Abstract
Bovine mastitis is the most important source of loss for the dairy industry. A rapid and specific test for the detection of the main pathogens of bovine mastitis is not actually available. Molecular probes reacting in PCR with bacterial DNA from bovine milk, providing direct and rapid detection of Escherichia coli, Staphylococcus aureus, Streptococcus agalactiae, Streptococcus dysgalactiae, Streptococcus parauberis, and Streptococcus uberis, have been developed. Two sets of specific primers were designed for each of these microorganisms and appeared to discriminate close phylogenic bacterial species (e.g., S. agalactiae and S. dysgalactiae). In addition, two sets of universal primers were designed to react as positive controls with all major pathogens of bovine mastitis. The sensitivities of the test using S. aureus DNA extracted from milk with and without a pre-PCR enzymatic lysis step of bacterial cells were compared. The detection limit of the assay was 3.125 × 102 CFU/ml of milk when S. aureus DNA was extracted with the pre-PCR enzymatic step compared to 5 × 103 CFU/ml of milk in the absence of the pre-PCR enzymatic step. This latter threshold of sensitivity is still compatible with its use as an efficient tool of diagnosis in bovine mastitis, allowing the elimination of expensive reagents. The two PCR tests avoid cumbersome and lengthy cultivation steps, can be performed within hours, and are sensitive, specific, and reliable for the direct detection in milk of the six most prevalent bacteria causing bovine mastitis.
Bovine mastitis (BM) is an inflammation of the mammary gland, usually due to a microbial infection (28), which causes North American dairy producers to lose billions of dollars every year. These losses are primarily due to lower milk yields, reduced milk quality, and higher production costs. BM often becomes chronic, and it is important to identify quickly the new clinical cases in order to control infection in the herd. The bacteria responsible for BM can be classified as environmental (Escherichia coli, Streptococcus dysgalactiae, Streptococcus parauberis, and Streptococcus uberis) or contagious (Staphylococcus aureus and Streptococcus agalactiae) depending of their primary reservoir (environment versus infected mammary gland quarter) (11, 25).
The suitability of a detection method for routine diagnosis depends on several factors, such as specificity, sensitivity, expense, amount of time, and applicability to large numbers of milk samples. The most common but unspecific method (2) to identify potential chronic infections is a somatic cell count: the California Mastitis Test in field conditions and the automated method in the diagnosis laboratory. Currently, the method of identification of the mammary gland pathogens is by in vitro culture, which provides the “gold standard”; however, this technique is labor-intensive and time-consuming. Two other problems can be encountered when these methods of identification are used: first, 2 to 3 days are required to grow, isolate, and identify the pathogen; second, some bacteria, like S. uberis and S. parauberis (S. uberis type II), cannot be distinguished by biochemical assays (16). It has been demonstrated that early detection procedures have been shown to enhance cure rates and reduce the time required to return to normal milk when coupled with appropriate antimicrobial therapy (20). It is important to identify the pathogen not only for antimicrobial therapy purposes but also to monitor and control the rate of infection at the farm level.
During the last 7 years, many tests have been developed for the diagnosis of BM. However, a rapid (less than 1 day), simple, and specific test for each kind of bacterium involved has not been achieved. Many tests for the detection of human pathogens already exist. Some of these tests have been applied to pathogens of a bovine origin, such as the Minitek Gram-Positive Set for Streptococcus (29), but with no success because of a lack of information on veterinary pathogens in the database. Enzyme-linked immunosorbent assay methods exist for S. aureus detection in cases of BM (10), but the antibody titer does not correlate with the amount of infecting bacteria (10, 15). Other enzyme-linked immunosorbent assays were developed to screen milk for contamination with Listeria organisms (1, 9). Most PCR and API methods used for the detection of microorganisms in milk or in other organic samples need a step of multiplying the bacteria in culture media (3, 18, 21, 22, 27, 30) and are, therefore, time-consuming. Rapid identification methods, in particular nucleic acid-based tests, have the potential to be extremely specific and can also discriminate between closely related organisms, such as S. parauberis and S. uberis. It has been previously shown that milk samples could serve as substrate for the amplification of specific DNA sequences using PCR (6, 17). For all these reasons, we selected a PCR technique as a method for identifying BM pathogens because of its precision, high limit of detection, and rapidity.
The aim of this study was to develop molecular tools to identify with rapidity, sensitivity, and specificity the major pathogens involved in intramammary infections in cows. To reach this objective, two sets of universal primers used as positive controls and two sets of specific primers for each bacterium species were developed to identify, by PCR, E. coli, S. aureus, S. agalactiae, S. dysgalactiae, S. parauberis, and S. uberis in milk samples inoculated with these bacteria. Finally, the sensitivity of two PCR assays performed on S. aureus DNA samples prepared with and without a pre-enzymatic lysis step in milk and in Tryptic Soy Broth (TSB) was analyzed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The organisms used in this study include E. coli DH5α, Staphylococcus epidermidis ATCC 35984, S. parauberis ATCC 13386, S. uberis ATCC 9927, and three clinical isolates from cases of BM, S. aureus 97-6609, S. agalactiae 91-0121, and S. dysgalactiae 97-6617, which were identified biochemically by the API Staph and Strep systems accordingly. All organisms were cultured in TSB (Difco Laboratories, Detroit, Mich.) at 37°C for about 17 h before DNA extraction. Cell numbers were determined by the preparation of serial dilutions of an overnight culture in phosphate-buffered saline (PBS) and plating on blood agar (Columbia agar base supplemented with 5% defibrinated sheep blood).
Preparation of bacterial DNA for PCR using the Dneasy Tissue kit from Qiagen.
Bacteria were grown overnight in 20 ml of TSB, and then 1.5 ml of this culture was centrifuged at 5,000 × g for 10 min. The Dneasy Tissue kit from Qiagen was used with some modifications to the protocol for gram-positive bacteria: the pellet was resuspended in 200 μl of enzyme incubation buffer (20 mM Tris-HCl, pH 8.0, 1.2% Triton, 20 μg of lysozyme/ml, and 200 μg of lysostaphin/ml) followed by an incubation period of 30 min at 37°C. Then, 20 μg of RNase H (GIBCO Life Technologies, Montreal, Canada)/ml was added with 200 μl of AL buffer (supplied with the kit) and incubated at 70°C for 10 min followed by the addition of 25 μl of proteinase K (supplied with the kit) and incubation at 70°C for 30 min. This mixture was transferred into columns and eluted with deionized autoclaved water by following the manufacturer's recommendation.
Preparation of bacterial DNA for the comparative study.
A culture of S. aureus (1.6 × 108 CFU/ml) in TSB was distributed in tubes and centrifuged at 5,000 × g for 15 min and the pellets were kept at −20°C until subsequent use. CFU per milliliter was determined by limit dilution and plating on blood agar (Columbia agar base supplemented with 5% defibrinated sheep blood). Thawed cells were resuspended in sterile bovine milk (ultra high temperature-treated [UHT] milk; Grand-Pré, Québec, Canada) or in TSB and submitted to twofold serial dilutions to reach concentrations of 4 × 104 to 3.125 × 102 CFU/ml. Aliquots of 1 ml of these dilutions were centrifuged for 5 min at 7,000 × g and submitted to four washes, and then the pellets were resuspended in 1 ml of 1× PBS and centrifuged at 5,000 × g for 5 min. PBS rather than water was used for washes to precipitate calcium ions from the milk, which is known to be a PCR inhibitor (8), and to protect the bacterial membranes from lysis during these steps. After the last centrifugation, the pellet was resuspended in 1 ml of autoclaved distilled water to allow the burst of bacterial cells. This preparation was directly used for PCR or treated with the Dneasy Tissue kit. At the end of this latter step, 600 μl of deionized autoclaved water was added to the DNA preparation to obtain a total volume of 1 ml ready for PCR use. Sample of 5 μl of these preparations were used for PCR.
PCR primers.
PCR primers were designed from highly divergent and species-specific regions of the DNA coding for 16S and 23S rRNA (16S or 23S rRNA) based on previously published sequence entries available in the GenBank database (E. coli, GI no. 42756; S. aureus, GI no. 288516; S. agalactiae, GI no. 2353759; S. dysgalactiae, GI no. 560494; S. parauberis, GI no. 433515; and S. uberis, GI no. 433703 and 2668550). Genes encoding rRNA were used as target sequences rather than genes encoding mRNA because of the signal enhancement due to the presence of several copies of genes encoding rRNA in the genome (4). The sequences, specificities, and G+C contents are summarized in Table 1. The primer combinations, the annealing temperatures, and the lengths of the amplified products are summarized in Table 2. These primers were synthesized by AlphaDNA (Montreal, Canada). All these primers were resuspended to a final concentration of 200 μM in deionized autoclaved water. The working concentration of the primers for PCR was 5 μM.
TABLE 1.
Oligonucleotide primer sequences used for PCR
| Primer | Specificity | Sequence(5′-3′) | G+C content (%) |
|---|---|---|---|
| Eco 223 | E. coli | ATC AAC CGA GAT TCC CCC AGT | 52 |
| Eco 455 | E. coli | TCA CTA TCG GTC AGT CAG GAG | 52 |
| Eco 2083 | E. coli | GCT TGA CAC TGA ACA TTG AG | 45 |
| Eco 2745 | E. coli | GCA CTT ATC TCT TCC GCA TT | 45 |
| Sau 234 | S. aureus | CGA TTC CCT TAG TAG CGG CG | 60 |
| Sau 1501 | S. aureus | CCA ATC GCA CGC TTC GCC TA | 60 |
| Sau 327 | S. aureus | GGA CGA CAT TAG ACG AAT CA | 45 |
| Sau 1645 | S. aureus | CGG GCA CCT ATT TTC TAT CT | 45 |
| Sag 40 | S. agalactiae | CGC TGA GGT TTG GTG TTT ACA | 48 |
| Sag 445 | S. agalactiae | CAC TCC TAC CAA CGT TCT TC | 50 |
| Sag 432 | S. agalactiae | CGT TGG TAG GAG TGG AAA AT | 45 |
| Sag 1018 | S. agalactiae | CTG CTC CGA AGA GAA AGC CT | 55 |
| Sdy 105 | S. dysgalactiae | AAA GGT GCA ACT GCA TCA CTA | 43 |
| Sdy 386 | S. dysgalactiae | GTC ACA TGG TGG ATT TTC CA | 45 |
| Sdy 519 | S. dysgalactiae | GGC TCA ACC ACT NTA CGC TT | 50 |
| Sdy 920 | S. dysgalactiae | ATC TCT AGA CCG GTC AGG AG | 55 |
| Spa 301 | S. parauberis | GCG ACG TGG GAT CAA ATA CT | 50 |
| Spa 1219 | S. parauberis | TAC CAT TAC CTC TAA AGG TA | 35 |
| Spa 2152 | S. parauberis | TTT CGT CTG AGG CAA TGT TG | 45 |
| Spa 2870 | S. parauberis | GCT TCA TAT ATC GCT ATA CT | 35 |
| Sub 302 | S. uberis | CGA AGT GGG ACA TAA AGT TA | 40 |
| Sub 396 | S. uberis | CTG CTA GGG CTA AAG TCA AT | 45 |
| Sub 1546 | S. uberis | TGA TGG GGA GCG AAA ATA AG | 45 |
| Sub 2170 | S. uberis | CCC AAC AAC GCC TCA AAC GA | 55 |
| Uni 678 | Universal | AGT GGA ATT CCA TGT GTA GC | 45 |
| Uni 888 | Universal | GAG TGC TTA ATG CGT TAG CT | 45 |
| Uni 1870 | Universal | TGG AAG GTT AAG AGG AGT GG | 50 |
| Uni 2308 | Universal | GCC TCC GTT ACC TTT TAG GA | 50 |
TABLE 2.
Primer conditions during PCR
| Forward primer | Reverse primer | Annealing temp (°C) | Size of product amplified (bp) |
|---|---|---|---|
| Eco 223 | Eco 455 | 64 | 232 |
| Eco 2083 | Eco 2745 | 57 | 662 |
| Sau 234 | Sau 1501 | 70 | 1,267 |
| Sau 327 | Sau 1645 | 64 | 1,318 |
| Sag 40 | Sag 445 | 60 | 405 |
| Sag 432 | Sag 1018 | 65 | 586 |
| Sdy 105 | Sdy 386 | 57 | 281 |
| Sdy 519 | Sdy 920 | 57 | 401 |
| Spa 301 | Spa 1219 | 57 | 918 |
| Spa 2152 | Spa 2870 | 57 | 718 |
| Sub 302 | Sub 396 | 56 | 94 |
| Sub 1546 | Sub 2170 | 59 | 624 |
| Uni 678 | Uni 888 | 56 | 210 |
| Uni 1870 | Uni 2308 | 58 | 438 |
PCR amplification.
PCR was performed in a GeneAmp PCR System 2400 (Perkin-Elmer). All reactions were carried out in a final volume of 100 μl. Volumes of 200 ng of extracted DNA template or 5 μl of bacterial preparation, 5 μM primer, 2.5 U of Taq DNA polymerase (GIBCO Life Technologies, Montreal, Canada), 10 μl of 10× PCR buffer minus Mg (GIBCO Life Technologies), 3 μl of 50 mM MgCl2, and all four deoxynucleotide triphosphates (supplied by GIBCO Life Technologies) were added to a 0.5-ml microcentrifuge tube. A pre-PCR step at 94°C for 2 min was applied. A total of 35 PCR cycles were run under the following conditions: denaturation at 94°C for 45 s, annealing (at the temperature in Table 2) for 1 min, and extension at 72°C for 2 min. After the final cycle, the preparation was kept at 72°C for 10 min to complete the reaction. The PCR products were stored in the thermocycler at 4°C until they were collected.
Detection of PCR products.
Twenty microliters of the PCR-amplified product was analyzed by electrophoresis on a 1.7% agarose gel stained with 0.5 μg of ethidium bromide/ml. Electrophoresis was carried out in 1× TAE (pH 8.0; 0.04 M Tris-acetate, 0.001 M EDTA) at 80 V for 2 h. The molecular size marker, a 100-bp, 1-kb DNA ladder (GIBCO Life Technologies), was run concurrently. Gels were visualized under UV illumination (Alphadoc; Alpha Innotech Corporation, San Leandro, Calif.) and photographed.
RESULTS
Specificity of primers.
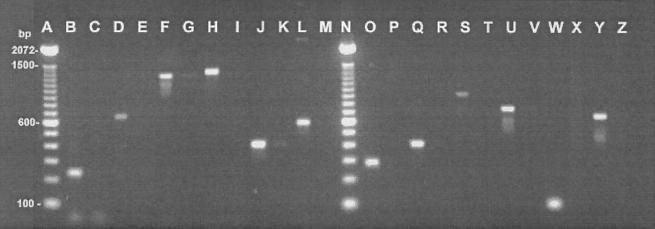
The specific oligonucleotide primer sequences designed to amplify and identify the main bacteria causing BM are presented in Table 1. Twelve pairs of primers were tested (2 pairs/bacterium). Prior to PCR, purified DNA was run on 0.8% agarose gel and quantified by UV absorbance at 260 and 280 nm to confirm its quantity and presence in PCR. The specificity of the primer pairs was confirmed by the positive amplification of the DNA from bacteria found in BM, whereas no DNA amplification was observed with the closest phylogenic bacteria tested, as demonstrated in Fig. 1. The assay was controlled as follows: Pseudomonas aeruginosa was tested as a negative control for E. coli, S. epidermidis was a negative control for S. aureus, S. dysgalactiae was a negative control for S. agalactiae, S. agalactiae was a negative control for S. dysgalactiae, S. uberis was a negative control for S. parauberis, and finally, S. parauberis was a negative control for S. uberis. Amplification of the DNA templates using proper primers produced the expected fragments (Fig. 1) between 94 and 1,318 bp as shown in Table 2.
FIG. 1.
Demonstration of the specificity of the molecular probes in PCR assay with purified bacterial DNA from different species. Amplification products of the different primer combinations were analyzed by electrophoresis on a 1.7% agarose gel. Lanes: A, 100-bp DNA ladder (GIBCO Life Technologies); B and C, primers Eco 223 and Eco 455 with E. coli (B) and P. aeruginosa (C); D and E, primers Eco 2083 and Eco 2745 with E. coli (D) and P. aeruginosa (E); F and G, primers Sau 234 and Sau 1501 with S. aureus (F) and S. epidermidis (G); H and I, primers Sau 327 and Sau 1645 with S. aureus (H) and S. epidermidis (I); J and K, primers Sag 40 and Sag 445 with S. agalactiae (J) and S. dysgalactiae (K); L and M, primers Sag 432 and Sag 1018 with S. agalactiae (L) and S. dysgalactiae (M); N, 100-bp DNA ladder (Life Technologies, Inc.); O and P, primers Sdy 105 and Sdy 386 with S. dysgalactiae (O) and S. agalactiae (P); Q and R, primers Sdy 519 and Sdy 920 with S. dysgalactiae (Q) and S. agalactiae (R); S and T, primers Spa 301 and Spa 1219 with S. parauberis (S) and S. uberis (T); U and V, primers Spa 2152 and Spa 2870 with S. parauberis (U) and S. uberis (V); W and X, primers Sub 302 and Sub 396 with S. uberis (W) and S. parauberis (X); and Y and Z, primers Sub 1546 and Sub 2170 with S. uberis (Y) and S. parauberis (Z).
Universal primer amplification.
Universal primer pairs Uni 678 plus Uni 888 and Uni 1870 plus Uni 2308 were tested against six bacterial species: E. coli, S. aureus, S. agalactiae, S. dysgalactiae, S. parauberis, and S. uberis (Fig. 2). A product of 210 bp was observed when Uni 678 and Uni 888 were tested, and a product of 438 bp was observed when Uni 1870 and Uni 2308 were used. Amplification was not observed in negative controls of PCR mix. The universal primers used in this study, Uni 678 plus Uni 888 and Uni 1870 plus Uni 2308, were able to detect all BM pathogens cited above. The intensity of each amplification was good with all the bacteria tested. The strongest signal was obtained with the primers Uni 678 and Uni 888 for S. uberis and Uni 1870 and Uni 2308 for S. aureus (Fig. 2). These primer sets were designed from DNA regions coding for 16S and 23S rRNA, respectively. Nucleotide sequence data comparison (BLASTN 2.0) showed that the primers Uni 678, Uni 888, Uni 1870, and Uni 2308 are conserved for all pathogens of BM.
FIG. 2.
PCR amplification of purified bacterial DNA by using two pairs of universal primers. Amplification products for the different primer combinations were analyzed by electrophoresis on a 1.7% agarose gel. Lanes: A, 100-bp DNA ladder (GIBCO Life Technologies); B to H, primers Uni 678 and Uni 888 with E. coli (B), S. aureus (C), S. agalactiae (D), S. dysgalactiae (E), S. parauberis (F), S. uberis (G), and a negative control without DNA (H); I to O, primers Uni 1870 and Uni 2308 with E. coli (I), S. aureus (J), S. agalactiae (K), S. dysgalactiae (L), S. parauberis (M), S. uberis (N), and a negative control without DNA (O); P, 100-bp DNA ladder (GIBCO Life Technologies).
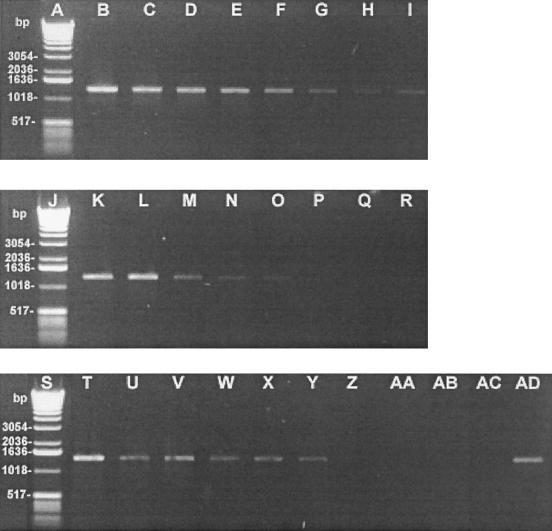
Sensitivity of PCR assay depending on pretreatment used.
In the aim to make the test as simple and cheap as possible, we tried to eliminate the pre-PCR enzymatic lysis step of bacterial cells. Secondly, in comparing milk with the TSB as support medium, it was possible to measure the inhibitory effect of milk, particularly calcium ions, on the PCR test. For control purposes, uninoculated samples of UHT milk and TSB media were also subjected to the sample preparation methods and to PCR. No amplification could be observed (Fig. 3c). Figure 3a shows the PCR results obtained following the pre-PCR enzymatic lysis step using the Qiagen kit: a very high level of sensitivity corresponding to 1.56 CFU/5 μl of PCR mixture or 3.125 × 102 CFU/ml of milk was obtained, as demonstrated by the twofold serial dilutions of the S. aureus samples. The sensitivity of the assay fell to 6.25 CFU/5 μl of PCR mixture or 1.25 × 103 CFU/ml of TSB in the absence of the pre-PCR enzymatic lysis step of the S. aureus cells (Fig. 3c). In milk, the elimination of the pre-PCR enzymatic lysis step of S. aureus gave a level of sensitivity of 25 CFU/5 μl of milk, or 5.0 × 103 CFU/ml of milk (Fig. 3b).
FIG. 3.
Sensitivity of the PCR assay in detecting DNA from two different media (milk and TSB) artificially inoculated with S. aureus 97-6609 with and without a pre-PCR enzymatic lysis step of bacterial cells. Primers Sau 327 and Sau 1645 were used. The amounts of bacteria used were 4 × 104 (lanes B, K, and T), 2 × 104 (C, L, U), 1 × 104 (D, M, V), 5 × 103 (E, N, W), 2.5 × 103 (F, O, X), 1.25 × 103 (G, P, Y), 6.25 × 102 (H, Q, Z), and 3.125 × 102 (I, R, AA) CFU. Lanes: A, J, and S, 1-kb DNA ladder (GIBCO Life Technologies); B to I, S. aureus samples resuspended in milk at the concentrations indicated above and extracted with the pre-PCR enzymatic step; K to R, S. aureus samples resuspended in milk at the concentrations indicated above and not subjected to the pre-PCR treatment; T to AA, S. aureus samples resuspended in TSB at the concentrations indicated above and not subjected to the pre-PCR treatment; AB, PCR mixture without bacteria in milk; AC, PCR mixture without bacteria in TBS; AD, positive control using 200 ng of S. aureus DNA.
DISCUSSION
An efficient vaccine against BM is not yet available, and prevention as a measure of control needs sensitive, rapid, and specific tests to identify the main bacteria that cause heavy losses in the dairy industry. Conventional procedures for the identification of BM pathogens are labor-intensive, and most of the commercial identification systems are not designed to identify important veterinary pathogens (16, 29). We aimed to develop a detection and identification test for BM pathogens that produced results in 1 day, did not need a culture step, and was as specific, sensitive, and cheap as possible. Molecular methods are very efficient tools for the development of newly improved diagnosis tests. When methods such as ribotyping are laborious, new methods using PCR based on the 16S or the 23S rDNA region sequences have been successfully applied for the identification of many bacteria (5, 7, 12, 19, 23, 24, 26, 31). The major advantages of PCR lay in the possibility of using only nanograms of nucleic acid samples, allowing the elimination of culture, rapidity, and easy analysis. Therefore, we selected PCR amplification of DNA regions coding for rRNA because of the presence of hypervariable regions, which facilitates the design of highly specific oligonucleotide probes (13) and common regions for the design of universal probes (14). Moreover, rDNA is present in many copies, which permits signal enhancement (4).
Specific primers described here (Table 1 and Fig. 1) were proven to be specific since on agarose gel only one band was observed for each set of primers and no signal was detected with negative controls. The primers for E. coli, S. aureus, S. parauberis, and S. uberis were designed based on a DNA sequence coding for 23S rRNA, and primers for S. agalactiae and S. dysgalactiae were based on DNA coding for 16S rRNA. All the signals were very obvious even though the bands produced by the primers Eco 2083 and Eco 2745 with E. coli and Spa 301 and Spa 1219 with S. parauberis were of lower intensity (Fig. 1). The difference of signal cannot be explained by the amount of DNA used for PCR because the same amount (200 ng) was used for all bacteria. Therefore, it is possible that the difference in the copy numbers of the coding regions for these particular probes could explain the phenomenon (4). Negative controls made with the closest phylogenic bacteria found in BM confirmed the specificity of each primer set. While the bacteria S. parauberis and S. uberis are genotypically and phenotypically close, PCR performed with the described primers made it possible to distinguish between them. The universal primer sets used as positive controls, Uni 678 plus Uni 888 and Uni 1870 plus Uni 2308, were designed from DNA regions coding for 16S and 23S rRNA, respectively. Even though the nucleotide sequence data comparison (BLASTN 2.0) showed that the four universal primers are conserved in all pathogens of BM, the sequences can vary by 1 or 2 nucleotides, therefore varying the signal intensity.
In the aim to evaluate the sensitivity level of the PCR test and to reduce the cost and the duration of the test, a study to compare the sensitivities of the test with and without a pre-PCR enzymatic lysis step of the bacterial cells was performed. It appeared that the limits of detection of the PCR test performed on infected milk samples treated with a pre-PCR enzymatic lysis step were 1.56 CFU/5 μl of PCR mixture and 3.12 × 102 CFU/ml of milk. Although these very high levels of sensitivity were diminished to 1.25 × 103 CFU per ml of TSB and 5 × 103 CFU per ml of milk (Fig. 3) in the absence of the pre-PCR enzymatic lysis step, these levels of the detection limit are sensitive enough to be used as a diagnosis tool for BM. The level of this detection limit was obtained by significantly reducing the quantity of calcium ions from milk by performing four washes with PBS. We think that it would be possible to increase the sensitivity level of CFU detection in milk in the absence of the pre-PCR enzymatic lysis step to the same level observed in TSB by improving the method to eliminate calcium ions. Even though PCR is less labor-intensive than bacterial culture and conventional methods of bacterial identification, the elimination of the pre-PCR enzymatic lysis step leads to a more important economy of expensive reagents and time.
In conclusion, a PCR-based assay for the detection of the major pathogens involved in BM is described here. The test, directly performed from milk samples without a culture step, is specific for E. coli, S. aureus, S. agalactiae, S. dysgalactiae, S. parauberis, and S. uberis. The primers were designed from the 16S and 23S rRNA sequences available in GenBank. The test can be performed with and without a pre-PCR enzymatic lysis step of the bacterial cells. The greatest advantage of the latter version is the elimination of expensive reagents. When used with the pre-PCR enzymatic lysis step, the test can be performed in 6 h and reaches a limit of detection of 3.12 × 102 CFU/ml of milk. The test without the pre-PCR enzymatic lysis step can be performed in 4.5 h and reaches a limit of detection of 5 × 103 CFU/ml of milk. Considering the level of detection obtained in TSB without the pre-PCR enzymatic lysis step, it could be possible to improve the detection limit of the latter assay in milk by using a better method to eliminate calcium ions. Secondly, it will be important to test in the near future the detection limit of the molecular probes in milk from actual mastitis cases. If the results comply with the present study, these PCR tests could be readily implemented in clinical veterinary microbiological laboratories and be of great value for promoting prevention of BM.
ACKNOWLEDGMENTS
This work was supported by the Fonds pour la formation de chercheurs et l'aide à la recherche (FCAR, Québec, Canada) and Novalait.
REFERENCES
- 1.Adams D S, McDonald J S, Hancock D, McGuire T C. Staphylococcus aureus antigens reactive with milk immunoglobulin G of naturally infected dairy cows. J Clin Microbiol. 1988;26:1175–1180. doi: 10.1128/jcm.26.6.1175-1180.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews A H, Blowey R W, Boyd H, Eddy R G. Bovine medicine: diseases and husbandry of cattle. London, United Kingdom: Blackwell Publishing Co.; 1992. pp. 289–300. [Google Scholar]
- 3.Andrews A T. Proteinases in normal bovine milk and their action on caseins. J Dairy Res. 1983;50:45–55. doi: 10.1017/s0022029900032519. [DOI] [PubMed] [Google Scholar]
- 4.Bentley R W, Leigh J A. Determination of 16S ribosomal RNA gene copy number in Streptococcus uberis, S. agalactiae, S. dysgalactiae and S. parauberis. FEMS Immunol Med Microbiol. 1995;12:1–7. doi: 10.1111/j.1574-695X.1995.tb00167.x. [DOI] [PubMed] [Google Scholar]
- 5.Bentley R W, Leigh J A. Development of PCR-based hybridization protocol for identification of streptococcal species. J Clin Microbiol. 1995;33:1296–1301. doi: 10.1128/jcm.33.5.1296-1301.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berri M, Laroucau K, Rodolakis A. The detection of Coxiella burnetii from ovine genital swabs, milk and fecal samples by the use of a single touchdown polymerase chain reaction. Vet Microbiol. 2000;72:285–293. doi: 10.1016/s0378-1135(99)00178-9. [DOI] [PubMed] [Google Scholar]
- 7.Bes M, Guerin-Faublee V, Meugnier H, Etienne J, Freney J. Improvement of the identification of Staphylococci isolated from bovine mammary infections using molecular methods. Vet Microbiol. 2000;71:287–294. doi: 10.1016/s0378-1135(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 8.Bickley J, Short J K, McDowell D G, Parkes H C. Polymerase chain reaction (PCR) detection of Listeria monocytogenes in diluted milk and reversal of PCR inhibition caused by calcium ions. Lett Appl Microbiol. 1996;22:153–158. doi: 10.1111/j.1472-765x.1996.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 9.Bourry A, Cochard T, Poutrel B. Serological diagnosis of bovine, caprine, and ovine mastitis caused by Listeria monocytogenes by using an enzyme-linked immunosorbent assay. J Clin Microbiol. 1997;35:1606–1608. doi: 10.1128/jcm.35.6.1606-1608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourry A, Poutrel B. Bovine mastitis caused by Listeria monocytogenes: kinetics of antibody responses in serum and milk after experimental infection. J Dairy Sci. 1996;79:2189–2195. doi: 10.3168/jds.S0022-0302(96)76595-5. [DOI] [PubMed] [Google Scholar]
- 11.Bramley, A., J. 1996. Current concepts of bovine mastitis. National Mastitis Council, Madison, Wis.
- 12.Forsman P, Tilsala-Timisjarvi A, Alatossava T. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S–23S rRNA spacer regions. Microbiology. 1997;143:3491–3500. doi: 10.1099/00221287-143-11-3491. [DOI] [PubMed] [Google Scholar]
- 13.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray M W, Sankoff D, Cedergren R J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in a small subunit ribosomal RNA. Nucleic Acids Res. 1984;12:5837–5852. doi: 10.1093/nar/12.14.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heisick J E, Harrell F M, Peterson E H, McLaughlin S, Wagner D E, Wesley I V, Bryner J. Comparison of four procedures to detect Listeria spp. in foods. J Food Prot. 1989;52:154–157. doi: 10.4315/0362-028X-52.3.154. [DOI] [PubMed] [Google Scholar]
- 16.Jayarao B M, Dore J J, Jr, Baumbach G A, Matthews K R, Oliver S P. Differentiation of Streptococcus uberis from Streptococcus parauberis by polymerase chain reaction and restriction fragment length polymorphism analysis of 16S ribosomal DNA. J Clin Microbiol. 1991;29:2774–2778. doi: 10.1128/jcm.29.12.2774-2778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipkin E, Shalom A, Khatib H, Soller M, Friedmann A. Milk as a source of deoxyribonucleic acid and as a substrate for the polymerase chain reaction. J Dairy Sci. 1993;76:2025–2032. doi: 10.3168/jds.S0022-0302(93)77536-0. [DOI] [PubMed] [Google Scholar]
- 18.Lorenz H, Jager C, Willems H, Baljer G. PCR detection of Coxiella burnetii from different clinical specimens, especially bovine milk, on the basis of DNA preparation with a silica matrix. Appl Environ Microbiol. 1998;64:4234–4237. doi: 10.1128/aem.64.11.4234-4237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendoza M, Meugnier H, Bes M, Etienne J, Freney J. Identification of Staphylococcus species by 16S–23S rDNA intergenic spacer PCR analysis. Int J Syst Bacteriol. 1998;48:1049–1055. doi: 10.1099/00207713-48-3-1049. [DOI] [PubMed] [Google Scholar]
- 20.Milner P, Page K L, Hillerton J E. The effects of early antibiotic treatment following diagnosis of mastitis detected by a change in the electrical conductivity of milk. J Dairy Sci. 1997;80:859–863. doi: 10.3168/jds.S0022-0302(97)76008-9. [DOI] [PubMed] [Google Scholar]
- 21.Queipo-Ortuno M I, Garcia-Ordonez M A, Colmenero J D, Morata P. Hydrogen peroxide improves the efficiency of a peripheral blood PCR assay for diagnosis of human brucellosis. BioTechniques. 1999;27:248–250. doi: 10.2144/99272bm06. , 252. [DOI] [PubMed] [Google Scholar]
- 22.Reale S, Maxia L, Vitale F, Glorioso N S, Caracappa S, Vesco G. Detection of Leishmania infantum in dogs by PCR with lymph node aspirates and blood. J Clin Microbiol. 1999;37:2931–2935. doi: 10.1128/jcm.37.9.2931-2935.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabat G, Rose P, Hickey W J, Harkin J M. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl Environ Microbiol. 2000;66:844–849. doi: 10.1128/aem.66.2.844-849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saruta K, Matsunaga T, Kono M, Hoshina S, Ikawa S, Sakai O, Machida K. Rapid identification and typing of Staphylococcus aureus by nested PCR amplified ribosomal DNA spacer region. FEMS Microbiol Lett. 1997;146:271–278. doi: 10.1111/j.1574-6968.1997.tb10204.x. [DOI] [PubMed] [Google Scholar]
- 25.Smith B P. Large animal internal medicine. 2nd ed. Boston, Mass: Mosby; 1996. pp. 1181–1193. [Google Scholar]
- 26.Straub J A, Hertel C, Hammes W P. A. 23S rDNA-targeted polymerase chain reaction-based system for detection of Staphylococcus aureus in meat starter cultures and dairy products. J Food Prot. 1999;62:1150–1156. doi: 10.4315/0362-028x-62.10.1150. [DOI] [PubMed] [Google Scholar]
- 27.Thomas E J, King R K, Burchak J, Gannon V P. Sensitive and specific detection of Listeria monocytogenes in milk and ground beef with the polymerase chain reaction. Appl Environ Microbiol. 1991;57:2576–2580. doi: 10.1128/aem.57.9.2576-2580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watts J L. Etiological agents of bovine mastitis. Vet Microbiol. 1988;16:41–66. doi: 10.1016/0378-1135(88)90126-5. [DOI] [PubMed] [Google Scholar]
- 29.Watts J L. Evaluation of the Minitek Gram-Positive Set for identification of streptococci isolated from bovine mammary glands. J Clin Microbiol. 1989;27:1008–1010. doi: 10.1128/jcm.27.5.1008-1010.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wernars K, Heuvelman C J, Chakraborty T, Notermans S H. Use of the polymerase chain reaction for direct detection of Listeria monocytogenes in soft cheese. J Appl Bacteriol. 1991;70:121–126. doi: 10.1111/j.1365-2672.1991.tb04437.x. [DOI] [PubMed] [Google Scholar]
- 31.Whitehead T R, Cotta M A. Development of molecular methods for identification of Streptococcus bovis from human and ruminal origins. FEMS Microbiol Lett. 2000;182:237–240. doi: 10.1111/j.1574-6968.2000.tb08901.x. [DOI] [PubMed] [Google Scholar]