Abstract
Growing evidence supports that N6‐methyladenosine (m6A) modification acts as a critical regulator involved in tumorigenesis at the mRNA level. However, the role of m6A modification at the noncoding RNA level remains largely unknown. We found that methyltransferase‐like 14 (METTL14) was significantly downregulated in renal cell carcinoma (RCC) tissues (n = 580). Gain‐of‐function and loss‐of‐function experiments revealed that METTL14 attenuated the proliferation and migration ability of RCC cells in vivo and in vitro. The methylated RNA immunoprecipitation experiments identified that METTL14 decreased the expression of long noncoding RNA nuclear enriched abundant transcript 1_1 (NEAT1_1) in an m6A‐dependent manner. Mechanistically, RNA pull‐down assay and RNA immunoprecipitation identified NEAT1_1 directly bound to m6A reader YTH N6‐methyladenosine RNA binding protein 2 (YTHDF2). Notably, YTHDF2 accelerated the degradation of NEAT1_1 by selectively recognizing METTL14‐mediated m6A marks on NEAT1_1. Multivariate analysis suggested that METTL14 downregulation was associated with malignant characteristics and predicted poor prognosis in RCC patients. In conclusion, our results uncover a newly identified METTL14‐YTHDF2‐NEAT1_1 signaling axis, which facilitates RCC growth and metastasis and provides fresh insight into RCC therapy.
Keywords: long noncoding RNA, m6A, METTL14, renal cell carcinoma, YTHDF2
Our results showed that N6‐methyladenosine (m6A) modification could mediate tumor progression and metastasis in renal cell carcinoma by regulating long noncoding RNA NEAT1, and that the METTL14‐YTHDF2‐m6A axis played an essential role in this process. These results could shed a light into the carcinogenic mechanism of renal cell carcinoma.
![]()
1. INTRODUCTION
Renal cell carcinoma (RCC) is one of the most frequently diagnosed malignancies of the genitourinary system, and the incidence has increased steadily. 1 , 2 The mortality of RCC is mainly due to a high rate of postoperative recurrence and metastasis. 3 , 4 The surgical and systemic therapy of RCC has progressed, but is less than satisfactory. 5 The carcinogenesis and development of RCC is a complex process caused by abnormal genetic and epigenetic changes. 6 Therefore, it is critical to explore the mechanism of carcinogenesis and the development of RCC.
In epigenetic regulation, similar to DNA or histone, there are over 100 kinds of posttranscriptional modifications in RNA levels. 7 N6‐methyladenosine (m6A) modification is the most common mRNA modification in eukaryotic cells. 8 The m6A modification is mainly mediated by “writers,” “readers,” and “erasers”, which can modulate RNA translation, stability, and localization by reversibly regulating the level of RNA m6A modification. 9 , 10 It has been reported that dysregulation of m6A modification is extensively related to tumorigenesis. 11 Methyltransferase‐like 14 (METTL14), as a critical member of the m6A methyltransferase complex, was recently reported to be essential for tumorigenesis and progression in several types of cancers, including pancreatic cancer, breast cancer, and colorectal cancer. 12 , 13 , 14 However, the regulatory mechanism for m6A modification in RCC remains unclear.
Long noncoding RNA (lncRNA) nuclear enriched abundant transcript 1 (NEAT1) is located on chromosome 11 (11q13.1), and plays an oncogenic role in a variety of solid tumors, in which it acts as a molecular sponge for microRNAs (miRNAs). 15 Knockdown of NEAT1 has been shown to be related to inhibition of proliferation and metastasis of tumor cells. For instance, NEAT1 acts as a competing endogenous RNA to reduce the expression of miRNA‐129‐5p, thereby promoting the malignant phenotype of hepatocellular carcinoma. 16 Similarly, it has been reported that NEAT1 also plays an oncogenic role in RCC. 17 Nuclear enriched abundant transcript 1 encodes a short variant (NEAT1_1, 3,756bp) and a long variant (NEAT1_2, 22,743bp). Notably, abundant m6A residues were found on the NEAT1_1 sequence in a recent study, 18 which suggested that m6A modification might influence the function of NEAT1_1.
Here, we found that METTL14 was expressed at lower levels in RCC than in normal tissues, and the downregulation of METTL14 expression portended poor prognosis of RCC. Moreover, we showed that METTL14 decreased the expression of oncogenic NEAT1_1 by upregulating the m6A level of NEAT1_1, thus inhibiting the proliferation and migration of RCC cells.
In this process, YTH N6‐methyladenosine RNA binding protein 2 (YTHDF2), as a specific reader of m6A, modified NEAT1_1 and accelerated the degradation of NEAT1_1. Overall, we emphasize the crucial role of RNA methyltransferase METTL14 in RCC, and suggest that METTL14 could be a new prognostic marker and therapeutic target for RCC.
2. MATERIALS AND METHODS
2.1. Clinical samples
The research protocol was reviewed and approved by the Ethics Committee of Changzheng Hospital of the Naval Medical University (Shanghai, China), and informed consent was obtained from all participants included in the study, in agreement with institutional guidelines. A total of 210 RCC tissues and 47 pairs of RCC and matched normal tissues were randomly selected and pathologically diagnosed as clear cell carcinoma. All the samples were taken from the tissue sample database of Changzheng Hospital. The samples were used for real‐time quantitative PCR and western blot analysis. The tissue microarray (TMA) of renal clear cell carcinoma was made by our group for immunohistochemical experiments.
2.2. Tissue microarray and immunohistochemistry analysis
The TMA contained 132 cases of RCC and normal tissues. Immunohistochemical analysis was undertaken with anti‐METTL14 (Cat#MA5‐24706; Thermo Fisher Scientific) or anti‐YTHDF2 Ab (1:1000, Cat#ab220163; Abcam). Briefly, the tissue chip was dipped in acetone solution for 5 minutes, dried, then soaked for 10 minutes in xylene solution. Next, the chip was immersed in ethanol solution and then 3% hydrogen peroxide solution. The chip was then blocked with goat serum. After incubation with targeted Ab and secondary Ab, the chip was dripped with DAB chromogenic solution and restained with hematoxylin. The results were analyzed by Aperio Image Scope software and the H‐score was calculated.
2.3. Western blot analysis
The tissue or cell was cleaved for 20 minutes with RIPA (Beyotime) lysate on ice. After centrifugation, the protein was quantified using a BCA protein quantitative kit (Beyotime). The protein concentration was determined by the protein standard curve, and the protein was adjusted into the proper concentration. Sample loading buffer (5×) (Beyotime) was added and the mixture was boiled at 100℃ for 5 minutes. The PAGE glue was prepared according to the molecular size of the corresponding protein and electrophoretic separation was performed after adding samples. The membrane was then transferred by constant current 270 mA for 90 minutes. We blocked the membrane with 5% milk, it was incubated with primary Ab then secondary Ab, and finally scanned and recorded with an Odyssey infrared scanner, semiquantified with internal reference GAPDH. Antibodies used in this experiment were as follows: anti‐GAPDH (1:5000, Cat# ab8245, Cat#ab9485; Abcam), anti‐METTL14 (1:1000, Cat#669602; BioLegend), anti‐YTHDF2 (1:1000, Cat#ab220163; Abcam), anti‐YTHDF1 (1:1000, Cat#17479‐1‐AP; Proteintech), anti‐YTHDF3 (1:1000, Cat#ab220161; Abcam), anti‐IGF2BP2 (1:1000, Cat#11601‐1‐AP; Proteintech), and anti‐IGF2BP3 (1:1000, Cat#14642‐1‐AP; Proteintech).
2.4. RNA isolation and real‐time PCR
The RNA of tissue or cell was extracted by an RNAsimple Total RNA Kit, DP419 (Tiangen) and the concentration of RNA was measured by Nanodrop. cDNA was synthesized by a reverse transcription kit (RR036A; Takara). The RT‐PCR reaction was carried out with a TB Green Premix Extra kit (RR420A; Takara). The Applied Bio Systems Step One Plus Real‐Time PCR System (Thermo Fisher Scientific) was used for reactions. All operations were carried out according to the product instructions. β‐Actin was used as the internal reference, and the relative expression of the target gene was calculated by 2−ΔΔCt, and the expression of the control group was determined as 1. These experiments were undertaken in triplicate. The primers used for quantitative RT‐PCR (qRT‐PCR) are listed in Table S1.
2.5. Cell lines, cell culture and transfection
The 786‐O and 769‐P cells were cultured in DMEM, HK‐2 was cultured in serum‐free medium (Keratinocyte‐SFM) under 37℃, 5% CO2, and saturated humidity. Cell transfections were carried out with siRNAs using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. The siRNAs and METTL14 overexpression and knockdown lentivirus were obtained from GenePharma Company, and the sequences are listed in Table S2. For transfection with lentivirus, the cells in logarithmic growth phase were washed twice with PBS, then 5 mL serum‐free medium containing 8 µg/mL polybrene (Beyotime) was added. Cells were cultured for 30 minutes and the virus was added according to MOI = 0.4. Cells were supplemented with medium and cultured for 3 days, then the liquid was changed, with medium added containing 1 µg/mL puromycin (Beyotime). The culture continued for 3 days, and the cells were collected.
2.6. Cell proliferation and migration assay
The cells were incubated into a 96‐well plate with 100 µL medium in each well. The culture medium was prepared with CCK‐8 (Dojindo Molecular Technologies) and added to a 96‐well plate. After incubation with CCK‐8 for 30 minutes, data were obtained in three independent experiments with five replicates over 72 hours. The migration ability was assessed according to the number of cells that crossed Transwell inserts. First, 5 × 104 cells were plated in the upper chamber with serum‐free medium, and the medium in the lower chamber contained 10% FBS. After 24 hours, the inserts were stained with 1% crystal violet and photographed. These experiments were carried out three times.
2.7. Animal experiments
The animal experiments in this study were approved by the Institutional Animal Care and Use Committee of the Naval Military Medical University. Mouse subcutaneous xenograft and lung metastasis experiments were carried out with six 4‐week‐old male BALB/c nude mice as we described previously. 19 For the subcutaneous xenograft, the size of subcutaneous nodules was recorded weekly, and mice were killed in the sixth week of the experiment. The H&E and immunofluorescent staining were undertaken on the dissected mouse tissues.
2.8. RNA immunoprecipitation and methylated‐RNA immunoprecipitation assay
A Magna RIP kit (#17‐700; Merck Millipore) was used in the RIP experiment. Renal cancer cells in 15‐cm plates were lysed and incubated with the magnetic beads prebound to METTL14, YTHDF2, or IgG Ab (5 µL) overnight at 4℃ with rotation. Then RNA was isolated from the binding protein by protease and purified. The experiment was divided into three groups: INPUT, IP, and IgG. The expression of purified RNA relative to INPUT was calculated by quantitative PCR. For methylated‐RNA immunoprecipitation assay (MeRIP), m6A Ab was used to immunoprecipitate methylated NEAT1_1. Antibodies used in this experiment were as follows: anti‐m6A (1:1000, Cat#202 003; Synaptic Systems), anti‐METTL14 (1:1000, Cat#669602; BioLegend), anti‐YTHDF2 (1:1000, Cat#ab220163; Abcam), and anti‐IgG (Cat#2729; Cell Signaling Technology).
2.9. N6‐methyladenosine quantification assay
The m6A levels in total RNAs were detected with the EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (P‐9005; Epigentek). Briefly, 200 ng RNAs (1‐8 µL) were bound to assay cells. According to the manufacturer’s instructions, diluted Capture Antibody solution and Detection Antibody solution were added successively into assay wells. The absorbance of each sample was measured at 450 nm and quantification of RNA m6A was carried out by using the standard curve.
2.10. RNA m6A dot blot assay
After the RNA extracted by TRIzol was treated with DNase I to remove DNA impurities, RNA (400 ng or 200 ng) was added to the nylon membrane. The membrane was then cross‐linked by UV light (254 nm), sealed for 1 hour, and incubated overnight with m6A Ab (1:1000, Cat# 202 003; Synaptic Systems) at 4°C. The membrane was washed three times and incubated with anti‐rabbit IgG Ab (Cat# 2729; Cell Signaling Technology) for 1 hour at room temperature. We scanned and photographed the nylon film with m6A point. The same 400 ng and 200 ng RNA was placed on the membrane, dyed with 0.2% methylene blue corrected by 0.3% sodium acetate (pH = 5.2) for 2 hours, then washed with RNase‐free water and photographed. These experiments were carried out three times.
2.11. RNA pull‐down assay
Nuclear enriched abundant transcript 1_1 was transcribed in vitro and biotinylated with biotinylated RNA labelling mix (Roche) and T7 RNA polymerase (Roche). Cell lysates (1 mg) were incubated with 3 μg biotin‐labelled NEAT1_1 at 25°C for 1 hour, and then the complexes were separated with streptavidin agarose beads (Invitrogen). After three washes, the protein present was retrieved from the pull‐down mix, and detected by immunoblotting analysis.
2.12. RNA FISH
Fluorescence‐conjugated NEAT1_1 or YTHDF2 probes were designed and purchased (GenePharma). Hybridization was carried out using DNA probe sets (GenePharma) following the manufacturer’s instructions. Cells were then observed and recorded.
2.13. RNA decay assays
RNA decay assays were undertaken to estimate the stability of RNA. Renal cell carcinoma cells were cultured in 6‐well plates and treated with 5 μg/mL actinomycin D (#15021; Cell Signaling Technology). Total RNAs of cells were isolated at 0, 4, 8 hours and RT‐qPCR was carried out to measure the relative RNA levels of NEAT1_1.
2.14. Statistical analysis
The classified variables in this experiment are expressed by frequency or percentage, and the continuous variables in the experiment are shown as mean ± SD. The data between the two groups were analyzed by Student’s t test, and multiple comparisons were analyzed by one‐way ANOVA or two‐way ANOVA. Statistical analyses were undertaken using SPSS 22.0.0 software.
3. RESULTS
3.1. Low expression of METTL14 is related to RCC progression and poor prognosis
To determine the function of m6A modification in RCC, we first determined the expression of major m6A methyltransferases, including METTL3 and METTL14 in RCC. As shown, the mRNA and protein expression of METTL14 in RCC tissues were significantly downregulated compared with paired normal tissues (Figure 1A,B). However, there was no significant difference in the expression level of METTL3 (Figure S1A). In addition, consistent expression patterns of METTL14 and METTL3 in RCC were validated in The Cancer Genome Atlas (TCGA) databases (Figures 1C and Figure S1B). Immunohistochemical staining was also used to analyze METTL14 expression in TMAs of RCC vs normal tissues (Figure 1D). Low METTL14 expression was observed in RCC (P < .001) (Figure 1E). Therefore, our results determined that METTL14 was significantly downregulated in RCC.
FIGURE 1.
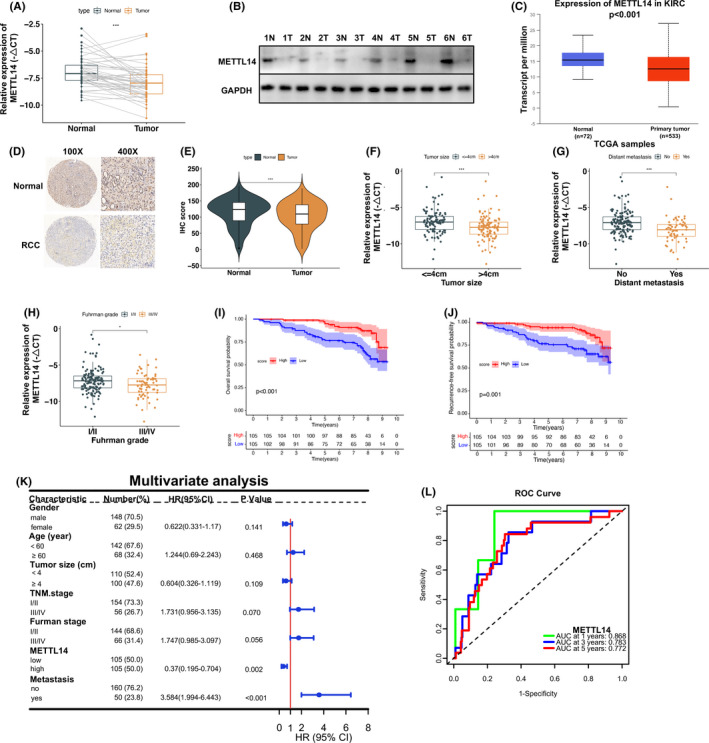
Low expression of methyltransferase‐like 14 (METTL14) is related to renal cell carcinoma (RCC) progression and poor prognosis. A, B, METTL14 mRNA (n = 47) and protein (n = 6) expression in RCC (T) and matched normal (N) tissues. C, METTL14 expression in KIRC from The Cancer Genome Atlas (TCGA) database (n = 533). D, E, Immunohistochemical (IHC) analysis of METTL14 in RCC tissue microarrays (n = 132). Magnification, 100×, 400×. F‐H, METTL14 expression in tumors >4 cm group (n = 100) vs tumors ≤4 cm group (n = 110), in the RCC with metastasis group (n = 50) vs RCC without metastasis group (n = 160), and in the Fuhrman I/II group (n = 144) vs Fuhrman III/IV group (n = 66). I, J, Kaplan‐Meier analysis of correlation between METTL14 and overall survival (OS) or recurrence‐free survival. K, Multivariate analysis of factors associated with OS in RCC patients (n = 210). L, Time‐dependent OS receiver operating characteristic (ROC) analysis. Error bars represent mean ± SD. Student’s t test (A, E‐H) and log‐rank test (I, J) were used. *P < .05, **P < .01, ***P < .001. AUC, area under the ROC curve; CI, confidence interval; HR, hazard ratio
To detect the correlation between METTL14 and clinicopathologic features, a cohort of 210 RCC cases was analyzed. We revealed that METTL14 expression was considerably lower in the group with tumors larger than 4 cm (P < .001), the group with distant metastasis (P < .001), those with higher pathological stage (P < .05), and those with higher TNM stage (Figures 1F‐H and S1C). Dividing the cohort into high and low METTL14 groups by median expression, based on qRT‐PCR results, we discovered that METTL14 expression negatively correlated with RCC tumor size (P < .001), pathological grade (P < .05), TNM stage (P < .01), and metastasis (P < .001) (Table S3). Accordingly, TCGA data also showed that METTL14 expression was lower in higher TNM stage (Figure S1D). An alluvial diagram was utilized to visualize the distribution of TNM stage, tumor grade, and METTL14 subgroup in the TCGA dataset (Figure S1E). The survival analysis indicated that overall survival and recurrence‐free survival were significantly worse in the low METTL14 group than in the high METTL14 group (Figures 1I,J and S1F,G). Furthermore, multivariate and time‐dependent receiver operating characteristic analyses identified METTL14 expression as an independent prognostic factor in RCC patients (Figures 1K,L and S1H).
3.2. Methyltransferase‐like 14 inhibits proliferation and migration of RCC in vitro
To assess the functional role of METTL14 and METTL3, specific siRNAs were transfected into 786‐O and 769‐P cell lines (Figure 2A,B). The CCK‐8 and colony formation assays showed that METTL14 knockdown significantly enhanced the proliferation of RCC cells (Figure 2C,D). Similarly, the Transwell migration assay showed that METTL14 knockdown significantly enhanced the migration of RCC cells (Figure 2E). However, CCK‐8 and Transwell migration assays for METTL3‐interfering RCC cells showed no significance, indicating that METTL14, rather than METTL3, played a prominent role in RCC (Figure S2). In addition, we constructed and transfected METTL14 overexpression (OE‐METTL14) and control lentivirus (OE‐NC) into RCC cell lines (Figure 2F,G). As expected, functional assays showed that METTL14 overexpression attenuated the proliferation and migration of RCC cells (Figure 2H‐J).
FIGURE 2.
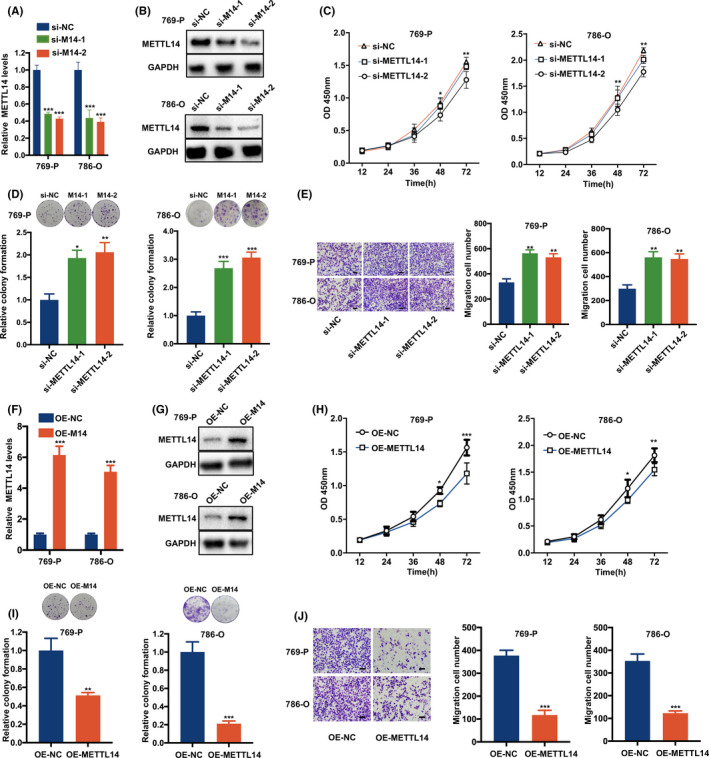
Methyltransferase‐like 14 (METTL14) inhibits the proliferation and migration of renal cell carcinoma (RCC) cells in vitro. A, B, METTL14 mRNA (A) and protein (B) levels in 769‐P and 786‐O cells transfected with METTL14 siRNAs (si‐M14‐1 and si‐M14‐2) or control (si‐NC). C‐E, CCK‐8 assay (C), clone formation assay (D), and Transwell migration assay (E) of 769‐P and 786‐O cells transfected with METTL14 siRNAs or control. F, G, METTL14 mRNA (F) and protein (G) levels in METTL14 overexpression (OE‐M14) and control (OE‐M14) RCC cells (P < .001). H‐J, CCK‐8 assay (H), cell clone formation assay (I), and Transwell migration assay (J) of METTL14 overexpression and control RCC cells. Error bars represent mean ± SD of at least three independent experiments. Two‐way ANOVA (C, H) and Student’s t test (A, D‐F, I, J) were used. *P < .05, **P < .01, ***P < .001
3.3. Methyltransferase‐like 14 suppresses tumor growth and metastasis in vivo
To assess the effects of METTL14 in vivo, we constructed OE‐METTL14 or OE‐NC 769‐P cells and injected them into mouse armpits to undertake the xenograft experiment (Figure 3A). At week 6 after injection, mice bearing OE‐METTL14 had lower tumor volumes and weights (Figure 3B,C). Moreover, immunohistochemical assays indicated that the expression levels of Ki‐67 were lower in the OE‐METTL14 group than in the OE‐NC group (Figure 3D). Nude mice were also injected intravenously with OE‐METTL14 and OE‐NC 769‐P cells to examine the impact of METTL14 on metastasis (Figure 3E). As a result, the mice injected with OE‐METTL14 cells had fewer pulmonary metastases (Figure 3F). Taken together, these results illustrated that higher METTL14 expression suppressed RCC tumor growth and metastasis in vivo.
FIGURE 3.
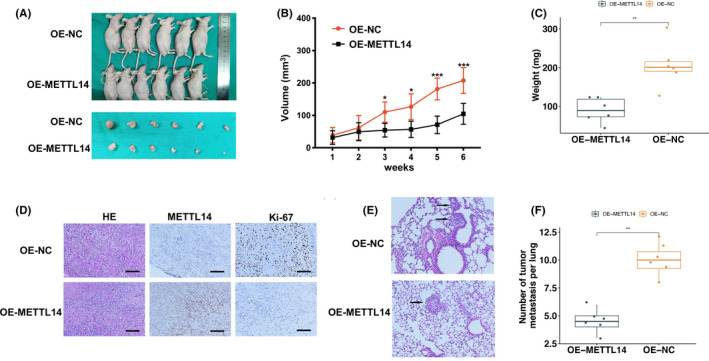
Methyltransferase‐like 14 (METTL14) suppresses renal cell carcinoma tumor growth and metastasis in vivo. A, Subcutaneous xenograft models were established with METTL14 overexpressing (OE‐METTL14) or control (OE‐NC) 769‐P cells (n = 6). B, C, Growth curve (B) and weight (C) of OE‐METTL14 tumors compared to control group (OE‐NC). D, Immunohistochemical assays showing METTL14 and Ki‐67 expression in each group. Scale bar, 200 μm. E, F, Representative metastatic lung nodules in each group and quantification of the number of metastatic nodules. Magnification, 400×. Error bars represent mean ± SD. Mann‐Whitney U test (C, F) and two‐way ANOVA test (B) were used. *P < .05, **P < .01, ***P < .001
3.4. Methyltransferase‐like 14 inhibits proliferation and migration of RCC cells by targeting lncRNA NEAT1_1
Next, we investigated the mechanism underlying the inhibitory effect of METTL14 on malignant behaviors of RCC cells. Previous investigations have shown that METTL14 facilitated the installation of m6A modification of lncRNAs and triggered downstream signaling pathways. 20 Potential target lncRNAs that might interact with METTL14 by m6A modification were identified through the RMBase version 2.0 database. 21 Subsequently, we verified these potential lncRNAs in the microarray data from the Gene Expression Omnibus database (GSE117890). It was found that lncRNA NEAT1 was upregulated in RCC (Figure 4A). In line with a recent study in prostate cancer, abundant m6A modification sites were located in NEAT1_1 through the SRAMP database (Figure 4B), which suggested that m6A methylation might play a pivotal part in regulating NEAT1_1. 18 , 22 In addition, expression of NEAT1_1 was almost 4‐5‐fold higher than NEAT1_2 in RCC cells (Figure S3A). By qRT‐PCR, NEAT1_1 was upregulated in METTL14‐interfering RCC cells (Figure 4C); NEAT1_1 was also reduced in RCC cells overexpressing METTL14 (Figure 4D). In addition, NEAT1_2 showed no significant difference between METTL14‐interfering RCC cells and control (Figure S3B). Thus, NEAT1_1 was selected for further study. It was found that NEAT1_1 expression was higher in RCC tissues than in normal tissues and negatively correlated with METTL14 expression (Figures 4E,F and S3C).
FIGURE 4.
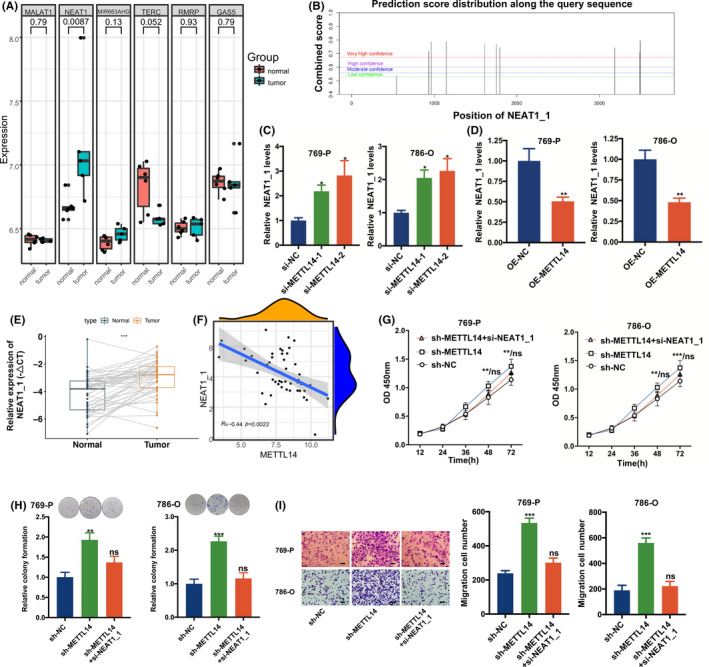
Methyltransferase‐like 14 (METTL14) inhibits the proliferation and migration of renal cell carcinoma (RCC) cells by targeting nuclear enriched abundant transcript 1_1 (NEAT1_1). A, Analysis of long noncoding RNA in RCC vs normal tissue from the Gene Expression Omnibus microarray. B, N6‐methyladenosine (m6A) sites on NEAT1_1 predicted by the SRAMP database. C, Relative NEAT1_1 expression in RCC cells treated with METTL14 siRNA or control (si‐NC). D, Relative expression of NEAT1_1 in METTL14 overexpression and control RCC cells. E, NEAT1_1 expression in RCC and matched normal tissues (n = 47). F, Linear correlation pattern between METTL14 and NEAT1_1 from our RCC samples (n = 47). G‐I, CCK‐8 assays, cell clone formation, and Transwell assays of RCC cells cotransfected with sh‐METTL14 and si‐NEAT1_1 or si‐NC. Error bars represent mean ± SD of at least three independent experiments. Mann‐Whitney U test (A), Student’s t test (C‐E, H, I), two‐way ANOVA test (G), and Pearson correlation analysis (F) were used. *P < .05, **P < .01, ***P < .001. ns, no significance relative to control
It was reported that NEAT1 played an oncogenic role through epithelial‐mesenchymal transition in RCC. 17 In our study, it was identified that NEAT1_1 interference attenuated proliferation and migration of RCC cells (Figure S3D‐H). We also found that NEAT1_1 interference attenuated the enhancement of cell proliferation and migration induced by METTL14 knockdown (Figures 4G‐I and S3I). These results indicated that METTL14 promoted proliferation and migration of RCC cells through decreasing NEAT1_1.
3.5. Methyltransferase‐like 14 negatively regulates NEAT1_1 in an m6A‐dependent manner
It was unknown whether METTL14 directly mediated the downregulation of NEAT1 in an m6A‐dependent manner. We examined the m6A expression profile between RCC and normal tissues by RNA m6A colorimetry, and found that m6A levels were lower in RCC than normal controls (Figure 5A,B). These data suggested the potential role of m6A modification in RCC. In addition, the m6A levels of RCC tissues were clearly correlated with the expression of METTL14, rather than METTL3 (Figures 5C and S3J). Through m6A dot blotting, we verified that silencing of METTL14 could decrease the m6A level of RCC cells, and overexpression of METTL14 could restore the reduction of m6A levels in RCC cells (Figure 5D). Methylated RNA immunoprecipitation was used to ascertain whether METTL14 regulates m6A levels of NEAT1_1. Compared with the control, the RNA abundance of m6A antibody was significantly increased in RCC cells overexpressing METTL14 (Figure 5E). These results confirmed that the m6A modification on NEAT1_1 could be transferred in large part by METTL14. Moreover, we dosed RCC cells with 3‐deazaadenosine (DAA), the global methylation inhibitor, and NEAT1_1 was decreased with the increase of DAA concentration (Figure 5F). However, no significant associations between the expression of NEAT1_1 and m6A level in RCC tissues were observed (Figure S3K), mainly due to heterogeneous components in clinical tumor tissues. These results indicated that METTL14 downregulated NEAT1_1 by promoting m6A methylation, inhibiting growth and metastasis of RCC.
FIGURE 5.
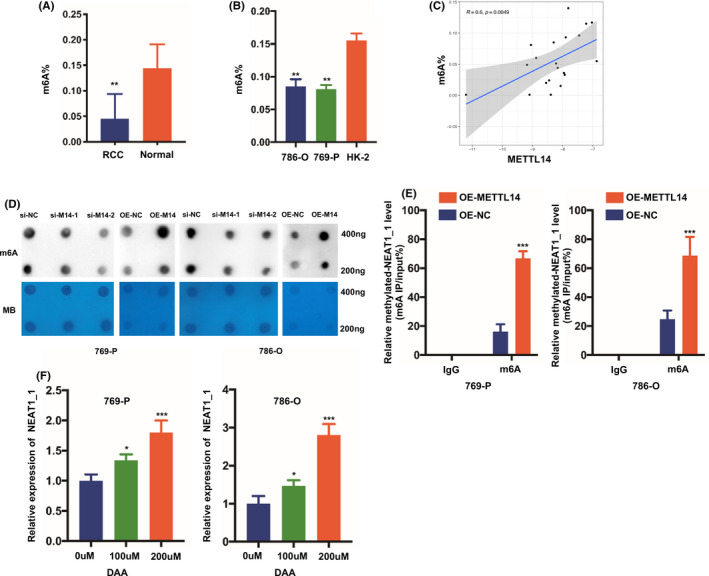
Methyltransferase‐like 14 (METTL14) negatively regulates nuclear enriched abundant transcript 1_1 (NEAT1_1) in an N6‐methyladenosine (m6A)‐dependent manner. A, m6A levels of total RNAs in tumor and matched normal tissues by RNA m6A colorimetry (n = 10). B, m6A levels of total RNAs in renal cell carcinoma (RCC) cell lines and HK‐2 cells by colorimetry. C, Correlation analysis between m6A and METTL14 in RCC samples (n = 20). D, m6A levels of METTL14 knockdown or overexpressing (OE) RCC cells by m6A dot blot assay. MB, methylene blue; NC, control. E, Methylated‐RNA immunoprecipitation (IP) assay‐quantitative PCR analysis of NEAT1_1 in METTL14 overexpressing (OE‐METTL14) RCC cells and the control (OE‐NC). F, NEAT1_1 expression in RCC cells treated with 3‐deazaadenosine (DAA) and the control. Error bars represent the mean ± SD. Student’s t test was used. *P < .05, **P < .01, ***P < .001
3.6. YTH N6‐methyladenosine RNA binding protein 2 inhibits growth and metastasis of RCC by recognizing and downregulating NEAT1
The m6A methylation is a labelling process that requires recognition by m6A readers of the downstream target RNAs. 23 We screened the NEAT1‐binding m6A reader proteins by RNA pull‐down assay, and found that biotin‐labelled NEAT1_1 significantly enriched YTHDF2, compared to other readers (Figures 6A and S4A). We further proved the combination of NEAT1_1 and YTHDF2 through RIP and double FISH assays (Figure 6B,C). It has been determined that YTHDF2 can promote the degradation of m6A‐dependent RNA. 24 Therefore, we hypothesized that YTHDF2 accelerated the decay of NEAT1_1 by recognizing m6A marks. Moreover, the mRNA and protein levels of YTHDF2 were lower in RCC than normal tissues (Figures 6D and S4B,C). Importantly, YTHDF2 expression was negatively correlated with NEAT1_1 (Figure 6E), which was verified in the TCGA database (Figure S4D). Furthermore, TCGA data also showed that YTHDF2 expression was negatively associated with the prognosis of RCC patients (Figure S4E,F). We undertook YTHDF2 knockdown experiments and found that YTHDF2 knockdown attenuated the degradation of NEAT1_1 (Figure 6F‐H), which indicated that YTHDF2 mediated the degradation of NEAT1_1. Next, we determined whether METTL14 inhibited RCC growth and migration through a YTHDF2‐dependent pathway. We transfected OE‐METTL14 RCC cells with si‐YTHDF2 or si‐NC, and we found that downregulation of NEAT1_1 induced by METTL14 overexpression could be reversed by YTHDF2 knockdown (Figure 6I). Functionally, CCK‐8 and Transwell migration assays showed that YTHDF2 knockdown could largely reverse the inhibitory effect of METTL14 overexpression (Figure 6J‐L).
FIGURE 6.
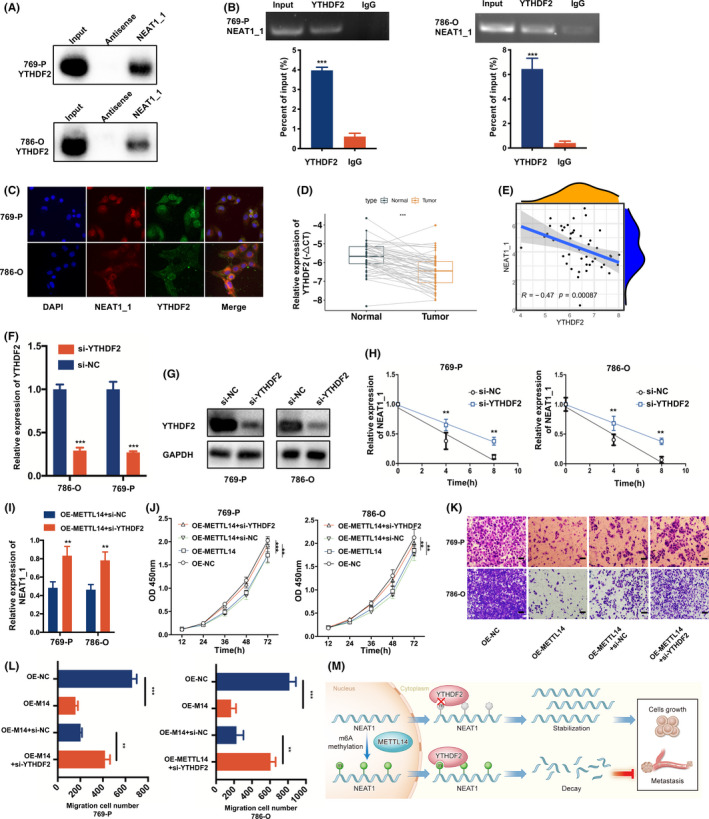
YTH N6‐methyladenosine RNA binding protein 2 (YTHDF2) inhibits growth and metastasis of renal cell carcinoma (RCC) by recognizing and downregulating nuclear enriched abundant transcript 1 (NEAT1). A, RNA pull‐down assays in RCC cells showing significant interaction between YTHDF2 and NEAT1_1. B, RIP assays in RCC cells showing the enrichment of NEAT1_1 on YTHDF2 relative to IgG. C, Double FISH experiment of YTHDF2 (green) and NEAT1_1 (red) showing the colocalization in RCC cells. D, YTHDF2 mRNA expression in RCC and matched normal tissues (n = 47). E, Negative linear correlation between YTHDF2 and NEAT1_1 from our samples (n = 47). F, YTHDF2 mRNA expression in RCC cells transfected with YTHDF2 siRNA or control. G, YTHDF2 protein expression in RCC cells transfected with YTHDF2 siRNA or control. H, Degradation rate of NEAT1_1 after YTHDF2 interference in RCC cells. I, NEAT1_1 expression in methyltransferase‐like 14 overexpressing (OE‐METTL14) RCC cells with or without YTHDF2 interference. J‐L, CCK‐8 assay (J) and Transwell assay (K, L) of OE‐METTL14 RCC cells with or without YTHDF2 interference at the indicated times. M, Graphic abstract of METTL14 regulating the malignant phenotype of RCC. Error bars represent mean ± SD. Student’s t test (B, D, F, I, K), Pearson correlation analysis (E), and two‐way ANOVA test (H, J) were used. *P < .05, **P < .01, ***P < .001
Taken together, these findings suggested that the METTL14‐induced m6A process suppressed the expression of NEAT1_1 through YTHDF2‐dependent RNA degradation (Figure 6M).
4. DISCUSSION
In this study, it was found that METTL14 was downregulated in RCC tissues and negatively associated with the prognosis of RCC patients. In addition, METTL14 inhibited the proliferation and migration of RCC by reducing NEAT1_1 in an m6A‐YTHDF2‐dependent manner.
As a key component of the N6‐methyltransferase complex, METTL14 has been reported to play a critical role in various tumors. 13 , 14 In the present study, we revealed that METTL14 was downregulated in RCC and low expression of METTL14 had a negative correlation with the prognosis of RCC patients, which was consistent with previous bioinformatics studies. 25 , 26 Although the TNM staging system has played a dominant role in prognostic prediction of RCC for decades, limitations remain. Thus, an m6A‐based scoring model could be valuable supporting indicator of prognosis for RCC patients. Larger prospective trials are required to validate whether METTL14 could be applied in clinical practice as a valuable prognostic biomarker. We confirmed that METTL14 could regulate m6A modification and impair the growth and metastasis of RCC cells in vitro and in vivo. However, the function of METTL14 varied in different tumors, as METTL14 played an oncogenic role in pancreatic cancer, promoting tumor growth and metastasis by decreasing PERP mRNA. 12 Similarly, recent studies reported that METTL3 and METTL14, which both serve as m6A writers, played oncogenic and antioncogenic roles, respectively, in hepatocellular carcinoma. 24 , 27 These results, which appear to be contradictory, implied that the functions of m6A in tumor progression is complicated, probably due to the differences of m6A modified sites on RNAs, different readers that recognize these m6A modifications, or different target genes regulating specific cellular processes. Thus, our results, along with other observations, showed that dysregulation of m6A methylation caused by METTL14 is closely associated with tumor progression. However, more research is needed to acquire all‐round and deep cognition of RNA m6A modification in tumorigenesis.
Increasing evidence shows that m6A plays a regulatory role for noncoding RNA (ncRNA), including lncRNA, and miRNA in various cancer types. 20 It was reported that METTL14 inhibited primary miRNA processing to facilitate hepatocellular carcinoma metastasis. 27 Another study stated that METTL14‐medicated m6A marks contribute in opposite fashion to lncRNA XIST in colorectal cancer. 28 In line with the previous study, NEAT1_1 was identified as an oncogene in RCC in our study, 17 and the expression of oncogenic NEAT1_1 negatively correlated with METTL14. As a highly abundant m6A‐modified transcript, we observed that the downregulation of NEAT1_1 in RCC induced by METTL14 depended on m6A modification. In accordance with previous studies, these results suggested that m6A mediated ncRNA methylation to regulate cancer progression.
The m6A modification is posttranscriptionally introduced by writers and reversed by erasers, including FTO and ALKBH5. 29 , 30 The m6A erasers also play an important role in tumor initiation and progression. For example, ALKBH5 mediated m6A demethylation of mRNA AURKB to enhance its stability, thus promoting cell proliferation in RCC. 31 In particular, ALKBH5 could upregulate NEAT1 by demethylation. 32 Although the specific m6A readers of NEAT1_1 and subsequent process were not elucidated in the study, these results, along with our data, indicated that upregulation of m6A levels might lead to downregulation of NEAT1_1 expression. However, whether ALKBH5 regulates m6A‐dependent lncRNA in synergy with METTL14 in RCC requires further investigation.
Previous studies had identified YTH and IGF2BP as prominent families of m6A readers that might control the fate of m6A‐modified mRNA. 33 , 34 Notably, m6A readers were reported to recognize the methylated lncRNA and determine further RNA processes. It was shown that IGF2BP2 facilitated stemness‐like properties and proliferation of pancreatic tumor by promoting the stability of lncRNA DANCR. 35 In addition, YTHDF2 recognized METTL14‐mediated m6A marks of lncRNA XIST, 28 accelerating decay of XIST. In the present study, YTHDF2, rather than the other m6A reader proteins, served as the specific m6A reader of NEAT1_1 and facilitated its degradation. Furthermore, YTHDF2 expression was negatively associated with NEAT1_1 and has a positive correlation with the prognosis of RCC patients. These results lead to the conclusion that the METTL14‐YTHDF2‐m6A signaling axis plays a regulatory role for lncRNAs. However, further research is needed to confirm the function of YTHDF2 in RCC.
The m6A modification is reportedly involved in regulation of lncRNA in various cancer types, such as MALAT1, XIST, NEAT1, DANCR, and PVT1. 18 , 28 , 35 , 36 , 37 Significantly, the cooperation of writer, eraser, and reader, rather than a single isolated gene, was proved to participate in regulating m6A‐modified lncRNA, which provided a better interpretation of the m6A regulatory network. We noticed that METTL14 and YTHDF2 regulated the expression of NEAT1_1 in synergy. However, the mechanism underlying this synergy effect and target specificity remains foggy. Moreover, in the present study, we did not locate the specific m6A sites on NEAT1_1 installed by METTL14 and the role of the m6A eraser in molecular mechanisms regulating the physiological functions of NEAT1_1 remain unclear. In addition, NEAT1_1 might be mediated by other regulators, such as transcription factors. Therefore, studies considering transcription factor regulatory mechanisms are required. Above all, these results imply that m6A‐modified lncRNA is an essential part of the complex m6A regulatory network.
In this study, we confirmed the specificity and efficacy of the METTL14‐YTHDF2‐NEAT1_1 signaling axis in regulating RCC progression and metastasis. Moreover, it was shown that METTL14 served as a prognostic marker for the prognosis of RCC patients. These results might shed light on future clinical treatment strategies in RCC, and more investigations are urgently needed to unveil the integral m6A regulatory networks.
DISCLOSURE
The authors declare no conflict of interest.
Supporting information
Fig S1‐S4
Table S1‐S3
ACKNOWLEDGMENTS
We would like to acknowledge the funding from the National Natural Science Foundation of China (81902560 and 81730073) and the Clinical Science and Technology Innovation Project of Shanghai Shenkang Hospital Development Centre (SHDC12018108).
Liu T, Wang H, Fu Z, et al. Methyltransferase‐like 14 suppresses growth and metastasis of renal cell carcinoma by decreasing long noncoding RNA NEAT1. Cancer Sci.2022;113:446–458. doi: 10.1111/cas.15212
Tao Liu, Hui Wang, and Zhibin Fu contributed equally to this work.
Funding information
National Natural Science Foundation of China, Grant/Award Number: 81902560, 81730073; Shanghai Shenkang Hospital Development Centre, Grant/Award Number: SHDC12018108
Contributor Information
Anbang Wang, Email: wanganbangcz@163.com.
Linhui Wang, Email: wanglinhui@smmu.edu.cn.
REFERENCES
- 1. Znaor A, Lortet‐Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67(3):519‐530. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Miller K, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. [DOI] [PubMed] [Google Scholar]
- 3. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rydzanicz M, Wrzesiński T, Bluyssen H, Wesoły J. Genomics and epigenomics of clear cell renal cell carcinoma: recent developments and potential applications. Cancer Lett. 2013;341(2):111‐126. [DOI] [PubMed] [Google Scholar]
- 5. Barata P, Rini B. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507‐524. [DOI] [PubMed] [Google Scholar]
- 6. Hsieh J, Le V, Oyama T, Ricketts C, Ho T, Cheng E. Chromosome 3p loss‐orchestrated VHL, HIF, and epigenetic deregulation in clear cell renal cell carcinoma. J Clin Oncol. 2018;36(36):3533‐3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cantara WA, Crain PF, Rozenski J, et al. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195‐D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet. 2014;15(5):293‐306. [DOI] [PubMed] [Google Scholar]
- 9. Wang X, Zhao B, Roundtree I, et al. N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X, Lu Z, Gomez A, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang H, Weng H, Chen J. mA modification in coding and non‐coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37(3):270‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang M, Liu J, Zhao Y, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peng F, Xu J, Cui B, et al. Oncogenic AURKA‐enhanced N‐methyladenosine modification increases DROSHA mRNA stability to transactivate STC1 in breast cancer stem‐like cells. Cell Res. 2021;31(3):345‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen X, Xu MU, Xu X, et al. METTL14‐mediated N6‐methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klec C, Prinz F, Pichler M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol Oncol. 2019;13(1):46‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye J, Lin Y, Yu Y, Sun D. LncRNA NEAT1/microRNA‐129‐5p/SOCS2 axis regulates liver fibrosis in alcoholic steatohepatitis. J Transl Med. 2020;18(1):445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ning L, Li Z, Wei D, Chen H, Yang C. LncRNA, NEAT1 is a prognosis biomarker and regulates cancer progression via epithelial‐mesenchymal transition in clear cell renal cell carcinoma. Cancer Biomark. 2017;19(1):75‐83. [DOI] [PubMed] [Google Scholar]
- 18. Wen S, Wei Y, Zen C, Xiong W, Niu Y, Zhao Y. Long non‐coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6‐methyladenosine. Mol Cancer. 2020;19(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Q, Liu T, Bao YI, et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR‐27a‐3p/TXNIP pathway. Cancer Lett. 2020;469:68‐77. [DOI] [PubMed] [Google Scholar]
- 20. Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction between N‐methyladenosine (mA) modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xuan J‐J, Sun W‐J, Lin P‐H, et al. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46:D327‐D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Y, Zeng P, Li Y, Zhang Z, Cui Q. SRAMP: prediction of mammalian N6‐methyladenosine (m6A) sites based on sequence‐derived features. Nucleic Acids Res. 2016;44(10):e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allis C, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487‐500. [DOI] [PubMed] [Google Scholar]
- 24. Chen M, Wei L, Law C, et al. RNA N6‐methyladenosine methyltransferase‐like 3 promotes liver cancer progression through YTHDF2‐dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254‐2270. [DOI] [PubMed] [Google Scholar]
- 25. Wang J, Zhang C, He W, Gou X. Effect of mA RNA methylation regulators on malignant progression and prognosis in renal clear cell carcinoma. Front Oncol. 2020;10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Q, Zhang H, Chen Q, Wan Z, Gao X, Qian W. Identification of METTL14 in kidney renal clear cell carcinoma using bioinformatics analysis. Dis Markers. 2019;2019:5648783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma J, Yang F, Zhou C, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N ‐methyladenosine‐dependent primary MicroRNA processing. Hepatology. 2017;65(2):529‐543. [DOI] [PubMed] [Google Scholar]
- 28. Yang X, Zhang S, He C, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down‐regulating oncogenic long non‐coding RNA XIST. Mol Cancer. 2020;19(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao XU, Yang Y, Sun B‐F, et al. FTO‐dependent demethylation of N6‐methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403‐1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang B, Yang Y, Kang M, et al. mA demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF‐1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Wang F, Wang Z, et al. ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an mA‐dependent manner. Ann Transl Med. 2020;8(10):646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo T, Liu D, Peng S, Xu A. ALKBH5 promotes colon cancer progression by decreasing methylation of the lncRNA NEAT1. Am J Transl Res. 2020;12(8):4542‐4549. [PMC free article] [PubMed] [Google Scholar]
- 33. Hsu P, Zhu Y, Ma H, et al. Ythdc2 is an N‐methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27(9):1115‐1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang H, Weng H, Sun W, et al. Recognition of RNA N‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu X, Peng W‐X, Zhou H, et al. IGF2BP2 regulates DANCR by serving as an N6‐methyladenosine reader. Cell Death Differ. 2020;27(6):1782‐1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jin D, Guo J, Wu Y, et al. mA mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1‐miR‐1914‐3p‐YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37. Chen S, Zhou L, Wang Y. ALKBH5‐mediated mA demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S4
Table S1‐S3


