Abstract
Purpose
To assess the potential of next-generation sequencing (NGS) technologies to characterize cases diagnosed with autosomal recessive (ar) or sporadic (s) macular dystrophies (ar/sMD) and describe their mutational spectrum.
Methods
A cohort of 1036 families was classified according to their suspected clinical diagnosis—Stargardt disease (STGD), cone and cone-rod dystrophy (CCRD) or other maculopathies (otherMD). Molecular studies included genotyping microarrays, Sanger sequencing, NGS, and sequencing of intronic regions of the ABCA4 gene. Clinical reclassification was done after the genetic study.
Results
At the end of the study, 677 patients (65%) had a confirmed genetic diagnosis, representing 78%, 63%, and 38% of STGD, CCRD, and otherMD groups of patients, respectively. ABCA4 is the most mutated gene in all groups, and a second pathogenic variant was found in 76% of STGD patients with one previously identified mutated ABCA4 allele. Autosomal dominant or X-linked mutations were found in 5% of cases together with not-MD genes (CHM, EYS, RHO, RPGR, RLBP1, OPA1, and USH2A among others) leading to their reclassification. Novel variants in the very rare genes PLA2G5 and TTLL5 revealed additional phenotypic associations.
Conclusions
This study provides for the first time a genetic landscape of 1036 ar/sMD families according to their suspected diagnosis. The analysis of >200 genes associated with retinal dystrophies and the entire locus of ABCA4 increase the rate of characterization, even regardless of available clinical and familiar data. The use of the suspected a priori diagnosis referred by the clinicians, especially in the past, could lead to clinical reclassifications to other inherited retinal dystrophies.
Keywords: Inherited macular dystrophies, genetics, next generation sequencing, clinical reclassification, autosomal recessive
Inherited macular dystrophies (MD) comprise a heterogeneous group of disorders characterized by bilateral central visual loss and atrophy of the macula and underlying retinal pigment epithelium (RPE). The hallmark of these diseases is a loss of visual acuity, that could affect people in every age. The genetic spectrum of MD is very heterogeneous. All patterns of inheritance have been associated with these diseases. The different forms of macular degeneration encompass a wide range of clinical and histological findings.1 Different types of MD include recessive juvenile Stargardt disease,2 dominant forms of Best disease,3 adult vitelliform macular disease,4 pattern dystrophies,5 X-linked (XL) juvenile retinoschisis,6 and age-related macular degeneration,7 among others.
Other inherited retinal dystrophies (IRD), such as cone and cone-rod dystrophies (CCRD), are commonly clinically related with MD because they also involve progressive degeneration or dysfunction of the central retina due to photoreceptor degeneration and lead to a similar vision loss.8 Full field electroretinography (ERG) recordings help to distinguish among these related diseases, with normal findings in MD, only reduced cone responses in cone dystrophies (CD) and both cone and rod responses reduced in cone-rod dystrophies (CRD).
Stargardt disease (STGD), the most frequent maculopathy, was described as a neuroepithelial disease affecting cones, RPE, and the underlying choroid.9 Autosomal recessive Stargardt disease (STGD1; OMIM no. 248200) is caused by biallelic mutations in the ABCA4 gene.10 This gene is also associated to CRD, depending on the severity of the variants found in patients.11–15 More than 1200 disease-causing ABCA4 variants have been described, including missense, nonsense, small indels, copy-number,16 and, more recently, non-canonical splice site and deep intronic variants that lead to splicing defects.17–22 This remarks the importance of sequencing noncoding regions in STGD patients and performing functional assays to unveil the pathogenicity of noncoding variants.
Next-generation sequencing (NGS) technologies allow studying a high number of genes and patients parallelly using exome-targeted approaches or customized gene panels to also include noncoding regions and identifying new candidate genes.23 In the pre-NGS era, genetic testing of IRD patients was performed using genotyping microarrays or Sanger sequencing, which were time-consuming and had low diagnostic rates due to failure to detect previously unknown mutations or new genes.
In this work we report the findings achieved before and after the implementation of NGS for the study of a large cohort of 1036 autosomal recessive or sporadic MD and CCRD (hereinafter, ar/sMD) families at the Genetic Department of the University Hospital Fundación Jimenez Diaz, an IRD reference laboratory testing the largest number of IRD patients in Spain.24 Almost 70% of ar/sMD patients had a molecular diagnosis at the end of this study, and we highlight the most frequent genes and describe mutations in very rare genes that had not been identified previously in the worldwide population.
Methods
Subjects and Samples
A cohort of 1036 unrelated families with ar/sMD was recruited at the Fundación Jiménez Díaz University Hospital (HUFJD, Madrid, Spain) since 1991 up to October 2020. Our laboratory receives internal and external referrals for patients with suspected IRD from different genetic and ophthalmologic services throughout Spain. DNA samples were collected from the HUFJD Biobank, and all participants or their legal guardians signed an informed consent before being included in this study. This study was performed in accordance with the tenets of the Helsinki Declaration and approved by the Research Ethics Committee of our institution.
Clinical Diagnosis and Molecular Characterization
Patients were included when they were referred to the Genetics Department of HUFJD with the following diagnosis and were classified according to it: (I) STGD cases, patients with a clinical suspicion of STGD or fundus flavimaculatus; (II) CCRD cases, patients with a clinical suspicion of CD or CRD; and (III) otherMD cases, patients referred with a suspicion of macular dystrophy, Best disease, vitelliform macular dystrophy, drusen maculopathy, pattern dystrophy, central areolar choroidal dystrophy, and retinoschisis.
In addition, they were classified according to the available data about the inheritance pattern in: (I) autosomal recessive (ar), when there were two or more affected individuals in the same generation (N = 283), (II) sporadic cases, in families with a single diagnosed patient reported (N = 707), or (III) unknown (N = 46), when clinicians did not report family information. Patients referred with an a priori dominant (AD) or XL pattern of inheritance were excluded.
A total of 1036 probands were studied by one or more genetic approaches (Supplementary Table S1). Until 2017, screening of patients with suspicion of ar/sMD was performed most commonly by genotyping microarrays and Sanger sequencing of coding regions of ABCA4 as first-tier testing approaches.25–28 Some unsolved patients were also screened by NGS when they gave their permission and DNA had enough quality, sequencing the genomic ABCA4 locus29 and using small NGS-targeted gene panels for up to 82 IRD genes.30 A few families were also analyzed by whole exome sequencing (WES).31,32
After the implementation of NGS in our laboratory in 2017, screening of new cases is routinely performed by clinical exome-targeted approaches prioritizing up to 229 IRD-associated genes.30,33–35 A few cases were sequenced for the ABCA4 gene (Asper Biogene, Tartu, Estonia). For most of the studied unsolved cases in which one pathogenic variant was found in ABCA4, several approaches were further applied depending on their availability: gene panels covering specific noncoding ABCA4 variants previously reported,36 the targeted sequencing of the complete coding or noncoding ABCA4 regions,22 or Sanger sequencing of specific ABCA4 deep intronic variants known to be prevalent in Spanish population (c.4253+43G>A, c.4539+2064C>T, and c.5196+1137G>A)35 (Supplementary Table S1). In all these analyses, variants were prioritized and their pathogenicity was established following the guidelines of the American College of Medical Genetics and Genomics,37 as previously described.34
Statistical Analysis
Different analyses were performed considering: (I) clinical groups (STGD, CCRD and otherMD); (II) genotype status: (a) biallelic ABCA4 patients; (b) other IRD-associated genes; (c) monoallelic ABCA4 patients; and (d) unsolved patients; and (III) period of their study: (before and after 2017, when implementing exome sequencing). McNemar test and Bonferroni correction for multiple comparisons were done using R version 4.0.3.
Results
Until October 2020, a total of 1036 ar/sMD cases had been studied, 744 recruited until 2017, and 292 new cases recruited during 2017–2020 period. Of them, 570, 223, and 243 presented a clinical suspected diagnosis of STGD, CCRD, and otherMD, respectively.
Molecular Screening and Diagnostic Yields
A total of 744 cases were recruited from 1991 until 2017. First approaches as genotyping microarrays or Sanger sequencing were used to study 523 patients. With these technologies, pathogenic mutations were found in 291 patients (56%), most of them in the ABCA4 gene.
Then, 213 patients were studied by gene panels and 8 patients by WES. In total, 89 patients were characterized (40%), 84 studied by gene panels, and five by WES (Supplementary Table S1).
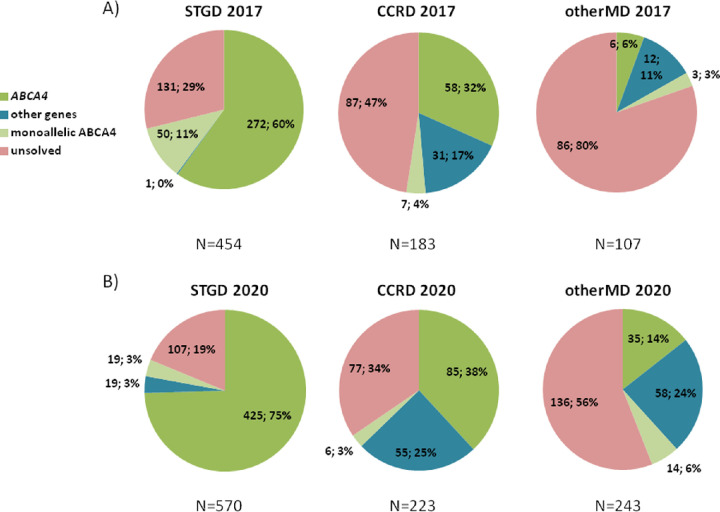
In summary, of the total of 744 cases studied, 364 patients were unsolved by the beginning of 2017. Figure 1A shows the diagnostic yields by genotype of STGD, CCRD, and otherMD patients in 2017 using these approaches.
Figure 1.
Diagnostic yields of patients with Stargardt, cone and cone-rod dystrophies, and other maculopathies regarding their genotype. (A) Before exome sequencing and screening of ABCA4 introns. A total of 744 patients were recruited until January 2017. STGD diagnosis patients presented biallelic mutations in the ABCA4 gene in 272 of the 454 cases (60%), and only one patient was characterized with mutations in other gene, PRPH2. The remaining patients were uncharacterized, with 50 (11%) of them carrying one pathogenic allele in the ABCA4 gene. This gene also explained one-third (58/183) and 5.7% (6/107) of the characterized CCRD patients and otherMD patients, respectively. Pathogenic variants in other genes were identified in 17% (31/183) and 11% (12/107) of CCRD and otherMD cases, respectively. Finally, 364 patients remained unsolved, including 60 that were monoallelic for ABCA4. (B) After exome sequencing and screening of ABCA4 introns. A total of 1036 ar/sMD cases were studied or restudied by the end of this study, October 2020. Three quarters of STGD patients (425/570) were characterized with biallelic ABCA4 mutations, 3% (19/570) presented a single pathogenic variant in this gene, and 3% (19/570) presented mutations in other IRD genes. The 19% (107/570) of studied patients remained genetically unsolved. Among the CCRD cases, a 38% (85/223) were found to carry mutations in ABCA4, and one-quarter (55/223) presented mutations in other IRD genes. In the case of otherMD patients, 14% (35/243) presented biallelic ABCA4 pathogenic variants, whereas 24% (58/243) carried mutations in other genes. CCRD and otherMD unsolved cases, including monoallelic ABCA4 patients, were 37% (83/223) and 62% (150/243) respectively. N, total of patients.
During 2017 to 2020, 223 of the 364 unsolved patients were restudied using NGS technologies (Supplementary Table S1) that allowed identification of the molecular cause of the disease in 136 patients (61%): 83 using clinical exome, 30 sequencing the entire ABCA4 gene, 19 using a gene panel, and four by WES. Remarkably, ABCA4 pathogenic variants were found in 41 and 14 patients studied by clinical exome or gene panel, respectively, in addition to the 30 screened for the entire gene. Segregation of variants was confirmed in 45% of restudied cases (61/136). In the case of ABCA4, variants were confirmed to be in trans in 52% of total ABCA4 restudied cases (44/85).
The diagnostic yields for these 744 arMD cases was compared before and after the implementation of NGS in the diagnostic routine in our lab for the 3 clinical groups, regarding their genotype (Table 1). The increase in the diagnostic yield underlying NGS was statistically significant for the clinical groups STGD and CCRD across (a) (ABCA4 solved cases) and (b) (solved other genes) genotype groups (P < 0.01 for all comparisons). In the case of otherMD group, there was a statistically significant increase in the (b) genotype group (solved other genes) (P < 0.001) but not in the case of (a) genotype group (ABCA4 solved cases). Furthermore, the reduction of unsolved cases (genotype d) showed statistically significant differences for all clinical groups across all genotype groups (P < 0.001 for all comparisons).
Table 1.
Diagnostic Yield Comparison Among Genotypes of the 744 Inherited Macular Dystrophy Families Recruited Until 2017, After the Restudy of Unsolved Cases
| Genotype Groups | ||||
|---|---|---|---|---|
| Period of Genetic Study | Solved ABCA4 | Solved Other Genes | Monoallelic ABCA4 | Unsolved |
| STGD diagnosis group (N = 454) | ||||
| Initial technologies | 272 (59.9%) | 1 (0.2%) | 50 (11%) | 131 (28.9%) |
| Restudy | 338 (74.4%) | 16 (3.5%) | 15 (3.3%) | 85 (18.7%) |
| P value | <0.001 | <0.001 | <0.001 | <0.001 |
| CCRD diagnosis group (N = 183) | ||||
| Initial technologies | 58 (31.7%) | 31 (16.9%) | 7 (3.8%) | 87 (47.5%) |
| Restudy | 73 (39.9%) | 43 (23.5%) | 3 (1.6%) | 64 (35%) |
| P value | <0.001 | 0.002 | NS | <0.001 |
| OtherMD diagnosis group (N = 107) | ||||
| Initial technologies | 6 (5.6%) | 12 (11.2%) | 3 (2.8%) | 86 (80.4%) |
| Restudy | 10 (9.3%) | 36 (33.6%) | 3 (2.8%) | 58 (54.2%) |
| P value | NS | <0.001 | NS | <0.001 |
NS, nonsignificant.
A second pathogenic ABCA4 variant was found in affected individuals for whom a single pathogenic ABCA4 variant had been previously identified using a complementary technique in 38/50 (76%) and 4/7 (57%) of STGD and CCRD cases, respectively. By contrast, two of the three cases with otherMD diagnosis and with only one previously discovered mutant ABCA4 allele presented mutations in a different gene (BEST1 and CERKL). Although for the STGD group the reduction of monoallelic ABCA4 cases was statistically significative (P < 0.001), this was not the case for the CCRD and otherMD groups.
Current Genetic Landscape of ar/sMD
From 2017 until the end of this study, 292 new cases were recruited and studied with a first approach of clinical exome (89%; 259/292) or by sequencing coding regions of the ABCA4 gene (33/292) (Supplementary Table S1). Solved patients were 134 and 18, respectively, resulting in a diagnostic yield of 52% (152/292). Segregation studies have been performed in 35% of them (54/152).
Twenty-seven of the new cases unsolved by clinical exome were further studied sequencing the entire ABCA4 gene due to the clinical suspicion of the patient or the presence of one pathogenic allele in this gene, identifying a second pathogenic allele in 9 of them (Supplementary Table S1) and confirming that both variants were in trans in the four patients in which the segregation analysis was performed (44%). Adding these 27 cases to the restudied cases recruited until 2017 described above, the total characterization rate of restudied cases was 58% (145/250) instead of 61% (136/223). The characterization yield obtained in both restudied and new cases groups according to their clinical diagnosis is summarized in Supplementary Figure S1.
To sum up, a total of 1036 ar/sMD cases were studied by the end of this study, October 2020. Of them, 677 (65%) were genetically characterized (Fig. 1B), in which 343 different pathogenic variants (Supplementary Table S2) were identified, 38 (11%) of them are novel.
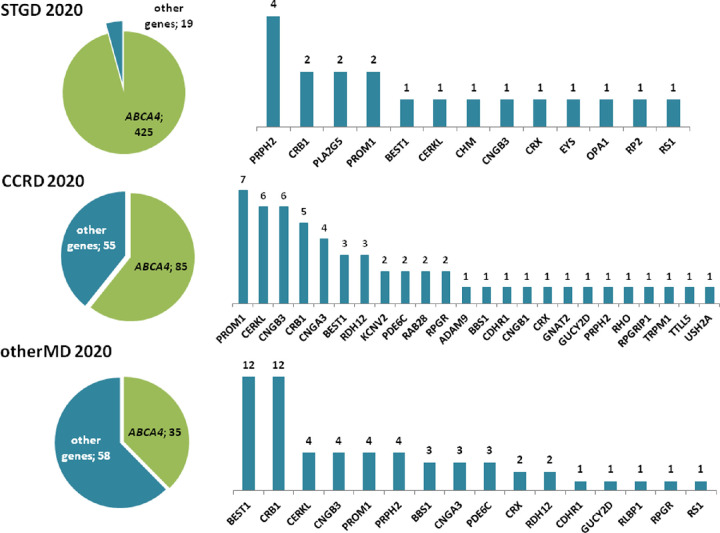
The genetic landscape of STGD, CCRD and otherMD cases at the end of the study, together with the genes mutated in each group, are shown in Figure 1B and Figure 2. Of the unsolved patients, 50% (53/107), 55% (46/83), and 21% (31/150) of STGD, CCRD and otherMD, respectively, could not be analyzed by NGS.
Figure 2.
Genetic landscape in characterized patients referred with Stargardt, cone and cone-rod dystrophies, and other maculopathies. ABCA4 gene was the most mutated gene in the three groups of patients, followed by PRPH2 in STGD patients; PROM1, CERKL and CNGB3 in CCRD patients; and BEST1 and CRB1 in otherMD patients.
The diagnostic yields for the 1036 ar/sMD cases were compared for each clinical and genotype groups (Table 2). The diagnostic yield for ABCA4 solved and unsolved cases showed statistically significant differences between STGD versus CCRD, STGD versus otherMD, and CCRD versus otherMD (P < 0.001). The increase in the diagnostic yield of patients solved with other genes was statistically significant when comparing CCRD and otherMD groups with the STGD group (P < 0.001). The diagnostic yield of monoallelic ABCA4 patients did not show statistically significant differences when comparing between the clinical groups of diagnosis, with a similar rate ranging from 3% to 6%. Around 65% (26/39) of monoallelic ABCA4 patients were unsuccessfully screened for noncoding variants.
Table 2.
Diagnostic Yield Comparison Among Genotypes and Clinical Groups at the End of the Study
| Genotype Groups | ||||
|---|---|---|---|---|
| End of the Study (Oct. 2020) | Solved ABCA4 | Solved Other Genes | Monoallelic ABCA4 | Unsolved |
| STGD vs. CCRD groups | ||||
| STGD (N = 570) | 425 (74.6%) | 19 (3.3%) | 19 (3.3%) | 107 (18.8%) |
| CCRD (N = 223) | 85 (38.1%) | 55 (24.7%) | 6 (2.7%) | 77 (34.5%) |
| P value | <0.001 | <0.001 | NS | <0.001 |
| STGD vs. OtherMD groups | ||||
| STGD (N = 570) | 425 (74.6%) | 19 (3.3%) | 19 (3.3%) | 107 (18.8%) |
| OtherMD (N = 243) | 35 (14.4%) | 58 (23.8%) | 14 (5.8%) | 136 (56%) |
| P value | <0.001 | <0.001 | NS | <0.001 |
| CCRD vs. OtherMD groups | ||||
| CCRD (N = 223) | 85 (38.1%) | 55 (24.7%) | 6 (2.7%) | 77 (34.5%) |
| OtherMD (N = 243) | 35 (14.4%) | 58 (23.8%) | 14 (5.8%) | 136 (56%) |
| P value | <0.001 | NS | NS | <0.001 |
NS, nonsignificant.
Genetic and Clinical Reclassification
A total of 677 families were genetically characterized after the molecular study. As expected, 95% of them carried recessive variants (647/677). Other pathogenic variants associated with an AD or XL inheritance patterns were found in 23 (4%) and seven (1%) families, respectively, both with an a priori ar inheritance pattern, sporadic, or without familiar data (Supplementary Table S2). In four AD-reclassified cases it was confirmed that one of the parents was actually affected. In three XL-reclassified cases, other unreported male relatives were also affected in the family. Segregation analysis was performed in eight cases, identifying incomplete penetrance in two families with reported unaffected relatives carrying previously reported CRX and PRPH2 pathogenic variants, respectively.
Regarding clinical reclassification, some mutations were in genes typically associated with diseases other than those that the respective patients had (Supplementary Table S2). In STGD group, although most of the patients carried biallelic ABCA4 variants, some patients presented pathogenic variants in genes causing STGD-like phenotypes (PRPH2 and PLA2G5) and other types of IRDs or eye diseases: CD (CNGB3), CRD or RP (CERKL, CRB1, CRX, EYS, PROM1 and RP2), Best disease (BEST1), choroideremia (CHM), optic atrophy (OPA1), and retinoschisis (RS1).
Within the CCRD group of patients, 11 patients presented variants in RP genes as CERKL, CNGB1, RHO, RPGR and USH2A, three patients were reclassified to Bestrophinopathy (BEST1), three to LCA (CRB1 and RPGRIP1) because of their early age at onset, and one patient presented congenital stationary night blindness with pathogenic variants in TRPM1. This last patient was diagnosed with retinal dystrophy at age one year.
Finally, in the group of otherMD patients we found 35 patients with ABCA4 variants that were classified to STGD1 or CRD, eight patients classified to CD (CNGB3, GUCYD2 and PDE6C), 36 patients with CRD (BBS1, CDHR1, CNGA3, CRB1, CRX, PROM1, PRPH2, RDH12) or RP (CERKL, RLBP1 and RPGR), 12 patients with BEST1 mutations, one with LCA (CRB1), and one with retinoschisis (RS1).
Novel Findings in Rare Genes
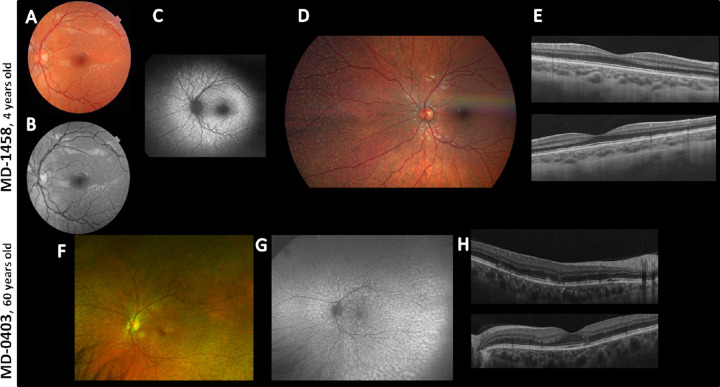
Two unrelated patients of our cohort referred as STGD diagnosis were homozygous for the PLA2G5 gene variant c.309C>A; p.(Cys103*) (rs571294470). Patient MD-1458 was diagnosed at four years of age and presented a best corrected visual acuity (BCVA) (decimal) of 0.5 in both eyes at that age. The fundus images revealed a discrete dotlike flecks in the peripheral fundus and within arcades without foveal involvement, and optic coherence tomography (OCT) showed RPE alterations in the macular area. Patient MD-0403 presented symptoms of visual acuity loss at age five years, and she was diagnosed at 46 years of age with fundus flavimaculatus. Her last ophthalmological examination, at age 60 years, revealed a BCVA (decimal) of 0.9 in both eyes, fundus and autofluorescence images showing macular and RPE atrophy with foveal-sparing, fine dotlike flecks in periphery and macula, and ERG with both scotopic a-waves and photopic b-waves slightly reduced in amplitude at age 55 years. Figure 3 shows ophthalmological findings of the two patients with the PLA2G5 variant.
Figure 3.
Ophthalmological images of patients MD-1458 and MD-0403 with the novel variant p.(Cys103*) in homozygosis in the PLA2G5 gene. (A and F) Fundus images of both patients revealed flecklike lesions perifoveally within arcades with foveal sparing (similar in both eyes, only left eye shown). (B) Infrared reflectance image showed hyporeflective lesions perifoveally within arcades in both eyes (left eye shown) that correspond with hypopigmented lesions in the color picture in MD-1458. (C) Autofluorescence showed hyperautofluorescent scatter lesions that correspond with the hypopigmented lesions of the color picture. (D) Color fundus photograph of the right eye showing nasal periphery disclosing scatter yellowish dots outside the arcades. (E) OCT images showed RPE alterations (up, right eye; down, left eye). (G) Ultra-widefield fundus autofluorescence revealed hyperautofluorescent scatter lesions that correspond with the hypopigmented lesions of the color picture in both eyes (left eye shown). (H) OCT images showed RPE alterations in the macula (up, right eye; down, left eye).
A family with two siblings who were diagnosed with a CD phenotype initially in 2005 were found by WES to have compound heterozygous, novel stop variants in TTLL5—previously associated to CD and CRD38—(NM_015072.4): c.211C>T; p.(Arg71*) (rs1439202144), and c.2029C>T; p.(Arg677*)(rs138370992). The ophthalmological examination of the proband at age 49 years showed a fundus appearance of a bull's-eye maculopathy with a hyperautofluorescent ring in macular area BCVA (decimal) of 0.6 in both eyes, dyschromatopsia, and cone-reduced responses in ERG.
Discussion
In this work, we report the genetic diagnosis of 1036 ar/sMD families studied for 30 years. The genetic testing options for studying patients with IRD are numerous and have progressed over time.39 First approaches used in our laboratory—genotyping microarrays and Sanger sequencing—before NGS allowed characterizing half of studied patients, most of them carrying ABCA4 pathogenic coding variants.
In recent years, NGS technologies have had a substantial impact in the molecular diagnosis of IRD patients.23 Here, the use of customized NGS-based panels covering >70 genes solved nearly 40% of ar/sMD cases, whereas their use for other IRD families of our cohort showed variable mutation rates of 27% and 57% in autosomal dominant and recessive cases, respectively.40,41 The restudy of unsolved cases by the implementation of clinical exome sequencing >200 IRD genes or noncoding regions of ABCA4 have allowed characterizing the 58% of the restudied cases. When clinical exome was used as the first-tier approach in the new cases, the characterization rate decreased to 52%. Other authors using NGS custom gene panels of >300 IRD-associated genes and noncoding regions reported a diagnosis yield of 85%.42 The differences in the characterization rate found between the restudied cases and the new cases can be explained by several reasons. First, both cohorts have been recruited in different periods of time and do not have the same number of patients in each clinical group. Although most of the restudied patients have a STGD diagnosis (52%), new cases included more otherMD diagnosis (46%) than STGD diagnosis (40%). As we have seen in this study, in this otherMD clinical group, there is the highest rate of unsolved cases. We suggest that in this miscellaneous group where most of the patients are referred with “macular dystrophy,” there may be nonhereditary cases. Second, additional regions studied by WES or the entire ABCA4 gene have been screened in restudied cases, representing the 2% and 32% of the total of characterized restudied cases, respectively. In 20 of these patients (8% of total restudied cases), we found pathogenic variants in genes not included in the clinical exome—TTLL5 case—or deep intronic variants in the ABCA4 gene. However, the remaining patients characterized after the restudy should have been solved if they had been sequenced by clinical exome as first approach. For all these reasons, the characterization rate obtained in this study is specific for each cohort and cannot be compared.
Three-quarters of patients referred with STGD diagnosis were biallelic for the ABCA4 gene, which is expected because Stargardt disease is due to mutations in this gene (STGD1).16 The ABCA4 gene was also the most frequent gene mutated in CCRD and otherMD groups, representing 38% and 14% of the total of studied patients, respectively.
It is well known that there are missing alleles in the ABCA4 gene in unsolved patients with one pathogenic allele.16 The fact that the monoallelic cases for the ABCA4 gene were only significantly reduced in the group of STGD diagnosis supports the necessity of sequencing the complete ABCA4 gene in those cases with a clear diagnosis of Stargardt disease to find a second pathogenic variant. In most cases, when all 50 coding exons have been already screened, this is likely located in deep-intronic regions22 or regulatory regions.21 At the end of our study, the frequency of monoallelic ABCA4 patients found in all groups of clinical diagnosis is between 3% to 6%. Pathogenic noncoding variants still could not be discarded in 54% of the total of monoallelic ABCA4 patients in which the complete sequencing of the ABCA4 locus has not been performed, including nine STGD patients, six of whom were unsuccessfully screened for the prevalent Spanish intronic variants. However, it is important to remark that only 1.6% of the mutated alleles in the ABCA4 gene are deep intronic pathogenic variants, as we described before.35 Nevertheless, our hypothesis is that most of these patients, specially CCRD and otherMD patients, could be merely carriers of a variant in the most frequent IRD mutated gene,43 as is also supported by previous studies reflecting a carrier rate of 6% for ABCA4 in Spanish population.28
Other genes different from ABCA4 known to cause other MD or IRD were identified in the group of suspected STGD cases. Mutations in PRPH2 have been associated with STGD-like phenotypes,16 but with an AD-inheritance pattern. This fact could be explained by the limited clinical information from relatives together with the reported incomplete penetrance and variable expressivity in this gene.44 Other genes associated to specific IRD-phenotypes with clinical features other than STGD were found in patients in whom no clinical information apart from their referred STGD diagnosis was available. Mutations in IRD-genes causing phenotypes different from STGD have also been reported when studying a priori STGD cases.45 Genes associated to RP, LCA, optic atrophy, retinoschisis and Best disease were found in the groups of CCRD and otherMD. This reflects that the clinical entity can be accurately defined when sequencing a large number of IRD-associated genes, as has been addressed before.42 The ideal scenario in those cases would be to collect ophthalmological information after the genetic study to provide a complete clinical reclassification. Moreover, in unsolved cases with a specific diagnosis, as the case of STGD patients, additional clinical examinations might reveal initial misdiagnosis, when both imaging technologies and the knowledge on retinal diseases was not as advanced as currently.
Remarkably, we identified a homozygous novel variant in the gene PLA2G5 in two unrelated probands with STGD-like phenotype. This gene was first associated with benign fleck retina,46 a condition that was added to the classification of the fleck disorders described by Krill and Folk,47 and where patients have no functional defect.48 Sergouniotis et al. 46 identified seven patients with yellow-white retinal lesions and no visual deficits carrying PLA2G5 mutations. No other patients have been reported to date to our knowledge, with our two patients being the eighth and ninth cases with mutations in this very rare gene. Our patients presented a fleck retina disorder involving the macula and different from the benign condition because they are symptomatic and have a mild functional dysfunction of vision. This may suggest that PLA2G5 gene could also been involved in retinal degeneration with functional disturbances on vision. The novel variant identified in the last exon of this gene leads to the truncation of the protein group V phospholipase A2 (PLA2G5) at amino acid 103, located before the aspartic acid amino acid at 111 position that is responsible for catalytic activity of the protein.49
The high-throughput genetic analysis by NGS could be successful, although the family history was ambiguous or the clinical manifestation was atypical.42 On the one hand, most of the patients included in this work were sporadic cases in which apparently the proband had not affected relatives, and, in few cases, there is no available information of the family members. As expected, 95% of these cases were confirmed to have an autosomal recessive pattern of inheritance after the identification of the mutated gene, and in 52% of them it was confirmed that each mutated allele was received from one progenitor. However, when variants in AD- or XL-associated genes were identified, as in the remaining 5% of patients, having clinical information of relatives and segregation studies are important to give accurate genetic counseling.
Finally, only 35% of the total ar/sMD patients were genetically unsolved; however, NGS screening has not been yet performed in a quarter. An extended analysis of noncoding and regulatory regions together with functional assays, and the identification of new genes causing IRD would be needed to characterize unsolved cases. Also, structural variants affecting genomic domains may be involved.50
This study provides a genetic landscape of 1036 ar/sMD families, giving a mutational spectrum of the genes involved in STGD, CCRD, and otherMD groups of patients according to their suspected diagnosis. We demonstrate the increase of the characterization diagnostic yield after the implementation of the exome sequencing even when no clinical and familiar data are available. Also, a new phenotypic association with a visual defect is described for the rare gene PLA2G5.
Supplementary Material
Acknowledgments
The authors thank Ignacio Mahillo Fernandez for statistical analysis, and Laura Cortazar and Alvaro Fernandez-Vega for ophthalmological pictures.
Supported by the Instituto de Salud Carlos III (ISCIII) of the Spanish Ministry of Health (FIS; PI16/00425 and PI19/00321), Centro de Investigación Biomédica en Red Enfermedades Raras (CIBERER, 06/07/0036), IIS-FJD BioBank PT13/0010/0012), Comunidad de Madrid (CAM, RAREGenomics Project, B2017/BMD-3721), European Regional Development Fund (FEDER), the Organización Nacional de Ciegos Españoles (ONCE), Fundación Ramón Areces, Fundación Conchita Rábago and the University Chair UAM-IISFJD of Genomic Medicine. M.C. is supported by the Miguel Servet Program (CPII17/00006) from ISCIII.
Disclosure: M. Del Pozo-Valero, None; R. Riveiro-Alvarez, None; I. Martin-Merida, None; F. Blanco-Kelly, None; S. Swafiri, None; I. Lorda-Sanchez, None; M.J. Trujillo-Tiebas, None; E. Carreño, None; B. Jimenez-Rolando, None; B. Garcia-Sandoval, None; M. Corton, None; A. Avila-Fernandez, None; C. Ayuso, None
References
- 1. Michaelides M. The genetics of inherited macular dystrophies. J Med Genet. 2003; 40: 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanna P, Strauss RW, Fujinami K, Michaelides M.. Stargardt disease: Clinical features, molecular genetics, animal models and therapeutic options. Br J Ophthalmol. 2017; 101: 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krämer F, White K, Pauleikhoff D, et al.. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. Eur J Hum Genet. 2000; 8: 286–292. [DOI] [PubMed] [Google Scholar]
- 4. Burgess R, Millar ID, Leroy BP, et al.. Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am J Hum Genet. 2008; 82: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rahman N, Georgiou M, Khan KN, Michaelides M.. Macular dystrophies: Clinical and imaging features, molecular genetics and therapeutic options. Br J Ophthalmol. 2020; 104: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forsius H, Vainio-Mattila B, Eriksson A.. X-linked hereditary retinoschisis. Br J Ophthalmol. 1962; 46: 678–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saksens NTM, Fleckenstein M, Schmitz-Valckenberg S, et al.. Macular dystrophies mimicking age-related macular degeneration. Prog Retin Eye Res. 2014; 39: 23–57. [DOI] [PubMed] [Google Scholar]
- 8. Birtel J, Eisenberger T, Gliem M, et al.. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci Rep. 2018; 8(1): 4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stargardt K. Über familiäre, progressive Degeneration in der Maculagegend des Auges. Albr von Graefes Arch für Ophthalmol. 1909; 71: 534–550. [Google Scholar]
- 10. Allikmets R, Singh N, Sun H, et al.. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997; 15: 236–246. [DOI] [PubMed] [Google Scholar]
- 11. Cremers FPMM, Van De Pol DJRR, Van Driel M, et al.. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt's disease gene ABCR. Hum Mol Genet. 1998; 7: 355–362. [DOI] [PubMed] [Google Scholar]
- 12. Bertelsen M, Zernant J, Larsen M, et al.. Generalized choriocapillaris dystrophy, a distinct phenotype in the spectrum of ABCA4-associated retinopathies. Invest Ophthalmol Vis Sci. 2014; 55: 2766–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verbakel SK, van Huet RAC, Boon CJF, et al.. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018; 66: 157–186. [DOI] [PubMed] [Google Scholar]
- 14. Tanaka K, Lee W, Zernant J, et al.. The rapid-onset chorioretinopathy phenotype of ABCA4 disease. Ophthalmology. 2018; 125: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martínez-Mir A, Paloma E, Allikmets R, et al.. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet. 1998; 18: 11–12. [DOI] [PubMed] [Google Scholar]
- 16. Cremers FPM, Lee W, Collin RWJ, Allikmets R.. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog Retin Eye Res. 2020; 79: 100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khan M, Cornelis SS, Khan MI, et al.. Cost-effective molecular inversion probe-based ABCA4 sequencing reveals deep-intronic variants in Stargardt disease. Hum Mutat. 2019; 40: 1749–1759. [DOI] [PubMed] [Google Scholar]
- 18. Fadaie Z, Khan M, Del Pozo-Valero M, et al.. Identification of splice defects due to noncanonical splice site or deep-intronic variants in ABCA4. Hum Mutat. 2019; 40: 2365–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sangermano R, Khan M, Cornelis SS, et al.. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018; 28: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sangermano R, Garanto A, Khan M, et al.. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet Med. 2019; 21: 1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bauwens M, Garanto A, Sangermano R, et al.. ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet Med. 2019; 21: 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khan M, Cornelis SS, Pozo-Valero M Del, et al.. Resolving the dark matter of ABCA4 for 1054 Stargardt disease probands through integrated genomics and transcriptomics. Genet Med. 2020; 22: 1235–1246. [DOI] [PubMed] [Google Scholar]
- 23. Broadgate S, Yu J, Downes SM, Halford S.. Unravelling the genetics of inherited retinal dystrophies: Past, present and future. Prog Retin Eye Res. 2017; 59: 53–96. [DOI] [PubMed] [Google Scholar]
- 24. Perea-Romero I, Gordo G, Iancu IF, et al.. Genetic landscape of 6089 inherited retinal dystrophies affected cases in Spain and their therapeutic and extended epidemiological implications. Sci Rep. 2021; 11(1): 1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aguirre-Lamban J, González-Aguilera JJ, Riveiro-Alvarez R, et al.. Further associations between mutations and polymorphisms in the ABCA4 gene: Clinical implication of allelic variants and their role as protector/risk factors. Invest Ophthalmol Vis Sci. 2011; 52: 6206–6212. [DOI] [PubMed] [Google Scholar]
- 26. Aguirre-Lamban J, Riveiro-Alvarez R, Maia-Lopes S, et al.. Molecular analysis of the ABCA4 gene for reliable detection of allelic variations in Spanish patients: Identification of 21 novel variants. Br J Ophthalmol. 2009; 93: 614–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riveiro-Alvarez R, Aguirre-Lamban J, Angel Lopez-Martinez M, et al.. Frequency of ABCA4 mutations in 278 Spanish controls: An insight into the prevalence of autosomal recessive Stargardt disease. Br J Ophthalmol. 2009; 93: 614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riveiro-Alvarez R, Lopez-Martinez MA, Zernant J, et al.. Outcome of ABCA4 disease-associated alleles in autosomal recessive retinal dystrophies: Retrospective analysis in 420 Spanish families. Ophthalmology. 2013; 120: 2332–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zernant J, ngel. Xie YA, Ayuso C, et al.. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum Mol Genet. 2014; 23: 6797–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin-Merida I, Avila-Fernandez A, Del Pozo-Valero M, et al.. Genomic landscape of sporadic retinitis pigmentosa. Ophthalmology. 2019; 126: 1181–1188. [DOI] [PubMed] [Google Scholar]
- 31. Corton M, Nishiguchi KM, Avila-Fernández A, et al.. Exome sequencing of index patients with retinal dystrophies as a tool for molecular diagnosis. PLoS One. 2013; 8(6): e65574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Riveiro-Álvarez R, Xie Y, López-Martínez MÁ, et al.. New mutations in the RAB28 gene in 2 Spanish families with cone-rod dystrophy. JAMA Ophthalmol. 2015; 133: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martin-Merida I, Aguilera-Garcia D, Fernandez-San Jose P, et al.. Toward the mutational landscape of autosomal dominant retinitis pigmentosa: A comprehensive analysis of 258 Spanish families. Invest Ophthalmol Vis Sci. 2018; 59: 2345–2354. [DOI] [PubMed] [Google Scholar]
- 34. Del Pozo-Valero M, Martin-Merida I, Jimenez-Rolando B, et al.. Expanded phenotypic spectrum of retinopathies associated with autosomal recessive and dominant mutations in PROM1. Am J Ophthalmol. 2019; 207: 204–214. [DOI] [PubMed] [Google Scholar]
- 35. Del Pozo-Valero M, Riveiro-Alvarez R, Blanco-Kelly F, et al.. Genotype-phenotype correlations in a Spanish cohort of 506 families with bi-allelic ABCA4 pathogenic variants. Am J Ophthalmol. 2020; 219: 195–204. [DOI] [PubMed] [Google Scholar]
- 36. Braun TA, Mullins RF, Wagner AH, et al.. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013; 22: 5136–5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richards S, Aziz N, Bale S, et al.. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sergouniotis PI, Chakarova C, Murphy C, et al.. Biallelic variants in TTLL5, encoding a tubulin glutamylase, cause retinal dystrophy. Am J Hum Genet. 2014; 94: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ratnapriya R, Swaroop A.. Genetic architecture of retinal and macular degenerative diseases: The promise and challenges of next-generation sequencing. Genome Med. 2013; 5(10): 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fernandez-San Jose P, Corton M, Blanco-Kelly F, et al.. Targeted next-generation sequencing improves the diagnosis of autosomal dominant retinitis pigmentosa in Spanish patients. Invest Ophthalmol Vis Sci. 2015; 56: 2173–2182. [DOI] [PubMed] [Google Scholar]
- 41. Perez-Carro R, Corton M, Sánchez-Navarro I, et al.. Panel-based NGS reveals novel pathogenic mutations in autosomal recessive retinitis pigmentosa. Sci Rep. 2016; 6: 19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gonzàlez-Duarte R, de Castro-Miró M, Tuson M, et al.. Scaling new heights in the genetic diagnosis of inherited retinal dystrophies. Adv Exp Med Biol. 2019; 1185: 215–219. [DOI] [PubMed] [Google Scholar]
- 43. Hanany M, Rivolta C, Sharon D.. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc Natl Acad Sci USA. 2020; 117: 2710–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boon CJF, den Hollander AI, Hoyng CB, et al.. The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog Retin Eye Res. 2008; 27: 213–35. [DOI] [PubMed] [Google Scholar]
- 45. Wolock CJ, Stong N, Ma CJ, et al.. A case–control collapsing analysis identifies retinal dystrophy genes associated with ophthalmic disease in patients with no pathogenic ABCA4 variants. Genet Med. 2019; 21: 2336–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sergouniotis PI, Davidson AE, MacKay DS, et al.. Biallelic mutations in PLA2G5, encoding group v phospholipase A 2, cause benign fleck retina. Am J Hum Genet. 2011; 89: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krill AE, Folk MR.. Retinitis punctata albescens. A functional evaluation of an unusual case. Am J Ophthalmol. 1962; 53: 450–455. [PubMed] [Google Scholar]
- 48. Sabel Aish SF, Dajani B.. Benign familial fleck retina. Br J Ophthalmol. 1980; 64: 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lambeau G, Gelb MH.. Biochemistry and physiology of mammalian secreted phospholipases A 2. Annu Rev Biochem. 2008; 77: 495–520. [DOI] [PubMed] [Google Scholar]
- 50. de Bruijn SE, Fiorentino A, Ottaviani D, et al.. Structural variants create new topological-associated domains and ectopic retinal enhancer-gene contact in dominant retinitis pigmentosa. Am J Hum Genet. 2020; 107: 802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.