ABSTRACT
The utility of Schlafen 11 (SLFN11) expression as a predictive biomarker for platinum‐based chemotherapy has been established for cancers from different histologies. However, the therapeutic relevance of SLFN11 in bladder cancer (BC) is unknown. Here, we examined the clinicopathologic significance of SLFN11 expression across 120 BC cases by immunohistochemistry. We divided the cases into two cohorts, one including 50 patients who received adjuvant or neoadjuvant platinum‐based chemotherapy, and the other including 70 BC patients treated by surgical resection without chemotherapy. In the cohort of 50 BC cases treated with platinum‐based chemotherapy, the SLFN11‐positive group (n = 25) showed significantly better overall survival than the SLFN11‐negative group (n = 25, P = .012). Schlafen 11 expression correlated significantly with the expression of luminal subtype marker GATA3. Multivariate analyses identified SLFN11 expression as an independent prognostic predictor (odds ratio, 0.32; 95% confidence interval, 0.11‐0.91; P = .033). Conversely, in the cohort of 70 BC cases not receiving platinum‐based chemotherapy, the SLFN11‐positive group (n = 29) showed significantly worse overall survival than the SLFN11‐negative group (n = 41, P = .034). In vitro analyses using multiple BC cell lines confirmed that SLFN11 KO rendered cells resistant to cisplatin. The epigenetic modifying drugs 5‐azacytidine and entinostat restored SLFN11 expression and resensitized cells to cisplatin and carboplatin in SLFN11‐negative BC cell lines. We conclude that SLFN11 is a predictive biomarker for BC patients who undergo platinum‐based chemotherapy and that the combination of epigenetic modifiers could rescue refractory BC patients to platinum derivatives by reactivating SLFN11 expression.
Keywords: biomarker, bladder cancer, chemotherapy, cisplatin, SLFN11
Schlafen 11 (SLFN11) is a predictive biomarker for bladder cancer patients who undergo platinum‐based chemotherapy. A combination of epigenetic modifiers could rescue bladder cancer patients who are refractory to platinum derivatives, by reactivating SLFN11 expression.

1. INTRODUCTION
Bladder cancer (BC) is one of the most frequently occurring cancers, with more than 430 000 men and women diagnosed worldwide every year. 1 Although 75% of BCs are non–muscle invasive cancers treated mainly by surgical resection, the remaining 25% develop muscle invasion and/or metastatic lesions (advanced BC) and require systemic therapies. 2 Even though several immune checkpoint inhibitors have recently been approved in the first‐ or second‐line setting for advanced BC by the US FDA, 3 70%‐80% of patients do not respond to those treatments. Hence, platinum‐based chemotherapy (PBC) has remained as a gold standard treatment for advanced BC for decades and up to the present. Platinum‐based chemotherapy for advanced BC patients typically consists of cisplatin and gemcitabine or carboplatin and gemcitabine. Although PBC initially provides high response in a subpopulation of advanced BC, recurrence frequently occurs within several years with acquired resistance, which results in an overall poor survival rate for advanced BC. Indeed, 80%‐90% of advanced BCs are refractory to PBC, and patient prognosis is unpredictable by current methodologies. 4 , 5 Hence, a major unmet need for the treatment of advanced BC patients is the development of novel strategies to overcome recurrent BC and the discovery of clinically available biomarkers to predict responders to PBC.
Schlafen 11 (SLFN11) was recently discovered to be a determinant of response to a broad type of DNA damaging agents (DDAs) including platinum‐derived drugs (cisplatin, carboplatin), DNA synthesis inhibitors (gemcitabine, cytarabine), poly (ADP‐ribose) polymerase inhibitors (olaparib, talazoparib), topoisomerase I inhibitors (camptothecin, topotecan), and topoisomerase II inhibitors (etoposide, doxorubicin) by investigating the NCI‐60 genomic and pharmacological databases. 6 The extremely high correlation between SLFN11 expression and drug sensitivity to topotecan was also reported through the analysis of the Cancer Cell Line Encyclopedia database. 7 , 8 Since these discoveries, highly significant correlations between SLFN11 expression and drug sensitivity to various DDAs have been reported in multiple tumor cell lines, tissue organoids, and xenograft models in mice. 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 Retrospective analyses of patient samples have shown that high SLFN11 expression correlates with enhanced responses to DDAs in breast cancers, 18 small cell lung cancers, 19 prostate cancers, 20 and esophageal cancers. 21 Moreover, our group first showed that SLFN11‐high gastric cancers showed a favorable response to PBC compared to SLFN11‐low gastric cancers 22 and proposed that evaluation of SLFN11 by immunohistochemistry (IHC) is able to predict response to PBC in clinical gastric cancer.
Mechanistically, SLFN11 induces a lethal replication block under DDA treatments through chromatin binding and chromatin remodeling, which in part explains the SLFN11‐dependent cell death occurring with DDA treatment. 12 , 14 , 23 , 24 , 25 Inactivation of SLFN11 expression is largely caused by epigenetic modulation of histones and DNA, which offers the opportunity to reactivate SLFN11 expression by epigenetic‐modifying drugs such as inhibitors of DNA methyltransferase (5‐azacytidine), 7 inhibitors of histone methyltransferase (EZH2 inhibitors), 26 and inhibitors of histone deacetylase (HDAC inhibitors). 17
Although the interest in SLFN11 is increasing, there is no information about SLFN11 in BC. In this study, we assessed the clinical and predictive values of SLFN11 in BC through the analysis of patient samples and multiple BC cell lines.
2. MATERIALS AND METHODS
2.1. Human tissues
Primary tumor samples were collected from 120 BC patients. Patients were treated at Hiroshima University Hospital or an affiliated hospital. In this cohort, 50 patients received adjuvant or neoadjuvant PBC. As histological examination was carried out to confirm the definitive diagnosis of malignancy, specimens included those from biopsy and transurethral resection. All samples were collected before chemotherapy. The chemotherapy regimen included cisplatin or carboplatin. Clinical outcomes were followed from the first day of chemotherapy initiation. Response to chemotherapy was decided clinically, according to RECIST. None of the 70 patients who were treated by radical cystectomy received adjuvant or neoadjuvant chemotherapy. The 7th TNM classification system was used for tumor staging. This study was approved by the Ethics Committee for Human Genome Research of Hiroshima University (No. E 326, Hiroshima, Japan) and conformed to the ethical guidelines of the Declaration of Helsinki.
2.2. Antibodies and IHC
Continuous 3‐µm‐thick sections were used for IHC. The Abs used in this study were as follows: mouse anti‐SLFN11 Ab (D‐2, #sc‐515071, 1:50 dilution for IHC and 1:500 dilution for western blot; Santa Cruz Biotechnology), mouse anti‐phospho‐Histone H2AX (Ser139) (γH2AX) Ab (JBW301, #DAM1493341, 1:200 dilution for immunofluorescence and 1:500 dilution for western blot; Sigma‐Aldrich), mouse anti‐β‐actin Ab (#127M4866V, 1:20 000 dilution; Sigma‐Aldrich), mouse anti‐p53 Ab (NCL‐L‐p53‐DO7, 1:200 dilution; Leica Biosystems), mouse anti‐cytokeratin 5/6 Ab (M7237, 1:200 dilution; Dako), mouse anti‐GATA3 Ab (ACR405B, 1:200 dilution; BIOCARE), rabbit anti‐ programmed death ligand 1 (PD‐L1) Ab (ab205921, 1:400 dilution; Abcam). The IHC procedures for SLFN11 and other Abs were described previously. 22 , 27 , 28 , 29 All staining was manually scored by two surgical pathologists (DT and NS) without the knowledge of clinical findings or patient outcome. Schlafen 11 was considered positive when at least 5% of the tumor cells were stained. P53, GATA3, and PD‐L1 were considered positive when at least 10%, 20%, and 1% of the tumor cells were stained, respectively. 30 Cytokeratin 5/6 (CK5/6) was considered positive when a full layer of the tumor cells was stained. Consensus regarding interpretation was made when there were discordant results.
2.3. Immunofluorescence
Procedures were the same as those for IHC until the second Ab application. For the secondary Ab, Alexa Fluor 488 donkey (#34330A, 1:100 dilution; Molecular Probes) was applied and incubated for 1 hour at room temperature. Nuclear staining was undertaken using DAPI (Vector Laboratories) for 10 minutes. Images were captured by an IX81 microscope (Olympus). Signal intensity in each cell was calculated by ImageJ software as described previously. 22
2.4. Cell lines and CRISPR‐Cas9
Seven urothelial carcinoma cell lines, including T24, UM‐UC13, UM‐UC3, 253JBV, KMBC2, RT112, and UM‐UC6, were used for in vitro experiments. T24 and KMBC2 were purchased from the Japanese Collection of Research Bioresources Cell Bank, and the other cell lines were kindly provided by Professor Peter C. Black (Department of Urologic Sciences, Vancouver Prostate Centre, University of British Columbia). Cells were cultured in phenol red‐containing minimum essential medium α (Fujifilm Wako Pure Chemical Corporation), supplemented with 10% FBS (BioWhittaker), 50 U/mL penicillin, and 50 µg/mL streptomycin in a humidified incubator with 5% CO2 at 37°C.
The SLFN11 KO cells were established in the T24, UM‐UC13, KMBC2, and RT112 cell lines using CRISPR‐Cas9 methods. Details were described previously. 12
2.5. Western blot analysis
Cell pellets were lysed with RIPA buffer (50 mmol/L Tris, pH 7.4, 125 mmol/L NaCl, 0.1% NP‐40, 5 mmol/L EDTA, and protease inhibitor cocktail [cOmplete; Roche]). Immunocomplexes were detected with an ECL Plus Western Blot Detection System (Amersham Biosciences). β‐Actin was used for internal control.
2.6. Drugs
Cisplatin (Pfizer), carboplatin (Nippon Kayaku), 5‐aza‐2′‐deoxycytidine (5‐aza; #SLBZ9636; Sigma Chemical), and entinostat (#14654; ChemScene) were used.
2.7. Viability assay
The viability of the cell lines was determined using an MTT assay. Three thousand cells were plated in each well of 96‐well plates. After 24 hours, the cells were continuously treated with various concentrations of the drugs. The culture medium was removed after another 72 hours, and 50 μL of a 0.5‐mg/mL solution of MTT (Sigma‐Aldrich) was added to each well. The plates were then incubated for 1 hour at 37°C. After the removal of the MTT solution, 50 μL DMSO (Wako) was added per well. For the combination assays, RT112 and 253JBV cell lines were pretreated with indicated concentrations of 5‐aza or entinostat for 2 days, washed, and then treated with the indicated concentrations of cisplatin or carboplatin for two additional days. Viability was measured by MTT assay 2 days after the cisplatin or carboplatin treatments. The absorbance at 540 nm was measured by an Envision 2104 Multilabel Reader (Perkin Elmer).
2.8. Cell growth assay
T24, UM‐UC13, and KMBC2 cell lines were treated for 4 hours with cisplatin at 400, 200, and 50 nmol/L, respectively. The cells were then washed and released into a drug‐free medium. Three thousand cells were plated per well in 96‐well plates. Cell number was checked at 1, 2, and 4 days after the cisplatin treatment by MTT assays.
2.9. Statistical methods
Associations between SLFN11 expression and clinicopathologic parameters and IHC results were examined by Fisher’s exact test and Student’s t test. The Kaplan‐Meier method was used to examine the overall survival (OS) of the patients. Overall survival was also analyzed using the log‐rank test and multivariate analysis based on the Cox proportional hazards method. The results are shown as the mean ± standard variance of triplicate measurements. A P value of less than .05 was considered to be statistically significant.
3. RESULTS
3.1. Opposite impact of SLFN11 on OS in BC patients receiving and not receiving PBC
To examine the prognostic impact of SLFN11 in BC, we evaluated SLFN11 expression by IHC using formalin‐fixed paraffin‐embedded samples obtained from 120 BC patients registered in the archives of Hiroshima University Hospital or an affiliated hospital. Expression of SLFN11 was observed exclusively in the nucleus as we reported previously 31 (Figure 1A). We scored SLFN11 expression by the averaged percentage of SLFN11‐positive tumor cells from multiple fields (Figure 1B). Among the 120 BC cases, 66 cases (55%) were totally negative for SLFN11 expression. To divide the population into two groups, we set a cut‐off value of 5% positivity as a threshold (Figure 1B). We found no significant correlation between SLFN11 expression and OS in the 120 BC cases (Figure 1C).
FIGURE 1.
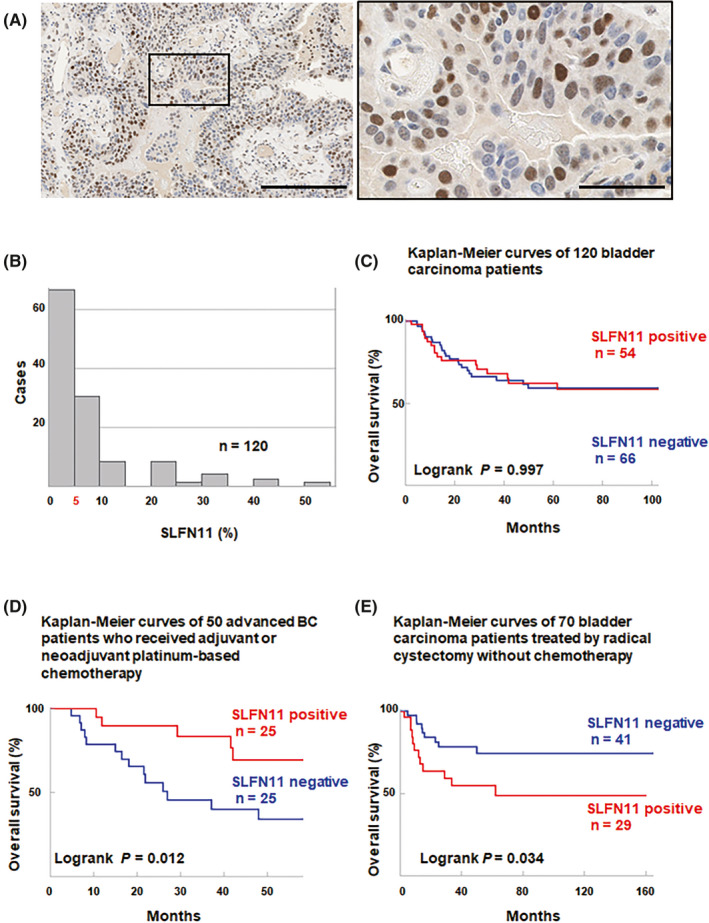
Immunohistochemical expression of Schlafen 11 (SLFN11) in bladder cancer (BC) and validation of the association between SLFN11 expression and clinical course. A, Representative immunohistochemical images of SLFN11 in BC. Scale bars, 200 µm (left) and 50 µm (right). B, Distribution of the immunohistochemical expression of SLFN11 in 120 BC cases, with 5% used as the cut‐off value. C, Correlation of the expression of SLFN11 protein with overall survival (OS) of 120 patients with BC. D, Correlation of the expression of SLFN11 protein with OS of 50 patients with unresectable locally advanced or metastatic BC treated with platinum‐based chemotherapy. E, Correlation of the expression of SLFN11 protein with OS of 70 patients with local BC treated by surgical resection without chemotherapy
Considering the function of SLFN11 as a sensitizer of cancer cells to platinum derivatives, we divided the 120 BC patients into two cohorts, one including 50 patients with clinically unresectable locally advanced or metastatic BC who received PBC (advanced BC with PBC) and the other including 70 local BC patients treated by surgical resection without chemotherapy (local BC without PBC; Table 1).
TABLE 1.
Association between Schlafen 11 (SLFN11) expression and clinicopathologic characteristics in bladder carcinoma (BC) patients (n = 120)
| Patient characteristics | SLFN11 expression, n (%) | P value | ||
|---|---|---|---|---|
| Positive | Negative | |||
|
Unresectable locally advanced or metastatic BC patients with platinum‐based chemotherapy (n = 50) | ||||
| Sex | Male | 17 (46) | 20 (54) | .333 |
| Female | 8 (62) | 5 (38) | ||
| Age, y | ≤70 | 10 (40) | 15 (60) | .157 |
| >70 | 15 (60) | 10 (40) | ||
| Cellular atypism classification | Low grade | 3 (50) | 3 (50) | 1.000 |
| High grade | 22 (50) | 22 (50) | ||
| Clinical TNM stage | Stage II | 7 (70) | 3 (30) | .329 |
| Stage III | 4 (44) | 5 (56) | ||
| Stage IV | 13 (43) | 17 (57) | ||
| First‐line chemotherapy regimen | GC or MVAC | 22 (49) | 23 (51) | .637 |
| GCa | 3 (60) | 2 (40) | ||
| Response to chemotherapy | SD or PD | 14 (48) | 15 (52) | .775 |
| PR or CR | 11 (52) | 10 (48) | ||
| BC patients treated by radical cystectomy without chemotherapy (n = 70) | ||||
| Sex | Male | 18 (36) | 32 (64) | .145 |
| Female | 11 (55) | 9 (45) | ||
| Age, y | ≤70 | 9 (33) | 18 (67) | .276 |
| >70 | 20 (47) | 23 (53) | ||
| Cellular atypism classification | Low grade | 1 (20) | 4 (80) | .313 |
| High grade | 28 (43) | 37 (57) | ||
| T classification | Ta/Tis/1 | 10 (36) | 18 (64) | .428 |
| T2/3/4 | 19 (45) | 23 (55) | ||
| Lymphatic invasion | Negative | 20 (41) | 28 (58) | .952 |
| Positive | 9 (41) | 13 (59) | ||
| Vascular invasion | Negative | 27 (42) | 36 (57) | .467 |
| Positive | 2 (29) | 5 (71) | ||
| N classification | N0 | 25 (44) | 32 (56) | .973 |
| N1/2/3 | 4 (44) | 5 (56) | ||
| TNM stage | 0/I/II | 14 (34) | 27 (66) | .141 |
| III/IV | 15 (52) | 14 (48) | ||
Abbreviations: CR, complete response; GC, gemcitabine and cisplatin; GCa, gemcitabine and carboplatin; MVAC, methotrexate, vinblastine, doxorubicin, and cisplatin; PD, progressive disease; PR, partial response; SD, stable disease; Ta, noninvasive papillary carcinoma; Tis, carcinoma in situ.
First, we examined the cohort of 50 advanced BC treated with PBC (Table 1). Twenty‐five of those 50 cases (50%) were SLFN11 positive. There was no significant association between SLFN11 expression and the clinicopathologic characteristics (Table 1). Kaplan‐Meier analysis revealed that the OS of the SLFN11‐positive group was significantly better than that of the SLFN11‐negative group (P = .012, Figure 1D). Univariate analyses revealed that SLFN11 expression was significantly associated with survival, and multivariate analyses identified clinical TNM stage and SLFN11 positivity as independent markers of better prognosis (Table 2).
TABLE 2.
Univariate and multivariate analysis of factors for prognosis of bladder cancer patients treated with platinum‐based chemotherapy (n = 50)
| Patient characteristic | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| Sex | Male | 1.000 | 1.000 | ||
| Female | 0.300 (0.069‐1.303) | .108 | 0.788 (0.163‐3.796) | .767 | |
| Age, y | ≤70 | 1.000 | 1.000 | ||
| >70 | 1.119 (0.454‐2.758) | .806 | 2.475 (0.863‐7.100) | .092 | |
| Cellular atypism classification | Low grade | 1.000 | 1.000 | ||
| High grade | 0.852 (0.248‐2.931) | .799 | 0.885 (0.208‐3.762) | .868 | |
| Clinical TNM stage | Stage II/III | 1.000 | 1.000 | ||
| Stage IV | 4.099 (1.351‐12.433) | .013 | 7.597 (1.913‐30.166) | .004 | |
| Visceral metastasis | Negative | 1.000 | 1.000 | ||
| Positive | 1.160 (0.337‐3.992) | .813 | 0.546 (0.131‐2.275) | .406 | |
| SLFN11 expression | Negative | 1.000 | 1.000 | ||
| Positive | 0.291 (0.104‐0.811) | .018 | 0.275 (0.094‐0.805) | .018 | |
Abbreviations: CI, confidence interval; OR, odds ratio; SLFN11, Schlafen 11.The significance of bold values are P values less than 0.05
We next analyzed the cohort of 70 local BC treated without PBC (Table 1). Twenty‐nine of the 70 cases (41%) were SLFN11 positive. There was no significant association between SLFN11 expression and the clinicopathologic characteristics (Table 1). Kaplan‐Meier analysis revealed that the OS of the SLFN11‐positive group was significantly worse than that of the SLFN11‐negative group (P = .034, Figure 1E). These results indicated that SLFN11 expression is an unfavorable prognostic marker for BC patients who do not receive PBC treatment, whereas SLFN11 expression can be a predictive marker of superior response to PBC for BC patients.
3.2. Schlafen 11 expression is associated with luminal subtype marker GATA3 but not with other subtype markers or an immune checkpoint marker
Bladder cancer can be classified into p53‐like, luminal, and basal subtypes. 32 , 33 To examine a possible association between these subtypes and SLFN11 expression, we undertook IHC staining for p53 (TP53), luminal marker GATA3, and basal marker CK5/6 in the 50 advanced BC with PBC (Figure 2). In addition, we undertook IHC staining for PD‐L1, a key molecule that determines the response to immune checkpoint inhibitors (Figure 2). Several cases could not be analyzed for technical reasons. We validated the positivity and negativity of the results according to previous reports 30 (see also Materials and Methods). Statistical analyses revealed that only GATA3 expression was significantly associated with SLFN11 expression (Table S1), implying that any drivers of luminal phenotype might activate SLFN11 expression.
FIGURE 2.
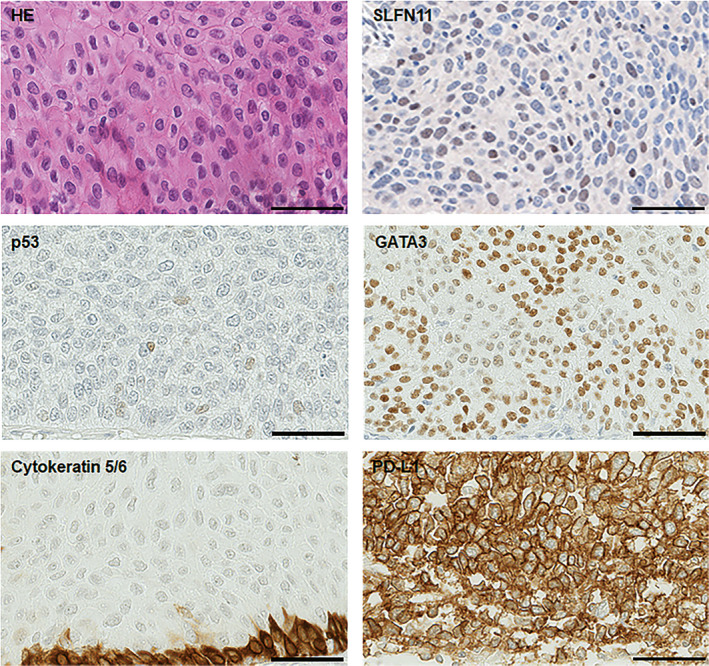
Representative images of H&E and immunohistochemical staining. Images of H&E, Schlafen 11 (SLFN11), p53, GATA3, cytokeratin 5/6, and programmed death ligand 1 (PD‐L1) staining in bladder cancer cells. Scale bars, 50 µm
3.3. Schlafen 11 expression is correlated with cisplatin response in BC cell lines and SLFN11 KO confers chemoresistance to cisplatin in BC cell lines
To validate our finding with the clinical BC samples by a genetic approach, we first used the CellMiner website (https://discover.nci.nih.gov/cellminercdb/) 34 with the Genomics of Drug Sensitivity in Cancer (GDSC) cancer cell line database, which includes 15 BC cell lines among the 849 cell lines tested with cisplatin (Figure 3A). As expected, 24 , 26 , 31 , 35 the cytotoxicity of cisplatin was found to be highly correlated with SLFN11 expression across the 849 cell lines (Figure 3A). Cisplatin ranked eighth in the top drugs, which included camptothecin, topotecan, talazoparib, LMP744, mitoxantrone, teniposide, and bendamustin. The 15 BC cell lines of GDSC tested with cisplatin also showed highly positive correlations between SLFN11 expression and response to cisplatin (Figure 3B).
FIGURE 3.
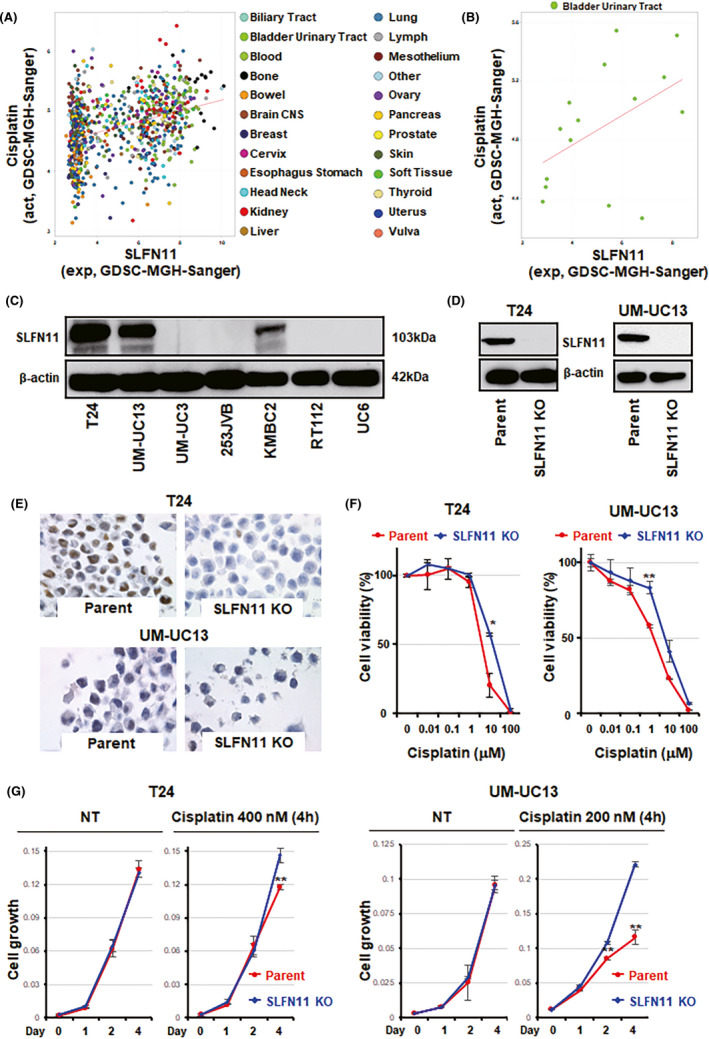
Inactivation of Schlafen 11 (SLFN11) induces resistance to cisplatin and inhibits cell death under replication stress in bladder cancer (BC) cell lines. A, Correlation between SLFN11 mRNA expression and cisplatin sensitivity among 849 cell lines, which includes 15 BC cell lines in the Genomics of Drug Sensitivity in Cancer (GDSC) cancer cell line database. Pearson correlation (r) = .36, P = 1.1e‐27. B, Correlation between SLFN11 mRNA expression and cisplatin sensitivity among 15 BC cell lines with the GDSC cancer cell line database. r = .47, P = .075. C, Western blot analysis of SLFN11 in BC cell lines and β‐actin as a loading control. D, Western blot analysis of SLFN11 KO cells generated by CRISPR‐Cas9 gene editing technology. β‐Actin was used as a loading control. E, Immunohistochemical images of parent and SLFN11 KO cells in T24 or UM‐UC13 cell lines. Scale bars, 100 µm. F, Dose‐dependent effects of cisplatin on the viability of T24 or UM‐UC13 cell lines with parent and SLFN11 KO cells. G, Cell growth curves of the indicated cell lines under normal conditions (NT) or treated with the indicated concentrations of cisplatin for 4 h and released into a drug‐free medium. *P < .05; **P < .01. act, drug activity (−log10 [IC50M]); exp, mRNA expression (log2); MGH, Massachusetts General Hospital; GDSC, Genomics of Drug Sensitivity in Cancer
Next, we undertook experiments in seven BC cell lines in our laboratory. High SLFN11 expression was observed in three of them: T24, UM‐UC13, and KMBC2 (Figure 3C), and we generated SLFN11 KO cells in those three cell lines by CRISPR‐Cas9 gene editing technology. Knockout of SLFN11 protein expression was confirmed by western blotting and IHC (Figures 3D,E and S1A,B). The SLFN11 KO cells acquired resistance to cisplatin after 48 hours of continuous treatment in all cases (Figures 3F and S1C). We also tested the impact of SLFN11 on cell growth after a brief exposure to cisplatin that mimics the clinical situation. Concentrations of cisplatin for the brief exposure tests were 10% of their IC50 values. 14 Although the presence of SLFN11 did not alter cell growth rate under the normal condition, treatment with a submicromolar concentration of cisplatin for 4 hours significantly delayed cell growth in the parental cells compared to the SLFN11 KO cells in the three BC lines (Figures 3G and S1D). Hence, SLFN11 sensitizes BC cells to cisplatin in response to either short or long treatment times (Figure 3F).
3.4. Knockout of SLFN11 does not alter the amount of DNA damage by cisplatin
To examine whether SLFN11 affects the amount of DNA damage, we treated T24 and KMBC2 cell sets (parental and the SLFN11 KO cells) with cisplatin for 4 hours. DNA damage was semiquantified by western blotting and immunofluorescence for γH2AX, a hallmark of histone modification under DNA damage. 36 We confirmed a comparable amount of DNA damage between the two cell sets (Figure S2). Taken together, these results indicate that the elevated cisplatin sensitivity in SLFN11‐expressing cells is not a result of increased DNA damage in BC cells.
3.5. Synergistic effect of epigenetic modifiers with platinum derivatives through SLFN11 reactivation
Schlafen 11 expression has been shown to be regulated by epigenetic modifications on DNA and/or histones. 7 , 22 , 24 Hence, it is feasible for SLFN11 to be reactivated by inhibitors of DNA methyltransferases, such as 5‐aza, or HDAC inhibitors, such as entinostat. 7 , 17 To examine this possibility, SLFN11‐negative RT112 and 253JBV cell lines were treated with the epigenetic modifiers (5‐aza or entinostat). Those treatments reactivated SLFN11 at the protein level (Figure 4A,B). To assess whether the reactivated SLFN11 can sensitize BC cells to platinum derivatives, we undertook combination assays. We chose a less toxic concentration of 5‐aza or entinostat delivered by a single agent, by which SLFN11 was reactivated (Figure S3). All combinations of 5‐aza or entinostat with a platinum derivative (cisplatin or carboplatin) sensitized RT112 and 253JBV cells significantly more compared to the single treatment of cisplatin or carboplatin (Figure 4C).
FIGURE 4.
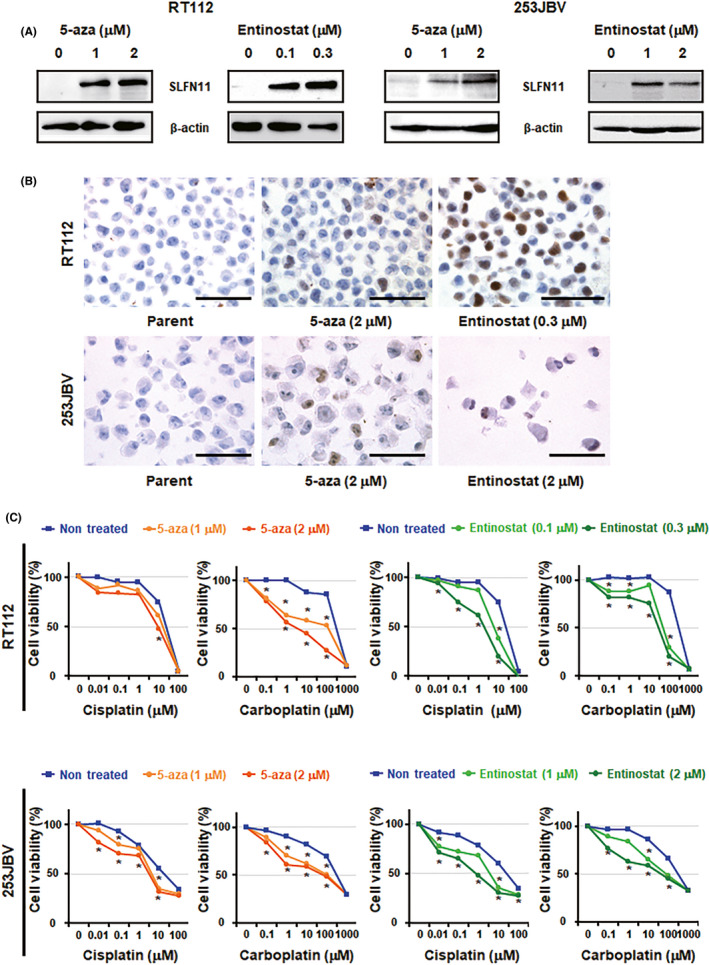
Epigenetic modulators reactivate Schlafen 11 (SLFN11) expression and sensitize bladder cancer cells to platinum agents. A, Western blot analysis of RT112 (left) and 253JBV (right) cell lines treated with the indicated concentrations of 5‐aza‐2′‐deoxycytidine (5‐aza) or entinostat for 2 d. β‐Actin was used as a loading control. B, Representative immunohistochemical images of SLFN11 in RT112 (upper) and 253JBV (lower) cell lines treated with the indicated concentrations of 5‐aza or entinostat for 2 d. Scale bars, 100 µm. C, Dose‐dependent effects of cisplatin or carboplatin on the viability of RT112 (upper) and 253JBV (lower) cell lines treated with the indicated concentrations of 5‐aza or entinostat
Because epigenetic modifiers alter the expression of multiple genes, we knocked out SLFN11 in the RT112 cell line to assess whether the synergistic effect was derived by SLFN11 reactivation. The absence of SLFN11 expression was validated by western blotting after the treatment with 5‐aza or entinostat (Figure S4A). We then tested the combination of 5‐aza or entinostat with cisplatin. The synergistic effects observed in the parental RT112 cells (Figure 4C) disappeared in the SLFN11 KO RT112 cells (Figure S4B). Moreover, such a synergistic effect was not observed in the T24 cell line, which has robust expression of SLFN11 under normal conditions (Figure S4C,D). The same results were obtained in our study on gastric cancer. 22 From these results, we concluded that reactivation of SLFN11 is a predominant factor for the synergistic effect of epigenetic modifiers with platinum derivatives. These results suggest a promising strategy for BC patients to overcome refractoriness to platinum derivatives.
4. DISCUSSION
To capture the tumor biology that is responsible for the PBC response in BCs, we investigated the correlation between SLFN11 expression and clinicopathologic characteristics. We found that positive expression of SLFN11 can predict better OS in advanced BC patients treated with PBC. Additionally, we observed that expression of SLFN11 was associated with worse OS in local BC patients who are treated without PBC. We report that resistance to cisplatin or carboplatin can be due to epigenetic silencing of SLFN11, and that epigenetic reactivation of SLFN11 restores sensitivity to platinum derivatives in SLFN11‐negative BC cell lines. Consequently, SLFN11 could serve as a predictive biomarker for PBC in advanced BC patients and evaluation of SLFN11 expression could potentially be a promising strategy to control refractory BC to PBC.
4.1. Clinical implications of SLFN11
We recently reported comprehensive analyses of SLFN11 expression across 16 human organs. 31 We found that no case of normal urothelial epithelium in bladder expressed SLFN11, whereas 68% of urothelial carcinomas expressed SLFN11 at various degrees, 31 which implies activation of SLFN11 expression in the process of tumorigenesis in urothelial carcinomas. In other words, any oncogenic factor of urothelial carcinomas could activate SLFN11 expression, which might be related to the worse OS in SLFN11‐positive compared to SLFN11‐negative local BC without PBC (Figure 1E).
Schlafen 11 is known to be induced by chronic inflammation 37 and cytokines such as interferon‐β. 38 Chronic inflammation and proinflammatory cytokines are known causes of the pathogenesis of BC. 39 , 40 In the present study, we found that the expression of GATA3, a luminal marker in breast and bladder cancers as well as a transcription factor of cytokines, correlated significantly with SLFN11 expression, which indicates that SLFN11 might be activated by GATA3 indirectly through the regulation of cytokines. Additionally, using the cancer cell line databases, we found that SLFN11 transcript expression was significantly correlated with the expression of the interleukin receptor‐associated kinase 1 in the 20 BC cell lines of the GDSC and in the 27 cell lines of Broad Cancer Cell Line Encyclopedia (r = .86 with P = 8.9e‐07 and r = .61 with P = .0007, respectively; Figure S1E,F). However, only four patients showed CK5/6 expression in our cohort, which implies that our data can be influenced by the biased cohort in terms of luminal/basal phenotype.
In the cohort of this study, nearly half of the BC cases (66/120) were totally negative for SLFN11 (Figure 1B). Hence, we applied the threshold of a 5% positive rate to divide the population into two groups. This situation is not always applicable to other cancers or different cohorts because the distribution of SLFN11‐positive cells varies by organs and tissue of origin. 24 Moreover, storage conditions of the formalin‐fixed paraffin‐embedded samples could affect staining sensitivity. 19 Accordingly, in our recent report on gastric cancers, we used 30% as the threshold based on two major populations at 0% and 30%. 22
For the approximately 50% of patients with SLFN11‐negative BC who are possibly refractory to platinums, we propose a combination strategy with epigenetic modifiers (Figure 4). One of the limitations of this study is that we did not examine the effect of combination therapy in the in vivo models, which will be necessary in additional studies to further establish the utility of combination therapy. In addition, we found no correlation between SLFN11 expression and response to chemotherapy, which might be due to the small number of advanced BC patients who received PBC.
4.2. Advantage of SLFN11 examination in the clinic
Cisplatin and carboplatin generate DNA adducts repaired by DNA damage response pathways. Defects in homology directed repair and transcription coupled nucleotide excision repair, which are key DNA repair pathways, have been established to be associated with elevated cisplatin response rates in urothelial carcinoma through whole and exon genome sequencing. 41 Although mutational analyses of these genes are valuable for precision medicine, such sequence analysis methods are currently applicable in limited institutions or hospitals at high sequencing analysis cost. By contrast, the evaluation of SLFN11 expression by IHC 22 , 31 is low‐cost and can be integrated with conventional methods as well as with RNA sequencing methods, which are becoming mainstream for precision medicine.
Although not stressed in this manuscript, we found that specimens obtained by biopsy or transurethral resection were adequate for the analysis of SLFN11 expression. Hence, patients with advanced BC whose tissue blocks are not available can also be stratified by the results of IHC.
The clinical study of SLFN11 is just beginning at many institutions. 11 , 22 , 37 , 42 , 43 , 44 , 45 Although the utility of SLFN11 as a predictive biomarker for DDAs has been established, regardless of the tissues of origin, most of the associated reports are limited to retrospective studies. Prospective studies should be planned to establish the clinical utility of SLFN11.
DISCLOSURE
The authors have no conflict of interest.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Table S1
ACKNOWLEDGMENTS
We thank Mr Shoichi Norimura for his excellent technical assistance. The present study was supported by Grants‐in‐Aid for Scientific Research (JP15H04713 and JP16K08691 to WY, JP16H06999 to NS, and JP19H03505 to JM) and Challenging Exploratory Research (26670175, JP16K15247 to WY) from the Japan Society for the Promotion of Science. The present study was also supported in part by research funds from the Yamagata prefectural government and the City of Tsuruoka. YP is supported by the Center for Cancer Research, the Intramural Program of the National Cancer Institute (Z01‐BC‐006150), NIH, Bethesda, MD.
Taniyama D, Sakamoto N, Takashima T, et al. Prognostic impact of Schlafen 11 in bladder cancer patients treated with platinum‐based chemotherapy. Cancer Sci.2022;113:784–795. doi: 10.1111/cas.15207
Funding information
Japan Society for the Promotion of Science, Grant/Award Number: JP15H04713, JP16K08691, JP16H06999, JP19H03505, 26670175, JP16K15247; Yamagata prefectural government; City of Tsuruoka; Center for Cancer Research, Grant/Award Number: Z01‐BC‐006150.
REFERENCES
- 1. Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71:96‐108. [DOI] [PubMed] [Google Scholar]
- 2. Smith AB, Deal AM, Woods ME, et al. Muscle‐invasive bladder cancer: evaluating treatment and survival in the National Cancer Data Base. BJU Int. 2014;114:719‐726. [DOI] [PubMed] [Google Scholar]
- 3. Massard C, Gordon MS, Sharma S, et al. Safety and efficacy of durvalumab (MEDI4736), an anti‐programmed cell death ligand‐1 immune checkpoint inhibitor, in patients with advanced urothelial bladder cancer. J Clin Oncol. 2016;34:3119‐3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Powles T, van der Heijden MS, Castellano D, et al. Durvalumab alone and durvalumab plus tremelimumab versus chemotherapy in previously untreated patients with unresectable, locally advanced or metastatic urothelial carcinoma (DANUBE): a randomised, open‐label, multicentre, phase 3 trial. Lancet Oncol. 2020;21:1574‐1588. [DOI] [PubMed] [Google Scholar]
- 5. von der Maase H, Sengelov L, Roberts JT, et al. Long‐term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602‐4608. [DOI] [PubMed] [Google Scholar]
- 6. Zoppoli G, Regairaz M, Leo E, et al. Putative DNA/RNA helicase Schlafen‐11 (SLFN11) sensitizes cancer cells to DNA‐damaging agents. Proc Natl Acad Sci USA. 2012;109:15030‐15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nogales V, Reinhold WC, Varma S, et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget. 2016;7:3084‐3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reinhold WC, Varma S, Sunshine M, et al. The NCI‐60 methylome and its integration into cell miner. Cancer Res. 2017;77:601‐612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allison Stewart C, Tong P, Cardnell RJ, et al. Dynamic variations in epithelial‐to‐mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget. 2017;8:28575‐28587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kang MH, Wang J, Makena MR, et al. Activity of MM‐398, nanoliposomal irinotecan (nal‐IRI), in Ewing's family tumor xenografts is associated with high exposure of tumor to drug and high SLFN11 expression. Cancer Res. 2015;21:1139‐1150. [DOI] [PubMed] [Google Scholar]
- 11. Moribe F, Nishikori M, Takashima T, et al. Epigenetic suppression of SLFN11 in germinal center B‐cells during B‐cell development. PLoS One. 2021;16:e0237554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murai J, Feng Y, Yu GK, et al. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget. 2016;7:76534‐76550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murai J, Pommier Y. PARP trapping beyond homologous recombination and platinum sensitivity in cancers. Annu Rev Cancer Biol. 2019;3:131‐150. [Google Scholar]
- 14. Murai J, Tang S‐W, Leo E, et al. SLFN11 blocks stressed replication forks independently of ATR. Mol Cell. 2018;69:371‐84.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rathkey D, Khanal M, Murai J, et al. Sensitivity of mesothelioma cells to parp inhibitors is not dependent on BAP1 but is enhanced by temozolomide in cells with high‐Schlafen 11 and low‐O6‐METHYLGUANINE‐DNA methyltransferase expression. J Thorac Oncol. 2020;15:843‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tang S‐W, Bilke S, Cao L, et al. SLFN11 is a transcriptional target of EWS‐FLI1 and a determinant of drug response in Ewing sarcoma. Clin Cancer Res. 2015;21:4184‐4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang S‐W, Thomas A, Murai J, et al. Overcoming resistance to DNA‐targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin Cancer Res. 2018;24:1944‐1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coussy F, El‐Botty R, Château‐Joubert S, et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple‐negative breast cancers. Sci Transl Med. 2020;12:eaax2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pietanza MC, Waqar SN, Krug LM, et al. Randomized, double‐blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed‐sensitive or refractory small‐cell lung cancer. J Clin Oncol. 2018;36:2386‐2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conteduca V, Ku SY, Puca L, et al. SLFN11 expression in advanced prostate cancer and response to platinum‐based chemotherapy. Mol Cancer Ther. 2020;19:1157‐1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kagami T, Yamade M, Suzuki T, et al. The first evidence for SLFN11 expression as an independent prognostic factor for patients with esophageal cancer after chemoradiotherapy. BMC Cancer. 2020;20:1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takashima T, Taniyama D, Sakamoto N, et al. Schlafen 11 predicts response to platinum‐based chemotherapy in gastric cancers. Br J Cancer. 2021;125(1):65‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mu Y, Lou J, Srivastava M, et al. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016;17:94‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murai J, Thomas A, Miettinen M, Pommier Y. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA‐targeting anti‐cancer therapies. Pharmacol Ther. 2019;201:94‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murai J, Zhang H, Pongor L, et al. Chromatin remodeling and immediate early gene activation by SLFN11 in response to replication stress. Cell Rep. 2020;30:4137‐4151.e4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2‐SLFN11 Axis. Cancer Cell. 2017;31:286‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Honda Y, Nakamura Y, Teishima J, et al. Clinical staging of upper urinary tract urothelial carcinoma for T staging: review and pictorial essay. Int J Urol. 2019;26:1024‐1032. [DOI] [PubMed] [Google Scholar]
- 28. Sekino Y, Han X, Kobayashi G, et al. BUB1B overexpression is an independent prognostic marker and associated with CD44, p53, and PD‐L1 in renal cell carcinoma. Oncology. 2021;99:240‐250. [DOI] [PubMed] [Google Scholar]
- 29. Shigematsu Y, Oue N, Sekino Y, et al. SEC11A expression is associated with basal‐like bladder cancer and predicts patient survival. Pathobiology. 2019;86:208‐216. [DOI] [PubMed] [Google Scholar]
- 30. Hayashi T, Sentani K, Ikeda K, et al. MP82‐03 Clinicopathological characteristics of upper tract urothelial cancer with loss of immunohistochemical expression of mismatch repair proteins in universal screening. J Urol. 2020;203(Supplement 4):e1246. [Google Scholar]
- 31. Takashima T, Sakamoto N, Murai J, et al. Immunohistochemical analysis of SLFN11 expression uncovers potential non‐responders to DNA‐damaging agents overlooked by tissue RNA‐seq. Virchows Arch. 2021;478(3):569‐579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi W, Porten S, Kim S, et al. Identification of distinct basal and luminal subtypes of muscle‐invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taber A, Christensen E, Lamy P, et al. Molecular correlates of cisplatin‐based chemotherapy response in muscle invasive bladder cancer by integrated multi‐omics analysis. Nat Commun. 2020;11:4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luna A, Elloumi F, Varma S, et al. Cell miner cross‐database (CellMinerCDB) version 1.2: exploration of patient‐derived cancer cell line pharmacogenomics. Nucleic Acids Res. 2021;49:D1083‐D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He T, Zhang M, Zheng R, et al. Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics. 2017;9:849‐862. [DOI] [PubMed] [Google Scholar]
- 36. Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957‐967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe S, Nishimura R, Shirasaki T, et al. Schlafen 11 is a novel target for mucosal regeneration in ulcerative colitis. J Crohns Colitis. 2021;15(9):1558‐1572. [DOI] [PubMed] [Google Scholar]
- 38. Li M, Kao E, Gao X, et al. Codon‐usage‐based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491:125‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Korac‐Prlic J, Degoricija M, Vilović K, et al. Targeting Stat3 signaling impairs the progression of bladder cancer in a mouse model. Cancer Lett. 2020;490:89‐99. [DOI] [PubMed] [Google Scholar]
- 40. Chen MF, Lin PY, Wu CF, Chen WC, Wu CT. IL‐6 expression regulates tumorigenicity and correlates with prognosis in bladder cancer. PLoS One. 2013;8:e61901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Börcsök J, Sztupinszki Z, Bekele R, et al. Identification of a synthetic lethal relationship between nucleotide excision repair (NER) deficiency and irofulven sensitivity in urothelial cancer. Clin Cancer Res. 2020;27:2011‐2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Coleman N, Zhang B, Byers LA, Yap TA. The role of Schlafen 11 (SLFN11) as a predictive biomarker for targeting the DNA damage response. Br J Cancer. 2021;124:857‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Knelson EH, Patel SA, Sands JM. PARP inhibitors in small‐cell lung cancer: rational combinations to improve responses. Cancers (Basel). 2021;13(4):727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mao S, Chaerkady R, Yu W, et al. Resistance to pyrrolobenzodiazepine dimers is associated with SLFN11 downregulation and can be reversed through inhibition of ATR. Mol Cancer Ther. 2021;20:541‐552. [DOI] [PubMed] [Google Scholar]
- 45. Winkler C, Armenia J, Jones GN, et al. SLFN11 informs on standard of care and novel treatments in a wide range of cancer models. Br J Cancer. 2021;124:951‐962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Table S1


