Abstract
The past decades witnessed a significant stride in deciphering the pathophysiology of inflammatory bowel disease, which further advanced drug development adding several new biologicals and small molecules to the arsenal of available therapies. Surprisingly, this wealth in therapeutic options did not yield the aspired high durable response rates. In addition, the increase in therapeutic availabilities ignited an increase in research toward biomarkers that could help assign therapies to patients with the highest probability of response. Luckily, major steps have been undertaken in this domain which resulted in the discovery of some interesting biomarkers that are still under validation. However, the pace in which this domain is progressing, the discordance between short-term endpoints in biomarker discovery studies and the ambition of the disease community in modifying disease course, and the uncertainties about the validity of discovered biomarkers highlight the need for a critical appraisal of research conduct in this domain. In this review, we shed light on areas of improvement in biomarker discovery studies that will help optimize the use of available therapies and break the current therapeutic ceiling.
Keywords: Biomarker discovery, Personalized medicine, Precision, Therapeutic ceiling, IBD, Omics
Abbreviations: CD, Crohn's disease; CRP, C reactive protein; FDA, Food and Drug Administration; IBD, inflammatory bowel diseases; MOA, mechanism of action; RCTs, randomized clinical trials; SES, simple endoscopic score; TDM, therapeutic drug monitoring; TNF, tumour necrosis factor; UC, ulcerative colitis; UST, ustekinumab
1. Introduction
Inflammatory bowel diseases (IBD) are chronic immune-mediated inflammatory disorders of the gastrointestinal tract entailing two main entities: Crohn's disease (CD) and ulcerative colitis (UC). Both diseases are characterized by a relapsing-remitting nature with heterogeneous phenotypes, disease course and complications (Roda et al., 2020; Kobayashi et al., 2020). Prevalence of IBD is forecast to increase in the coming years, with a particular rise in incidence in recently industrialized areas (Ng et al., 1474; Kaplan and Windsor, 1759). This epidemiologic expansion will further compound the economic and social burden of IBD (Kaplan, 2015). Although the exact pathophysiology of IBD remains elusive, available evidence suggests that a dysregulated immune response towards the microbiome in genetically and environmentally susceptible individuals induces and maintains tissue damage (Chang, 2020). The widespread availability of high throughput analytical techniques has led to a better understanding of IBD pathogenesis and resulted in an ever-expanding therapeutic armamentarium with many new compounds in late-stage development (Al-Bawardy et al., 2021). This increasing availability in therapeutic options is indeed embraced by IBD physicians, but unfortunately not yet optimally utilized as evidenced by the striking ceiling in remission rates (20%–30%) observed also with new therapies (Alsoud et al., 2021a). Currently, assigning therapies is based on clinical features, co-morbidities, side effects and patient preference on mode of delivery of the drug and its speed of action. Furthermore, many clinical and laboratory variables have shown an association with (non-)response to available therapies such as disease duration, previous exposure to anti-tumor necrosis factor alpha (anti-TNF-a) biologicals, baseline laboratory values (such as albumin, C reactive protein (CRP) and calprotectin), increased weight and previous bowel resection (Kopylov and Seidman, 2016). While these features may be informative to some extent, none of them could predict the likelihood for a certain drug to induce disease remission in a certain patient at a certain time with reliable accuracy (Verstockt and Ferrante, 2020). Luckily, the aforementioned complexity of the current therapeutic landscape in IBD was accompanied by a shift in stance of IBD investigators and triggered an endeavor aiming to optimize the use of available medications in a more efficient personalized approach. The early steps in this research domain delivered several promising biomarkers that are being currently further assessed for validity and clinical utility (Verstockt et al., 2021a). However, despite the widespread belief that predictive biomarkers are of high priority, this domain has been facing many challenges complicating discovery and validation of biomarkers. In this review, we outline challenges and pitfalls in this infant field.
1.1. Defining the suitable outcome for an effective biomarker
Presently, there is a lack of consensus on the ideal outcomes for patients included in IBD randomized clinical trials (RCTs), real-life studies, and biomarker discovery studies. Biomarker discovery studies have aimed for a variety of endpoints, including normalization of inflammatory markers, clinical or endoscopic response, or endoscopic remission (Peyrin-Biroulet et al., 2015; Turner et al., 2021). This simple and unidimensional approach currently used in assessing the outcome of therapies in IBD does not appreciate the various and complex phenotypes of the disease, nor serve the ambition of the IBD community in modifying disease course and restoring the quality in all aspects of a patient's life. For instance, a patient with fistulizing CD necessitating recurrent surgical interventions and complicating daily life activities, but with only modest luminal disease, cannot be counted as a real remitter in a biomarker study considering solely a decrease in luminal endoscopic indices (such as SES-CD) while the fistulizing component is not equally healed. Such scenarios will lead to an overestimation of the real value of investigated biomarkers. Predictive biomarker studies recruiting IBD patients with heterogeneous phenotypes, are more likely to benefit from a holistic and multi-dimensional assessment of therapy effect, which can be a composite of several unidimensional outcome measures. The recent SPIRIT initiative to establish a consensus on outcome measures for disease modification could serve as a starting point towards an objective, holistic and patient-centered outcome assessment that can be further adapted to serve the aims of biomarker studies (Le Berre and Peyrin-Biroulet, 2021). Alternatively, objectivity in assessing the reliability of discovered biomarkers can be ensured by designing studies including subgroups of patients with homogenous phenotypes, where therapy effect can be adequately assessed in each subgroup using the most suitable simple outcome measure. Besides the need to identify the most suitable outcomes for IBD patients, the heterogeneity in outcome definitions is an additional major limitation, especially in biomarker development. For example, various definitions are being used for endoscopic remission, including the absence of ulcerations or specific endoscopic score cut-offs (Dulai et al., 2015). Standardizing outcomes definitions across IBD studies is vital to ensure robustness in the discovery and validation of new biomarkers, and to facilitate the comparison between results obtained in different studies. Currently used endoscopic and histologic indices have been constructed and validated as continuous variables and proven to be correlated with other inflammatory biomarkers (CRP and calprotectin). However, their categorization into binary or ordinal variables is still questionable. Dichotomizing continuous indices leads to an important loss of information, and ignores within-category information as all patients below or above a certain cut-off are treated as equal (responder vs non-responder, with partial responders often categorized as non-responders) (Fedorov et al., 2009). This information loss jeopardizes the reliability of machine learning models used to select the ideal panel of biomarkers as the explained-variance significantly drops (Altman and Royston, 2006). Therefore, additional studies are required to determine the clinically relevant minimal change in outcome indices, investigate the non-inferiority of categorized outcomes, and define the optimal cut-offs.
1.2. Aiming for biomarkers predicting primary non-response and long-term response
To date, all studies investigating biomarkers for the prediction of therapy outcome in IBD have been aiming for short-term response (6 months–1 year). Such biomarkers will undoubtedly be of a great importance in assigning therapies and lowering rates of primary non-response. While there are currently very limited long-term real-life reports on non-anti-TNF-a drugs, the rates of loss of response are high for anti-TNF-a biologicals (Roda et al., 2016). This reality forms a major clinical challenge and make it reasonable to question the durability of certain biomarkers as follow-up reports on accuracy performance of these biomarkers in predicting long-term outcomes are lacking. Without the guarantee to predict long-term benefits of a certain drug, the cost-effectiveness of biomarkers-based therapy assignment in comparison with the current random trial-and-error approach might be limited to some extent. However, it is presently not clear from the available evidence whether the same biomarkers predicting initial (non-)response would also be capable of predicting the maintenance of a stable long-term remission, as pathways involved in these processes may be different. Therefore, researchers are encouraged to also report on the long-term predictive performance of biomarkers, and to investigate molecular processes underlying “loss of response” phenomenon and IBD flares. These parallel efforts are necessary to ultimately break the impasse of cycling biologicals which a considerable proportion of patients is currently facing. Predicting (non-)response to biologicals and small molecules will undoubtedly bring IBD community many steps closer to realizing precision medicine. However, due to the cost of these therapies and the preference of many patients to withdraw therapy if feasible, one should explore the potential of biomarkers to predict disease recurrence upon treatment discontinuation in patients in deep remission. Randomized (unblinded) trials have demonstrated that a subset of patients does indeed not experience relapse after anti-TNF withdrawal (Kobayashi et al., 2021). A post-hoc analysis of the STORI trial identified a potential proteomic signature predictive of disease recurrence (Pierre et al., 2020), though validation in the SPARE trial (NCT02177071) and other large datasets is required prior to implementation in daily clinical practice (Verstockt et al., 2021b; Louis, 2022). On top, longitudinal (sequential) assessment of biomarkers holds even large promise to identify those patients who can harmlessly discontinue their treatment. This approach should yet be investigated in IBD, as promising results are emerging from other fields (Kameda et al., 2021).
1.3. Ensuring sufficient drug exposure among included patients
So far, only few predictive biomarker studies provided information on drug exposure (Verstockt et al., 2019a). Ideally, all patients should be offered an equal chance to attain a sufficient drug exposure. This can be achieved through implementing therapeutic drug monitoring (TDM) to exclude a pharmacokinetic (i.e. underexposure), rather than a pharmacodynamic, reason for therapy failure (Papamichael et al., 2015; Dreesen, 2021). Consequently, in many previous predictive biomarker studies, it cannot be ruled out that patients who were classified as “non-responders” would have been labeled as “responders”, if TDM would have been embedded in the study. Failing to discriminate between pharmacodynamic (mechanistic) non-responders and those non-responding as a result of underexposure could jeopardize the reliability of biomarker studies.
1.4. Confounding baseline inflammation
Non-specific measures of baseline inflammation in blood (C RP), feces (calprotectin) or intestinal tissue (endoscopic indices and histologic scorings) have proven to be correlated with disease response to therapy (Kopylov and Seidman, 2016; Narula et al., 2020). As a result, to ascertain its added value, any biomarker should exhibit an ability to predict therapy response better than these non-specific measures. Furthermore, inflammatory state will undoubtedly impact tissue architecture, gene expression, gene translation and cellular functions. As a result, a putative biomarker, even with a validated high accuracy, might be a surrogate of inflammation and not specifically related to the mechanism of action (MOA) of the drug whose response this biomarker is claimed to predict. To eliminate this suspicion, functional studies should ensue to investigate the molecular mechanisms through which this biomarker is implicated in predicting therapy response to a certain drug irrespective of baseline inflammatory status (Prins et al., 2021).
1.5. Confounding baseline concomitant therapies
Conventional therapies such as steroids still hold an important position in the therapeutic arsenal of IBD, either as a first option to ameliorate symptoms in newly diagnosed patients or as a bridging option while awaiting the effect of an initiated biological or small molecule. Despite their positive role in controlling IBD, these concomitant therapies may confound results of predictive biomarker studies. For instance, the exact processes and molecular changes mediating the effects of therapeutic steroids are, till now, not yet entirely understood. Broadly, it is reported that steroids could exert these effects through genomic or non-genomic pathways (Ramamoorthy and Cidlowski, 2016). Hence, these unpredictable effects that steroids (and plausibly other concomitant therapies) may have on molecular signatures, could lead to false conclusions pertaining the predictive role of certain biomarkers and hamper their validation (Alsoud et al., 2021b).
1.6. Unknown effect of disease duration
Data from both RCTs and real-life studies clearly showed that therapy outcomes in IBD are better in patients with shorter disease duration (Ungaro et al., 2020; Solitano et al., 2020). The exact underlying mechanisms are not yet elucidated, but it is conceivable to hypothesize that pathological networks are dynamic and thus change over time, generally leading to an increase in disease refractoriness. These changes could be intrinsic and programmed in the genetic component of IBD pathophysiology or could result from extrinsic effects of environmental, pharmaceutical or dietary factors on epigenetics and intestinal immunology, or could be a combination thereof (Renz et al., 2011). As a result, it's plausible that IBD disease biology in a newly diagnosed patient is entirely different from the biology the same patient could have after 10 or 20 years. Until this phenomenon is fully understood, one could question whether a putative biomarker that is claimed to predict response to a certain drug would exhibit the same predictive performance across patients with different disease durations. Additionally, investigators should carefully ascertain that therapy outcome in biomarker studies is not solely driven by confounding differences in disease duration among study subjects (which is currently thought to be correlated with disease refractoriness). In such instances, the putative biomarker could be indicative for accumulation of molecular changes caused by disease progression over time, rather than being indicative for response probability to a specific therapy and its specific MOA. Ideally, the accuracy of discovered biomarkers should be validated across patients' groups with different disease durations in order to ensure a wide utility in real-life clinical practice.
1.7. Unknown effect of previous exposures
Beside the effect of disease duration on therapy outcome, lower response rates in anti-TNF exposed patients is a repetitive finding in many RCTs and real-life studies that yet require underpinning (Rosario et al., 2017; Verstockt et al., 2020). Given the lack of evidence that these lower rates were driven by alterations in drug clearance (Liefferinckx et al., 2019), it's plausible that the introduction of one drug may trigger processes that eventually lead to increased refractoriness to (some) future drugs. This hypothesis was evidenced by molecular studies using intestinal tissue and serum samples from ustekinumab (UST) phase 3 trials. In these studies, only anti-TNF naïve patients showed significant normalization in CD expression profiles as a result of UST induction and maintenance therapy (Li et al., 2017; Monast et al., 2017). In addition, more inflammatory serum proteins elevated at baseline normalized in the anti-TNF naïve group during maintenance phase (Li et al., 2017). Hence, a careful consideration should be made regarding previous drug exposures while conducting predictive biomarker studies. This is important to avoid drawing inaccurate conclusions by attributing the predictive character of a certain biomarker to its implication in (non-)response to a specific drug, while being in reality a non-specific parameter for increased disease refractoriness in general.
1.8. Need for a shift from knowledge-based to data-driven biomarker discovery
Until now, many predictive biomarker studies have been driven by theories based on earlier knowledge or assumptions about the MOA of the targeted drugs and mechanisms of (non-)response to them. For instance, to predict response to a drug that is presumed to intercept mucosal inflammation through its role in blocking lymphocyte trafficking, the most intuitable approach is to search for biomarkers among chemokines, binding ligands or integrins known to be implicated in the mucosal immune system. Such a narrow approach could lead to pursue futile research as previous knowledge or speculations may appear to be inaccurate or at least incomplete (Zeissig et al., 2019). Instead, “data-driven research” holds substantial promise to advance biomarker discovery in IBD. This latter approach has become recently more feasible through the increasing availability of high-throughput analytical techniques whose data output can be further interrogated and modelled by numerous unbiased artificial intelligence algorithms (Seyed Tabib et al., 2020). Additionally, several molecular layers (proteomics, transcriptomics, metagenomic, epigenetic, metabolomic and genomics) can presently be easily and simultaneously analyzed from patients’ bio-samples and further integrated in multi-omic models. 41As opposed to the former knowledge-based research, unprejudiced data-driven research relies entirely on data to infer molecular components (i.e. biomarkers) implicated in (non-)response to a certain drug. Using currently available biologic networks databases, researchers can further investigate how the identified biomarkers are involved in pathways related to mucosal inflammation and help set new theories for future functional studies (Fiocchi and Iliopoulos, 2021). Such approaches will likely result in predictive signatures, rather than single gene/protein biomarkers, and hence might have a more robust predictive accuracy. Furthermore, integrating multi-omic data from biomarker studies of several drugs and coalescing new insights on mechanisms of actions and (non-)response may guide physicians towards evidenced-based combinations of available therapies that could yield optimal synergic effects (Stalgis et al., 2021). This innovative approach is still not widely adopted in IBD studies, mainly due to scarcity of resources and lack of standardized methodologies tailored to the purpose of advancing personalized medicine in IBD. However, IBD professional organization and research consortia can play a vital role in formulating clear methodologies and secure sufficient fundings to help anchor this approach in IBD research. This robust and highly predictive potential of multi-omic biomarkers is accompanied with a tradeoff in increased complexity, as it is impractical to collect many different biosamples and apply different technologies in the clinical setting. Therefore, IBD researchers should translate these complex single or multi-omic signatures into easy-to-implement clinical tools. This translation is not a hypothetical aspiration, but has already been applied with the development of a whole blood easy-to-use qPCR-based prognostic classifier as surrogate for a CD 8 T cell gene expression signature (Biasci et al., 2019).
1.9. Collaboration and data sharing
It is well known that oncology has been the leading field in medicine in delivering personalized therapy to patients in the past years. Although the complexity of cancer is different than IBD, this success could never have been achieved without the early realization of all actors in the field that the delivery of personalized medicine is a “team sport”, and that working together is the key to overcome obstacles, standardize methodologies and advance the field (Rodriguez and Pennington, 2018). Recently, biomarker discovery research in IBD has witnessed the start of some promising collaborations among academic research groups, and also between academia and pharmaceutical industry (e.g. COLLIBRI consortium, IMI programs including 3 TR and Immuniverse). Further augmenting this collaborative mindset is essential to deliver the long-awaited breakthroughs and offer the optimum in personalized therapy to IBD patients. Besides collaborative efforts on certain projects, the public availability of omics data generated from predictive biomarker studies is paramount to expedite the advances in the field. These publicly available omics data, along with standardized and detailed reporting on phenotypes, characteristics and therapy outcomes of patients in whom the data were generated and code scripts used to run the bioinformatic processing, offer several advantages: allow ing an objective judgment of the design of the studies and their results; enabling other researchers to correctly validate previous results using their own new datasets; providing the suitable materials bioinformaticians and statisticians need to tailor bioinformatic tools to the pathology of IBD; and offering the chance for other investigators to apply newly developed bioinformatic algorithms that may refine previous findings yielding new insights, and even more reliable biomarkers.
1.10. Validation and clinical application
Before biomarkers can be clinically implemented, their clinical usefulness in guiding therapeutic decisions to achieve improved outcomes should ideally be determined. Many aspects of this step are still debated, including whether determining this clinical utility is always necessary, what the sufficient sample size would be and whether this step should solely be performed through RCTs. Although not being a therapeutic predictive biomarker, the widely used TPMT testing, assessed before initiating thiopurines, has never undergone an RCT assessment of clinical utility. Furthermore, large RCTs may not be needed to determine the clinical utility of a predictive biomarker: the United States Food and Drug Administration (FDA) approved microsatellite instability testing, predictive for the response to pembrolizumab in patients with colorectal cancer, based on data from only 149 patients (Marcus et al., 2019). In addition, high quality evidence can also be generated from studies other than RCTs: the prospective-specimen-collection, retrospective-blinded-evaluation (PRoBE) design has been proposed and well described to avoid complexities of RCTs, while ensuring the statistical integrity of biomarker evaluation in real-life clinical settings (Pepe et al., 2008).
Another consideration is the importance of validating discovered biomarkers in diverse populations, to explore a world-wide future clinical utility. As different geographic and ethnic populations have different genetic predisposition status, different diets and different environmental exposures, it is reasonable to assume the existences of analogous differences in processes underlying the development of the disease, its flares, and mechanisms leading to (non-) response to drugs. These differences may be the reason for the inconsistent findings pertaining the whole-blood TREM1 biomarker generated from different cohorts, in addition to differences in outcome definitions (Verstockt et al., 2019a; Gaujoux et al., 2019; Verstockt et al., 2019b; Verstockt, 2022).
Evidence-based analytical validity and clinical utility are not the only catalysts to guarantee a high uptake of biomarkers in clinical practice. Regulatory and financial aspects are also issues that should pragmatically be solved by regulators with clear and relevant guidelines. Currently, a large share of IBD expenditure is attributed to a small group of patients who have very refractory forms of IBD, requiring frequent switching of biologicals and multiple surgical interventions that are obviously associated with high need for in-patient care and lost productive life years (Alulis et al., 2021; Park et al., 2019). Those patients are most likely to benefit from personalized therapy approach using developed biomarkers. It is the responsibility of IBD biomarker researchers and interested health economists to convey the benefits of biomarker-driven therapeutic decisions in improving population health and alleviating scarcity in financial resources to policy makers and regulators. Based on the collective (basic, translational and economic) evidence, regulators can then impose the implementation of predictive biomarkers in therapeutic decision in order to be entitled to insurance coverage. Costs of utilizing properly developed and robust biomarkers would eventually be negligible compared to the overall financial gain.
3. Conclusion
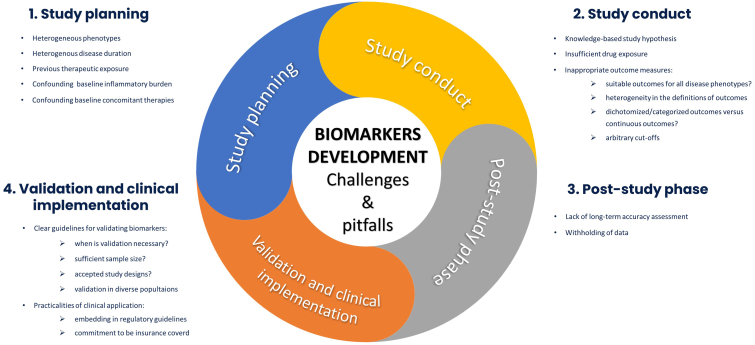
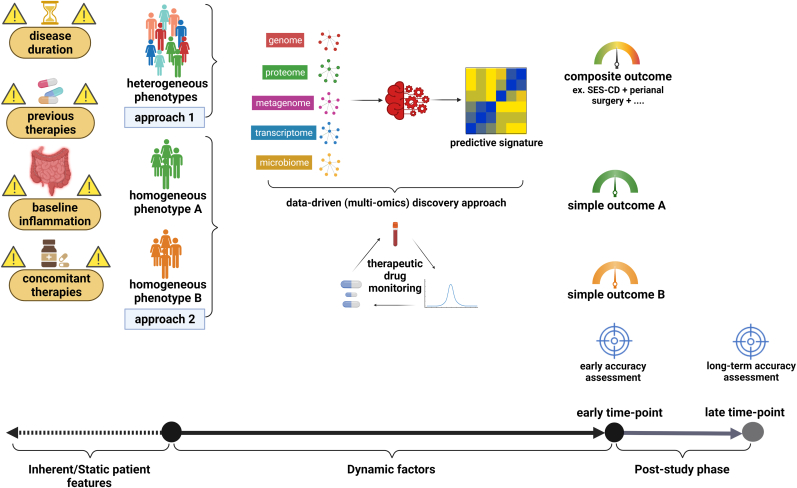
Biomarker discovery to predict therapy response in IBD is an area of prime importance, driven by the pressing need to break the current therapeutic ceiling. Although initial steps have been taken in this direction, it is paramount to critically assess current design of predictive biomarker studies to identify methodological weaknesses and optimize the use of currently scarce resources. Cautious conduct of biomarker studies taking into account the various challenges (Fig. 1) and alternatives (Fig. 2) we highlighted in this article is required in order to deliver reliable predictive biomarkers that are direly needed to increase the effectiveness of available therapies and raise hope for IBD patients and their treating physicians in the decade ahead.
Fig. 1.
Challenges and pitfalls in predictive biomarker studies in inflammatory bowel disease.
Fig. 2.
Alternative design of predictive biomarker studies to deal with various challenges. Precaution is required for heterogenous patients' characteristics (including phenotype, disease duration, baseline inflammation, previous and concomitant therapies). Therapeutic drug monitoring is ideally implemented to ensure sufficient drug exposure. Baseline bio-samples are analyzed yielding several molecular layers which are then interrogated through artificial intelligence algorithms to confer signatures that are most predictive for disease outcomes. Disease outcomes are assessed using indices/measures that are compatible with disease phenotypes. Accuracy of identified signatures is reported in predicting early and long-term disease outcomes.
Contributors
DA: literature search, creating figures and drafting the manuscript. SV and BV: critical revision of the manuscript. All authors agreed on the final manuscript.
Declaration of competing interest
B Verstockt reports financial support for research from Pfizer; lecture fees from Abbvie, Biogen, Chiesi, Falk, Ferring, Galapagos, Janssen, MSD, Pfizer, R-Biopharm, Takeda and Truvion; consultancy fees from Applied Strategic, Atheneum, Bristol Myers Squibb, Guidepoint, Ipsos, Janssen, Progenity, Sandoz, Sossei Heptares and Takeda. S Vermeire reports financial support for research: MSD, AbbVie, Galapagos, Takeda, Pfizer, J&J; Lecture fees from MSD, AbbVie, Takeda, Ferring, Centocor, Hospira, Pfizer, J&J, Genentech/Roche; consultancy fees from MSD, AbbVie, Takeda, Ferring, Centocor, Hospira, Pfizer, J&J, Genentech/Roche, Celgene, Mundipharma, Celltrion, SecondGenome, Prometheus, Shire, Prodigest, Gilead, Galapagos. D Alsoud declares no conflicts of interest.
Acknowledgments
SV and BV received research grant from the Leona M. and Harry B. Helmsley Charitable Trust, and DA is funded by this grant. DA received research grant from the IBD Patient's Association Flanders (CCV VZW). BV supported by the Clinical Research Fund (KOOR) at the University Hospitals Leuven, Belgium.
References
- Al-Bawardy B., Shivashankar R., Proctor D.D. Novel and emerging therapies for inflammatory bowel disease. Front. Pharmacol. 2021;12:569. doi: 10.3389/fphar.2021.651415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoud D., Verstockt B., Fiocchi C., Vermeire S. Breaking the therapeutic ceiling in drug development in ulcerative colitis. Lancet Gastroenterol. Hepatol. 2021;6(7):589–595. doi: 10.1016/S2468-1253(21)00065-0. [DOI] [PubMed] [Google Scholar]
- Alsoud D., Verstockt S., Sabino J., et al. P062 Effects of exposure to steroids on the PredictSURE whole blood prognostic assay in Inflammatory Bowel Disease. J. Crohn's Colitis. 2021;15(Supplment_1):S168–S. [Google Scholar]
- Altman D.G., Royston P. The cost of dichotomising continuous variables. Br. Med. J. 2006;332(7549):1080. doi: 10.1136/bmj.332.7549.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alulis S., Vadstrup K., Olsen J., et al. The cost burden of Crohn's disease and ulcerative colitis depending on biologic treatment status – a Danish register-based study. BMC Health Serv. Res. 2021;21(1):836. doi: 10.1186/s12913-021-06816-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasci D., Lee J.C., Noor N.M., et al. A blood-based prognostic biomarker in IBD. Gut. 2019;68(8):1386. doi: 10.1136/gutjnl-2019-318343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.T. Pathophysiology of inflammatory bowel diseases. N. Engl. J. Med. 2020;383(27):2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- Dreesen E. New tools for therapeutic drug monitoring: making big things out of small pieces. J. Crohn's Colitis. 2021 doi: 10.1093/ecco-jcc/jjab137. [DOI] [PubMed] [Google Scholar]
- Dulai P.S., Levesque B.G., Feagan B.G., D'Haens G., Sandborn W.J. Assessment of mucosal healing in inflammatory bowel disease: review. Gastrointest. Endosc. 2015;82(2):246–255. doi: 10.1016/j.gie.2015.03.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov V., Mannino F., Zhang R. Consequences of dichotomization. Pharmaceut. Stat. 2009;8(1):50–61. doi: 10.1002/pst.331. [DOI] [PubMed] [Google Scholar]
- Fiocchi C., Iliopoulos D. IBD systems biology is here to stay. Inflamm. Bowel Dis. 2021;27(6):760–770. doi: 10.1093/ibd/izaa343. [DOI] [PubMed] [Google Scholar]
- Gaujoux R., Starosvetsky E., Maimon N., et al. Cell-centred meta-analysis reveals baseline predictors of anti-TNFα non-response in biopsy and blood of patients with IBD. Gut. 2019;68(4):604–614. doi: 10.1136/gutjnl-2017-315494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda H., Hirata A., Katagiri T., et al. Prediction of disease flare by biomarkers after discontinuing biologics in patients with rheumatoid arthritis achieving stringent remission. Sci. Rep. 2021;11(1):6865. doi: 10.1038/s41598-021-86335-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G.G. The global burden of IBD: from 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015;12(12):720–727. doi: 10.1038/nrgastro.2015.150. [DOI] [PubMed] [Google Scholar]
- Kaplan GA-O, Windsor JA-O. The Four Epidemiological Stages in the Global Evolution of Inflammatory Bowel Disease. (1759-5053 (Electronic)). [DOI] [PMC free article] [PubMed]
- Kobayashi T., Siegmund B., Le Berre C., et al. Ulcerative colitis. Nat. Rev. Dis. Prim. 2020;6(1) doi: 10.1038/s41572-020-0205-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Motoya S., Nakamura S., et al. Discontinuation of infliximab in patients with ulcerative colitis in remission (HAYABUSA): a multicentre, open-label, randomised controlled trial. Lancet Gastroenterol. Hepatol. 2021;6(6):429–437. doi: 10.1016/S2468-1253(21)00062-5. [DOI] [PubMed] [Google Scholar]
- Kopylov U., Seidman E. Predicting durable response or resistance to antitumor necrosis factor therapy in inflammatory bowel disease. Ther. Adv. Gastroenterol. 2016;9(4):513–526. doi: 10.1177/1756283X16638833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Berre C., Peyrin-Biroulet L. Selecting end points for disease-modification trials in inflammatory bowel disease: the SPIRIT consensus from the IOIBD. Gastroenterology. 2021;160(5):1452–1460. doi: 10.1053/j.gastro.2020.10.065. e21. [DOI] [PubMed] [Google Scholar]
- Li K., Hayden K., Wadman E., et al. Molecular response to ustekinumab in moderate-to-severe Crohn's disease by serum protein and biopsy gene expression analysis: results from Ustekinumab phase 3 studies. J. Crohn's Colitis. 2017;11(Suppl. 1):S57–S58. [Google Scholar]
- Liefferinckx C., Verstockt B., Gils A., et al. Impact of first-line infliximab on the pharmacokinetics of second-line vedolizumab in inflammatory bowel diseases. U. Eur. Gastroenterol. J. 2019;7(6):750–758. doi: 10.1177/2050640619841538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E., et al. OP01 Withdrawal of infliximab or anti-metabolite therapy in Crohn's Disease patients in sustained remission on combination therapy: a randomized unblinded controlled trial (SPARE) J. Crohn's Colitis. 2022 [Google Scholar]
- Marcus L., Lemery S.J., Keegan P., Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin. Cancer Res. 2019;25(13):3753–3758. doi: 10.1158/1078-0432.CCR-18-4070. [DOI] [PubMed] [Google Scholar]
- Monast C., Li K., Myshkin E., Brodmerkel C., Friedman J., Baribaud F. Molecular surrogates of histologic activity in Crohn's disease. U. Eur. Gastroenterol. J. 2017;5(5 Suppl. 1):A523. [Google Scholar]
- Narula N., Wong E.C.L., Aruljothy A., et al. Ileal and rectal ulcer size affects the ability to achieve endoscopic remission: a post hoc analysis of the SONIC trial. Am. J. Gastroenterol. 2020;115(8):1236–1245. doi: 10.14309/ajg.0000000000000617. [DOI] [PubMed] [Google Scholar]
- Ng SC, Shi HY, Hamidi N, et al. Worldwide Incidence and Prevalence of Inflammatory Bowel Disease in the 21st Century: a Systematic Review of Population-Based Studies. (1474-547X (Electronic)). [DOI] [PubMed]
- Papamichael K., Gils A., Rutgeerts P., et al. Role for therapeutic drug monitoring during induction therapy with TNF antagonists in IBD. Inflamm. Bowel Dis. 2015;21(1):182–197. doi: 10.1097/MIB.0000000000000202. [DOI] [PubMed] [Google Scholar]
- Park K.T., Ehrlich O.G., Allen J.I., et al. The cost of inflammatory bowel disease: an initiative from the Crohn's & colitis foundation. Inflamm. Bowel Dis. 2019;26(1):1–10. doi: 10.1093/ibd/izz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe M.S., Feng Z., Janes H., Bossuyt P.M., Potter J.D. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J. Natl. Cancer Inst. 2008;100(20):1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrin-Biroulet L., Sandborn W., Sands B.E., et al. Selecting therapeutic targets in inflammatory bowel disease (STRIDE): determining therapeutic goals for treat-to-target. Off. J. Am. Coll. Gastroenterol. 2015;110(9):1324–1338. doi: 10.1038/ajg.2015.233. [DOI] [PubMed] [Google Scholar]
- Pierre N., Baiwir D., Huynh-Thu V.A., et al. Discovery of biomarker candidates associated with the risk of short-term and mid/long-term relapse after infliximab withdrawal in Crohn's patients: a proteomics-based study. Gut. 2020 doi: 10.1136/gutjnl-2020-322100. [DOI] [PubMed] [Google Scholar]
- Prins M.M., Verstockt B., Ferrante M., Vermeire S., Wildenberg M.E., Koelink P.J. Monocyte TREM-1 levels associate with anti-TNF responsiveness in IBD through autophagy and fcγ-receptor signaling pathways. Front. Immunol. 2021;12(750) doi: 10.3389/fimmu.2021.627535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy S., Cidlowski J.A. Corticosteroids: mechanisms of action in health and disease. Rheum. Dis. Clin. N. Am. 2016;42(1):15–vii. doi: 10.1016/j.rdc.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz H., von Mutius E., Brandtzaeg P., Cookson W.O., Autenrieth I.B., Haller D. Gene-environment interactions in chronic inflammatory disease. Nat. Immunol. 2011;12(4):273–277. doi: 10.1038/ni0411-273. [DOI] [PubMed] [Google Scholar]
- Roda G., Jharap B., Neeraj N., Colombel J.-F. Loss of response to anti-TNFs: definition, epidemiology, and management. Clin. Transl. Gastroenterol. 2016;7(1):e135–e. doi: 10.1038/ctg.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roda G., Chien Ng S., Kotze P.G., et al. Crohn's disease. Nat. Rev. Dis. Prim. 2020;6(1) doi: 10.1038/s41572-020-0156-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez H., Pennington S.R. Revolutionizing precision oncology through collaborative proteogenomics and data sharing. Cell. 2018;173(3):535–539. doi: 10.1016/j.cell.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario M., French J.L., Dirks N.L., et al. Exposure-efficacy relationships for vedolizumab induction therapy in patients with ulcerative colitis or crohn's disease. J. Crohn's Colitis. 2017;11(8):921–929. doi: 10.1093/ecco-jcc/jjx021. [DOI] [PubMed] [Google Scholar]
- Seyed Tabib N.S., Madgwick M., Sudhakar P., Verstockt B., Korcsmaros T., Vermeire S. Big data in IBD: big progress for clinical practice. Gut. 2020;69:1520–1532. doi: 10.1136/gutjnl-2019-320065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solitano V., D’amico F., Zacharopoulou E., Peyrin-Biroulet L., Danese S. Early intervention in ulcerative colitis: ready for prime time? J. Clin. Med. 2020;9(8):1–12. doi: 10.3390/jcm9082646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalgis C., Deepak P., Mehandru S., Colombel J.F. Rational combination therapy to overcome the plateau of drug efficacy in inflammatory bowel disease. Gastroenterology. 2021;161(2):394–399. doi: 10.1053/j.gastro.2021.04.068. [DOI] [PubMed] [Google Scholar]
- Turner D., Ricciuto A., Lewis A., et al. STRIDE-II: an update on the selecting therapeutic targets in inflammatory bowel disease (STRIDE) initiative of the international organization for the study of IBD (IOIBD): determining therapeutic goals for treat-to-target strategies in IBD. Gastroenterology. 2021;160(5):1570–1583. doi: 10.1053/j.gastro.2020.12.031. [DOI] [PubMed] [Google Scholar]
- Ungaro R.C., Aggarwal S., Topaloglu O., Lee W.-J., Clark R., Colombel J.-F. Systematic review and meta-analysis: efficacy and safety of early biologic treatment in adult and paediatric patients with Crohn's disease. Aliment. Pharmacol. Ther. 2020;51(9):831–842. doi: 10.1111/apt.15685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstockt B., Ferrante M. Editorial: a clinical decision tool to identify patients who might benefit most from intensified dosing in the biological era-getting nearer? Aliment. Pharmacol. Ther. 2020;51(7):737–738. doi: 10.1111/apt.15634. [DOI] [PubMed] [Google Scholar]
- Verstockt B., Verstockt S., Blevi H., et al. TREM-1, the ideal predictive biomarker for endoscopic healing in anti-TNF-treated Crohn's disease patients? Gut. 2019;68(8):1531–1533. doi: 10.1136/gutjnl-2018-316845. [DOI] [PubMed] [Google Scholar]
- Verstockt B., Verstockt S., Dehairs J., et al. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine. 2019;40:733–742. doi: 10.1016/j.ebiom.2019.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstockt B., Mertens E., Dreesen E., et al. Influence of drug exposure on vedolizumab-induced endoscopic remission in anti-tumour necrosis factor [TNF] naïve and anti-TNF exposed IBD patients. J. Crohn's Colitis. 2020;14(3):332–341. doi: 10.1093/ecco-jcc/jjz151. [DOI] [PubMed] [Google Scholar]
- Verstockt B., Noor N.M., Marigorta U.M., Pavlidis P., Deepak P., Ungaro R.C. Results of the Seventh Scientific Workshop of ECCO: precision medicine in IBD - disease outcome and response to therapy. J. Crohn's Colitis. 2021 doi: 10.1093/ecco-jcc/jjab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstockt B., Abreu N., Torres J. Selecting the ideal candidate for anti-TNF discontinuation in crohn's disease, dream or reality? Gastroenterology. 2021;161(1):353–355. doi: 10.1053/j.gastro.2021.05.005. [DOI] [PubMed] [Google Scholar]
- Verstockt B., et al. DOP81 Baseline whole-blood gene expression of TREM1 does not predict clinical or endoscopic outcomes following adalimumab treatment in patients with Ulcerative Colitis or Crohn's Disease in the SERENE studies. J. Crohn's Colitis. 2022 doi: 10.1093/ecco-jcc/jjad170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeissig S., Rosati E., Dowds C.M., et al. Vedolizumab is associated with changes in innate rather than adaptive immunity in patients with inflammatory bowel disease. Gut. 2019;68(1):25–39. doi: 10.1136/gutjnl-2018-316023. [DOI] [PubMed] [Google Scholar]