments might help understand the effect of LcS comprehensively and would be an interesting topic of further study.
In conclusion, LcS can attenuate age‐related sarcopenia through the gut–muscle axis, and oral LcS supplementation may be a potential strategy to prevent age‐related sarcopenia.
Funding
This work was supported in part by Ministry of Science and Technology, Taiwan (MOST‐109‐2320‐B‐038‐058‐MY3).
Conflicts of interest
The authors declare no conflicts of interest.
Supporting information
Table S1. Criteria for grading score of senescence in mice.
Table S2. Primer sequences.
Acknowledgements
We would like to thank Dr Ming‐Fu Wang provided SAMP8 mice.
Chen L.‐H., Chang S.‐S., Chang H.‐Y., Wu C.‐H., Pan C.‐H., Chang C.‐C., Chan C.‐H., and Huang H.‐Y. (2022) Probiotic supplementation attenuates age‐related sarcopenia via the gut–muscle axis in SAMP8 mice, Journal of Cachexia, Sarcopenia and Muscle, 13, 515–531, 10.1002/jcsm.12849
Li‐Han Chen and Shy‐Shin Chang contributed equally to the study.
Contributor Information
Li‐Han Chen, Email: lihan.h.chen@gmail.com.
Hui‐Yu Huang, Email: maggieh323@hotmail.com.
References
- 1. Marzetti E, Calvani R, Tosato M, Cesari M, Di Bari M, Cherubini A, et al. Sarcopenia: an overview. Aging Clin Exp Res 2017;29:11–17. [DOI] [PubMed] [Google Scholar]
- 2. Picca A, Fanelli F, Calvani R, Mule G, Pesce V, Sisto A, et al. Gut dysbiosis and muscle aging: searching for novel targets against sarcopenia. Mediators Inflamm 2018;2018:7026198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ticinesi A, Nouvenne A, Cerundolo N, Catania P, Prati B, Tana C, et al. Gut microbiota, muscle mass and function in aging: a focus on physical frailty and sarcopenia. Nutrients 2019;11:e1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lustgarten MS. The role of the gut microbiome on skeletal muscle mass and physical function: 2019 update. Front Physiol 2019;10:1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lahiri S, Kim H, Garcia‐Perez I, Reza MM, Martin KA, Kundu P, et al. The gut microbiota influences skeletal muscle mass and function in mice. Sci Transl Med 2019;11:eaan5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ren GX, Zhang JP, Li MH, Tang ZC, Yang ZN, Cheng GY, et al. Gut microbiota composition influences outcomes of skeletal muscle nutritional intervention via blended protein supplementation in posttransplant patients with hematological malignancies. Clin Nutr 2021;40:94–102. [DOI] [PubMed] [Google Scholar]
- 7. Munukka E, Rintala A, Toivonen R, Nylund M, Yang BR, Takanen A, et al. Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high‐fat fed mice. ISME J 2017;11:1667–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li BL, Evivie SE, Lu JJ, Jiao YH, Wang CF, Li ZY, et al. Lactobacillus helveticus KLDS1.8701 alleviates d‐galactose‐induced aging by regulating Nrf‐2 and gut microbiota in mice. Food Funct 2018;9:6587–6599. [DOI] [PubMed] [Google Scholar]
- 9. Bindels LB, Beck R, Schakman O, Martin JC, De Backer F, Sohet FM, et al. Restoring specific Lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. Plos One 2012;7:e37971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cervantes‐Tolentino M, Beltran‐Campos V, Padilla‐Raygoza N. Influence of Lactobacillus casei Shirota strain on body composition: a review. Biomed Pharmacol J 2020;13:89–99. [Google Scholar]
- 11. Finamore A, Ambra R, Nobili F, Garaguso I, Raguzzini A, Serafini M. Redox role of Lactobacillus casei Shirota against the cellular damage induced by 2,2′‐azobis (2‐amidinopropane) dihydrochloride‐induced oxidative and inflammatory stress in enterocytes‐like epithelial cells. Front Immunol 2018;9:e1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macnaughtan J, Figorilli F, Garcia‐Lopez E, Lu H, Jones H, Sawhney R, et al. A double‐blind, randomized placebo‐controlled trial of probiotic Lactobacillus casei Shirota in stable cirrhotic patients. Nutrients 2020;12:e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joseph N, Vasodavan K, Saipudin NA, Yusof BNM, Kumar S, Nordin SA. Gut microbiota and short‐chain fatty acids (SCFAs) profiles of normal and overweight school children in Selangor after probiotics administration. J Funct Foods 2019;57:103–111. [Google Scholar]
- 14. Matsumoto K, Takada T, Shimizu K, Moriyama K, Kawakami K, Hirano K, et al. Effects of a probiotic fermented milk beverage containing Lactobacillus casei strain Shirota on defecation frequency, intestinal microbiota, and the intestinal environment of healthy individuals with soft stools. J Biosci Bioeng 2010;110:547–552. [DOI] [PubMed] [Google Scholar]
- 15. Brunetti D, Bottani E, Segala A, Marchet S, Rossi F, Orlando F, et al. Targeting multiple mitochondrial processes by a metabolic modulator prevents sarcopenia and cognitive decline in SAMP8 mice. Front Pharmacol 2020;11:1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okubo H, Sakoda H, Kushiyama A, Fujishiro M, Nakatsu Y, Fukushima T, et al. Lactobacillus casei strain Shirota protects against nonalcoholic steatohepatitis development in a rodent model. Am J Physiol Gastrointest Liver Physiol 2013;305:G911–G918. [DOI] [PubMed] [Google Scholar]
- 17. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber‐deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016;167:1339–1353, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang SY, Chen LH, Wang MF, Hsu CC, Chan CH, Li JX, et al. Lactobacillus paracasei PS23 delays progression of age‐related cognitive decline in senescence accelerated mouse prone 8 (SAMP8) mice. Nutrients 2018;10:e894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen LH, Huang SY, Huang KC, Hsu CC, Yang KC, Li LA, et al. Lactobacillus paracasei PS23 decelerated age‐related muscle loss by ensuring mitochondrial function in SAMP8 mice. Aging‐US 2019;11:756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen LH, Wang MF, Chang CC, Huang SY, Pan CH, Yeh YT, et al. Lacticaseibacillus paracasei PS23 effectively modulates gut microbiota composition and improves gastrointestinal function in aged SAMP8 mice. Nutrients 2021;13:e1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marttinen M, Ala‐Jaakkola R, Laitila A, Lehtinen MJ. Gut microbiota, probiotics and physical performance in athletes and physically active individuals. Nutrients 2020;12:e2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012;488:178–184. [DOI] [PubMed] [Google Scholar]
- 23. Liao X, Wu M, Hao Y, Deng H. Exploring the preventive effect and mechanism of senile sarcopenia based on “gut‐muscle axis”. Front Bioeng Biotechnol 2020;8:590869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suzuk TA, Phifer‐Rixey M, Mack KL, Sheehan MJ, Lin DN, Bi K, et al. Host genetic determinants of the gut microbiota of wild mice. Mol Ecol 2019;28:3197–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo WL, Xiang QR, Mao BY, Tang X, Cui SM, Li XF, et al. Protective effects of microbiome‐derived inosine on lipopolysaccharide‐induced acute liver damage and inflammation in mice via mediating the TLR4/NF‐kappa B pathway. J Agric Food Chem 2021;69:7619–7628. [DOI] [PubMed] [Google Scholar]
- 26. Koh A, De Vadder F, Kovatcheva‐Datchary P, Backhed F. From dietary fiber to host physiology: short‐chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–1345. [DOI] [PubMed] [Google Scholar]
- 27. Falony G, Vlachou A, Verbrugghe K, De Vuyst L. Cross‐feeding between Bifidobacterium longum BB536 and acetate‐converting, butyrate‐producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 2006;72:7835–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsumoto S, Hara T, Hori T, Mitsuyama K, Nagaoka M, Tomiyasu N, et al. Probiotic Lactobacillus‐induced improvement in murine chronic inflammatory bowel disease is associated with the down‐regulation of pro‐inflammatory cytokines in lamina propria mononuclear cells. Clin Exp Immunol 2005;140:417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mankhong S, Kim S, Moon S, Kwak HB, Park DH, Kang JH. Experimental models of sarcopenia: bridging molecular mechanism and therapeutic strategy. Cells‐Basel 2020;9:e1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guedj A, Volman Y, Geiger‐Maor A, Bolik J, Schumacher N, Kunzel S, et al. Gut microbiota shape ‘inflamm‐ageing’ cytokines and account for age‐dependent decline in DNA damage repair. Gut 2020;69:1064–1075. [DOI] [PubMed] [Google Scholar]
- 31. Fu D, Li P, Sheng QF, Lv ZB. Beta‐arrestin‐2 enhances intestinal epithelial apoptosis in necrotizing enterocolitis. Aging‐US 2019;11:8294–8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Le WD. Autophagy and ubiquitin‐proteasome system. Autophagy: Biology and Diseases: Basic Science 2019;1206:527–550. [DOI] [PubMed] [Google Scholar]
- 33. Zhou Z, Sun B, Yu DS, Bian M. Roles of tRNA metabolism in aging and lifespan. Cell Death Dis 2021;12:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo AY, Leung KS, Siu PM, Qin JH, Chow SK, Qin L, et al. Muscle mass, structural and functional investigations of senescence‐accelerated mouse P8 (SAMP8). Exp Anim 2015;64:425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spangler EL, Patel N, Speer D, Hyman M, Hengemihle J, Markowska A, et al. Passive avoidance and complex maze learning in the senescence accelerated mouse (SAM): age and strain comparisons of SAM P8 and R1. J Gerontol A Biol Sci Med Sci 2002;57:B61–B68. [DOI] [PubMed] [Google Scholar]
- 36. Onishi S, Ishino M, Kitazawa H, Yoto A, Shimba Y, Mochizuki Y, et al. Green tea extracts ameliorate high‐fat diet‐induced muscle atrophy in senescence‐accelerated mouse prone‐8 mice. PLoS ONE 2018;13:e0195753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andreani C, Bartolacci C, Guescini M, Battistelli M, Stocchi V, Orlando F, et al. Combination of coenzyme Q10 intake and moderate physical activity counteracts mitochondrial dysfunctions in a SAMP8 mouse model. Oxid Med Cell Longev 2018;2018:8936251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang CH, Xue SJ, Wang Y, Yu D, Hua LM, Guo CH, et al. Oral administration of Lactobacillus casei Shirota improves recovery of hand functions after distal radius fracture among elder patients: a placebo‐controlled, double‐blind, and randomized trial. J Orthop Surg Res 2019;14:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Watanabe T, Nishio H, Tanigawa T, Yamagami H, Okazaki H, Watanabe K, et al. Probiotic Lactobacillus casei strain Shirota prevents indomethacin‐induced small intestinal injury: involvement of lactic acid. Am J Physiol‐Gastr L 2009;297:G506–G513. [DOI] [PubMed] [Google Scholar]
- 40. Tsilingiri K, Barbosa T, Penna G, Caprioli F, Sonzogni A, Viale G, et al. Probiotic and postbiotic activity in health and disease: comparison on a novel polarised ex‐vivo organ culture model. Gut 2012;61:1007–1015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Criteria for grading score of senescence in mice.
Table S2. Primer sequences.


