Abstract
At present, there is no effective noninvasive method for the accurate diagnosis of early‐stage lung adenocarcinoma (LUAD). This study examined the profile of plasma extracellular vesicle (EV)‐delivered microRNAs (miRNAs) in patients with invasive stage I LUAD. In this study, a total of 460 participants were enrolled, including 254 patients with LUAD, 76 patients with benign pulmonary nodules (BPNs), and 130 healthy control patients (HCs). miRNA sequencing was used to analyze the EV miRNA profile of the patient plasma samples (n = 150). A diagnostic signature (d‐signature) was identified by applying a stepwise logistic regression algorithm, and a single‐center training cohort (n = 150) was tested, followed by a multicenter validation cohort (n = 100). A d‐signature comprising four EV‐derived miRNAs (hsa‐miR‐106b‐3p, hsa‐miR‐125a‐5p, hsa‐miR‐3615, and hsa‐miR‐450b‐5p) was developed for the early detection of LUAD. The d‐signature had high precision with area under the curve (AUC) values of 0.917 and 0.902 in the training and test cohorts, respectively. Moreover, the d‐signature could recognize patients with adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA) with AUC values of 0.846 and 0.92, respectively. To sum up, our study detailed the plasma EV–derived miRNA profile in early LUAD patients and developed an EV‐derived miRNA d‐signature to detect early LUAD.
Keywords: biomarker, diagnostic, extracellular vesicles, lung adenocarcinoma, microRNA
This study provides the largest genome‐wide analysis of extracellular vesicle (EV) microRNA (miRNA) in plasma from early lung adenocarcinoma (LUAD) patients, suggesting the feasibility of identifying cancer biomarkers based on EV miRNA profiling. We developed an EV miRNA–based diagnostic signature (d‐signature) that showed high accuracy for the diagnosis of early LUAD based on multicentric validation. The d‐signature was able to detect adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA) as readily as invasive stage I invasive adenocarcinoma (IAC).
1. INTRODUCTION
Lung cancer, including non–small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), is one of the most common solid tumors worldwide and the main cause of cancer‐associated deaths. 1 , 2 Lung adenocarcinoma (LUAD) is the most common histological subtype of NSCLC and accounts for about 40% of lung malignancies 3 with a 5‐year survival rate of less than 20%. 4 However, if the disease is diagnosed at stage IA, the survival rates increase from 6% to nearly 82%. 5 Moreover, patients with adenocarcinoma in situ (AIS) are categorized as TisN0M0 at stage 0, while minimally invasive adenocarcinoma (MIA) patients are categorized as T1a (mi) and stage IA LUAD. 6 The 5‐year survival rate of these two early types of LUAD has been evaluated as 100%. 6 Low‐dose computed tomography (LDCT) screening is recommended for the early investigation of lung cancer and can decrease the mortality of lung cancer patients by 20%. 7
Nevertheless, it can be difficult to distinguish lung cancer from benign pulmonary nodules (BPNs). 8 The application of LDCT is also limited by high costs and the need for repeated scanning. 8 Moreover, the diagnostic performance of commonly used lung cancer biomarkers, including carcinoembryonic antigen (CEA), squamous cell carcinoma (SCC) antigen, and cytokeratin 19 fragment antigen 21‐1 (CYFRA21‐1), is also limited due to unsatisfactory sensitivity and specificity for early LUAD. 9 Thus, it is imperative to identify novel effective early diagnostic markers for lung cancer patients.
MicroRNAs (miRNAs) are a class of small noncoding RNAs (approximately 18‐22 nucleotides in length) that participate in a wide range of cancer biology processes that promote tumor growth, apoptosis, progression, metastasis, and immune evasion. 10 , 11 , 12 Current evidence suggests that miRNAs can be effective diagnostic markers and therapeutic targets for common solid tumors, including lung cancer. 13 , 14 However, the biological functions of these circulating miRNAs remain unclear.
Extracellular vesicles (EVs), such as exosomes and microvesicles, are lipid membrane vesicles that contain various cargoes, including proteins, lipids, and RNAs. 15 , 16 EVs can be used to mobilize miRNAs, prevent RNA enzyme degradation, and influence cell‐cell communication via the transportation of their contents to target cells in the lung cancer microenvironment. Thus, EVs may be ideal circulating biomarkers. 17 , 18 Numerous studies have suggested that circulating EV miRNAs may play a role in early lung cancer. 18 , 19 , 20 , 21 Jin et al reported significant changes in plasma levels of EV miR‐181‐5p, miR‐30a‐3p, miR‐30e‐3p, and miR‐361‐5p in LUAD patients and further revealed that EV miRNAs could distinguish stage I LUAD patients from healthy individuals. 22 Unfortunately, these studies were limited by selection bias inherent to retrospective studies, analysis of single‐center cohorts, a small number of enrolled cases, and less rigorous identification methods. 23 Additionally, only one study reported the potential value of circulating EV miRNAs in stage I LUAD. Furthermore, there have been no reports directly comparing the differences in circulating EV miRNAs in AIS, MIA, and noncancerous (NC) healthy individuals to the best of our knowledge.
This multicenter, prospective study examined plasma samples from 460 subjects, including early LUAD patients, BPN patients, and healthy participants. EV miRNA–sequencing and quantitative reverse‐transcription polymerase chain reaction (RT‐qPCR) was performed to establish and validate an EV miRNA–based diagnostic signature (d‐signature) with high sensitivity and specificity to detect early LUAD.
2. MATERIALS AND METHODS
2.1. Patient cohort
This research enrolled 460 participants, including 254 patients diagnosed with LUAD, 76 patients with BPNs, and 130 healthy controls (HCs). Among them, 323 participants recruited from the National Cancer Center (NCC) were divided into a discovery cohort (n = 150), a screening cohort (n = 60), and a training cohort (n = 150). One hundred participants from the Peking Union Medical College Hospital and China‐Japan Friendship Hospital were recruited as the test cohort. The patient clinical characteristics, including age, gender, tumor size, and tumor stage, are displayed in Table 1.
TABLE 1.
Patients' characteristics for the training and validation cohorts
| Characteristic |
Discovery cohort (n = 150) |
Screen cohort (n = 60) |
Training cohort (n = 150) |
Validation cohort (n = 100) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Percent | Number | Percent | Number | Percent | Number | Percent | ||
| Age | Mean ± SD | 54.08 ± 9.40 | – | 49.12 ± 10.30 | – | 54.07 ± 10.76 | – | 53.79 ± 12.23 | – |
| Gender | Female | 84 | 56% | 38 | 63.33% | 55 | 36.67% | 39 | 39% |
| Male | 66 | 44% | 22 | 36.67% | 95 | 63.33% | 61 | 61% | |
| Smoking history | Yes | 114 | 76% | 42 | 70% | 114 | 76% | 79 | 79% |
| No | 36 | 24% | 18 | 30% | 36 | 24% | 21 | 21% | |
| Family history | Yes | 33 | 22% | 17 | 28.33% | 31 | 20.67% | 26 | 26% |
| No | 117 | 78% | 43 | 71.67% | 119 | 79.33% | 74 | 74% | |
| Nodule size | Mean ± SD | 1.13 ± 0.49 | – | 1.20 ± 0.52 | – | 1.29 ± 0.57 | – | 1.41 ± 0.58 | – |
| Location | RUL | 48 | 40% | 15 | 37.5% | 18 | 17.65% | 11 | 16.18% |
| RML | 11 | 9.17% | 4 | 10% | 20 | 19.61% | 15 | 22.06% | |
| RLL | 27 | 22.5% | 8 | 20% | 21 | 20.59% | 18 | 26.47% | |
| LUL | 24 | 20% | 8 | 20% | 12 | 11.76% | 8 | 11.76% | |
| LLL | 10 | 8.33% | 5 | 12.5% | 31 | 30.39% | 16 | 23.53% | |
| Type | LUAD | 91 | 60.67% | 30 | 50% | 80 | 53.33% | 53 | 53% |
| NC | 59 | 39.33% | 30 | 50% | 70 | 46.67 | 47 | 47% | |
| Pathology | HC | 30 | 20% | 20 | 33.33% | 48 | 32% | 32 | 32% |
| BPN | 29 | 19.33% | 10 | 16.67% | 22 | 14.67% | 15 | 15% | |
| AIS | 31 | 20.67% | 10 | 16.67% | 14 | 9.33% | 9 | 9% | |
| MIA | 29 | 19.33% | 10 | 16.67% | 13 | 8.67% | 9 | 9% | |
| IAC | 31 | 20.67% | 10 | 16.67% | 53 | 35.33% | 35 | 35% | |
Abbreviations: AIS, adenocarcinoma in situ; BPN, benign pulmonary nodule; HC, healthy control; IAC, invasive carcinoma; LLL, left lower lobe; LUAD, lung adenocarcinoma; LUL, left upper lobe; MIA, minimally invasive adenocarcinoma; NC, noncancerous control; RLL, right lower lobe; RML, right middle lobe; RUL, right upper lobe; SD, standard deviation.
2.2. Plasma sample collection
Peripheral blood samples were collected from individuals in EDTA tubes and centrifuged at 2000 g for 10 minutes at 4°C. Following aspiration, the plasma was stored at −80°C until use.
2.3. Isolation of EVs
The isolation of EVs via ultracentrifugation (UC) was performed as previously described. 24 , 25 After thawing at 37°C, plasma samples were centrifuged at 3000 g for 15 minutes to remove cell debris. The resultant supernatant was diluted with an eightfold volume of phosphate‐buffered saline (PBS) and centrifuged at 13 000 g to remove large particles. The supernatant was then ultracentrifuged in a P50A72‐986 rotor (CP100NX; Hitachi) at 150 000 g for 4 hours at 4°C to pellet the exosomes. The pellet was resuspended in PBS and centrifuged again at 150 000 g for 2 hours at 4°C. After washing with PBS, the exosome pellet was resuspended in 200 µL PBS.
2.4. Nanoparticle tracking analysis (NTA)
The vesicle suspensions were diluted to concentrations ranging from 1 × 107 to 1 × 109 particles/mL. Nanoparticle tracking was performed using the ZetaView PMX 110 (Particle Metrix) configured with a 405‐nm laser to determine the size and number of particles separated. A video of 60 seconds duration was shot at a frame rate of 30 frames/s, and NTA software was used to analyze particle movement (ZetaView 8.02.28).
2.5. Transmission electron microscopy (TEM)
A total of 10 µL exosome solution was placed on a copper mesh and incubated at room temperature for 1 minute. The exosome samples were then washed with sterile distilled water, stained with uranyl acetate solution for 1 minute and then dried under incandescent light for 2 minutes. Samples were viewed under a transmission electron microscope (H‐7650; Hitachi Ltd.), and photos were captured.
2.6. Western blot analysis
The exosome samples were denatured in 5× sodium dodecyl sulfonate (SDS) buffer before Western blot analysis as previously described. Briefly, 50 µg protein was loaded onto a 10% SDS‐polyacrylamide gel for electrophoresis. The rabbit polyclonal antibodies applied were CD63 (sc‐5275), CD9 (60232‐I‐Ig; Proteintech), HSP90 (60318‐I‐Ig; Proteintech), Alix (sc‐53540), TSG101 (sc‐13611), and calnexin (10427‐2‐AP; Promega). Protein bands were visualized using the Tanon 4600 automatic chemiluminescence image analysis system (Tanon).
2.7. ExoRNA isolation and RNA analyses
According to the manufacturer's instructions, the extraction and purifying of total RNA from plasma exosomes was performed using the miRNeasy® mini kit (Qiagen, cat. No. 217 004). Samples were run on 1.5% agarose gels to assess RNA degradation and contamination and DNA contamination. RNA concentration and purity were assessed using the RNA Nano 6000 assay kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies).
2.8. Library preparation and sequencing
According to the manufacturer's recommendations, the sequencing libraries were generated by applying 5 ng of exosome samples to the QIAseq miRNA library kit (Qiagen). Index codes were put to property sequences for every sample. The quality of the library was assessed using the Agilent Bioanalyzer 2100 and qPCR. The clustering of the index‐coded samples was assessed on the acBot cluster generation system applying TruSeq PE Cluster Kitv3‐cBot‐HS (Illumina) based on the manufacturer's instructions. After generating clusters, the sequencing of library preparations was performed on an Illumina NovaSeq platform, and paired‐end reads were produced.
2.9. Quantification and analysis of the differentially expressed miRNA (DEmiRs)
Bowtie tools, Clean Reads, Silva database, GtRNAdb database, Rfam database, and Repbase database sequence alignments were used to filter ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and other noncoding RNAs (ncRNAs) and repeats. Known miRNAs and new miRNAs were detected using the remaining reads, and known miRNAs were compared with miRbase and the human genome (GRCh38). Read counts for every miRNA were obtained from the mapping outcomes, and the transcripts per million (TPM) were calculated. The determination of the DEmiRs between NC and LUAD samples was performed using the Mann‐Whitney U‐test with a cutoff P‐value <.05 and average TPM >10. The DEmiRs were further screened with a P‐value <.01 or mean TPM >100. Finally, a total of 30 DEmiRs were identified for further analysis.
2.10. Sequence similarity filtering
The miRNAs with highly similar sequences showed the same expression trend. The Clustal omega tool was used to analyze the sequence similarity for the 30 DEmiRs filtered by P‐value and fold change. For the DEmiRs clusters whose similarity is less than 0.1, one miRNA was retained for qPCR verification. After sequence similarity filtering, a total of 22 candidate miRNAs were identified.
2.11. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
Gene Ontology enrichment analysis of the target genes of the DEmiRs was carried out using the topGO R packages. The KEGG pathway enrichment analysis (http://www.genome.jp/kegg/) was applied to determine the enriched biological systems associated with the genes of the DEmiRs. The statistical significance of the enrichment pathways was determined using the KOBAS software in KEGG pathways. 26
2.12. Selection of the normalization candidates
Cel‐39‐3p was used as an exogenous reference control in the qPCR analysis as previously reported. 27 Analysis of variance (ANOVA) in the RNA‐seq data showed that miR‐142‐5p had the smallest coefficient of variation, and thus, this was also used as an internal reference gene.
2.13. RNA isolation and RT‐qPCR
The Caenorhabditis elegans cell‐39‐3p miRNA was spiked into each EV sample as an external calibration before RNA extraction from exosomes using the miRNeasy® mini kit (Qiagen, cat. No. 217 004) according to the manufacturer's protocol. PrimeScript™ RT reagent kit (Perfect Real Time) (TAKARA, RR037A) was applied for the reverse transcription of total RNA to synthesize cDNA. For real‐time qPCR, 2 µL of cDNA was used to detect the abundance of the target gene. Table S1 shows the sequence of the primers and probes used in this study.
2.14. Quantitative PCR screening data set
A screening data set of 60 samples, including 30 NC samples and 30 LUAD samples, was used for the preliminary screening of miRNA biomarkers. qPCR verification was performed for 22 candidate miRNAs. Four targets were undetectable and were eliminated. Thus, a total of 18 miRNAs were obtained from the qPCR results of the screening data set. Among the 18 miRNAs, eight miRNAs with the highest single‐marker area under the curve (AUC) were selected for subsequent analysis.
2.15. Model training and validation
To verify the performance of 10 candidate miRNAs, 150 samples were included in the training set to train the logistic model. Another 100 samples were used as the test set to verify the model. In the training set, there were 70 NC samples and 80 LUAD samples. In the test set, there were 47 NC and 53 LUAD samples. With the 150 samples of the training set, all 10 miRNAs were incorporated into the model. In the model training process, stepwise regression was performed to eliminate the markers using the Akaike information criterion (AIC) as the standard to evaluate the goodness of fit. The lower the AIC, the better the goodness of fit. When any miRNA in the current model is eliminated, and the AIC of the model no longer declines, that miRNA will be the final biomarker. Otherwise, the miRNA that reduces the model's AIC the most will be removed. After data training with the 150 samples, four miRNAs (hsa‐miR‐106b‐3p, hsa‐miR‐125a‐5p, hsa‐miR‐3615, and hsa‐miR‐450b‐5p) were included in the final logistic model with Z = (1.021343 + 1.648991 × hsa‐miR‐106b‐3p + 0.795234 × hsa‐miR‐125a‐5p + −0.731803 × hsa‐miR‐3615 + −0.426805 × hsa‐miR‐450b‐5p). The model probability =EXP(Z)/(1+EXP(Z)). The cutoff of the model probability was set to 0.576112, and samples with a predicted value greater than the cutoff were classified as the cancer group.
2.16. Statistical analysis
The R 3.2.3 software was used for statistical analysis. The Wilcoxon rank‐sum test and the Mann‐Whitney U‐test were used to compare dependent and independent samples, respectively. The glm and step functions were used for stepwise logistic regression analysis. In the ANOVA, the Shapiro test was used to assess whether the data followed a normal distribution. Bartlett's test was used to assess the homogeneity of the data. When the above tests were satisfied, the aov function for ANOVA or the Kruskal test for statistical testing was applied. Receiver‐operating characteristic (ROC) curves were applied to test candidate miRNAs' diagnostic accuracy or combinations, and the AUC was calculated.
3. RESULTS
3.1. Characterization of plasma EVs
Characterization of plasma EVs using TEM (Figure 1A‐B) and NTA (Figure 1C) revealed that the isolated EVs were cup‐shaped, double‐membrane–bound vesicle‐like structures with a size between 75 nm to 200 nm. Western blot analysis showed that the EV markers CD9, CD63, and Tsg101 were detected in the separated EVs (Figure 1D). Calnexin is an intracellularly enriched protein on the endoplasmic reticulum and was used as a negative control protein marker for EV recognition. As expected, calnexin was not detected (Figure 1D), suggesting that the isolated EVs mainly consisted of exosomes.
FIGURE 1.
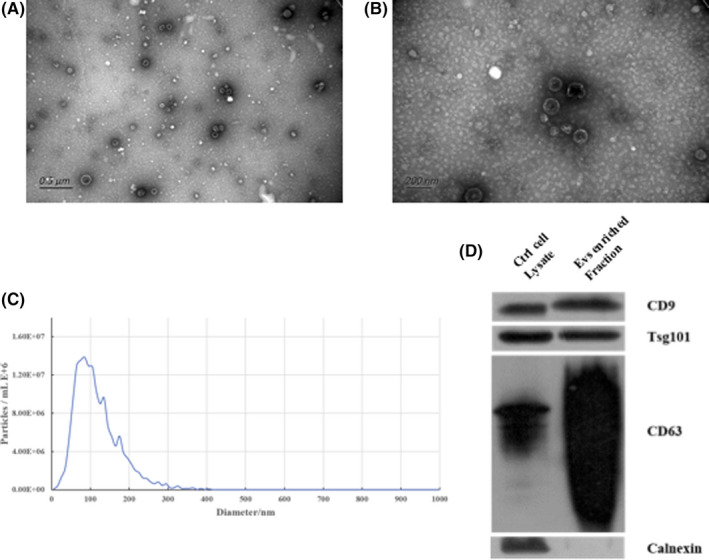
Characterization of plasma extracellular vesicles. A, Electron microscopy wide‐field image of extracellular vesicles (EVs) (bar = 0.5 μm). B, Electron microscopy close‐up image of EVs (bar = 200 nm). C, Size distribution measurements of isolated EVs. D, Western blot analysis of unenriched and exosomes‐enriched proteins in isolated EVs
3.2. Plasma EV miRNA profiling in the discovery cohort
A total of 91 early LUAD patients and 59 NC controls patients were recruited in the discovery cohort. The early LUAD group consisted of 31 AIS patients, 29 MIA patients, and 31 stage I invasive adenocarcinoma (IAC) patients. The NC group consisted of 29 patients with BPN and 30 HCs. Table 1 summarizes the demographic and clinical characteristics of the patients, including age, gender, and tumor size.
An improved strategy for EV miRNA‐seq analysis using human plasma (Figure S1) was adopted. The EV miRNA‐seq obtained a median read count of 10.55 million mapped reads per sample. Although there were wide mapped reads, about 1000 miRNAs (Figure 2A) were consistently detected. The numbers of detected miRNAs did not significantly differ among early LUAD samples, BPN samples, and HC samples (Figure 2B). Principal component analysis (PCA) showed significant differences in the EV miRNA profiles among LUAD patients, healthy individuals, and patients with BPN (Figure 2C). A total of 44 EV miRNAs were shown to be differentially expressed in LUAD samples compared with NC samples (BPN and HC) (P <.05; TPM >10). A clear separation of LUAD and NC samples (Figure 2D) was observed with unsupervised hierarchical clustering. KEGG pathway analysis revealed that the differentially expressed EV miRNAs (DEmiRs) were enriched for some cancer‐related pathways, including the PI3K‐Akt signaling pathway, platelet activation, and extracellular matrix (ECM)‐receptor interaction (Figure S2). In addition, GO analysis showed that these 44 DEmiRs were mainly enriched in ECM disassembly, axon guidance, cytoplasm, and ATP binding (Figure S3A‐C). These results suggested that EV miRNAs may be potential biomarkers for LUAD. From the 44 DEmiRs, 30 were selected for further analysis (Mann‐Whitney U‐test, P <.01, mean TPM >100; Figure S4).
FIGURE 2.
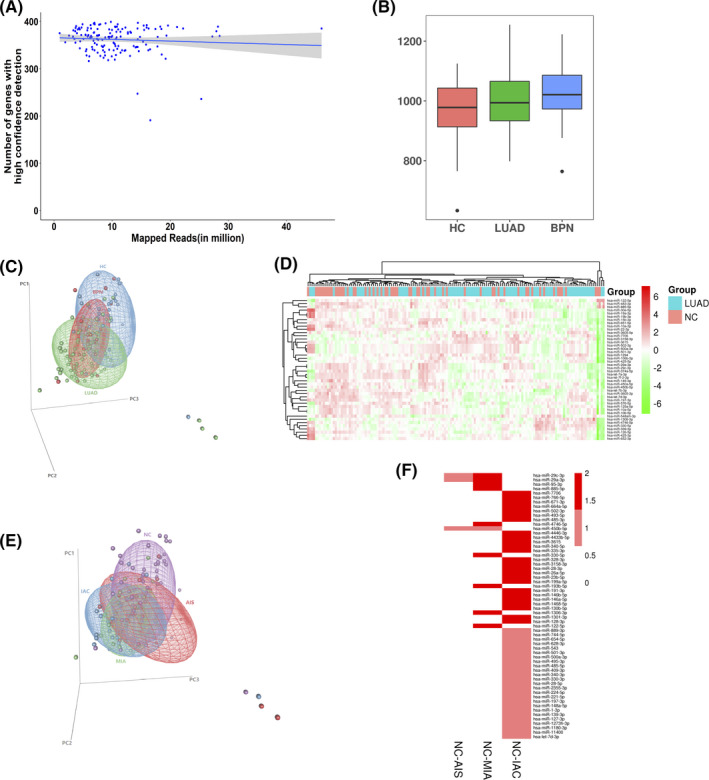
Plasma extracellular vesicle microRNA (miRNA) sequencing results. A, Distribution of the total mapped reads to the annotated genes with high confidence detection. B, Distribution of extracellular vesicle (EV) miRNAs per sample among lung adenocarcinoma (LUAD) patients, benign pulmonary nodule (BPN) patients, and healthy controls (HCs). C, Principal component analysis (PCA) for the differential extracellular vesicle (EV) miRNA profiles of LUAD patients compared with BPN patients and HCs. D, Heatmap of unsupervised hierarchical clustering of the differentially expressed EV miRNAs (DEmiRs) between LUAD patients and noncancerous controls (NC). E, PCA for the differential EV miRNA profiles of patients from the NC group. F, ANOVA analysis identified 60 DEmiRs in adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), and invasive adenocarcinoma (IAC) groups compared with the NC group
To further investigate the differences between the various stages of early LUAD, the LUAD cases were separated into the AIS group, the MIA group, and the IAC group. PCA revealed that the EV miRNA profiles of IAC patients did not differ from those of AIS patients and MIA patients (Figure 2E). Furthermore, ANOVA analysis identified 60 DEmiRs in these LUAD subgroups compared with the NC group (P <.05; Figure 2F). KEGG pathway analysis demonstrated that these DEmiRs were enriched in the PI3K‐Akt signaling pathway, cancer‐related pathways, Notch signaling pathway, miRNAs in cancer, Hippo signaling pathway, and basal cell carcinomas (Figure S5). These results showed that the IAC group is more dissimilar to the NC group than either AIS or MIA.
3.3. Candidate EV miRNA selection through the screening cohort
qPCR was used to evaluate the 30 DEmiRs using a screening cohort comprising 30 LUAD samples and 30 NC controls (20 healthy volunteer samples and 10 BPN samples). Four undetectable targets were eliminated. Another eight targets were eliminated due to sequence similarity (Figure S6). The expression levels of the remaining 18 EV miRNAs are shown in Figure 3. There were 14 miRNAs with upregulated expression in LUAD patients compared with NC controls (P <.05). ROC curve analysis examining sensitivity and specificity (Table S2) identified eight miRNAs (hsa‐mir‐106b‐3p, hsa‐mir‐10a‐5p, hsa‐mir‐125a‐5p, hsa‐mir‐30e‐5p, hsa‐miR‐3615, hsa‐miR‐450b‐5p, hsa‐miR‐4746‐5p, and hsa‐miR‐502‐3p; Figure 4) for further analysis. In addition to these eight candidate miRNAs, two other EV miRNAs (hsa‐miR‐181a‐5p and hsa‐miR‐361‐5p), which have been previously reported as LUAD biomarkers, were also selected for further analysis.
FIGURE 3.
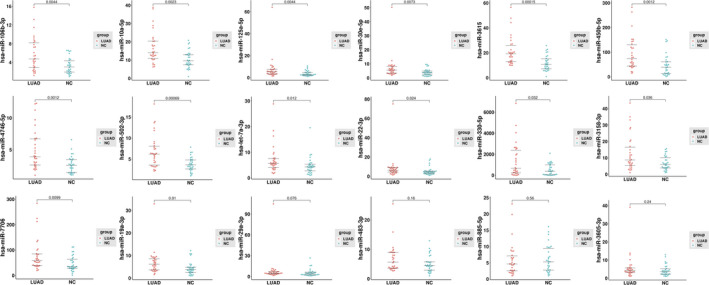
Quantitative reverse‐transcription polymerase chain reaction (RT‐qPCR) validation of the 18 candidate extracellular vesicle microRNAs (miRNAs) in the screening cohort (n = 60). The data were normalized to cel‐miR‐39 as an internal control
FIGURE 4.
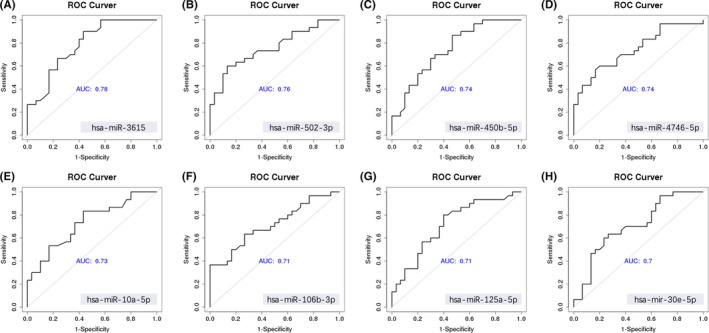
Receiver‐operating characteristic (ROC) curve analysis of the screening cohort's top eight candidate extracellular vesicle microRNAs (miRNAs) (n = 60)
3.4. Setting up an EV miRNA d‐signature for LUAD
Figure S7 shows the workflow design for identifying an EV miRNA d‐signature for the detection of early LUAD. After analysis of the discovery cohort and the screening cohort, 10 EV miRNAs were selected. A training cohort consisting of 70 NC participants (BPN and healthy individuals) and 80 LUAD patients were recruited. For qPCR analysis, two reference genes, cel‐39 and miR‐142‐5p, were used for normalizing EV miRNA expression levels. Stepwise logistic regression was used to reduce the number of variables. Finally, four EV miRNA markers, namely hsa‐miR‐106b‐3p, hsa‐miR‐125a‐5p, hsa‐miR‐3615, and hsa‐miR‐450b‐5p, were selected and used to set up a LUAD classifier. A diagnostic model was established using the logistic regression algorithm, and four miRNA d‐signatures were generated for LUAD. The cel‐39 gene was used as a control. The d‐signature distinguished LUAD patients from NC patients with an AUC of 0.917 (95% confidence interval [CI]: 0.874 to 0.96), a sensitivity of 83.8% (95% CI: 75% to 91.2%), and a specificity of 87.1% (95% CI: 78.6% to 94.3%) in the training cohort. The diagnostic precision was 0.853 (Figure 5A and B; Table 2). To determine if the d‐signature had comparable diagnostic value in different populations, 100 samples from three different centers were recruited for the validation cohort. The d‐signature distinguished LUAD patients from the NC group with an AUC of 0.902 (95% CI: 0.846‐0.959), a sensitivity of 84.9% (95% CI: 75.5%‐94.3%), a specificity of 80.9% (95% CI: 70.2%‐91.5%), and a diagnostic precision of 0.83 (Figure 5A and B; Table 2). In addition, the performance of the d‐signature was maintained even when the reference gene was changed to miR‐142‐5p (Figure S8, Table S3).
FIGURE 5.
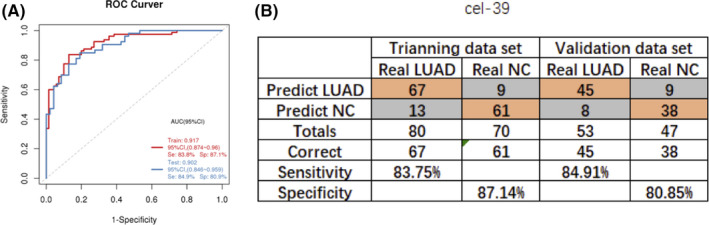
Establishment and validation of the extracellular vesicle (EV) microRNAs (miRNA) diagnostic signature (d‐signature) for lung adenocarcinoma (LUAD) (cel‐miR‐39 as reference gene). A, Receiver‐operating characteristic (ROC) curve analysis for the performance of the EV miRNA d‐signature in the training cohort and the validation cohort. B, Confusion table for the performance of the EV miRNA d‐signature in the training cohort and validation cohort
TABLE 2.
Performance of d‐signature in the diagnosis of early LUAD (Cel‐39 as reference gene)
| Training cohort (n = 150) | Validation cohort (n = 100) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | AUC | Sensitivity | Specificity | Accuracy | N | AUC | Sensitivity | Specificity | Accuracy | |
| LUAD vs NC | ||||||||||
| LUAD | 80/70 | 0.917 (0.874‐0.96) | 0.838 (0.75‐0.912) | 0.871 (0.786‐0.943) | 0.853 | 53/47 | 0.902 (0.846‐0.959) | 0.849 (0.755‐0.943) | 0.809 (0.702‐0.915) | 0.830 |
| AIS | 14/70 | 0.895 (0.813‐0.977) | 0.714 (0.429‐0.929) | 0.871 (0.786‐0.943) | 0.845 | 9/47 | 0.792 (0.639‐0.945) | 0.556 (0.222‐0.889) | 0.809 (0.702‐0.915) | 0.768 |
| MIA | 13/70 | 0.915 (0.84‐0.991) | 0.769 (0.538‐1) | 0.871 (0.786‐0.943) | 0.855 | 9/47 | 0.924 (0.826‐1) | 0.889 (0.667‐1) | 0.809 (0.702‐0.915) | 0.821 |
| IAC | 53/70 | 0.923 (0.875‐0.972) | 0.887 (0.792‐0.962) | 0.871 (0.786‐0.943) | 0.878 | 35/47 | 0.925 (0.872‐0.979) | 0.914 (0.8‐1) | 0.809 (0.702‐0.915) | 0.854 |
| LUAD vs HC | ||||||||||
| LUAD | 80/48 | 0.918 (0.87‐0.967) | 0.838 (0.75‐0.912) | 0.875 (0.771‐0.985) | 0.852 | 53/32 | 0.909 (0.849‐0.968) | 0.849 (0.755‐0.943) | 0.812 (0.687‐0.938) | 0.835 |
| AIS | 14/48 | 0.902 (0.822‐0.981) | 0.714 (0.429‐0.929) | 0.875 (0.771‐0.985) | 0.839 | 9/32 | 0.806 (0.647‐0.964) | 0.556 (0.429‐0.929) | 0.812 (0.687‐0.938) | 0.756 |
| MIA | 13/48 | 0.915 (0.84‐0.991) | 0.769 (0.538‐1) | 0.875 (0.771‐0.985) | 0.852 | 9/32 | 0.927 (0.829‐1) | 0.889 (0.667‐1) | 0.812 (0.687‐0.938) | 0.829 |
| IAC | 53/48 | 0.923 (0.875‐0.972) | 0.887 (0.792‐0.962) | 0.875 (0.771‐0.985) | 0.881 | 35/32 | 0.93 (0.874‐1) | 0.914 (0.8‐1) | 0.812 (0.687‐0.938) | 0.866 |
| LUAD vs BPN | ||||||||||
| LUAD | 80/22 | 0.914 (0.857‐0.971) | 0.838 (0.75‐0.912) | 0.864 (0.727‐1) | 0.843 | 53/15 | 0.889 (0.796‐0.982) | 0.849 (0.755‐0.943) | 0.8 (0.6‐1) | 0.838 |
| AIS | 14/22 | 0.88 (0.768‐0.991) | 0.714 (0.429‐0.929) | 0.864 (0.727‐1) | 0.806 | 9/15 | 0.763 (0.565‐0.961) | 0.556 (0.222‐0.889) | 0.8 (0.6‐1) | 0.708 |
| MIA | 13/22 | 0.916 (0.821‐1) | 0.769 (0.538‐1) | 0.864 (0.727‐1) | 0.829 | 9/15 | 0.919 (0.801‐1) | 0.889 (0.667‐1) | 0.8 (0.6‐1) | 0.833 |
| IAC | 53/22 | 0.923 (0.863‐0.983) | 0.887 (0.792‐0.962) | 0.864 (0.727‐1) | 0.880 | 35/15 | 0.914 (0.826‐1) | 0.914 (0.8‐1) | 0.8 (0.6‐1) | 0.880 |
Abbreviations: AUC, area under the curve; AIS, adenocarcinoma in situ; BPN, benign pulmonary nodule; HC, healthy control; IAC, invasive carcinoma; LUAD, lung adenocarcinoma; MIA, minimally invasive adenocarcinoma; NC, noncancerous control; SD, standard deviation.
The clinical value of a tumor biomarker relies on its ability to detect cancer at an early stage. The study found that the LUAD group showed a high median d‐signature score compared with the HC group (P <.001; Figure 6A) and the BPN group (P <.001; Figure 6A). When comparing the two cohorts, the d‐signature could identify early LUAD patients from NC controls with an AUC of 0.91 (95% CI: 0.875 to 0.944; Figure 6B). The d‐signature score was not correlated with the tumor stage of early LUAD (AIS, MIA, and IAC) (Figure 6C), indicating that the diagnostic characteristics of the d‐signature did not depend on tumor burden and thus can be an excellent tool for early diagnosis. Evaluation of the diagnostic performance of this d‐signature for patients with AIS and MIA demonstrated that the AUCs for AIS and MIA compared with NC samples were 0.895 and 0.792, respectively, in the training cohort, and 0.915 and 0.924, respectively, in the validation cohort (Table 2). In the integrated two cohorts, the d‐signature was able to identify AIS and MIA patients from NC participants with AUCs of 0.846 (95% CI: 0.764 to 0.944) and 0.92 (95% CI: 0.862 to 0.978), respectively (Figure 6D). Therefore, the d‐signature can also be adopted for the high‐precision diagnosis of AIS and MIA.
FIGURE 6.
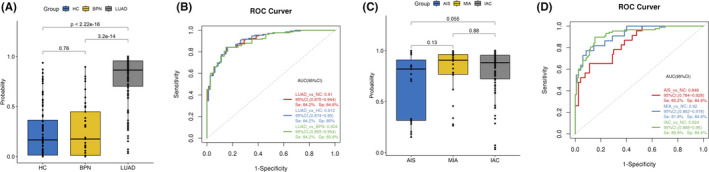
ExLR d‐signature for the diagnosis of stage I lung adenocarcinoma (LUAD). A, Extracellular vesicle (EV) microRNA (miRNA) diagnostic signature (d‐signature) in healthy control (HC) (n = 80), benign pulmonary nodule (BPN) patients (n = 38), and LUAD patients (n = 133). B, Receiver‐operating characteristic (ROC) curve for the performance of the EV miRNA d‐signature in LUAD in the combined cohorts. C, EV miRNA d‐signature score in adenocarcinoma in situ (AIS) patients (n = 23), minimally invasive adenocarcinoma (MIA) patients (n = 22), and invasive adenocarcinoma (IAC) patients (n = 90). D, ROC curve for the performance of the EV miRNA d‐signature for AIS, MIA, and IAC in the combined cohorts
3.5. Gene targeting analysis
The target mRNAs for the DEmiRs were predicted using the Miranda database and the RNAhybrid database. The screening standard for the Miranda database was set as score ≥150 and energy ≤−25. The screening standard for the RNAhybrid database was set as energy ≤−25 and P‐value <.05. In total, 2396 genes were targeted by these four candidate miRNAs (Figure S9A). GO analysis for the selected target genes revealed that they are greatly enriched in mRNA processing, collagen catabolic process, proteinaceous ECM, and ATP binding. Moreover, KEGG pathway analysis showed that the genes are mostly involved in the Notch signaling pathway, Rap1 signaling pathway, ECM‐receptor interaction, and miRNAs in cancer (Figure S9B‐D).
4. DISCUSSION
In this research, EV miRNA‐seq expression profiles were obtained from 150 human plasma EV samples. This is the biggest miRNA‐seq expression profile library from human plasma EVs to the best of our knowledge. The differences in EV miRNA expression among patients with early LUAD, patients with BPN, and healthy individuals were compared. A d‐signature with high accuracy was established and validated to distinguish LUAD patients and NC control participants.
Recent research has strongly highlighted a role for plasma EV miRNAs as diagnostic biomarkers for malignancies, including lung cancer. 17 , 28 In earlier work, Cazzoli et al reported that EV miRNAs (miR‐151a‐5p, miR‐154‐3p, miR‐200b‐5p, miR‐629, miR‐100, and miR‐30a‐3p) could differentiate LUAD patients from patients suffering from lung granulomas. 21 Jin et al demonstrated that plasma EV miRNAs (miR‐10b‐5p, miR‐15b‐5p, and miR‐320b) could differentiate LUAD from normal individuals with an AUC of 0.936. 22 Zhou and colleagues identified a panel of six EV miRNAs (miR‐19b‐3p, miR‐21‐5p, miR‐221‐3p, miR‐584‐5p, miR‐425‐5p, and miR‐409‐3p), which could distinguish LUAD patients from healthy volunteers with AUC values of 0.72, 0.74, and 0.84 in the training, testing, and verification cohorts, respectively. 29 Furthermore, Zhong et al observed significant variations in plasma EV miR‐520c‐3p and miR‐1274b levels between LUAD patients and HCs. The latter study also assessed the accuracy of these two miRNAs and reported AUC values of 0.857 and 0.845, respectively, for distinguishing NSCLC and NSCLC stage I patients from NC controls. 30
Unfortunately, these previous reports were limited by their selection bias and relatively small sample sizes. In the current prospective study, the EV miRNA profiles of consecutive plasma samples from patients with early LUAD, patients with BPN, and healthy individuals were examined using high‐throughput sequencing. Significant differences were observed between the EV miRNA profile of LUAD patients and that of the NC group. According to KEGG pathway analysis, these DEmiRs were enriched in several cancer‐related pathways. Therefore, EV miRNAs may be potential diagnostic biomarkers for the detection of LUAD. In addition, differences in EV miRNA levels between patients with AIS, MIA, and IAC, and NC controls were examined. The results showed that the IAC group was the most dissimilar to the NC group, while the AIS and MIA groups were similar to the NC group. This suggested that it is more difficult to distinguish AIS and MIA patients from NC individuals.
A total of eight newly identified EV miRNA markers and two previously published markers were further analyzed. Using a single‐center training cohort, a d‐signature was constructed including four EV miRNAs (hsa‐miR‐106b‐3p, hsa‐miR‐125a‐5p, hsa‐miR‐3615, and hsa‐miR‐450b‐5p). The d‐signature could distinguish early LUAD patients from NC controls with an AUC of 0.917, a sensitivity of 83.8%, and a specificity of 87.1%. The current results are promising for clinical application based on the following findings. First, in the test cohort, patients were recruited from our center and two other centers. The results showed that the d‐signature could differentiate early LUAD patients from the NC group with an AUC of 0.902, a sensitivity of 84.9%, and a specificity of 80.9%. Second, in the LUAD group, several cases were in very early stages of AIS and MIA. Tumor burden exerted little impact on our d‐signature differentiation of patients, indicating that the EV miRNA d‐signature may be conducive to detecting LUAD at a very early stage. Indeed, the d‐signature could distinguish AIS and MIA patients from NC controls in the integrated cohorts with an AUC of 0.846 and 0.92, respectively. Identification of the AIS and MIA cases with a noninvasive tool may facilitate the overall prognostic process and 5‐year survival rate of LUAD. Therefore, the four EV miRNA signatures have good diagnostic value and stability.
Before EV miRNAs can be used as biomarkers in cancer, we must better understand their role and function. Previous studies have shown that the EV miR‐106b level was much higher in the serum of lung cancer patients than in healthy volunteers. 31 Mechanistically, EV‐derived miR‐106b could strengthen lung cancer cells' migratory and invasive capability and enhance matrix metalloproteinase (MMP)‐2 and MMP‐9 expression. 31 Zhong et al revealed that miR‐125a‐5p acts as a tumor suppressor in lung carcinoma cells by targeting STAT3 directly. 32 Naidu and colleagues also reported that miR‐125a‐5p could control lung tumorigenesis by targeting various elements of the KRAS and NF‐kB pathways. 33 MiR‐450‐5p has also been shown to possess a tumor‐inhibiting function in lung SCC and hepatocellular carcinoma. 34 , 35 However, its clinical significance in LUAD remains unclear. MiR‐3615 is positively related to TNM stage, alpha‐fetoprotein (AFP) levels, and Ki‐67 levels in hepatocellular carcinoma 36 ; however, little is known regarding miR‐3615 in lung cancer. Indeed, certain miRNAs such as serum EV miR‐106b and circulating miR‐125a‐5p have been investigated for their potential role in diagnosing and prognosis of LUAD. 31 , 37 , 38 , 39 , 40 Collectively, these studies demonstrate that there are many miRNAs in plasma EVs that may act as potential biomarkers for the detection of LUAD.
There were several limitations to this investigation. First, although the EV miRNA d‐signature is a noninvasive diagnostic method with the potential for screening, its true value is detecting early LUAD before diagnosis by imaging. This requires longitudinal cohort studies that apply samples from biobanks based on large populations. Second, although multicentric validation research was conducted involving various centers in Beijing, validating the EV miRNA d‐signature in other regions of China is required. Other ethnic populations or other states may improve the effectiveness and stability of this diagnostic approach. Third, there was insufficient prognostic data, as all cases were newly diagnosed. Therefore, the prognostic value of this signature could not be evaluated in this study.
In conclusion, the current report demonstrated that LUAD patients present a particular plasma EV miRNA profile in early‐phase LUAD compared with noncarcinoma control individuals. The special EV miRNA d‐signature comprising four EV miRNA markers (hsa‐miR‐106b‐3p, hsa‐miR‐125a‐5p, hsa‐miR‐3615, and hsa‐miR‐450b‐5p) exhibits relatively good sensitivity and specificity. This d‐signature is a promising noninvasive biomarker for the early detection and routine screening of patients with LUAD.
DISCLOSURE
The authors declare no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All patients provided written informed consent. The study was approved by the Clinical Research Ethics Committee of the National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences (approval number: 20/370‐2155).
Supporting information
Fig S1
Table S1
Gao S, Guo W, Liu T, et al. Plasma extracellular vesicle microRNA profiling and the identification of a diagnostic signature for stage I lung adenocarcinoma. Cancer Sci.2022;113:648–659. doi: 10.1111/cas.15222
Shugeng Gao and Wei Guo contributed equally to this study.
Clinical trial registration information: The study was also registered on the Chinese Clinical Trial Registry (ChiCTR) on May 28, 2020 (ChiCTR2000033339).
Contributor Information
Shugeng Gao, Email: gaoshugeng@vip.sina.com.
Yibo Gao, Email: gaoyibo@cicams.ac.cn.
Jie He, Email: prof.jiehe@gmail.com.
DATA AVAILABILITY STATEMENT
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCE
- 1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐132. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7‐33. [DOI] [PubMed] [Google Scholar]
- 3. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong K‐K. Non‐small‐cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535‐546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446‐454. [DOI] [PubMed] [Google Scholar]
- 5. Zhang J‐T, Qin H, Man Cheung FK, et al. Plasma extracellular vesicle microRNAs for pulmonary ground‐glass nodules. J Extracell Vesicles. 2019;8:1663666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39‐51. [DOI] [PubMed] [Google Scholar]
- 7. Snowsill T, Yang H, Griffin ED, et al. Low‐dose computed tomography for lung cancer screening in high‐risk populations: a systematic review and economic evaluation. Health Technol Assess. 2018;22(69):1‐276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Silva M, Pastorino U, Sverzellati N. Lung cancer screening with low‐dose CT in Europe: strength and weakness of diverse independent screening trials. Clin Radiol. 2017;72:389‐400. [DOI] [PubMed] [Google Scholar]
- 9. Villalobos P, Wistuba II. Lung cancer biomarkers. Hematol Oncol Clin North Am. 2017;31:13‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uddin A, Chakraborty S. Role of miRNAs in lung cancer. J Cell Physiol. 2018:1‐10. doi: 10.1002/jcp.26607 [DOI] [PubMed] [Google Scholar]
- 11. Kasinski AL, Slack FJ. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321‐333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bottani M, Banfi G, Lombardi G. Circulating miRNAs as diagnostic and prognostic biomarkers in common solid tumors: focus on lung, breast, prostate cancers, and osteosarcoma. J Clin Med. 2019;8:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang S, Li S, Liu T, et al. MicroRNAs: Emerging oncogenic and tumor‐suppressive regulators, biomarkers and therapeutic targets in lung cancer. Cancer Lett. 2021;502:71‐83. doi: 10.1016/j.canlet.2020.12.040 [DOI] [PubMed] [Google Scholar]
- 15. Van Niel G, d'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213. [DOI] [PubMed] [Google Scholar]
- 16. Andaloussi SE, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discovery. 2013;12:347‐357. [DOI] [PubMed] [Google Scholar]
- 17. Guo W, Gao Y, Li N, et al. Exosomes: new players in cancer. Oncol Rep. 2017;38:665‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu J, Shen Z. Exosomal miRNAs as biomarkers for diagnostic and prognostic in lung cancer. Cancer Med. 2020;9:6909‐6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Yin Z, Fan J, Zhang S, Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct Target Ther. 2019;4:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8:1156‐1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin X, Chen Y, Chen H, et al. Evaluation of tumor‐derived exosomal miRNA as potential diagnostic biomarkers for early‐stage non–small cell lung cancer using next‐generation sequencing. Clin Cancer Res. 2017;23:5311‐5319. [DOI] [PubMed] [Google Scholar]
- 23. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7(3):789‐804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30(1):21‐23. [DOI] [PubMed] [Google Scholar]
- 25. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316‐W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Madadi S, Soleimani M. Comparison of miR‐16 and cel‐miR‐39 as reference controls for serum miRNA normalization in colorectal cancer. J Cell Biochem. 2019;120(4):4802‐4803. [DOI] [PubMed] [Google Scholar]
- 28. Fortunato O, Gasparini P, Boeri M, Sozzi G. Exo‐miRNAs as a new tool for liquid biopsy in lung cancer. Cancers. 2019;11:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X, Wen W, Shan X, et al. A six‐microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhong Y, Ding X, Bian Y, et al. Discovery and validation of extracellular vesicle‐associated miRNAs as noninvasive detection biomarkers for early‐stage non‐small‐cell lung cancer. Mol Oncol. 2021;15(9):2439‐2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xue X, Wang C, Xue Z, et al. Exosomal miRNA profiling before and after surgery revealed potential diagnostic and prognostic markers for lung adenocarcinoma. Acta Biochim Biophys Sin. 2020;52:281‐293. [DOI] [PubMed] [Google Scholar]
- 32. Zhong L, Sun S, Shi J, et al. MicroRNA‐125a‐5p plays a role as a tumor suppressor in lung carcinoma cells by directly targeting STAT3. Tumor Biol. 2017;39:1010428317697579. [DOI] [PubMed] [Google Scholar]
- 33. Naidu S, Shi L, Magee P, et al. PDGFR‐modulated miR‐23b cluster and miR‐125a‐5p suppress lung tumorigenesis by targeting multiple components of KRAS and NF‐kB pathways. Sci Rep. 2017;7:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Shen S, Chen X, Ren Z, Li Z, Yu Z. miR‐450b‐5p loss mediated KIF26B activation promoted hepatocellular carcinoma progression by activating PI3K/AKT pathway. Cancer Cell Int. 2019;19:1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ye P, Lv X, Aizemaiti R, Cheng J, Xia P, Di M. H3K27ac‐activated LINC00519 promotes lung squamous cell carcinoma progression by targeting miR‐450b‐5p/miR‐515‐5p/YAP1 axis. Cell Prolif. 2020;53:e12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan X, Zhang Y, Yu Z. Expression and clinical significance of miR‐3615 in hepatocellular carcinoma. J Int Med Res. 2021;49:0300060520981547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang P, Yang D, Zhang H, et al. Early detection of lung cancer in serum by a panel of microRNA biomarkers. Clin Lung Cancer. 2015;16:313–319. e311. [DOI] [PubMed] [Google Scholar]
- 38. Wang R‐J, Zheng Y‐H, Wang P, Zhang J‐Z. Serum miR‐125a‐5p, miR‐145 and miR‐146a as diagnostic biomarkers in non‐small cell lung cancer. Int J Clin Exp Pathol. 2015;8:765. [PMC free article] [PubMed] [Google Scholar]
- 39. Lin Q, Mao W, Shu Y, et al. A cluster of specified microRNAs in peripheral blood as biomarkers for metastatic non‐small‐cell lung cancer by stem‐loop RT‐PCR. J Cancer Res Clin Oncol. 2012;138:85‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu W‐Y, Luo B, An J‐Y, et al. Differential expression of miR‐125a‐5p and let‐7e predicts the progression and prognosis of non‐small cell lung cancer. Cancer Invest. 2014;32:394‐401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Data Availability Statement
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.


