Abstract
Background
Pulmonary metastasis (M1-PUL) as first site of dissemination in pancreatic ductal adenocarcinoma (PDAC) is a rare event and may define a distinct biological subgroup.
Patients and methods
Arbeitsgemeinschaft Internistische Onkologie-Young Medical Oncologists-Pankreas-0515 study (AIO-YMO-PAK-0515) was a retrospective German multicenter study investigating clinical and molecular characteristics of M1-PUL PDAC patients; 115 M1-PUL PDAC patients from 7 participating centers were included. Clinical characteristics and potential prognostic factors were defined within the M1-PUL cohort. Archival tumor samples were analyzed for Her2/neu, HNF1A and KRT81 expression. Additionally, messenger RNA (mRNA) expression analysis (using a 770-gene immune profiling panel) was carried out in the M1-PUL and in a control cohort (M1-ANY).
Results
Median overall survival in the entire M1-PUL cohort was 20 months; the most favorable prognosis (median survival: 28 months) was observed in the subgroup of 66 PDAC patients with metachronous lung metastases after previous curative-intent surgery. The number of metastatic lesions, uni- or bilateral lung involvement as well as metastasectomy were identified as potential prognostic factors. Her2/neu expression and PDAC subtyping (by HNF1A and KRT81) did not differ between the M1-PUL and the M1-ANY cohort. mRNA expression analysis revealed significant differentially expressed genes between both cohorts: CD63 and LAMP1 were among the top 20 differentially expressed genes and were identified as potential mediators of organotropism and favorable survival outcome of M1-PUL patients.
Conclusion
M1-PUL represents a clinically favorable cohort in PDAC patients. Site of relapse might already be predetermined at the time of surgery and could potentially be predicted by gene expression profiling.
Key words: chemotherapy, gene expression, lung metastasis, pancreatic cancer, prognosis
Highlights
-
•
The retrospective multicenter AIO-YMO-PAK-0515 study defines M1-PUL as a clinically favorable subgroup in PDAC.
-
•
The number of metastatic lesions, bilateral lung involvement and surgical metastasectomy may serve as prognostic factors.
-
•
Immune-related gene expression differs between patients with isolated pulmonary relapse versus other sites of relapse.
Introduction
The last decade saw an impressive increase of available therapies that harness the immune system or tackle specific genomic alterations of solid cancers. Yet, combination chemotherapy remains the mainstay of treatment for most patients with pancreatic ductal adenocarcinoma (PDAC).1 As seen for other cancer entities, data on genetic alterations in PDAC have been rapidly growing in recent years.2, 3, 4 PDAC thereby often is defined by an oncogenic KRAS mutation, followed by mutations in SMAD4, TP53 and CDKN2A. Other mutations are much less frequent and show large interindividual heterogeneity.4 Tumors with microsatellite instability or mutations in genes that code for DNA damage repair (most prominently BRCA1/2) are rare exceptions for actionable genetic alterations in PDAC.1 The clinical and molecular characterization of prognostically exceptional subgroups could thus be a key element in developing efficient, tailored treatment approaches.5
Liver metastasis represents the first site of dissemination in >80% of metastatic PDAC patients.6 Pulmonary metastasis (M1-PUL) as first site of dissemination is a rare event and has been suggested to define a unique clinical subgroup of PDAC patients with a favorable prognosis in different single-center studies.7, 8, 9, 10 Thus far, only insufficient knowledge on biological determinates for M1-PUL in PDAC exists. As one putative mechanism for organotropism in PDAC, overexpression of Her2/neu has been described.11 Furthermore, prognosis of PDAC patients has been consistently reported to correlate with three distinct biological subtypes as determined by gene expression profiling and more recently by immunohistochemistry (HNF1A and KRT81): quasi-mesenchymal (KRT81+), classical (KRT81 and HNF1A negative) and exocrine-like (HNF1A+).2,3,12, 13, 14 Additionally, it has been suggested that metastatic organotropism could also be immune-mediated.15
Here we report the final results of a retrospective multicenter study conducted at seven large academic cancer centers of the Arbeitsgemeinschaft Internistische Onkologie (AIO) study group in Germany. The aim of this study was to confirm previous single-center observations regarding outcome and to define prognostic factors for PDAC patients with isolated M1-PUL. Additionally, we sought to elucidate potential molecular predictors of isolated M1-PUL in PDAC. Our group thereby focused on a potential relationship between isolated M1-PUL and an overexpression of Her2/neu, previously defined molecular PDAC subtypes and the expression of immune-related genes.
Patients and methods
Patient population and treatment
Patients who had been diagnosed and/or treated with PDAC at one of the participating study centers were identified retrospectively and included into a central patient database at Ludwig-Maximilians-University of Munich. For the current study, medical records and correlating computed tomography (CT) findings were retrospectively analyzed for all included PDAC patients with isolated lung metastases (M1-PUL cohort). The following data were evaluated: patient and tumor characteristics including age, sex, tumor–node–metastasis (TNM) stage, grading, date of initial PDAC diagnosis, date of first appearance of M1-PUL, treatment (surgery, radiotherapy, first- to third-line chemotherapeutic regimens), as well as size, number and site of M1-PUL upon initial diagnosis and follow-up. Occurrence of M1-PUL had to be confirmed by histology or retrospective review of serial CT scans, showing enlarging pulmonary nodules over time. To rule out synchronous extrapulmonary dissemination, abdominal CT scans were reviewed for the presence of extrapulmonary metastases. Survival status was determined by (i) review of medical records at the respective institution, (ii) consultation of patient’s primary care physician or (iii) consultation of patient’s civil registrar office.
The multicenter Arbeitsgemeinschaft Internistische Onkologie-Young Medical Oncologists-Pankreas-0515 study (AIO-YMO-PAK-0515) study was developed within the ‘Young Medical Oncologists (YMO)’ group of the German AIO study group and supported by the working group ‘Pancreatic cancer’ of the AIO. The project was approved by all local ethics committees at the respective study centers after initial approval of this multicenter study project by Ludwig-Maximilians-University of Munich (approval number 134-15).
Existing cohorts of patients with resected PDAC from Ludwig-Maximilians-University of Munich who developed metastases at any other site (excluding patients with M1-PUL) were used as controls for immunohistochemical analyses: 47 patients from an available dataset with recurrent PDAC with liver metastases, peritoneal carcinomatosis or local recurrence (M1-ANY). For messenger RNA (mRNA) expression analysis, an M1-ANY subcohort (n = 29) with well-established clinical characteristics (derived from a prospectively maintained database) was utilized as the control group.
Immunohistochemical detection and evaluation of Her2/neu, HNF1A and KRT81
Four-micrometer-thick sections from formalin-fixed, paraffin-embedded (FFPE) tissue were dewaxed and stained as follows: Her2/neu expression was detected using the Ventana ready-to-use Her2 kit (rabbit clone 4B5) on an automated slide stainer (Ventana Benchmark ULTRA, Ventana Medical Systems, Tucson, AZ) strictly following the manufacturer’s instructions. Her2/neu expression was examined and scored as four-tire score as described previously.11 KRT81 and HNF1A expression was detected by manual staining (anti-KRT81, clone 3B10-5B10, dilution 1 : 120, LS Bio, Seattle, WA; polyclonal rabbit anti-HNF1A, dilution 1 : 100, Atlas antibodies, Stockholm, Sweden). Briefly, tumors displaying strong and specific staining in the respective cellular compartment (membrane and cytoplasm for KRT81; nucleus for HNF1A) were scored as positive and otherwise as negative. Appropriate positive and negative control tissue was included in each staining run.
RNA expression analysis
For tumor RNA extraction, tumor cell content and tissue suitability for nucleic acid extraction was assessed by a board-certified pathologist (SO) on routine hematoxylin–eosin-stained slides and tumor areas were determined. Tumor-containing tissue areas with at least 50% tumor cell content were extracted from subsequent unstained FFPE tissue slides by microscopically controlled tissue dissection and RNA was isolated using RNeasy FFPE mini kits (Qiagen, Hilden, Germany). A total of 250 ng of total RNA was used in the gene expression analysis on the nCounter® FLEX Analysis System (NanoString Technologies Inc., Seattle, WA) using the nCounter® PanCancer Immune Profiling Panel. Raw counts were quality controlled, background subtracted and normalized using 18 housekeeping genes in nSolver Analysis Software (v4.0) (NanoString Technologies Inc.). Expression values were log2-transformed for statistical analyses.
Statistical analyses
Overall survival from the time of first diagnosis of M1-PUL to the time of death from any cause was selected as the primary study endpoint. Patients who were alive at their last follow-up were censored. Estimated overall survival was calculated using the Kaplan–Meier method; differences in overall survival according to size, number and site of M1-PUL were compared using the log-rank test. Differences in immunohistochemical expression of Her2/neu, KRT81 and HNF1A were determined using the chi-square test. Statistical analyses for mRNA expression analysis were carried out using the nSolver 4.0 software (NanoString Technologies Inc.). SPSS PASW 25.0 (SPSS Inc., Chicago, IL) was used for the remaining statistical analyses. If applicable, a P value of ≤0.05 was considered statistically significant.
Results
Patient characteristics
A total of 115 PDAC patients from 7 German AIO study centers with isolated M1-PUL were included. Participating centers were: Ludwig-Maximilians-University of Munich (n = 43), Charité Berlin (n = 30) and the university hospitals of Essen (n = 12), Ulm (n = 12), Frankfurt (n = 10), Wuerzburg (n = 5) and Rostock (n = 3). Median age was 69 years and the majority (57%) of patients presented with an isolated pulmonary relapse after initial resection of the pancreatic primary in curative intent (n = 66) (Table 1). Database lock was carried out on 31 May 2017 and at that time point 78 patients had died. The estimated median overall survival for all included PDAC patients with isolated M1-PUL was 20 months; patients with metachronous lung metastases after pancreatic resection had a numerically longer median overall survival compared to patients with synchronous metastatic PDAC with isolated lung involvement (28 versus 19 months; see Table 1).
Table 1.
Patient characteristics of the AIO-YMO-PAK-0515 cohort
| Median overall survival,a months (95% CI) | ||
|---|---|---|
| All patients, n (%) | 115 (100) | 20.0 (16.3-23.7) |
| Median age (range),b years | 69 (41-84) | |
| Gender, n (%) | ||
| Male | 54 (47) | |
| Female | 61 (53) | |
| Diagnosis of pulmonary metastases, n (%) | ||
| Radiographic | 84 (73) | |
| Histological | 31 (27) | |
| Stage at initial diagnosis, n (%) | ||
| Resectable PDAC | 66 (57) | 28.0 (17.7-38.3) |
| Locally advanced PDAC | 12 (10) | 20.0 (17.2-22.7) |
| Metastatic PDAC | 37 (32) | 19.0 (13.6-24.4) |
CI, confidence interval; PDAC, pancreatic ductal adenocarcinoma.
Median overall survival was calculated from the time of initial diagnosis of pulmonary metastases (i.e. time of pulmonary relapse for patients with resectable PDAC at initial diagnosis/time of progression with pulmonary metastases for patients with locally advanced PDAC).
Age was not available for patients from Charité Berlin.
Tumor samples of the included patients were collected between 2016 and 2017; however, PDAC patients included in this study were diagnosed between 2002 and 2016. FFPE tumor tissue from resected primaries was available from 34 of the included PDAC patients and was analyzed centrally at the Ludwig-Maximilians-University of Munich. Presliced FFPE tumor sections from resected primaries with sufficient tumor content were available from an additional 13 patients.
Prognostic variables
Our group previously reported that patients with metachronous M1-PUL, <10 M1-PUL and/or unilateral lung involvement may have a favorable prognosis.10 To facilitate multicenter analysis of CT images, we defined a slightly different cut-off for number (>3 or ≤3) of M1-PUL. Metachronous M1-PUL, three or less metastases and unilateral lung involvement were confirmed as favorable prognostic variables in our cohort (Tables 1 and 2). Additionally, we retrospectively evaluated survival of patients who underwent resection of pulmonary metastases in our cohort: the estimated median overall survival in PDAC M1-PUL patients who underwent surgical metastasectomy was 33.0 versus 19.0 months for patients who did not (P = 0.029) (see Table 2).
Table 2.
Prognostic clinical factors for patients with isolated pulmonary metastases
| n | Median OS (months) | 95% CI (months) | P value | |
|---|---|---|---|---|
| Number of pulmonary metastases | ||||
| ≤3 | 33 | 33.0 | 27.1-38.9 | 0.003 |
| >3 | 71 | 19.0 | 15.1-22.9 | |
| Size of pulmonary metastases, mm | ||||
| ≤5 | 22 | 24.0 | 7.5-40.5 | 0.365 |
| >5 | 75 | 19.0 | 15.0-23.0 | |
| Location of pulmonary metastases | ||||
| Unilateral | 35 | 30.0 | 23.7-36.3 | 0.004 |
| Bilateral | 61 | 18.0 | 13.4-22.5 | |
| Resection of pulmonary metastases | ||||
| Yes | 24 | 33.0 | 22.2-43.8 | 0.029 |
| No | 90 | 19.0 | 16.0-22.0 | |
Statistical significant values are given in bold.
CI, confidence interval; OS, overall survival.
Her2/neu expression
Immunohistochemical analysis of Her2/neu receptor protein expression was carried out in primary tumor tissue of 47 resected PDAC patients who subsequently developed isolated M1-PUL versus 47 resected patients from a control cohort who developed relapse of the disease at other sites (excluding patients with M1-PUL, M1-ANY). Scoring of Her2/neu expression was carried out as described for PDAC previously, based on the scoring system initially developed for breast and gastric cancer.11,16 Her2/neu positivity (score: 3+) was observed in only one patient in the M1-PUL as well as in the M-ANY group, while equivocal Her2/neu expression was slightly more frequent in M1-PUL patients (differences not statistically significant; Table 3).
Table 3.
Her2/neu expression in the cohorts M1-PUL (n = 47) and M1-ANY (n = 47)
| Her2/neu expression (IHC score) |
||||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| M1-PUL, n (%) | 21 (45) | 15 (32) | 10 (21) | 1 (2) |
| M1-ANY (controla), n (%) | 27 (57) | 14 (30) | 5 (11) | 1 (2) |
Chi-square test: P = 0.484.
IHC, immunohistochemistry; M1-PUL, pulmonary metastasis; M1-ANY, any metastases excluding lung-only involvement.
Patients with recurrence to liver, peritoneal carcinomatosis or local recurrence.
PDAC subtyping by KRT81 and HNF1A expression
To evaluate whether isolated M1-PUL correlate with known transcriptional PDAC subtypes (quasi-mesenchymal, exocrine-like and classical subtype), we used a previously described immunohistochemical assessment of KRT81 and HNF1A.14 Staining was technically successful in 38 of 47 PDAC samples from primary tumors of resected patients who subsequently developed isolated M1-PUL. A majority of tumors (50%) were classified as ‘classical’-type PDAC, with 29% and 21% of patient samples being classified as ‘quasi-mesenchymal’ or ‘exocrine-like’, respectively. A similar distribution of PDAC subtypes was observed in the control cohort of patients with relapse at other sites (excluding patients with M1-PUL, M1-ANY; see Table 4).
Table 4.
Subtyping by KRT81 and HNF1A expression in the cohorts M1-PUL (n = 47) and M1-ANY (n = 47)a
| Quasi-mesenchymal (KRT81+) | Exocrine-like (HNF1A+) | Classical (double negative) | |
|---|---|---|---|
| M1-PUL, n (%) | 11 (29) | 8 (21) | 19 (50) |
| M1-ANY (controlb), n (%) | 8 (24) | 5 (15) | 20 (61) |
Chi-square test: P = 0.707.
M1-PUL, pulmonary metastasis; M1-ANY, any metastases excluding lung-only involvement.
Staining technically successful for 38 of 47 PDAC samples from M1-PUL patients and 33 of 47 control samples.
Patients with recurrence to liver, peritoneal carcinomatosis or local recurrence.
mRNA expression analysis
To evaluate whether gene expression of immune-related genes in primary tumor samples of resected patients with a subsequent development of isolated M1-PUL differs from a general PDAC population, we carried out mRNA expression analysis of 770 genes using the nCounter® PanCancer Immune Profiling Panel. nCounter analysis was carried out on tumor samples from 62 resected PDAC patients. Results of six patients had to be removed for the following reasons: three patients were still relapse-free at the date of last follow-up and thus inapt to answer our research question. Another three patients had to be excluded for technical reasons (amount of mRNA below critical threshold, n = 2; binding density above critical threshold, n = 1). The clinical characteristics of the successfully analyzed samples from 56 patients of the M1-PUL and the M1-ANY control cohort are summarized in Table 5: gender, age, tumor stage and grading at initial diagnosis were well balanced between the M1-PUL and M-ANY cohorts. Of note, more patients in the M1-PUL cohort had nodal-positive disease at surgery (91% versus 59%) and received chemoradiotherapy (CRT) as adjuvant treatment (27% versus 3%). Among the 56 patients included in the mRNA expression analysis set, survival was significantly longer for patients who relapsed with isolated M1-PUL versus patients who did relapse with local recurrence, liver or peritoneal metastases (M1-ANY; 28 versus 15 months, P = 0.034; see Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2021.100363).
Table 5.
Clinical characteristics of the cohorts M1-PUL and M1-ANY analyzed for mRNA expression (n = 56)
| M1-PUL (n = 27) | M1-ANY (n = 29) | |
|---|---|---|
| Gender, n (%) | ||
| Male | 13 (48) | 16 (55) |
| Female | 14 (52) | 13 (45) |
| Age | ||
| Median age (range), years | 69 (46-82) | 69 (53-83) |
| pT stage, n (%) | ||
| pT1 | — | — |
| pT2 | — | 1 (3) |
| pT3 | 24 (100) | 28 (97) |
| Not documented | 3 | — |
| pN stage, n (%) | ||
| pN0 | 2 (9) | 12 (41) |
| pN1 | 22 (91) | 17 (59) |
| Not documented | 3 | — |
| Grading, n (%) | ||
| G1 | 1 (5) | 0 (0) |
| G2 | 12 (50) | 12 (41) |
| G3 | 10 (46) | 17 (59) |
| Not documented | 4 | — |
| Adjuvant treatment, n (%) | ||
| None | 2 (8) | 4 (14) |
| Gemcitabine | 17a (65) | 24b (83) |
| Chemoradiotherapy | 7 (27) | 1 (3) |
| Not documented | 1 | — |
| First site of recurrence, n (%) | ||
| Lung | 27 (100) | — |
| Liver | — | 14 (48) |
| Peritoneal carcinomatosis | — | 4 (14) |
| Local recurrence | — | 11 (38) |
M1-PUL, pulmonary metastasis; M1-ANY, any metastases excluding lung-only involvement; mRNA, messenger RNA.
Two patients were treated with gemcitabine plus erlotinib within a clinical trial.
One patient was treated with gemcitabine plus cisplatin in combination with regional hyperthermia within a clinical trial.
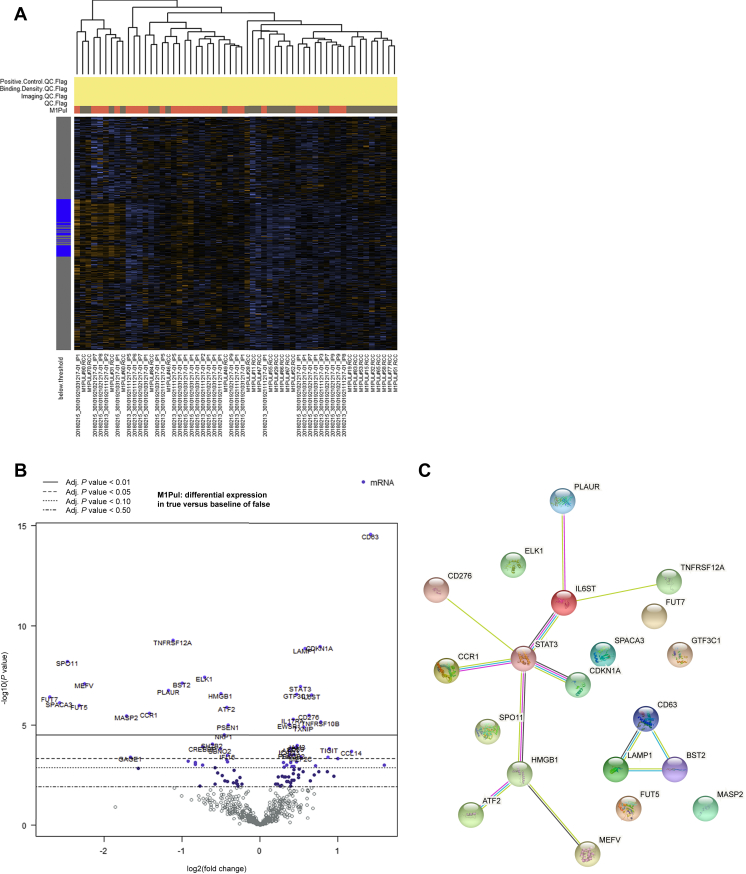
The heatmap of mRNA expression with automated clustering according to M1-PUL versus M1-ANY is shown in Figure 1A. Differentially expressed genes are depicted in Figure 1B. Statistical significance in differential gene expression—after adjusting for multiplicity testing—was determined by the nSolver algorithm. The top 20 gene results were confirmed using the limma software package (for details see Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100363).17 While there was a noticeable proportion of differentially expressed genes that are involved in chemokine regulation, there was no clear enrichment for a specific immunological gene set. Networks of gene products from the top 20 differentially expressed genes were visualized using the STRING database (see Figure 1C).18 Genes in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100363, are colored red and blue according to their affiliation to one of the two protein networks. Genes without affiliated protein network are marked gray in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100363. Among the top 20 differentially expressed genes summarized in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100363, mRNA expression was up-regulated in 7 and reduced in 13 genes (Figure 1B). A literature research for all 20 genes revealed that of the 7 up-regulated genes, 3 had been previously described to be associated with favorable outcomes in PDAC (CD63 and LAMP1) or breast cancer (GTF3C1). Of note, CD63 and LAMP1 were found to have multiple interactions in the STRING database analysis. As visualized using the STRING database, there is also a strong interaction between CDKN1A, STAT3 and IL6ST (Figure 1C). Of the 13 genes whose expression was reduced, 7 are part of predefined immunological gene sets in the nCounter® advanced analysis [tumor necrosis factor (TNF) superfamily, regulation, chemokines, cytokines, cancer testis antigen, cell functions, leukocyte functions, natural killer and T-cell functions]. Three of the 13 genes whose expression was reduced have been previously implicated in metastasis formation, cancer progression and/or epithelial–mesenchymal transition of PDAC. Furthermore, 8 out of these 13 genes have been implicated in metastasis formation, cancer progression and/or epithelial–mesenchymal transition of other cancer types (TNFRSF12A, PLAUR, FUT5, FUT7, BST2, ELK1, CCR1, MASP2).
Figure 1.
Results of the mRNA expression analysis of 770 genes using the nCounter® PanCancer Immune Profiling Panel.
nCounter analysis was successfully carried out on tumor samples from 56 resected PDAC patients (27 patients from the M1-PUL and 29 patients from the M1-ANY cohort). (A) Heatmap of mRNA expression with automated clustering according to M1-PUL versus M1-ANY cohorts. (B) Differentially expressed genes depicted as a volcano plot. (C) Networks of gene products form the top 20 differentially expressed genes visualized by using the STRING database.
Adj., adjusted; M1-PUL, pulmonary metastasis; M1-ANY, any metastases excluding lung-only involvement; mRNA, messenger RNA; PDAC, pancreatic ductal adenocarcinoma.
Discussion
AIO-YMO-PAK-0515 was a retrospective multicenter study of 115 PDAC patients with isolated lung metastasis from 7 large German cancer centers. Its aim was to improve our understanding of the clinical characteristics and the prognosis of this unique PDAC subgroup. As already hypothesized by single-center analyses on this topic,7, 8, 9, 10 and recently summarized within a systematic review and meta-analysis,19 our study confirms the favorable prognosis of M1-PUL PDAC patients. The median overall survival estimate for the whole cohort was 20 months. A particularly favorable prognosis was observed in patients who develop lung metastases after PDAC surgery (median overall survival: 28 months). Of note, more than half of the M1-PUL patients (n = 66/115) included in this AIO cohort study underwent surgical PDAC resection in curative intent before experiencing a relapse with isolated lung involvement. A similar observation was recently reported by Zheng and co-workers from a Chinese single-center study of 24 M1-PUL patients with relapse after surgery.20 A Surveillance, Epidemiology, and End Results (SEER) database analysis of 13 233 patients with stage IV PDAC carried out by Oweira et al. also found that patients with isolated lung metastases had a better overall and pancreatic cancer-specific survival compared to patients with isolated liver metastases.21 Very recently, a pooled analysis from three large randomized German trials (CONKO-001, CONKO-005 and CONKO-006) defined an isolated pulmonary recurrence as an independent favorable prognostic factor in 689 PDAC patients who experienced relapse after receiving adjuvant chemotherapy.22 In our M1-PUL multicenter cohort, the number of metastases as well as their localization (uni- or bilateral) were identified as potential prognostic factors for survival. Furthermore, the subgroup of 24 patients who underwent surgical metastasectomy for PDAC lung metastases seemed to have improved outcome compared to patients who did not undergo surgery.
Our group also made an effort to retrospectively collect tumor tissue from patients included in the clinical M1-PUL cohort, and we succeeded in obtaining tissue samples from 47 of the 115 recruited patients. A control cohort of PDAC patients without lung metastases (M1-ANY) was set up based on a prospective patient registry at the Ludwig-Maximilians-University of Munich. For mRNA expression analysis, only M1-ANY patients with well-defined clinical characteristics were used. We were thereby able to carry out a more detailed characterization of clinical-pathological factors of 56 patients from the M1-PUL and M1-ANY cohorts included in the gene expression analyses set (see Table 5). Of note, within the M1-PUL cohort, we observed an increased number of patients with nodal-positive disease at the time of surgery and a trend toward a more frequent use of adjuvant CRT in this subgroup. If both variables (pN+, use of CRT) might serve as pre-disposition factors for the subsequent development of lung metastases upon relapse remains unclear and should be analyzed within an extern validation cohort and in future studies, respectively.
Within the translational study, we were not able to confirm our hypotheses that the appearance of lung metastases is influenced either by the Her2/neu status or by any of the molecularly defined prognostic subgroups for PDAC (using KRT81 and HNF1A as markers for previously defined molecular subgroups). Recent studies suggest that the metastatic organotropism to the lung (in PDAC and other cancers) might be related not only to tumor-associated factors (like genetic alterations in DNA repair) but also to immune features (like an ‘inflammatory phenotype’) or to mechanisms of epithelial plasticity.15,23, 24, 25 The process of metastasis and organotropism in PDAC thus might depend on complex tumor and host factors that are not fully understood yet. By using mRNA expression analysis applying a commercial immune profiling panel, we were able to identify genes with significant differential expression in resected pancreatic primaries from M1-PUL versus M1-ANY patients. This suggests that a gene expression signature—that is already present at surgery—might influence the site of a subsequent relapse. If confirmed by other studies, such a gene signature may have important implications in management of patients after PDAC resection in curative intent (e.g. prognostic biomarker for a favorable prognosis, routine and regular CT imaging of the lung during structured aftercare). Additionally, this gene expression signature could inform further translational studies on the—to date still poorly understood—mechanism of organotropism in PDAC as well as other cancer types.
The main limitations of the current study arise from its retrospective nature and an expected bias potentially resulting from the local identification process of M1-PUL patients (especially those with a prolonged survival). Thus, when comparing subgroups in the current study (e.g. if metastasectomy or previous CRT was carried out), one must keep in mind that a potential reporting and selection bias may confound the results from these analyses. Another potential bias arises from the fact that the included patients were diagnosed within a wide time frame (2002-2016) and that we did not receive data on the applied chemotherapy regimens upon relapse or for advanced disease, respectively. Archival tumor tissue was available from 47 of the included 115 patients (41%) only; thus, from more than half of the recruited patients no translational data are available. These limitations clearly illustrate the challenges that investigators are faced with when carrying out research on rare subgroups in a disease like PDAC—in which also tissue acquisition represents a challenge since decades. Of course, a prospective study of M1-PUL PDAC cases would help to overcome many of these limitations, but such a project has—at least to our knowledge—not been reported up to now. Additionally, due to the use of a targeted mRNA gene expression panel, we might have missed further important genes involved in pulmonary organotropism of pancreatic cancer. For example, our panel did not include protein kinase D1, a protein that has recently been shown to have reduction of expression in pancreatic primaries of patients with M1-PUL.26 Nevertheless, at least to our knowledge, the current study is one of the largest multicenter datasets on the M1-PUL topic in PDAC to date and also represents a unique approach as we were able to collect archival tumor samples from a significant number of the enrolled study patients for additional translational work-up.
In conclusion, AIO-YMO-PAK-0515 confirmed that the M1-PUL patient population represents a favorable prognostic PDAC subgroup with specific clinical characteristics that may serve as determinants for developing lung metastases and as prognostic factors in PDAC patients with a pulmonary involvement only. The definition of candidate genes for prediction of isolated M1-PUL could help to inform further research on potential biomarkers for organotropism and translational studies aiming at a better understanding of the tumor biology of this interesting subgroup. Further research (e.g. by reverse translation) is strongly recommended for this specific disease entity.
Acknowledgments
Funding
This work was supported by a grant from the Friedrich-Baur-Stiftung to SO (Reg. Nr. 14/15). AR and SFK were supported by the Else-Kröner Fresenius Stiftung. SK is supported by the international doctoral program ‘i-Target: Immunotargeting of cancer’ funded by the Elite Network of Bavaria (no grant number); the Marie-Sklodowska-Curie Program Training Network for Optimizing Adoptive T Cell Therapy of Cancer funded by the H2020 Program of the European Union [grant number 955575]; the European Research Council Starting Grant [grant number 756017]; the DFG (no grant number); the Fritz-Bender-Foundation (no grant number); the José-Carreras Foundation (no grant number); and the Hector Foundation (no grant number).
Disclosure
CBW received personal and speakers’ fees, reimbursement for travel and accommodation and honoraria for participation in advisory boards from Bayer, BMS, Celgene, GSK, Ipsen, MedScape, Merck, MSD, Rafael Pharmaceuticals, RedHill, Roche, Servier, Shire/Baxalta, SirTex and Taiho and scientific grant support from Roche. JK received honoraria and reimbursement for travel and accommodation for participation in advisory boards and speaker’s bureau from AstraZeneca, Novartis, Quality Initiative in Pathology (QuIP) and Roche Pharma. OW received personal and speakers’ fees, reimbursement for travel and accommodation and honoraria for participation in advisory boards from Abbvie, Amgen, Bayer, BMS, Celgene, Eisai, Incyte, Ipsen, Merck Serono, MSD, Novartis, Roche, Servier and Shire. SFK is a full-time employee of MSD Sharp & Dohme GmbH; his work on this manuscript was carried out independently from his position at MSD: until 31 December 2020 as a full-time employee at LMU Munich (clinician scientist and medical oncologist at LMU) and from 1 January 2021 as a guest researcher and lecturer at LMU Munich. The remaining authors have declared no conflicts of interest.
Supplementary data
References
- 1.Ducreux M., Seufferlein T., Van Laethem J.L., et al. Systemic treatment of pancreatic cancer revisited. Semin Oncol. 2019;46(1):28–38. doi: 10.1053/j.seminoncol.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Waddell N., Pajic M., Patch A.M., et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey P., Chang D.K., Nones K., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 4.Lowery M.A., Jordan E.J., Basturk O., et al. Real-time genomic profiling of pancreatic ductal adenocarcinoma: potential actionability and correlation with clinical phenotype. Clin Cancer Res. 2017;23(20):6094–6100. doi: 10.1158/1078-0432.CCR-17-0899. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Aharon I., Elkabets M., Pelossof R., et al. Genomic landscape of pancreatic adenocarcinoma in younger versus older patients: does age matter? Clin Cancer Res. 2019;25(7):2185–2193. doi: 10.1158/1078-0432.CCR-18-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yachida S., Iacobuzio-Donahue C.A. The pathology and genetics of metastatic pancreatic cancer. Arch Pathol Lab Med. 2009;133(3):413–422. doi: 10.5858/133.3.413. [DOI] [PubMed] [Google Scholar]
- 7.Deeb A., Haque S.U., Olowokure O. Pulmonary metastases in pancreatic cancer, is there a survival influence? J Gastrointest Oncol. 2015;6(3):E48–E51. doi: 10.3978/j.issn.2078-6891.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wangjam T., Zhang Z., Zhou X.C., et al. Resected pancreatic ductal adenocarcinomas with recurrence limited in lung have a significantly better prognosis than those with other recurrence patterns. Oncotarget. 2015;6:36903–36910. doi: 10.18632/oncotarget.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downs-Canner S., Zenati M., Boone B.A., et al. The indolent nature of pulmonary metastases from ductal adenocarcinoma of the pancreas. J Surg Oncol. 2015;112(1):80–85. doi: 10.1002/jso.23943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruger S., Haas M., Burger P.J., et al. Isolated pulmonary metastases define a favorable subgroup in metastatic pancreatic cancer. Pancreatology. 2016;16:593–598. doi: 10.1016/j.pan.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Chou A., Waddell N., Cowley M.J., et al. Clinical and molecular characterization of HER2 amplified-pancreatic cancer. Genome Med. 2013;5(8):78. doi: 10.1186/gm482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collisson E.A., Sadanandam A., Olson P., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noll E.M., Eisen C., Stenzinger A., et al. CYP3A5 mediates basal and acquired therapy resistance in different subtypes of pancreatic ductal adenocarcinoma. Nat Med. 2016;22(3):278–287. doi: 10.1038/nm.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muckenhuber A., Berger A.K., Schlitter A.M., et al. Pancreatic ductal adenocarcinoma subtyping using the biomarkers hepatocyte nuclear factor-1A and cytokeratin-81 correlates with outcome and treatment response. Clin Cancer Res. 2018;24(2):351–359. doi: 10.1158/1078-0432.CCR-17-2180. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura T., Qian B.Z., Pollard J.W. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15(2):73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann M., Stoss O., Shi D., et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology. 2008;52(7):797–805. doi: 10.1111/j.1365-2559.2008.03028.x. [DOI] [PubMed] [Google Scholar]
- 17.Ritchie M.E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D., Gable A.L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerra F., Barucca V., Coletta D. Metastases or primary recurrence to the lung is related to improved survival of pancreatic cancer as compared to other sites of dissemination. Results of a systematic review with meta-analysis. Eur J Surg Oncol. 2020;46(10 Pt A):1789–1794. doi: 10.1016/j.ejso.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Zheng B., Ohuchida K., Yan Z., Okumura T., Ohtsuka T., Nakamura M. Primary recurrence in the lung is related to favorable prognosis in patients with pancreatic cancer and postoperative recurrence. World J Surg. 2017;41(11):2858–2866. doi: 10.1007/s00268-017-4068-6. [DOI] [PubMed] [Google Scholar]
- 21.Oweira H., Petrausch U., Helbling D., et al. Prognostic value of site-specific metastases in pancreatic adenocarcinoma: a Surveillance Epidemiology and End Results database analysis. World J Gastroenterol. 2017;23(10):1872–1880. doi: 10.3748/wjg.v23.i10.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurreck A., Weckwerth J., Modest D.P., et al. Impact of completeness of adjuvant gemcitabine, relapse pattern, and subsequent therapy on outcome of patients with resected pancreatic ductal adenocarcinoma - a pooled analysis of CONKO-001, CONKO-005, and CONKO-006 trials. Eur J Cancer. 2021;150:250–259. doi: 10.1016/j.ejca.2021.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Reichert M., Bakir B., Moreira L., et al. Regulation of epithelial plasticity determines metastatic organotropism in pancreatic cancer. Dev Cell. 2018;45(6):696–711.e8. doi: 10.1016/j.devcel.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Mulero S., Alonso M.H., Pardo J., et al. Lung metastases share common immune features regardless of primary tumor origin. J Immunother Cancer. 2020;8(1) doi: 10.1136/jitc-2019-000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson M.D., Dong L., Wan J., et al. Molecular alterations associated with DNA repair in pancreatic adenocarcinoma are associated with sites of recurrence. J Gastrointestinal Cancer. 2019;50(2):285–291. doi: 10.1007/s12029-018-0073-8. [DOI] [PubMed] [Google Scholar]
- 26.Armacki M., Polaschek S., Waldenmaier M., et al. Protein kinase D1, reduced in human pancreatic tumors, increases secretion of small extracellular vesicles from cancer cells that promote metastasis to lung in mice. Gastroenterology. 2020;159(3):1019–1035.e22. doi: 10.1053/j.gastro.2020.05.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.