Abstract
Species are facing strong selection pressures to adapt to inhospitable high-altitude environments. Yaks are a valuable species and an iconic symbol of the Qinghai-Tibet Plateau. Extensive studies of high-altitude adaptation have been conducted, but few have focused on metabolism. In the present study, we determined the differences in the serum metabolomics between yaks and the closely related species of low-altitude yellow cattle and dairy cows. We generated high-quality metabolite profiling data for 36 samples derived from the three species, and a clear separation trend was obtained between yaks and the other animals from principal component analysis. In addition, we identified a total of 63 differentially expressed metabolites among the three species. Functional analysis revealed that differentially expressed metabolites were related to the innate immune activation, oxidative stress-related metabolism, and energy metabolism in yaks, which indicates the important roles of metabolites in high-altitude adaptation in yaks. The results provide new insights into the mechanism of adaptation or acclimatization to high-altitude environments in yaks and hypoxia-related diseases in humans.
Keywords: Yak, High-altitude adaptation, Metabolomics, Cattle, Dairy cow
INTRODUCTION
The Qinghai-Tibetan Plateau (QTP), characterized by low temperature and hypobaric hypoxia, is the highest plateau in the world, with an average altitude > 4,000 m above sea level. Species are facing strong selection pressure to adapt to inhospitable high-altitude environments [1]. The yak (Bos grunniens) is an important domesticated ruminant. It is the only large mammal inhabiting the QTP and is an iconic symbol of the QTP [2]. Yaks living at high altitudes more than 7,000 years, and must adapt to the stress of decreased oxygen availability [3]. Yaks have numerous special morphological and physiological mechanisms for life at high altitudes, e.g., blunted hypoxic pulmonary vasoconstriction [4], increased foraging ability [5], enhanced glucose uptake and aerobic respiration [6], and improved bioenergy metabolism than mammals living in the plains [7]. Genome analysis identified an expansion in yak of gene families related to sensory perception and energy metabolism comparied with cattle [8], and differentially expressed miRNAs have also been found to be enriched in hypoxia-related pathways [9]. In addition, to reduce the risk of infection and disease, the activation of innate immunity was higher in yaks than in other cattle [10]. These findings partially reveal the adaptive mechanisms of yaks due to natural selection in a high-altitude and hypoxic environment, but few investigations have focused on the role of metabolites.
High-altitude hypoxia continuously affects the physical performance of people and animals [11]. Survival in high-altitude hypoxia requires a profound adaptive shift in metabolic processes [12]. In addition, hypoxia is related to homeostasis and the metabolic rate in adult tissues [13]. Organic metabolites are the reactants, intermediates or products of enzymatic reactions and represent the final products of cellular processes. The trend of contemporary scientific development is to follow systems biology. Investigation into the metabolome in response to genetic modification or physiological stimulus is a part of systems biology [14]. Identifying metabolic pathways has the potential to improve the understanding of physiological mechanisms [15]. The metabolome can reveal total metabolic profile changes in biological phenotypes and silent phenotypes [16]. In this study, the serum metabolites of yaks (B. grunniens), yellow cattle (Bos taurus) and China Holstein dairy cows (Bos taurus) were analyzed using a nontargeted metabolomics approach based on ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF-MS). Comparing yaks, yellow cattle and China Holstein dairy cows may contribute to understanding evolutionary adaptation and provide meaningful data for survival at high altitudes.
MATERIALS AND METHODS
Sample collection
Blood samples were collected between 9:00 and 10:00 am by jugular venous puncture using vacuum tubes from 12 white yaks (Qilian Township, 37°41’6”N, 102°26’24”E, altitude: 3,600 m), 12 local yellow cattle (Tanshanling Town, 37°4’38”N, 102°24’14”E, altitude: 2,200 m), and 12 China Holstein dairy cows (Anyuan Town, 37°8’24”N, 102°37’48”E, altitude: 1,700 m) in Tianzhu County on the edge of QTP, Gansu Province, China (Fig. 1). After the blood was left to stand for 30 min, it was centrifuged at 1,000 ×g for 5 min at 4°C. Then, the serum was extracted, immediately frozen in liquid nitrogen and stored until analysis was carried out. All animals are female, and about three years old. Yaks and yellow cattle graze the natural grassland throughout the year without supplementary feed and housing. Holstein dairy cows (milk production: 27.1 ± 0.85 kg/day, parity: 2, days in milk [DIM]: 91.6 ± 7.5 days) were fed the total mixed rations (TMR) diets ad libitum, the basal diet was formulated based on the Feeding Standards of Dairy Cattle in China. The three species had similar physical characteristics, and the characteristics enrolled yak, cattle, and Holstein dairy cows are shown in Table 1. The three pasture sites are traditionally used by local herders for grazing, with similar environment and climatic conditions (temperature: 19.2 ± 1.1°C, relative humidity: 65.0 ± 2.2%), except altitude. In order to minimize the controlling variables of feeding and environmental factors among the three species, the blood was collected in August, 2021. The animal experiment was approved, and the animals received humane care according to the Ethical Committee rules of Lanzhou University (RIB21110301).
Fig. 1. The geographic distribution of the sampling locations for white yak, yellow cattle and dairy cow on the edge of Qinghai-Tibetan Plateau, China.
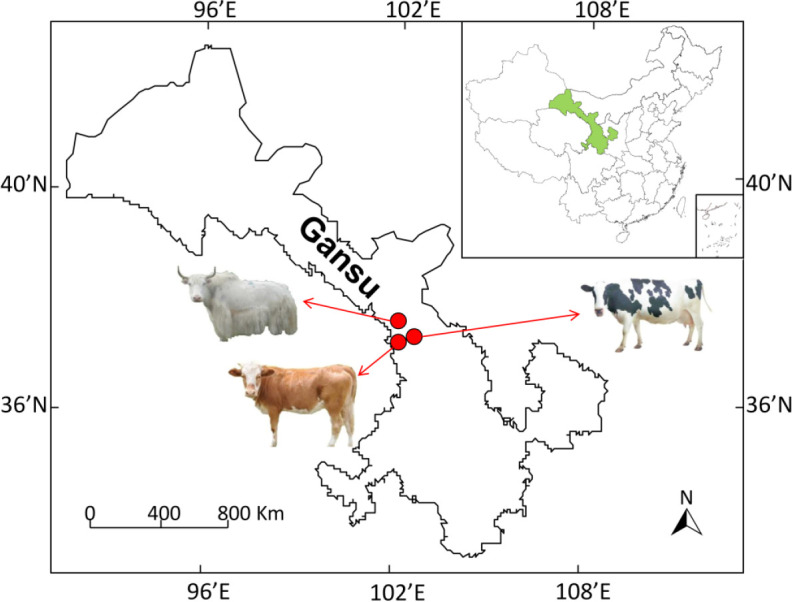
Table 1. The characteristics of enrolled yak, cattle, and Holstein dairy cows.
| Group | Body condition score | Parity | Age (y) | Milk production (kg/d) | Days in milk (d) |
|---|---|---|---|---|---|
| Yak | 3.5 ± 0.4 | 2 | 3 | - | 89.6 ± 10.5 |
| Cattle | 3.3 ± 0.5 | 2 | 3 | - | 87.5 ± 9.6 |
| Holstein dairy cows | 3.2 ± 0.3 | 2 | 3 | 27.1 ± 0.85 | 91.6 ± 7.5 |
Metabolite extraction
The collected samples were thawed at 4°C, and 100 μL of sample was mixed with 400 μL of precooled methanol/acetonitrile (1:1, v/v). It was incubated at room temperature for 10 min and then centrifuged. The supernatants were collected, dried, and then resuspended in 30 μL water/acetonitrile (98:2, v/v) for MS analysis.
Liquid chromatography conditions
First, a UPLC system (SCIEX, Framingham, MA, USA) was used for chromatographic separations. Reversed-phase separation was performed using an ACQUITY UPLC T3 column. Solvent A (ultrapure water, 0.1% formic acid) and solvent B (acetonitrile, 0.1% formic acid) comprised the mobile phase.
Quadrupole time-of-flight mass spectrometry conditions
The metabolites were detected using a tandem mass spectrometer (TripleTOF5600plus, SCIEX). The details of the Q-TOF mass spectrometry conditions were based on our previous publication [17].
Processing of metabolomics data
LC−MS raw data files were processed by the CAMERA and XCMS packages of R software. Retention time (RT) and m/z data were used to identify each ion. The metabolites were annotated using the human metabolome database (HMDB) database and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. MetaX was used to further preprocess the intensity of the peak data. The “50% rule” was applied to remove the systematic bias or technical variation by normalizing the data according to our previous publication [17], and the results showed a normal distribution after normalization processing. Outlier detection and batch effects were evaluated by principal component analysis (PCA). False discovery rate (FDR) and supervised partial least squares-discriminant analysis (PLS-DA) were conducted to adjust the p value. The important features were selected based on a variable importance in the projection (VIP) cutoff value of 1.0.
Determination of inflammatory cytokines and antioxidant enzymes
The levels of interleukin-2 (IL-2), interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) in serum were determined using ELISA kits. The levels of malondialdehyde (MDA), total antioxidant capacity (T-AOC), and glutathione peroxidase (GSH-Px) in serum were measured by chemical colorimetry. All kits were purchased from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). The detailed principles and methods for the detection of MDA, T-AOC and GSH-Px have been described in our previous publication [18].
Statistical analysis
GraphPad Prism 9 was employed to perform statistical analyses by single factor analysis of variance (one-way ANOVA). P values less than 0.05 indicated a significant difference.
RESULTS
Serum metabolite analysis
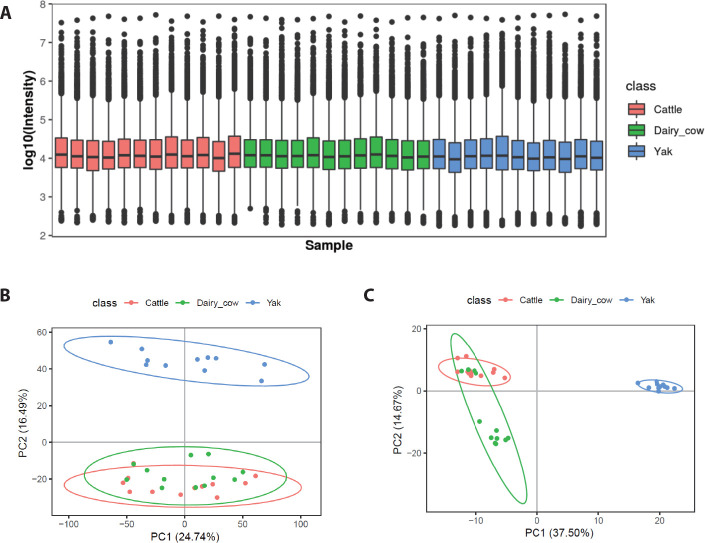
Box plots were used to analyze the identified serum metabolites in yaks, yellow cattle and dairy cows. All samples showed a similar range of metabolite levels (Fig. 2A). Serum metabolomic analysis was used to determine whether the metabolic profiles of yaks are separable from those of dairy cows and yellow cattle, we used PCA for visualization. Based on the serum metabolic profiles, the score plots of the PCA model discriminating yaks, dairy cows and yellow cattle are presented in Figs. 2B and C. PCA showed that the positive mode of the total variance data was 41.23%, represented by the first two principal components (Fig. 2A), and the negative mode was 52.17% (Fig. 2B). The plot revealed that the serum metabolic profiles of yellow cattle were closely related to those of dairy cows and were not obviously changed. However, the profiles of yaks showed a clear separation trend from those of and yellow cattle and dairy cows.
Fig. 2. Comparison of distribution of serum metabolite levels among yak, dairy cow and yellow cattle.
(A) Box plots represent the distribution of metabolite peak intensity measurements from serum samples across all subjects. PCA scores plots of serum metabolomic profiles derived from UPLC-Q-TOF-MS spectra showing separation between yak and yellow cattle and dairy cow in the positive mode (B) and negative mode (C). PC, principal component; PCA, principal component analysis; UPLC-Q-TOF-MS, ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry.
Identification of differentially expressed serum metabolites among yaks, yellow cattle and dairy cows
From the 1,815 detected metabolites, we investigated 63 differentially expressed metabolites (Table 2), including L-glutamine, L-glutamic acid, α-linolenic acid, tauroursodeoxycholic acid, and LysoPC (ratio ≥ 5.0 or ≤ 0.2, p < 0.01 and VIP ≥ 1). The relative concentrations of 23 metabolites were significantly higher, while 30 were significantly lower in yaks than in yellow cattle (Table 2). The relative concentrations of 11 metabolites were significantly higher and 15 were significantly lower in yaks than in dairy cows. The relative concentrations of 11 metabolites were significantly lower in yellow cattle than in dairy cows. These metabolites were carbohydrates, amino acids, lipids and their metabolites, suggesting that these metabolic pathways were different among yaks, yellow cattle, and dairy cows. Furthermore, metabolic pathways were significantly different between yaks and yellow cattle and between yaks and dairy cows, but there were similarities between yellow cattle and dairy cows based on the very few differential metabolic profiles (Table 2).
Table 2. List of serum differential metabolites among yak, yellow cattle and dairy cow (n = 12).
| NO. | Metabolites | Yak vs cattle | Yak vs dairy cow | Cattle vs dairy cow | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ratio | p-value | VIP | Ratio | p-value | VIP | Ratio | p-value | VIP | ||
| 1 | L-Acetopine | 9.287 | 1.17E-03 | 2.237 | 5.183 | 4.55E-03 | 2.082 | |||
| 2 | LysoPC (18:0) | 7.211 | 7.94E-06 | 2.558 | 7.621 | 3.30E-06 | 2.906 | |||
| 3 | 7-Ketodeoxycholic acid | 6.780 | 2.93E-03 | 2.358 | 0.161 | 5.11E-04 | 3.567 | |||
| 4 | LysoPC (16:0) | 6.379 | 1.09E-04 | 2.311 | 0.131 | 6.89E-06 | 2.572 | |||
| 5 | LysoPC (18:1) | 6.219 | 3.65E-05 | 2.401 | 5.979 | 3.69E-05 | 2.539 | |||
| 6 | Ganoderol A | 6.042 | 1.81E-05 | 2.564 | 5.110 | 4.73E-05 | 2.264 | |||
| 7 | α-Linolenic acid | 0.179 | 1.93E-05 | 2.342 | 0.181 | 1.73E-07 | 2.816 | |||
| 8 | Stearoylcarnitine | 0.141 | 4.00E-13 | 2.782 | 0.103 | 7.26E-14 | 3.428 | |||
| 9 | Taurocholic acid | 0.099 | 5.33E-06 | 3.151 | 0.186 | 7.83E-05 | 2.477 | |||
| 10 | TG (14:0) | 0.098 | 4.37E-08 | 2.979 | 0.081 | 8.98E-10 | 1.936 | |||
| 11 | Heme O | 0.089 | 3.83E-07 | 3.345 | 0.081 | 1.98E-07 | 3.623 | |||
| 12 | Tauroursodeoxycholic acid | 0.084 | 3.71E-06 | 3.393 | 0.133 | 2.61E-05 | 3.119 | |||
| 13 | Ganodermic acid TQ | 0.063 | 3.96E-07 | 3.500 | 0.147 | 2.16E-05 | 2.721 | |||
| 14 | L-Glutamine | 41.331 | 2.93E-10 | 2.094 | 50.738 | 3.27E-09 | 2.243 | |||
| 15 | L-Glutamic acid | 15.615 | 1.84E-07 | 1.764 | 16.328 | 3.30E-06 | 2.096 | |||
| 16 | Daucic acid | 8.296 | 2.66E-05 | 1.410 | 6.906 | 5.44E-05 | 1.351 | |||
| 17 | L-Malic acid | 5.635 | 2.69E-09 | 1.320 | 8.920 | 1.45E-08 | 1.798 | |||
| 18 | Pterosin N | 0.095 | 1.69E-06 | 1.474 | 0.073 | 5.82E-17 | 1.882 | |||
| 19 | Linoleic acid | 0.078 | 2.03E-06 | 1.744 | 0.060 | 2.18E-06 | 1.826 | |||
| 20 | Pyrocatechol sulfate | 0.031 | 9.83E-11 | 2.130 | 0.106 | 1.62E-04 | 1.167 | |||
| 21 | Methyl levulinate | 0.007 | 2.59E-17 | 2.578 | 0.019 | 3.04E-05 | 1.582 | |||
| 22 | LysoPC (22:6) | 8.040 | 1.68E-09 | 3.084 | ||||||
| 23 | Lactapiperanol D | 7.664 | 1.46E-08 | 3.178 | ||||||
| 24 | Octadecyl fumarate | 6.929 | 1.33E-07 | 2.797 | ||||||
| 25 | 27-Norcholestanehexol | 6.225 | 1.89E-07 | 3.014 | ||||||
| 26 | Lysyl-Threonine | 5.756 | 4.46E-07 | 2.940 | ||||||
| 27 | 5-Hydroxy-tryptophol | 5.682 | 1.08E-05 | 2.663 | ||||||
| 28 | LysoPC (22:4) | 5.665 | 4.70E-08 | 2.611 | ||||||
| 29 | beta-Elemonic acid | 5.555 | 1.21E-07 | 2.704 | ||||||
| 30 | L-Carnitine | 5.539 | 7.49E-08 | 2.715 | ||||||
| 31 | Malonyl-Carnitin | 5.461 | 1.78E-07 | 2.920 | ||||||
| 32 | Cortisol | 5.210 | 2.99E-05 | 2.455 | ||||||
| 33 | Polyporusterone B | 5.058 | 1.78E-05 | 2.896 | ||||||
| 34 | Acetaminophen glucuronide | 0.199 | 5.80E-03 | 2.147 | ||||||
| 35 | Eicosatetraenoic acid | 0.195 | 4.34E-06 | 2.348 | ||||||
| 36 | 12-Ketodeoxycholic acid | 0.191 | 1.81E-04 | 2.932 | ||||||
| 37 | Pterosin O | 0.149 | 4.72E-06 | 2.921 | ||||||
| 38 | Phenylalanyl-Tryptophan | 0.144 | 9.62E-08 | 2.968 | ||||||
| 39 | N-Heptanoylglycine | 0.137 | 1.79E-09 | 2.834 | ||||||
| 40 | Hexaethylene glycol | 0.125 | 4.52E-04 | 2.333 | ||||||
| 41 | Glycyrrhizin | 0.107 | 1.10E-06 | 3.200 | ||||||
| 42 | Campesteryl linoleate | 0.103 | 1.07E-04 | 3.643 | ||||||
| 43 | Psychosine sulfate | 0.091 | 1.75E-09 | 3.191 | ||||||
| 44 | D-Xylose | 0.084 | 3.42E-03 | 2.262 | ||||||
| 45 | Cellulose triacetate | 0.076 | 2.61E-06 | 3.463 | ||||||
| 46 | 6a-Hydroxy-paclitaxel | 0.070 | 6.90E-13 | 3.368 | ||||||
| 47 | TG (18:1) | 0.063 | 7.34E-10 | 3.311 | ||||||
| 48 | Paradol | 0.053 | 2.32E-15 | 3.789 | ||||||
| 49 | Torvonin A | 5.959 | 1.10E-10 | 1.534 | ||||||
| 50 | Allantoin | 0.168 | 2.66E-04 | 1.632 | ||||||
| 51 | Ethyl pyruvate | 0.154 | 8.04E-05 | 1.661 | ||||||
| 52 | 2-Hydroxybutyric acid | 0.148 | 5.76E-07 | 1.539 | ||||||
| 53 | Galactitol | 0.078 | 8.58E-04 | 1.423 | ||||||
| 54 | Cystathionine ketimine | 9.393 | 2.68E-04 | 1.256 | ||||||
| 55 | L-Phenylalanine | 5.220 | 1.86E-08 | 1.467 | ||||||
| 56 | PC (16:0) | 7.816 | 8.68E-03 | 2.112 | ||||||
| 57 | LysoPC (20:3) | 0.196 | 4.88E-05 | 2.219 | ||||||
| 58 | TG (15:0) | 0.014 | 4.52E-18 | 4.611 | 0.139 | 9.69E-05 | 2.035 | |||
| 59 | Heliantriol A1 | 0.183 | 1.00E-03 | 1.682 | ||||||
| 60 | Corbisterol | 0.183 | 4.19E-06 | 5.027 | ||||||
| 61 | Diacetoxypropyl stearate | 0.176 | 9.65E-04 | 4.455 | ||||||
| 62 | Famprofazone | 0.164 | 4.15E-07 | 5.073 | ||||||
| 63 | Leontogenin | 0.147 | 2.01E-06 | 5.420 | ||||||
The ratio is > 5.0 or < 0.2.
LysoPC, lysophosphatidylcholine; TG, triglycerides.
Metabolic Kyoto encyclopedia of genes and genomes pathway analysis
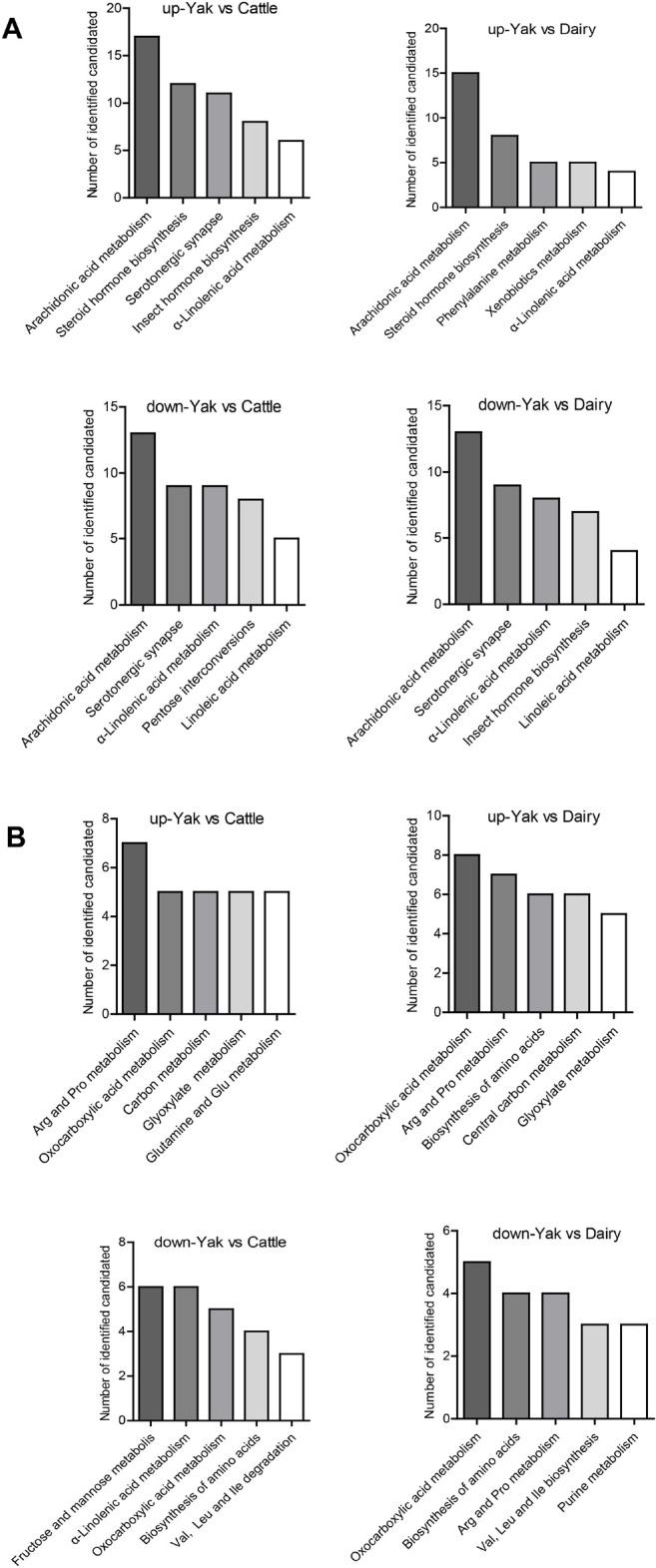
KEGG analysis was used to predict metabolic pathways for all differential metabolites. Fig. 3 shows the functional enrichment of the top 5 different pathways. The most enriched functional pathways among yak, yellow cattle and dairy cow belonged to metabolic pathways: amino acid metabolism (e.g., phenylalanine, arginine, proline, glycine, valine, leucine, isoleucine and glutamine), phospholipid metabolism (lysophosphatidylcholines [LysoPCs]), and fatty acid metabolism (arachidonic acid metabolism, α-linolenic acid and linolenic acid metabolism).
Fig. 3. The top 5 different pathways for differential metabolites (up and down) among yak, yellow cattle, and dairy cow in positive (A) and negative (B) ion modes.
Inflammatory cytokines and antioxidant levels
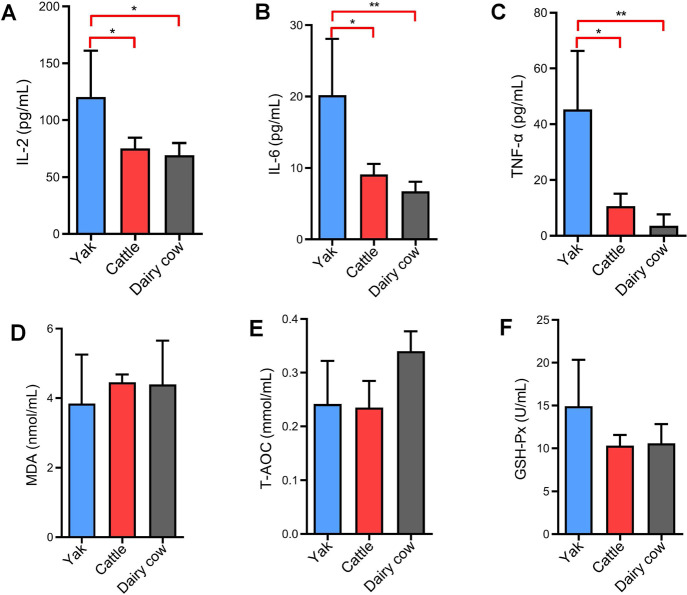
The levels of the inflammatory cytokines IL-2, IL-6, and TNF-α in yak serum were significantly higher than those in yellow cattle and dairy cows (p < 0.05 or p < 0.01) (Figs. 4A, B and C). Oxidative stress is an imbalance of reactive oxygen species (ROS) generation and elimination. High altitude-associated hypobaric hypoxia stress induces ROS production [19]. To determine if the elevated peripheral inflammatory cytokines were accompanied by reactive oxygen production or oxidative damcage [20], serum levels of MDA, T-AOC and GSH-Px were measured by chemical colorimetry. The results showed that there was no significant differences in MDA, T-AOC and GSH-Px levels among yaks, yellow cattle and dairy cows (p > 0.05) (Figs. 4D, E, and F).
Fig. 4. The inflammatory cytokines of IL-2 (A), IL-6 (B), and TNF-α (C) were determined using ELISA kits, and MDA level (D), T-AOC (E) and GSH-Px activity (F) related to antioxidant defense system in serum were measured by chemical colorimetry.
*p < 0.05, **p < 0.01. IL, interleukin; TNF, tumor necrosis factor; MDA, malondialdehyde; T-AOC, total antioxidant capacity; GSH-Px, glutathione peroxidase.
DISCUSSION
Yaks are an iconic symbol of QTP and can be used as a model to elucidate the mechanisms of hypoxia adaptation. The three specises of yak, yellow cattle and Holstein dairy cow belong to subtribe Bovina [21]. Holstein and yellow cattle should be probably separated from yak about 4.4 to 5.3 million years ago [10,22]. Systems biology is the trend of contemporary scientific development [23]. Comparative transcriptome sequencing revealed that the innate immunity were more activated in yak lung than low-altitude cattle (Sanjiang and Holstein cattle) [10,24]. Proteomics of skeletal muscle mitochondria showed that the significantly affected pathway in yaks and cattle was oxidative phosphorylation [7]. Identification of metabolic pathways using metabolomics comparisons between closely related species has the potential to provide insights into the basis of mammalian divergence and adaptation. To understand differences in the global metabolic profiles and relevant metabolic pathways of yaks, yellow cattle and dairy cows during acclimatization to high altitude, we utilized UPLC-Q-TOF-MS to determine the serum metabolite profiles of the three breeds.
We detected a clear separation trend between yaks and yellow cattle and dairy cows. A total of 63 different metabolites were obtained in serum. An integrative view plot of the metabolic changes among white yaks, yellow cattle and dairy cows was prepared (Fig. 3). The major perturbed metabolic patterns and plausible pathways are involved in amino acid metabolism, phospholipid metabolism, and fatty acid metabolism, which are associated with hypobaric hypoxia.
Amino acid metabolism
Glutamine is a key metabolite in the alanine, aspartate and glutamate metabolism pathways [25]. A previous study reported that high-altitude exposure leads to lower glutamate levels due to decreased activity of glutamine synthetase [26]. In contrast, we found significantly elevated levels of glutamic acid and glucogenic amino acids that produce pyruvic acid, α-ketoglutaric acid, and oxaloacetic acid in the serum of yaks in comparison with yellow cattle and dairy cows [27] (Table 2). The results of this study indicate that yaks have the highest levels of protein catabolism and amino acid mobilization. Therefore, the mobilization of yak muscle protein may be a metabolic adaptation to hypobaric hypoxia. Pathway analysis also showed improved energy metabolism and promoted acclimatization to high altitude by increasing the metabolism of phenylalanine, arginine, proline and glutamine to meet the energy requirements in yaks (Fig. 3). This result is consistent with the previous finding that hypobaric hypoxia exposure can enhance glucose and amino acid metabolism [28].
Phospholipid metabolism
Phospholipids play a role as a cellular bilayer with membrane proteins, and they are involved in the maintenance of hepatic lipid metabolism [29]. Our results show that almost all LysoPCs, including LysoPC (18:0), LysoPC (16:0), LysoPC (18:1), LysoPC (22:6) and LysoPC (22:4), were markedly increased in yaks compared with yellow cattle and dairy cows (Table 2). LysoPCs participate in the inflammatory response by mediating cell signaling pathways in monocytes and macrophages [30,31]. To verify the increased LysoPCs, the serum levels of cytokines IL-2, IL-6, and TNF-α were detected. The results showed that the levels of IL-2, IL-6, and TNF-α were significantly higher in yaks than in cattle and dairy cows (Figs. 4A, B, and C), which is consistent with the increased LysoPCs. Environmental factors such as hypobaric hypoxia, cold and UV exposure at high altitude can suppress the immune system [32]. Tumor necrosis factors and interleukins can mediate innate immunity signaling. Xin et al. [10] reported that the immune system was more activated and the genes related to immune were up-regulated in yak compared with Sanjiang and Holstein cattle [10]. A significant elevation of LysoPCs and cytokines (IL-2, IL-6, and TNF-α) might be responsible for yaks being more tolerant to hypoxia at high altitudes than yellow cattle and dairy cows by activating innate immunity system.
Fatty acids metabolism
Hypoxia is associated with an increase in the generation of reactive oxygen species (ROS), and an excessive load of ROS generated may result in cell injury and dysfunction [33]. Excessive ROS can lead to lipid peroxidation, MDA can reflect the level of lipid peroxidation. ROS are balanced by natural antioxidant compounds such as GSH-Px, superoxide dismutase (SOD) and catalase (CAT) [34]. We found that serum MDA levels, T-AOC and GSH-Px activity were not significantly changed in yak in comparison with yellow cattle and Holstein dairy cows (Figs. 4D, E, and F), based on they had similar physical characteristics (Table 1). The results demonstrate that yaks adapt to hypoxia-induced oxidative stress at high altitudes do not by increasing antioxidant enzyme levels. Free fatty acids (FFAs) are risk factors for cardiovascular diseases and are closely related to metabolic syndromes [35]. FFAs are significant sources of ROS [36], mainly through the activation of NADPH oxidase [37]. There was a dose-dependent increase in ROS in monocytes exposed to FFAs [38]. Polyunsaturated fatty acids (PUFAs) are a favorable target for ROS [39]. Oxidative breakdown of PUFAs may affect lipid metabolism and the expression of genes and proteins related to cell differentiation [40]. α-Linolenic acid and linoleic acid are PUFAs. Linoleic acid contains unsaturated double bonds that are highly vulnerable to ROS [41], and has been linked to red blood cell damage by promoting redox reactions [42]. The ROS production was greater in bovine mammary epithelia cells treated with linoleic acid and α-linolenic acid [43]. Arachidonic acid has been demonstrated to promote inflammatory responses by activating the mitogen-activated protein kinase (MAPK) and c-Jun N-terminal kinases (JNK) pathways by increasing TNF-α levels [44,45]. Arachidonic acid-derived metabolites also can propagate inflammation and oxidative stress [46]. Arachidonic acid suppressed the cell growth of hepatic cells by dose‐dependently inducing the production of ROS [47]. In the present work, FFAs (α-linolenic acid, linoleic acid and arachidonic acid) were significantly decreased in yaks compared with yellow cattle and Holstein dairy cows (Table 2). We speculate that yaks can decrease the level of FFAs (α-linolenic acid, linoleic acid and arachidonic acid) in serum induced by hypoxia and that the decreased FFAs can attenuate cell injury and hypoxia dysfunction by inhibiting oxidative stress.
CONCLUSION
A clear separation trend between the serum metabolic profiles of yaks and yellow cattle and dairy cows was demonstrated by PCA. In addition, a total of 63 differentially expressed metabolites were identified among the three species. Functional analysis revealed that differentially expressed metabolites were related to the innate immune activation (elevation of LysoPCs and cytokines), oxidative stress-related metabolism (arachidonic acid metabolism, α-linolenic acid metabolism, and linoleic acid metabolism) and energy metabolism (fatty acid metabolism and amino acid metabolism) in yaks, which indicates the important roles of metabolites in high-altitude adaptation in yaks.
Acknowledgements
Not applicable.
Competing interests
No potential conflict of interest relevant to this article was reported.
Funding sources
National Natural Science Foundation of China (No. 31802256) and Gansu Province Science Fund for Distinguished Young Scholars (No. 20JR5RA579).
Availability of data and material
Upon reasonable request, the datasets of this study can be available from the corresponding author.
Authors’ contributions
Conceptualization: Wang H.
Data curation: Huang M, Wang H.
Formal analysis: Huang M.
Methodology: Zhang X, Yan W, Liu J.
Software: Huang M.
Validation: Wang H.
Investigation: Yan W.
Writing - original draft: Wang H.
Writing - review & editing: Huang M, Zhang X, Yan W, Liu J, Wang H.
Ethics approval and consent to participate
The animal experiment was approved and received humane care according to Ethical Committee rules of Lanzhou University, China (RIB21110301).
REFERENCES
- 1.Tang Q, Gu Y, Zhou X, Jin L, Guan J, Liu R, et al. Comparative transcriptomics of 5 high-altitude vertebrates and their low-altitude relatives. Gigascienc. 2017;6:gix105. doi: 10.1093/gigascience/gix105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui GX, Yuan F, Degen AA, Liu SM, Zhou JW, Shang ZH, et al. Composition of the milk of yaks raised at different altitudes on the Qinghai–Tibetan Plateau. Int Dairy J. 2016;59:29–35. doi: 10.1016/j.idairyj.2016.02.046. [DOI] [Google Scholar]
- 3.Qiu Q, Wang L, Wang K, Yang Y, Ma T, Wang Z, et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat Commun. 2015;6:10283. doi: 10.1038/ncomms10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolt KS, Mishra MK, Karar J, Baig MA, Ahmed Z, Pasha MAQ. cDNA cloning, gene organization and variant specific expression of HIF-1α in high altitude yak (Bos grunniens) Gene. 2007;386:73–80. doi: 10.1016/j.gene.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Shao B, Long R, Ding Y, Wang J, Ding L, Wang H. Morphological adaptations of yak (Bos grunniens) tongue to the foraging environment of the Qinghai-Tibetan Plateau. J Anim Sci. 2010;88:2594–603. doi: 10.2527/jas.2009-2398. [DOI] [PubMed] [Google Scholar]
- 6.Xin JW, Chai ZX, Zhang CF, Zhang Q, Zhu Y, Cao HW, et al. Signature of high altitude adaptation in the gluteus proteome of the yak. J Exp Zool B Mol Dev Evol. 2020;334:362–72. doi: 10.1002/jez.b.22995. [DOI] [PubMed] [Google Scholar]
- 7.Long L, Zhu Y, Li Z, Zhang H, Liu L, Bai J. Differential expression of skeletal muscle mitochondrial proteins in yak, dzo, and cattle: a proteomics-based study. J Vet Med Sci. 2020;82:1178–86. doi: 10.1292/jvms.19-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44:946–9. doi: 10.1038/ng.2343. [DOI] [PubMed] [Google Scholar]
- 9.Guan J, Long K, Ma J, Zhang J, He D, Jin L, et al. Comparative analysis of the microRNA transcriptome between yak and cattle provides insight into high-altitude adaptation. PeerJ. 2017;5:e3959. doi: 10.7717/peerj.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xin JW, Chai ZX, Zhang CF, Zhang Q, Zhu Y, Cao HW, et al. Transcriptome profiles revealed the mechanisms underlying the adaptation of yak to high-altitude environments. Sci Rep. 2019;9:7558. doi: 10.1038/s41598-019-43773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julian CG, Wilson MJ, Moore LG. Evolutionary adaptation to high altitude: a view from in utero. Am J Hum Biol. 2009;21:614–22. doi: 10.1002/ajhb.20900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Brien KA, Griffin JL, Murray AJ, Edwards LM. Mitochondrial responses to extreme environments: insights from metabolomics. Extrem Physiol Med. 2015;4:7. doi: 10.1186/s13728-015-0026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Araldi E, Schipani E. Hypoxia, HIFs and bone development. Bone. 2010;47:190–6. doi: 10.1016/j.bone.2010.04.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer U, Heinemann M, Zamboni N. Getting closer to the whole picture. Science. 2007;316:550–1. doi: 10.1126/science.1142502. [DOI] [PubMed] [Google Scholar]
- 15.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–4. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40:387–426. doi: 10.1039/B906712B. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Liu Z, Wang S, Cui D, Zhang X, Liu Y, et al. UHPLC-Q-TOF/MS based plasma metabolomics reveals the metabolic perturbations by manganese exposure in rat models. Metallomics. 2017;9:192–203. doi: 10.1039/C7MT00007C. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Liu Z, Huang M, Wang S, Cui D, Dong S, et al. Effects of long-term mineral block supplementation on antioxidants, immunity, and health of Tibetan sheep. Biol Trace Elem Res. 2016;172:326–35. doi: 10.1007/s12011-015-0593-z. [DOI] [PubMed] [Google Scholar]
- 19.Bakonyi T, Radak Z. High altitude and free radicals. J Sports Sci Med. 2004;3:64–9. [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Cao G, Chen DQ, Wang M, Vaziri ND, Zhang ZH, et al. Metabolomics insights into activated redox signaling and lipid metabolism dysfunction in chronic kidney disease progression. Redox Biol. 2016;10:168–78. doi: 10.1016/j.redox.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Lenstra JA, Zhang S, Liu W, Liu J. Evolution and domestication of the Bovini species. Anim Genet. 2020;51:637–57. doi: 10.1111/age.12974. [DOI] [PubMed] [Google Scholar]
- 22.Gu Z, Zhao X, Li N, Wu C. Complete sequence of the yak (Bos grunniens) mitochondrial genome and its evolutionary relationship with other ruminants. Mol Phylogenet Evol. 2007;42:248–55. doi: 10.1016/j.ympev.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 23.Liao WT, Liu B, Chen J, Cui JH, Gao YX, Liu FY, et al. Metabolite modulation in human plasma in the early phase of acclimatization to hypobaric hypoxia. Sci Rep. 2016;6:22589. doi: 10.1038/srep22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan D, Xiong X, Ji W, Li J, Mipam TD, Ai Y, et al. Transcriptome profile and unique genetic evolution of positively selected genes in yak lungs. Genetica. 2018;146:151–60. doi: 10.1007/s10709-017-0005-8. [DOI] [PubMed] [Google Scholar]
- 25.Guo L, Tan G, Liu P, Li H, Tang L, Huang L, et al. Three plasma metabolite signatures for diagnosing high altitude pulmonary edema. Sci Rep. 2015;5:15126. doi: 10.1038/srep15126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radák Z, Asano K, Fu Y, Nakamura A, Nakamoto H, Ohno H, et al. The effect of high altitude and caloric restriction on reactive carbonyl derivatives and activity of glutamine synthetase in rat brain. Life Sci. 1998;62:1317–22. doi: 10.1016/S0024-3205(98)00065-4. [DOI] [PubMed] [Google Scholar]
- 27.Lapierre H, Lobley GE, Doepel L, Raggio G, Rulquin H, Lemosquet S. Triennial Lactation Symposium: mammary metabolism of amino acids in dairy cows. J Anim Sci. 2012;90:1708–21. doi: 10.2527/jas.2011-4645. [DOI] [PubMed] [Google Scholar]
- 28.Horscroft JA, Murray AJ. Skeletal muscle energy metabolism in environmental hypoxia: climbing towards consensus. Extrem Physiol Med. 2014;3:19. doi: 10.1186/2046-7648-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh HA, Lee H, Park S, Lim Y, Kwon O, Kim JY, et al. Analysis of plasma metabolic profiling and evaluation of the effect of the intake of Angelica keiskei using metabolomics and lipidomics. J Ethnopharmacol. 2019;243:112058. doi: 10.1016/j.jep.2019.112058. [DOI] [PubMed] [Google Scholar]
- 30.Duong CQ, Bared SM, Abu-Khader A, Buechler C, Schmitz A, Schmitz G. Expression of the lysophospholipid receptor family and investigation of lysophospholipid-mediated responses in human macrophages. Biochim Biophys Acta Mol Cell Biol Lipids. 2004;1682:112–9. doi: 10.1016/j.bbalip.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Kabarowski JH. G2A and LPC: regulatory functions in immunity. Prostaglandins Other Lipid Mediat. 2009;89:73–81. doi: 10.1016/j.prostaglandins.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra KP, Ganju L. Influence of high altitude exposure on the immune system: a review. Immunol Invest. 2010;39:219–34. doi: 10.3109/08820131003681144. [DOI] [PubMed] [Google Scholar]
- 33.Clanton TL. Hypoxia-induced reactive oxygen species formation in skeletal muscle. J Appl Physiol. 2007;102:2379–88. doi: 10.1152/japplphysiol.01298.2006. [DOI] [PubMed] [Google Scholar]
- 34.Wu JQ, Kosten TR, Zhang XY. Free radicals, antioxidant defense systems, and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2013;46:200–6. doi: 10.1016/j.pnpbp.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Boden G. Obesity and free fatty acids. Endocrinol Metab Clin North Am. 2008;37:635–46. doi: 10.1016/j.ecl.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh A, Gao L, Thakur A, Siu PM, Lai CWK. Role of free fatty acids in endothelial dysfunction. J Biomed Sci. 2017;24:50. doi: 10.1186/s12929-017-0357-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cury-Boaventura MF, Curi R. Regulation of reactive oxygen species (ROS) production by C18 fatty acids in Jurkat and Raji cells. Clin Sci. 2005;108:245–53. doi: 10.1042/CS20040281. [DOI] [PubMed] [Google Scholar]
- 38.Zhang WY, Schwartz E, Wang Y, Attrep J, Li Z, Reaven P. Elevated concentrations of nonesterified fatty acids increase monocyte expression of CD11b and adhesion to endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:514–9. doi: 10.1161/01.ATV.0000200226.53994.09. [DOI] [PubMed] [Google Scholar]
- 39.Behn C, Araneda OF, Llanos AJ, Celedón G, González G. Hypoxia-related lipid peroxidation: evidences, implications and approaches. Respir Physiol Neurobiol. 2007;158:143–50. doi: 10.1016/j.resp.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Bork CS, Baker EJ, Lundbye-Christensen S, Miles EA, Calder PC. Lowering the linoleic acid to alpha-linoleic acid ratio decreases the production of inflammatory mediators by cultured human endothelial cells. Prostaglandins Leukot Essent Fatty Acids. 2019;141:1–8. doi: 10.1016/j.plefa.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Fedor DM, Adkins Y, Mackey BE, Kelley DS. Docosahexaenoic acid prevents trans-10, cis-12–conjugated linoleic acid-induced nonalcoholic fatty liver disease in mice by altering expression of hepatic genes regulating fatty acid synthesis and oxidation. Metab Syndr Relat Disord. 2012;10:175–80. doi: 10.1089/met.2011.0113. [DOI] [PubMed] [Google Scholar]
- 42.Yuan T, Fan WB, Cong Y, Xu HD, Li CJ, Meng J, et al. Linoleic acid induces red blood cells and hemoglobin damage via oxidative mechanism. Int J Clin Exp Pathol. 2015;8:5044–52. [PMC free article] [PubMed] [Google Scholar]
- 43.Basiricò L, Morera P, Dipasquale D, Tröscher A, Bernabucci U. Comparison between conjugated linoleic acid and essential fatty acids in preventing oxidative stress in bovine mammary epithelial cells. J Dairy Sci. 2017;100:2299–309. doi: 10.3168/jds.2016-11729. [DOI] [PubMed] [Google Scholar]
- 44.Balsinde J, Winstead MV, Dennis EA. Phospholipase A2 regulation of arachidonic acid mobilization. FEBS Lett. 2002;531:2–6. doi: 10.1016/S0014-5793(02)03413-0. [DOI] [PubMed] [Google Scholar]
- 45.Saito Y, Watanabe K, Fujioka D, Nakamura T, Obata J, Kawabata K, et al. Disruption of group IVA cytosolic phospholipase A2 attenuates myocardial ischemia-reperfusion injury partly through inhibition of TNF-α-mediated pathway. Am J Physiol Heart Circ Physiol. 2012;302:H2018–30. doi: 10.1152/ajpheart.00955.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rink C, Khanna S. Significance of brain tissue oxygenation and the arachidonic acid cascade in stroke. Antioxid Redox Signal. 2011;14:1889–903. doi: 10.1089/ars.2010.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin XY, Lu J, Cai M, Kojima S. Arachidonic acid suppresses hepatic cell growth through ROS-mediated activation of transglutaminase. FEBS Open Bio. 2018;8:1703–10. doi: 10.1002/2211-5463.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]