Summary
Microfluidic dielectrophoresis (DEP) technology has been applied to many devices to perform label-free target cell separation. Cells separated by these devices are used in laboratories, mainly for medical research. The present study designed a microfluidic DEP device to fabricate a rapid and semiautomated cell separation system in conjunction with microscopy to enumerate the separated cells. With this device, we efficiently segregated bacterial cells from liquid products and enriched one cell type from two mixed eukaryotic cell types. The device eliminated sample pretreatment and established cell separation by all-in-one operation in a lab-on-chip, requiring only a small sample volume (0.5–1 mL) to enumerate the target cells and completing the entire separation process within 30 min. Such a rapid cell separation technique is in high demand by many researchers to promptly characterize the target cells.
Subject areas: Biological sciences, Biological sciences research methodologies, Biology experimental methods, Biotechnology, Microbiological method
Graphical abstract
Highlights
-
•
A new all-in-one microfluidic dielectrophoresis integrated chip is fabricated
-
•
Simultaneous operation of buffer exchange and continuous cell separation on a chip
-
•
Chip’s cell separation performance is evaluated with bacterial and eukaryotic cells
Biological sciences; Biological sciences research methodologies; Biology experimental methods; Biotechnology; Microbiological method
Introduction
Cell separation from living organisms and environmental samples is essential for research, medicinal, and industrial purposes. For example, separating circulating tumor cells (CTCs) from the bloodstream, which contains various cell types, such as small platelets and sizable white blood cells, is vital to provide accurate diagnostics and prognostics (Allard et al., 2004; Allard and Terstappen, 2015; Balasubramanian et al., 2017). Separating stem cells and their derivatives is also crucial for developing stem cell therapies since these cells can repair and replace damaged cells after injury and disease (Hwang et al., 2009; Song et al., 2015).
Cell separation methods have traditionally been developed based on three principles: size, density, and antigens possessed by the cells. Cell sorting by size generally uses a filter with various pore sizes and materials, exemplified by leukocyte removal from whole blood (Dzik, 1993). A significant percentage of healthy blood donors carry Chlamydia pneumoniae in their blood, and polyester fiber-based filtration has been successfully used for the removal of C. pneumoniae from red blood cell units (Ikejima et al., 2005). Low cost and simplicity of operation are advantages for size-based cell separation, but the limitations are a lack of specificity and accuracy in separating target cells.
Cell sorting by density, such as density gradient centrifugation, provides higher accuracy than size-based cell separation; however, it requires a centrifuge and skilled laboratory personnel. This technique can successfully separate nucleated white blood cells from anucleated red blood cells (English and Andersen, 1974). The usefulness of density separation methods was also shown for CTC enrichment from whole blood (Rosenberg et al., 2002; Gascoyne and Shim, 2014) and neutrophil isolation from peripheral blood (Degel and Shokrani, 2010).
Antigen-based cell separation recognizes target cells explicitly to separate the target cells, and there are currently two major techniques, fluorescence-activated cell sorting and magnetic-activated cell sorting. These methods have been used in a variety of applications owing to their many advantages, including high antibody specificity and the ability to simultaneously separate multiple markers (Nicoletti et al., 1991; Miltenyi et al., 1990; Soeth et al., 1996; Wolff et al., 2003; Ferrari Belinda et al., 2004; Applegate et al., 2004). However, the cells obtained from these techniques are difficult to use in therapeutics because the antibody must be removed, requiring a complicated procedure that increases the chance of damaging the target cells and running up costs (Lee et al., 2016). Although some cell sorters with devised flow path shapes and separation techniques are commercially available, these devices have not yet resolved the issues caused by markers (On-chip Sort/On-chip Biotechnologies Co., Ltd., Cell Sorter MA900/Sony Biotechnology Inc.). Flow cytometry can sort cells with high sensitivity and accuracy, but it also requires fluorescent biomarkers and skilled personnel (Yamaguchi et al., 2011).
More easily operated devices for cell separation and quantification without using invasive markers such as antibodies are preferable. Thus, the concept of dielectrophoresis (DEP) came into place to satisfy these criteria. Dielectrophoresis, a phenomenon that induces motion of a suspended particle by applying a nonuniform electric field, has been studied aside from cell separation purposes (Hughes, 2016). The concept attracted attention in the cell separation field because it does not require biochemical labels or direct contact with the cell surfaces during the operation, reducing the chances of damaging cells (Sakamoto et al., 2005; Pethig, 2010). Instead, it uses the cells' capacitance to separate multicomplex cell groups in heterogeneous populations based on their unique dielectric properties. Since a DEP-based device was demonstrated to be able to separate dead and live yeast cells, the first separated biological cells (Pohl and Hawk, 1966; Pohl and Crane, 1972), DEP device manipulation has been exploited considerably as a noninvasive cell separation technique. Subsequently, a DEP-based device successfully separated cell chloroplasts and bacteria in the 1980s (Ting et al., 1971; Dimitrov et al., 1984), and more recently, a microfluidic DEP separated human mesenchymal stem cells and their differentiation progenies, osteoblasts (Adams et al., 2020; Song et al., 2015). Following this, Tada et al. (2017) separated live and dead human epithermal breast cells in a cell-separation DEP flow chamber. Thus, DEP-based devices have shown broad separation applications for mammalian cells, blood cells, cancer cells, human leukocytes, neural cells, and CTCs (Markx et al., 1996; Stephens et al., 1996; Becker et al., 1995; Morgan et al., 1999; Shim et al., 2013).
The drawbacks of current DEP-based cell sorting devices are that they require presample treatments such as buffer exchange because the sample cells are generally in a liquid with high conductivity, such as biological medium and saline, and these suspensions disrupt DEP operation (Faraghat et al., 2017). In addition, the currently available DEP-based devices cannot handle large volumes. Thus, the challenge is to improve DEP-based devices for rapid, easy, and scale-up cell sorting operations without damaging the cells.
The present study aimed to develop a microfluidic DEP device for cell sorting: a label-free and one-step operation, a lab-on-chip. This device uses an engineered microchannel integrated chip that operates the entire cell sorting process, including sample preparation, buffer exchange, and cell collection with an assigned volume. Because the system does not require sample pretreatment, such as buffer exchange, the time to process the cell sorting is significantly reduced. The one-step operation performs continuous cell separation, increasing the rate of rare cell collection. The device is designed to sort cells by size, which can simultaneously remove undesired matter, such as bacteria mixed in with the eukaryotic samples in the different fractions. Because the device is a label-free and one-step operation, cell damage during the operation is minimized. Our device is similar to what Park et al. (2019) introduced, a microfluidic DEP capable of replacing buffer by an integrated DLD (deterministic lateral displacement) module. The two devices differ in cell sorting systems, “batch” or “continuous” separation (Hughes, 2016). The device by Park et al. (2019) uses the former, where positive DEP traps one cell type while the other is repelled and collected, requiring a second step to recover the trapped cells (Hughes, 2016). I device by Park et al. is good to capture a single target cell using microwells, whereas our device aimed at continuous cell separation and collection using DEP. This system is more complicated than the former because the microfluidic flow must be adjusted for target cells to be directed to one of the multiple outputs (Hughes, 2016). However, this device can subsequently collect target cells from a larger volume since trapping cells is avoided. Furthermore, cell separation by size and electricity are simultaneously performed by a connected flow path on one plane so that the device can be easily assembled, which leads to an advantage in the manufacturing process and in terms of costs. We validated our device for accurate continuous cell separation using three different bacterial cells and two eukaryotic cells by (1) performance testing of size separation by using microparticles, (2) confirmation of the separation efficiency by using cultured bacteria, and (3) checking the electrical character of the eukaryotic cells and their separation from a cell mixture.
Working principles and chip fabrication
Working principles
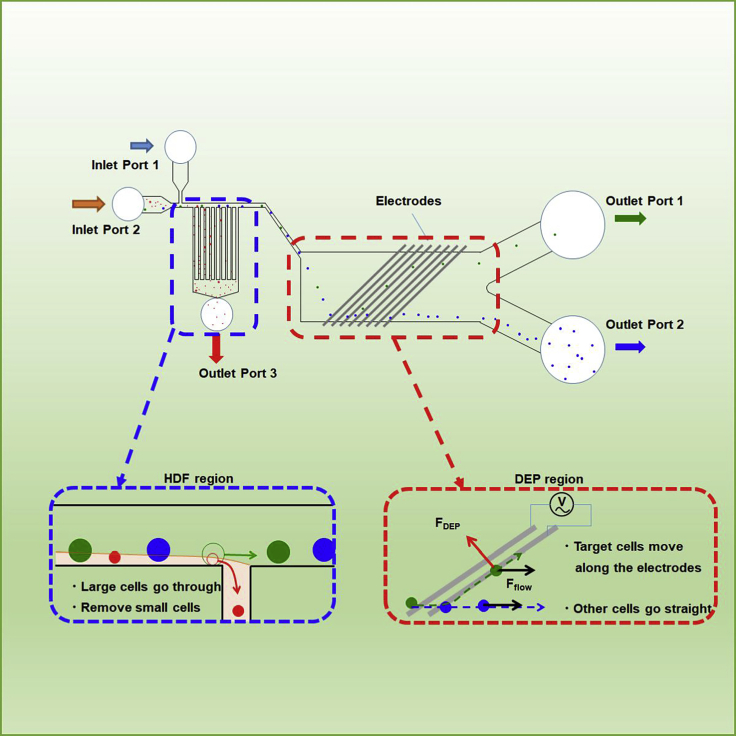
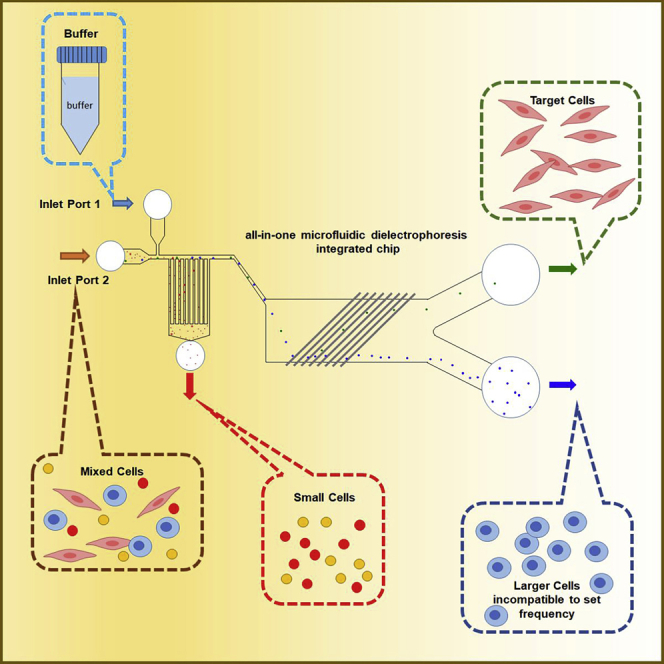
The schematic microfluidic dielectrophoresis integrated chip is shown in Figure 1. The chip contains two inlets, Inlet Port 1 and Inlet Port 2, in which DEP buffer and samples, respectively, are pumped in by a syringe at a constant flow rate (Figure 2). The chip consists of two cell sorting regions, the hydrodynamic filtration (HDF) region and the dielectrophoresis (DEP) region. In the HDF region, smaller cells and sample solvent move into Outlet Port 3, and larger cells and DEP buffer move to the DEP region from the main channel. The channel dimension of this region was designed as shown in Figure 3 based on the theory of hydrodynamics in microfluidic channel by Yamada and Seki (2005 and 2006). The branch channel width was determined by the ratio of the particle radius to the width of main channel. A certain percentage of sample solution and small particles are removed into a branch channel. By repeating this flow splitting, all sample fluid and small particles are removed into Outlet Port 3 in the principle. A series of inclined comb-shaped electrodes were placed in the DEP region. Because the DEP buffer replaces the sample solvent in the HDF region, the dielectrophoretic force can efficiently operate on cell sorting in the DEP region at a stable conductivity. When an AC voltage with a specific frequency for target cells is applied to the electrodes, the electrodes capture the target cells flowing from the HDF region into the DEP region by dielectrophoretic force and they are discharged to Outlet Port 1. The cells that do not respond to a specific frequency are not captured by the electrodes and are discharged to Outlet Port 2. The channels to Outlet Ports 1 and 2 are designed symmetrically. This structure delivers the same fluid volume into Outlet Port 1 and Outlet Port 2. Connecting the HDF and DEP regions in one flow path allows continuous separation of three or more different types of cells.
Figure 1.
Schematic function of the designed microfluidic dielectrophoresis integrated chip
The DEP buffer and sample are injected into Inlet Ports 1 and 2, respectively, while applying constant flow. The mixture of sample and DEP buffer flows into the HDF region where small particles are separated into Outlet Port 3. Larger cells move to the DEP region where the target cells are separated by DEP force into Outlet Port 1. The other large cells go through the electrodes and move into Outlet Port 2. The channels into Outlet Ports 1 and 2 from the DEP region were designed symmetrically to deliver the same fluid volume into Outlet Port 1 and Outlet Port 2.
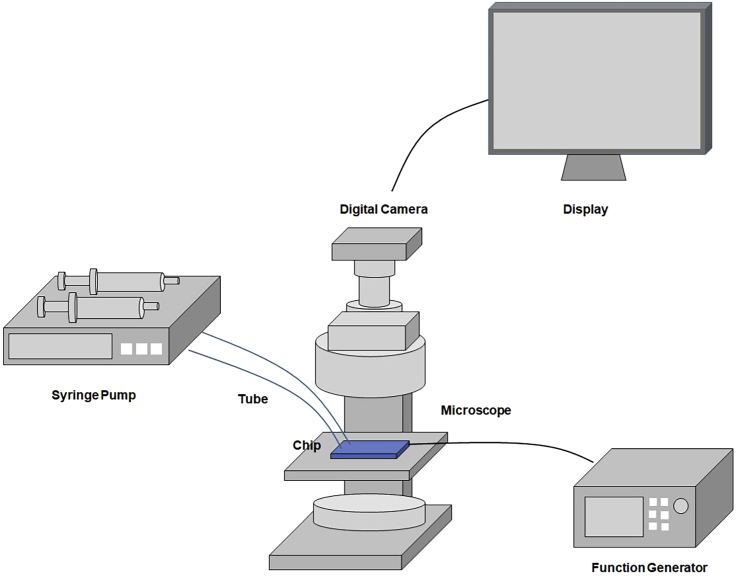
Figure 2.
Diagram of the microfluidic dielectrophoresis integrated chip for the separation experiments
Syringe pumps are used for the sample and buffer injections. These syringes are connected to Inlet Ports 1 and 2 of the designed chip. The function generator provides AC voltage to the electrode on the chip. The chip is observed by microscopy and the display. The movies are recorded by a digital camera to count the separated cell numbers at Outlet Ports 1 and 2.
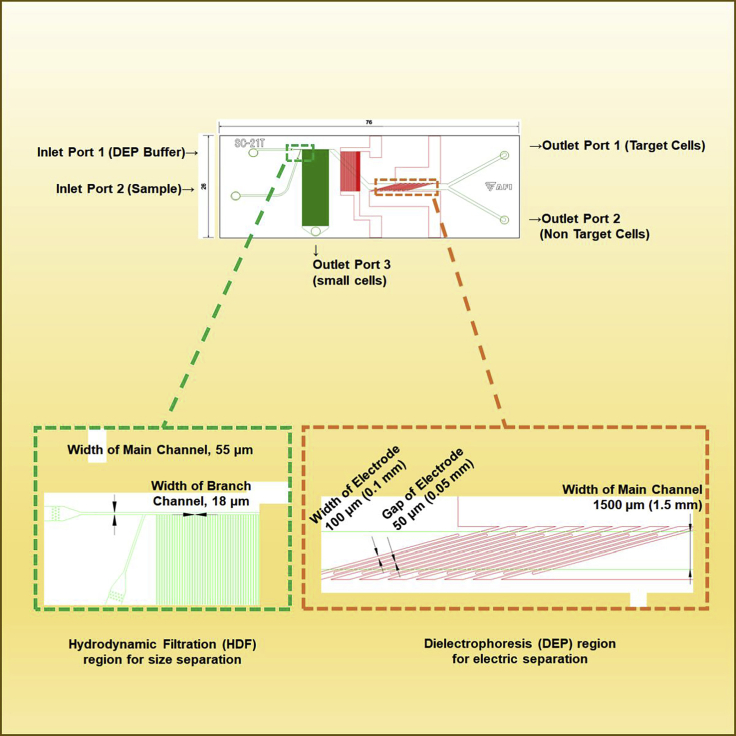
Figure 3.
Dimensions of the microfluidic dielectrophoresis integrated chip
The microfluidic channel is shown as a green line. The electrode pattern is shown as a red line. The minimum width of the main channel is 55 μm in the HDF region, and the maximum width is 1.5 mm in the DEP region. The branch channels (100 lines, 18 μm width) are connected to the main channel of the HDF region and Outlet Port 3. There are eight pairs of electrodes (0.1 mm width), and the gap of the electrodes is 50 μm in the DEP region. The electrodes incline 15° to the flow direction.
Chip fabrication
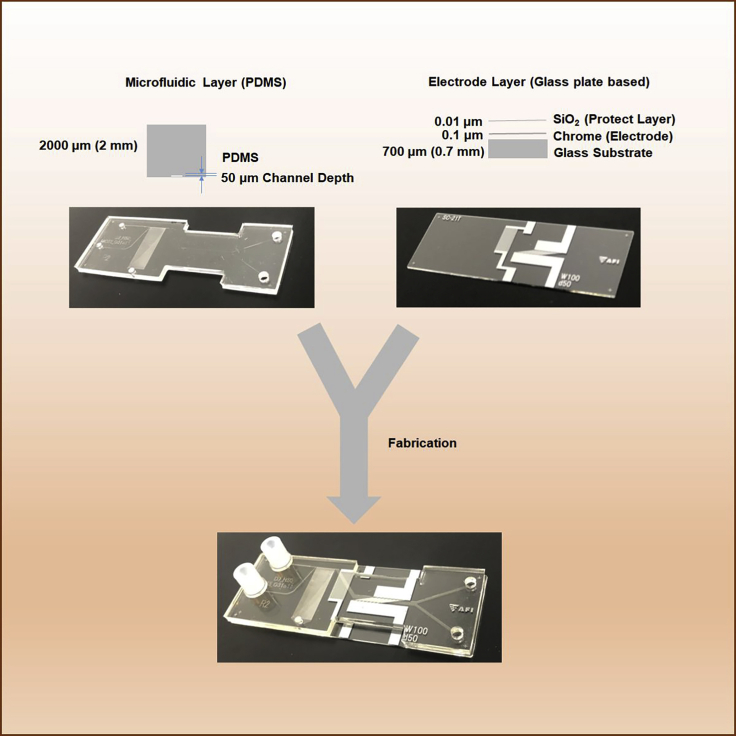
The microfluidic dielectrophoresis integrated chip was designed and manufactured for this study (Figure 4). The designed chip comprises two layers: the microfluidic channel layer and the electrode layer. The microfluidic layer was made of poly(dimethylsiloxane) with 2 mm thickness and 50 μm microchannel depth as follows: SU-8 was spin coated on a silicon wafer to a thickness of 50 μm. A channel mold was prepared by UV exposure and a channel-patterned photomask. The layer was prepared using poly(dimethylsiloxane) cast from the mold. The inlet and outlet in the layer were drilled at φ2 mm or φ3 mm, respectively. The electrode chip comprised three layers and was fabricated as follows: chromium (0.1 μm thick) was deposited by sputtering on a 0.7-mm-thick glass substrate. The electrode pattern was prepared by a standard photolithography method and wet etching. Silicon dioxide (0.01 μm thick) was sputtered as an electrode protection layer. The electrode chip and the microfluidic channel were treated with vacuum plasma equipment and bonded together.
Figure 4.
Structure of the microfluidic dielectrophoresis integrated chip
The chip comprises a microfluidic layer and an electrode layer. The microfluidic layer is made of poly(dimethylsiloxane) with 2 mm thickness and 50 μm microchannel depth. The electrode layer comprises three layers. On top of the glass substrate (0.7 mm thick), a 0.1-μm-thick electrode made of chromium is aligned, and the glass substrate and the chromium electrode are covered by a silicon dioxide protection layer (0.01 μm thick).
Results
Performance of size separation and solution replacement
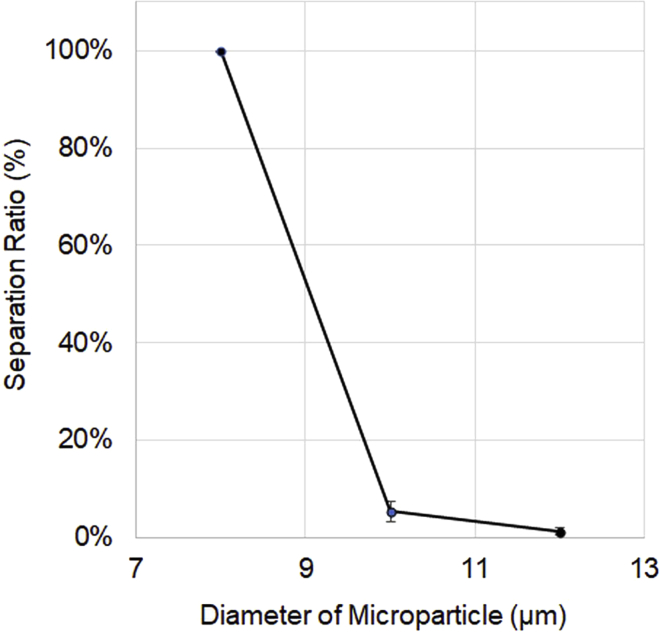
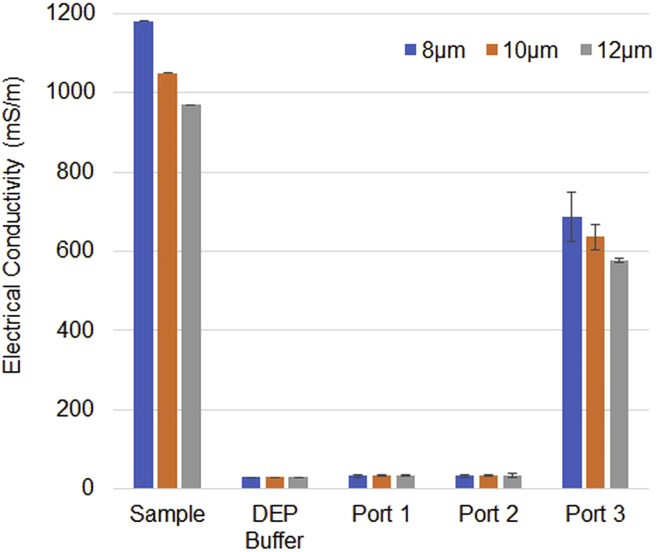
The performance of the size separation and buffer exchange in the HDF region was tested using polystyrene particle standards (diameter of 8, 10, 12 μm). More than 99.7% of the 8-μm-diameter particles were eluted from Outlet Port 3 (Figure 5). In contrast, only 3.46%–7.64% of the 10-μm particles and even fewer of the 12-μm particles (0.75%–1.96%) were eluted from Outlet Port 3, and most of them passed through the HDF region and were detected in the DEP region. The conductivity of these standards (polystyrene particle standard diameters of 8, 10, and 12 μm in PBS) was approximately 1,000 mS/m, which was reduced to the level of the DEP buffer conductivity (33.3–34.3 and 34.0–35.3 mS/m) at Outlet Port 1 and Outlet Port 2, respectively, showing no significant difference in conductivity at both Outlet Ports and the DEP buffer (Figure 6).
Figure 5.
Polystyrene particle standard separation at Outlet Port 3
Three different diameters of polystyrene particles (8, 10, 12 μm) were applied to the developed microfluid DEP cell separation system. The numbers show the percentage of particle counts that passed through Outlet Port 3 (n = 3).
Figure 6.
Performance of buffer replacement in the HDF region
The sample was a polystyrene microparticle suspension of three different diameters (8, 10, 12 μm). Each bar shows the average of the triplicate conductivity measurements with the SD at Outlet Ports 1, 2, and 3 compared with that of the sample suspension and the DEP buffer (n = 3).
Bacterial separation
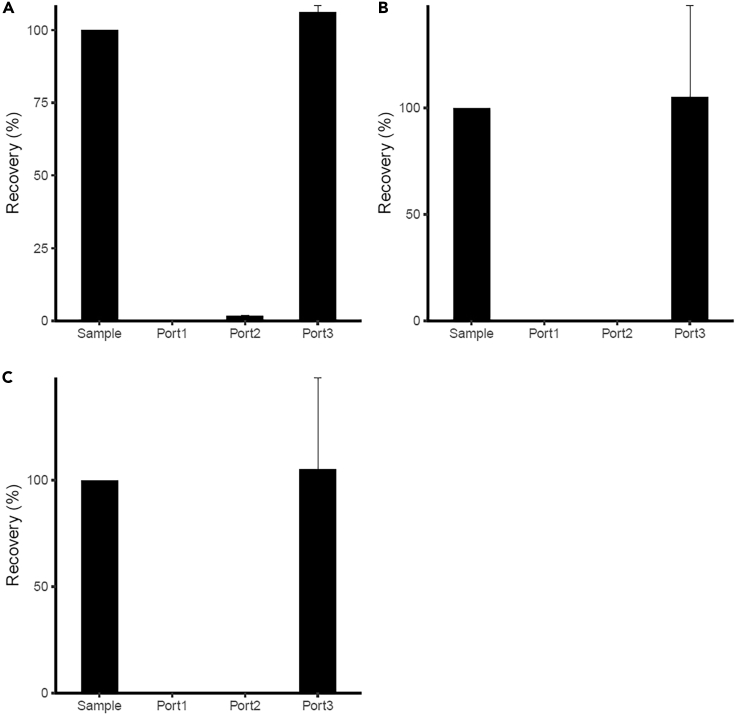
Each bacterial sample, Bacillus cereus, Staphylococcus aureus, and Escherichia coli, was individually applied to the microfluid DEP cell separation system, and the cell numbers in the starting sample and that eluted from the three outlets were estimated by colony forming units (CFUs) (Figure 7). For all three bacterial types, most cells eluted from Outlet Port 3. For B. cereus, the eluate from Outlet Port 1 did not grow colonies, whereas that from Outlet Port 2 formed 2% of colonies compared with the starting sample. For S. aureus and E. coli, no colony was formed from the eluate collected from Outlet Ports 1 and 2.
Figure 7.
Performance of bacteria separation in the HDF and DEP region
The bacterial cell numbers were estimated by colony forming units (CFU) from the sample from Outlet Ports 1, 2, and 3.
(A–C) (A) Bacillus cereus (n = 3), (B) Staphylococcus aureus (n = 4), and (C) Escherichia coli (n = 6).
Observation of separation in the HDF region
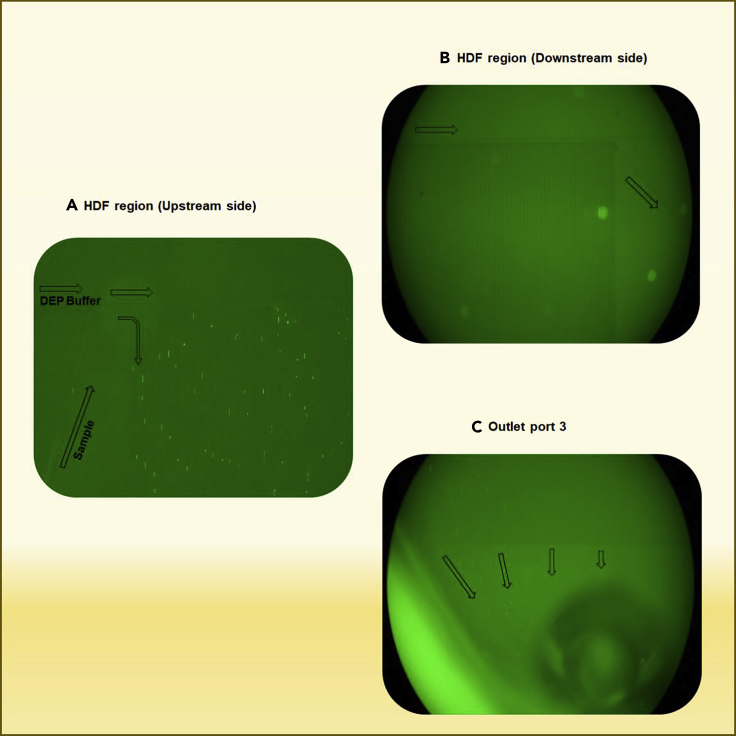
An image of E. coli separating into Outlet Port 3 via the HDF region is shown in Figure 8 and the movie clip (Video S1). When the E. coli sample was injected in Inlet Port 1, many cells immediately moved to the branch channels in the HDF region, which comprises 100 branch channels, to remove the small particles (Figure 8A). The E. coli cells passed through the branch channels and moved to Outlet Port 3 (Figure 8C). No cells were observed in the channel connected to the DEP region (Figure 8B).
Figure 8.
Separation of E. coli in the HDF region
(A–C) (A) bacteria flow into the branch channel on the upstream side of the HDF region, (B) no bacterial cells were observed in the channel connected to the DEP region, and (C) bacteria collected into Outlet Port 3 from the branch channels.
Eukaryotic cell separation
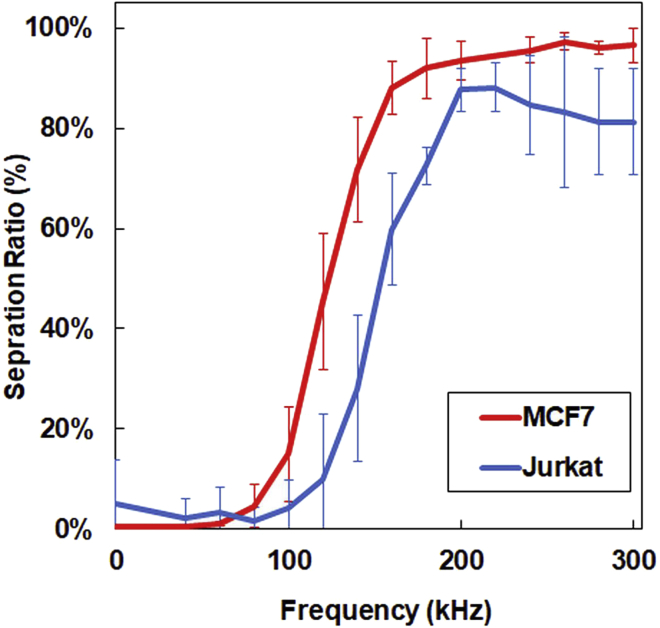
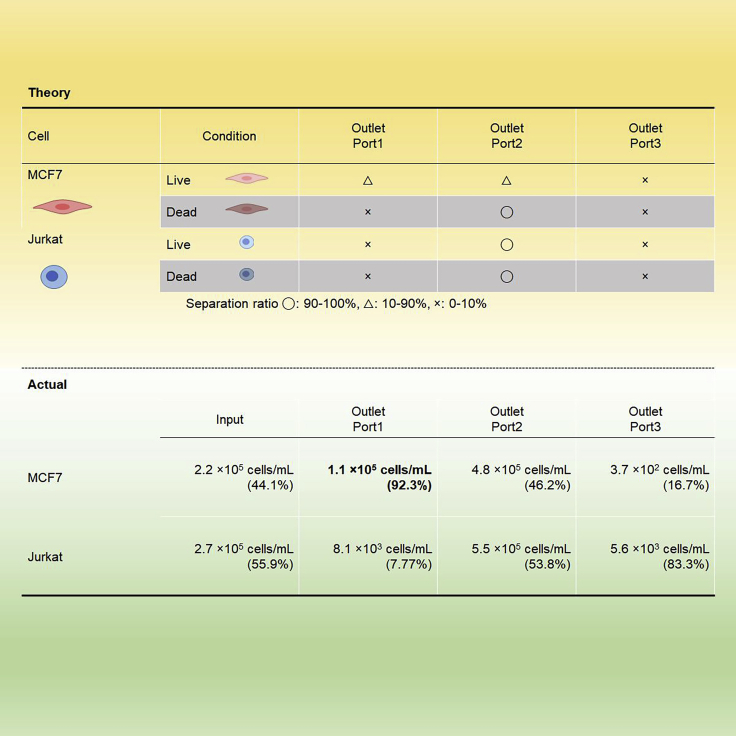
For both MCF7 and Jurkat cells, the higher the frequency of the AC voltage, the more the target cells that were collected (Figure 9). The separation rate for MCF7 started increasing at 80 kHz, reached 45% separation at 120 kHz, and plateaued at approximately 180 kHz. For Jurkat cells, only 10% separation was observed at 120 kHz, although its separation reached a plateau also at approximately 180 kHz. The mixture of MCF7 and Jurkat cells at similar counts (2.2 × 105 and 2.7 × 105 cells/mL, respectively) was processed with 14 Vpp and 120 kHz. In theory, MCF7 cells respond to 120 kHz and 45% of the total injected MCF7 cells should elute from Outlet Port 1. As Jurkat cells have poor response to 120 kHz, most Jurkat cells should elute in Outlet Port 2 (Figure 10, upper half). The result showed, of all cells eluted in Outlet Port 1, 92.3% (86.3%–97.2%) was MCF7 and 7.77% was Jurkat cells (Figure 10). The actual cell concentration of MCF7 at Outlet Port 1 was 1.1 × 105 cells/mL, the half of input.
Figure 9.
Separation of MCF7 and Jurkat cells at various frequencies of AC voltage
Separation ratio (%) = {Outlet Port 1/(Outlet Port 1 + 2)}× 100 (n = 3).
Figure 10.
Eukaryotic cell separation at 14 Vpp and 120 kHz
A mixture of equal counts of MCF7 and Jurkat cells was passed through the device. The upper half describes hypothetical separation based on Figure 9. The lower half shows actual cell concentration at each outlet port (n = 3). The numbers in the parentheses are the cell purity of each port.
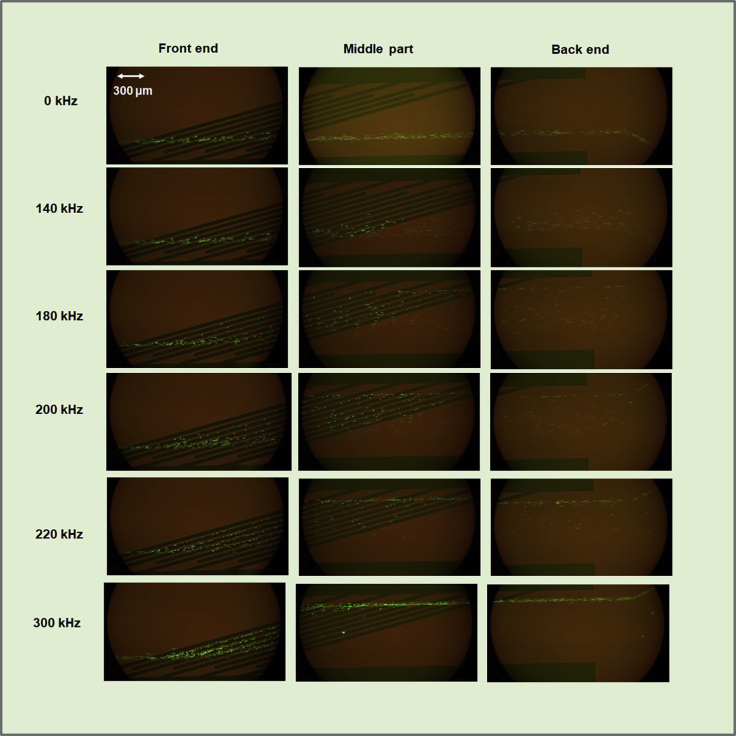
Observation of the separation in the DEP region
An image of Jurkat cells separated in the DEP region is shown in Figure 11 and in the movie clips (Videos S2, S3, and S4). When the frequency was 0 kHz (no voltage), the cells flowed straight into Outlet Port 2. When a frequency over 140 kHz was applied, the cells reacted to the frequency and moved to the electrodes. At a lower frequency (i.e., 140 kHz), the cells stopped moving at the middle point of the electrodes. When a higher frequency was applied, more cells were transferred into Outlet Port 1.
Figure 11.
Separation of Jurkat cells in the DEP region
The frequency was changed from 0 to 300 kHz at 14 Vpp. The cells were stained with fluorescent markers for observation. Scale bars are 300 μm.
Discussion
The present study fabricated a newly designed microfluidic DEP device to perform rapid and semiautomated cell separation and enumeration and it showed successful separation of polystyrene particle standards, bacterial cells, and eukaryotic cells. The study also demonstrated a selective separation of one type of eukaryotic cell from the binary cell culture by manipulating the frequency. The highlight of our chip is its ability of simultaneous operation of buffer exchange and continuous cell separation on a chip using DEP force. Each one of these technologies has been introduced in the past, but the combination of all in one is new. We believe that our chip can provide new values to users: not requiring pre-centrifugation of samples, reducing damages to samples, collecting a larger volume of separated cells, rapid cell enrichment, and all in one operation.
The present device installed the HDF region before the DEP region to elute small cells such as bacteria and erythrocytes in the former and transfer cells larger than 10 μm in the latter. The study confirmed the cell separation efficiency of each region. In the HDF region, the standard polystyrene particles with the smallest diameter (8 μm) were eluted from Outlet Port 3. All three bacterial cells, B. cereus, S. aureus, and E. coli, were mostly eluted from the HDF region to Outlet Port 3; all bacteria are smaller than 10 μm, B. cereus and E. coli are rod shaped and S. aureus is round (Turnbull 1981; Drobniewski, 1993). In the DEP region, standard polystyrene particles with 10 and 12 μm diameters were mostly transferred and eluted from Outlet Ports 1 and 2. When a small amount of larger particles enters the HDF region where the path is narrow, aggregation at the entrance of branch channels can occur, especially with poorly dispersed cell samples. Although we did not encounter this issue during this study, such cell stacking can be avoided by adequately distributing cells. Both mammalian cells (MCF7 and Jurkat) with average diameters of over 20 and 11 μm, respectively (Rosenbluth et al., 2006), were eluted from Outlet Ports 1 and 2. The port in the DEP region where these larger cells elute depends on the frequency of AC voltage applied. The large cells eluted from Outlet Port 2 when no voltage was applied, while the cells charged with dielectrophoretic force changed their migration direction in the electrodes toward Outlet Port 1. Therefore, this device can also be used for separating dead cells, which do not respond to electric charge, from live cells. By manipulating the frequency and using cell capacitance, this device is promising for broader applications, for example, removing small cells such as erythrocytes, bacteria, and toxins from whole blood cells.
Of all ingenuities our DEP device has, the best feature is one-step cell separation without the need for sample pretreatment (Table 1). Several label-free cell separation DEP devices have been previously developed (i.e., Song et al., 2015; Yoshioka et al., 2018). In fact, ApoStream (ApoCell, Inc., Houston, TX, USA) is a commercialized DEP-based cell separation product (Gupta et al., 2012). However, buffer exchange prior to sample loading is required for these devices. In pretreatment, the sample cells are centrifuged and suspended in osmotic-adjusted DEP buffer in a low conductivity medium, and thus, there is concern regarding how the physical force of centrifugation and the prolonged exposure to these buffers would affect the cell quality. Similar to Park et al. who first introduced a concept of buffer exchange on a chip in 2019, our device replaced sample media by buffer in the HDF region on the chip, showing equal osmolality for the buffer and the elution at Outlet Port 1 and Outlet Port 2, whereas the elute from Outlet Port 3 remained at the original sample’s osmolality. The difference in the two devices is that we aimed continuous cell separation using DEP, whereas Park et al. focused on capturing a single target cell by batch separation method. Because the present device is an all-in-one device, the time required for cells to migrate from the HDF region to the DEP region was only seconds, minimizing cell damage by shortening the buffer exchange duration. In fact, the entire cell separation process took only 30 min from sample loading to cell collection.
Table 1.
Comparison of Microfluidic dielectrophoresis integrated chip systems
| Manual buffer exchange | Separation by size | Cell position focusing | DEP force | Continuous separation | Novelty | Advantage | Disadvantage | |
|---|---|---|---|---|---|---|---|---|
| Vahey and Voldman (2008) | Required | No | Not required | Slanted (pDEP, nDEP) | Yes | Conductivity gradient is created in the chip. Cells are separated by slanted electrodes | Possible continuous separation of many types of cells | Samples need to be preadjusted to constant conductivity. Separation by size cannot be performed |
| Moon et al. (2011) | Required | Yes (DEP-MOFF) | Requirted | Slanted (pDEP) | Yes | MOFF using inertial force and its combination with slanted electrodes | Size separation | Samples need to be preadjusted to constant conductivity |
| Song et al. (2015) | Required | No | Not required | Slanted (pDEP) | Yes | Electricity is applied intermittently. Cell are separated by slanted electrodes | Possible continuous separation | Samples need to be preadjusted to constant conductivity. Separation by size cannot be performed |
| Park et al. (2019) | Not required | Yes (DEP-DLD) | Not required | Microwell array | No by batch | Separation with DEP array after adjusting the conductivity in the chip | Not requiring buffer exchange and size separation | Continuous separation cannot be performed |
| This study (2021) | Not required | Yes (DEP-HDF) | Not required | Slanted (pDEP) | Yes | Continuous separation with slanted electrodes after adjusting the conductivity in the chip | Not requiring buffer exchange and continuous size separation | Sample throughput need to be improved. HDF region need to be designed for adopting to another cell size |
Another feature of this device is its diagonal electrode alignment designed downflow the HDF region. In the DEP region, the voltage-charged cells change their migration direction in the diagonal electrodes and rapidly elute to Outlet Port 1. As a result, the time for cell exposure to the electric field is shortened. The importance and novelty of using slanted electrodes in microfluidic dielectrophoresis integrated chip systems was previously introduced by several groups (Vahey and Voldman, 2008; Moon et al., 2011; Song et al., 2015). Vahey and Voldman separated cells using p-DEP and n-DEP with conductivity gradient on the slanted electrodes. The slanted electrode by Moon et al. (2011) consists of multi-orifice flow fractionation (MOFF) for size separation and two DEP electrodes to ensure that cells flow to the focusing region before separation. Our device separates cells using only p-DEP in constant low conductivity without requiring focusing region before DEP separation, and these features are accommodated with automated buffer exchange in the HDF region. With the batch separation method, the trapped cells are exposed to electricity at all times, which may increase a chance of damaging the cells (Yoshioka et al., 2018).
An open outlet port is another advantage of this device. The cells are continuously and noninvasively collected from the outlet ports, and the collected cells can be used for characterization, diagnostics, medical use, and preparing cultures. The sample concentrations of both mammalian cells after injection were increased almost 2-fold at Outlet Port 2, indicating that the samples were successfully enriched through the device. In addition, target cells were successfully separated by manipulating the frequency.
Limitations of the study
The cell count recovery after passing through the device varied and sometimes fell below 50%, indicating a loss of target cells in the system. This is a current limitation of this device and is a future challenge for improvement. A possible cause for this phenomenon is cell adsorption on the surface of the flow path, perhaps due to the silicone used for the sample injection tube. The other cause may be cell stacking in the gap of flow path. We observed stacked cells at seams between the tube and inlet ports of chip. The tube material compatibility must be tested, and alternatives, low adsorption material tubes, or blocking reagents should be considered. The seams in chip also may be redesigned for considering for cell loss. Once the cell count recovery is improved, better dense-target cells can be obtained with this device. Throughput is another limitation for this device. In this study, MCF-7 of 2.2 × 105 cells/mL was delivered at 30 μL/min, accounting for a separation rate of 1.1 × 102 cells/s. A maximum concentration of 1 × 106 cells/mL of MCF-7 and Jurkat cells (separation rate of 5 × 102 cells/s) was tested without encountering a problem (data not shown). This throughput does not reach industrial standard, namely, the commercial product ApoStream system can separate 1 × 104 cells/s (6 × 105 cells/min) using the DEP system. No higher concentrations were tested in this study, and it is one of the subjects to be incorporated in the next generation of our chip. Addition of active pumping at ports may be one way to better control the flow rate.
Conclusions
The DEP-based cell separation device developed in this study is a rapid, label-free, all-in-one continuous cell separation device. This study verified that the device successfully functioned as a cell separation and concentration tool. We believe that our chip can provide new values to users and have a high potential contribution for study areas that require cell separation and concentration techniques such as cell research, medicine, and industrial purposes.
Declaration of patent
JP6441838 B2 (PCT/JP2017/002943)
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Bacillus cereus | ATCC | ATCC7004 |
| Staphylococcus aureus | ATCC | ATCC29213 |
| Escherichia coli | ATCC | ATCC25922 |
| Biological samples | ||
| MCF7 (human breast cancer cells) | ATCC | HTB-22 |
| Jurkat (human T cell leukemia) | RIKEN BioResouece Research Center, Ibaraki, Japan | RCB3052 |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 medium | Thermo Fisher Scientific, Waltham, MA | C11875500BT |
| 10% fetal bovine serum, | Thermo Fisher Scientific, Waltham, MA | 10437 |
| Penicillin-Streptomycin solution (×100) | FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan | 168-23191 |
| D-PBS(-) | FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan | 045-29795 |
| CROSSORTERTM Buffer | AFI Corporation, Kyoto, Japan | ECB-101 ECB-102 |
| CellTracker™ Red CMTPX | Thermo Fisher Scientific, Waltham, MA | C34552 |
| 4Na-EDTA solution | Fuji Film Wako Chemical Corporation, Osaka, Japan | 202–16931 |
| SYBR Green I | Lonza, Switzerland | 50513 |
| Other | ||
| SU-8 | Nippon Kayaku Co., Ltd., Tokyo, Japan | Resist 3050 |
| Poly (dimethylsiloxane), PDMS | Dow Corning Toray, Co. Ltd., Tokyo | Silpot 184 |
| nonalkali glass substrate | Nippon Electric Glass Co., Ltd., Shiga, Japan | OA-10G |
| vacuum plasma equipment | SAKIGAKE-Semiconductor Co., Ltd., Kyoto, Japan | YHS-R |
| Polystyrene microparticles | Thermo Fisher Scientific K.K., Tokyo, Japan | 4208A 8 μm 4210A 10 μm 4212A 12 μm, |
| syringe pumps | KD Scientific Inc., MA, USA | Legato111 |
| Arbitrary Function Generator | Tektronix, Tokyo, Japan | AFG1022 |
| optical microscope | Olympus, Tokyo, Japan | BX53 |
| digital camera | Sony, Tokyo, Japan | α7 |
| Syringe | Terumo, Tokyo, Japan | SS-01T |
| conductivity meter | HORIBA, Kyoto, Japan | B-771 |
| LUNA Automated Cell Counter | Logos Biosystems, South Korea | L10001 |
Resource availability
Lead contact
Further information and other requests should be directed to and will be fulfilled by the lead contact, Fumito Maruyama (fumito@hiroshima-u.ac.jp).
Materials availability
The developed microfluidic dielectrophoresis integrated chip in this study is available from AFI Corporation, Kyoto, Japan. CROSSORTERTM Buffer ECB-101 and ECB-102 are available from AFI Corporation, Kyoto, Japan
Experimental model and subject details
Polystyrene microparticles were used as standards to evaluate the performance of the HDF region. The particle concentration was adjusted to 106 particles/mL with Phosphate-Buffered Saline (PBS). The selectivity of the bacterial separation was evaluated using three types of bacteria with different sizes and shapes: Bacillus cereus (ATCC7004), Staphylococcus aureus (ATCC29213, MSSA), and Escherichia coli (ATCC25922), which were purchased from American Tissue Culture Collection. These bacteria were cultured on standard agar medium at 35 °C. The colonies were suspended in PBS at 105 ∼ 106 cells/mL. For evaluation of eukaryotic cell separation, MCF7 (human breast cancer cells) and Jurkat (human T cell leukemia) cells were used. MCF7 (HTB-22) was purchased from American Tissue Culture Collection, and Jurkat cells (RCB3052) was purchased from RIKEN BioResouece Research Center (Ibaraki, Japan). These cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in a 5% CO2 atmosphere. The sample concentration was adjusted to approximately 106 cells/mL with culture medium.
Method details
Separation setup
The cell sorting experiments used the designed chip, syringe pumps, Arbitrary Function Generator, optical microscope, and a digital camera. Two types of CROSSORTERTM Buffer were used as the DEP Buffer. Two syringes, one filled with 1 mL DEP buffer and the other with 0.5–1 mL sample, were prepared for Inlet Port 1 and Inlet Port 2, respectively. Each syringe was connected to the inlet through a polytetrafluoroethylene (PTFE) tube, and the flow path was filled with DEP buffer to keep out air. An arbitrary function generator was connected to the electrodes of the designed chip, and an AC voltage of 8∼20 Vpp and a frequency of 100 kHz–10 MHz were applied. Both the sample and DEP buffer were delivered to the designed chip at a flow rate of 30 μL/min using a syringe pump. The cell separation stage was monitored with an optical microscope, and the number of cells moving to Outlet Ports 1 and 2 was measured using a video captured by a digital camera. The number of cells separated into Outlet Port 3 was measured using a hemocytometer. The conductivity of the liquid collected from each port was measured with a conductivity meter.
Performance of size separation in the HDF region
Polystyrene microparticle standards (diameter of 8–12 μm) were used as samples. The DEP buffer (ECB-101) and the sample were loaded on the systems through Inlet Ports 1 and 2, respectively. The flow was set at 30 μL/min for 15 minutes without applying a voltage to the electrodes. The number of separated particles was counted by the LUNA Automated Cell Counter.
Bacterial separation
Each of the three types of bacteria was used independently as a sample. The DEP buffer (ECB-102) and 500 μL of the sample were loaded on the systems through Inlet Ports 1 and 2, respectively. The flow was set at 30 μL/min for 16.7 minutes with an AC voltage of 12 Vpp and 2,000 kHz frequency (the preliminary testing selected these conditions). The liquid flow-through from all three Outlet Ports was diluted with PBS at an appropriate ratio and cultured on standard agar medium at 35 °C for 20 hours. The number of colonies was counted to calculate the number of living bacteria.
Eukaryotic cell separation
MCF7 and Jurkat cells were independently applied to the designed chip. The suitable frequency to be used in the DEP region for each cell was examined. DEP buffer (ECB-101) and 500–1000 μL of the sample were loaded on the systems through Inlet Ports 1 and 2, respectively. The flow was set at 30 μL/min with a constant AC voltage of 14 Vpp and a variable frequency between 0 and 300 kHz (intervals 20 kHz). Separation by the device was conducted for 1 minute at each frequency. The cell separation ratios at Outlet Ports 1 and 2 were monitored by video. The cell separation ratio was calculated by the cell numbers eluted into Outlet Port 1 over the total number of cells applied to the system at each frequency.
Cell separation from binary eukaryotic cell cultures
MCF7 and Jurkat cells were stained first. The final concentration of 1 μM CellTracker™ Green CMFDA was added to the Jurkat cell culture and incubated for 20 min at 37 °C. After washing the cells twice with medium, the stained cells were measured by a LUNA Automated Cell Counter. A final concentration of 1 μM CellTracker™ Red CMTPX was added to the MCF7 cell culture and incubated for 20 min at 37 °C. After washing the cells with PBS, 100 μL of 0.05 w/v% trypsin and 0.53 mmol/L 4Na-EDTA solution were added and incubated for 5 min at 37 °C. The cells were dispersed into 1 mL medium for cell counting. Each cell concentration was adjusted to 2.5 × 105 cells/mL by the respective media. Binary cell culture was prepared from stained MCF7 and Jurkat cells. DEP buffer and binary cell culture were applied to the respective inlet ports with a flow rate of 30 μL/min, and cell separation was performed with a constant AC voltage of 14 Vpp and a frequency of 120 kHz. After separation, the cell concentration of each outlet port was recorded.
Observation of cell movement in the HDF and DEP regions
Escherichia coli (ATCC25922) at 108 cells/mL in PBS was stained with SYBR Green Ⅰ. After staining for 2 hours at room temperature, the cells were washed twice and diluted to 106 cells/mL with PBS. A flow rate of 30 μL/min was too fast to observe cell movement in the HDF region, and thus, a flow rate of 0.3 μL/min was used. Stained Jurkat cells were used to observe the cell movement in the DEP region. A cell concentration of 5 × 104 cells/mL, flow rate of 30 μL/min, and a voltage of 14 Vpp were used for Jurkat cells.
Quantification and statistical analysis
The number of separated polystyrene microparticle standards was counted by the LUNA Automated Cell Counter. The number of separated bacteria was counted by colony count on agar plates. Eukaryotic cells were stained first and then the separated cell were counted by by the LUNA Automated Cell Counter. Cell separation ratio was calculated by the cell numbers eluted into Outlet Port 1 over the total number of cells applied to the system. All experiments were performed triplicate or above replicates and mean and standard deviation were calculated using EXCEL calculation function.
Acknowledgment
The final manuscript was edited by a professional editor from American Journal Experts (AJE). The work done by K.Y., S.F., K.K., and F.M. was supported by a grant (JPMJSA1705) for a study on Science and Technology Research Partnership for Sustainable Development-Monitoring Algae in Chile (SATREPS-MACH).
Author contributions
All authors reviewed the manuscript and approved the final article. In addition, each author undertook the following responsibilities: K.O., Y.W., M.T., and T.I. designed and engineered the device. K.O. performed the experimental design, data collection, and writing of the manuscript. H.O. contributed by supervising K.O. K.K. and T.I. performed the laboratory experiments. K.Y. and S.F. contributed to the experimental design, manuscript writing, and editing. F.M. is the project leader who supervised the project.
Declaration of interests
We have no financial interest to declare. It should be noted that the authors K.O., Y.W., M.T., and T.I. are employees of AFI Corporation.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103776.
Data and code availability
-
•
All data produced in this study are included in the published article and its supplemental information, or are available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams T.N.G., Jiang A.Y.L., Mendoza N.S., Ro C.C., Lee D.H., Lee A.P., Flanagan L.A. Label-free enrichment of fate-biased human neural stem and progenitor cells. Biosens. Bioelectron. 2020;152:111982. doi: 10.1016/j.bios.2019.111982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard W.J., Matera J., Miller M.C., Repollet M., Connelly M.C., Rao C., Tibbe A.G.J., Uhr J.W., Terstappen L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- Allard W.J., Terstappen L.W.M.M. CCR 20th anniversary commentary: paving the way for circulating tumor cells. Clin. Cancer Res. 2015;21:2883–2885. doi: 10.1158/1078-0432.CCR-14-2559. [DOI] [PubMed] [Google Scholar]
- Applegate R.W., Squier J., Vestad T., Oakey J., Marr D.W.M. Optical trapping, manipulation, and sorting of cells and colloids in microfluidic systems with diode laser bars. Opt. Express. 2004;12:4390–4398. doi: 10.1364/OPEX.12.004390. [DOI] [PubMed] [Google Scholar]
- Balasubramanian P., Kinders R.J., Kummar S., Vishal Gupta V., Hasegawa D., Menachery A., Lawrence S.M., Wang L., Ferry-Galow K., Davis D., et al. Antibody-independent capture of circulating tumor cells of non-epithelial origin with the ApoStream® system. PLoS One. 2017;12:e0175414. doi: 10.1371/journal.pone.0175414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker F.F., Wang X.B., Huang Y., Pethig R., Vykoukal J., Gascoyne P.R. Separation of human breast cancer cells from blood by differential dielectric affinity. Proc. Natl. Acad. Sci. U S A. 1995;92:860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degel J., Shokrani M. Validation of the efficacy of a practical method for neutrophils isolation from peripheral blood. Clin. Lab Sci. 2010;23:94–98. doi: 10.29074/ascls.23.2.94. [DOI] [PubMed] [Google Scholar]
- Dimitrov D.S., Tsoneva J., Stoicheva N., Zhelev D. An assay for dielectrophoresis: applications to electromagnetically induced membrane adhesion and fusion. J. Biol. Phys. 1984;12:26–30. doi: 10.1007/BF01857654. [DOI] [Google Scholar]
- Drobniewski F.A. Bacillus cereus and related species. Clin. Microbiol. Rev. 1993;6:324–338. doi: 10.1128/cmr.6.4.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzik S. Leukodepletion blood filters: filter design and mechanisms of leukocyte removal. Transfus. Med. Rev. 1993;7:65–77. doi: 10.1016/s0887-7963(93)70125-x. [DOI] [PubMed] [Google Scholar]
- English D., Andersen B. Single-step separation of red blood cells, granulocytes and mononuclear leukocytes on discontinuous density gradients of Ficoll-Hypaque. J. Immunol. Methods. 1974;5:249–252. doi: 10.1016/0022-1759(74)90109-4. [DOI] [PubMed] [Google Scholar]
- Faraghat S.A., Hoettges K.F., Steinbach M.K., van der Veen D.R., Brackenbury W.J., Henslee E.A., Labeed F.H., Hughes M.P. High-throughput, low-loss, low-cost, and label-free cell separation using electrophysiology-activated cell enrichment. Proc. Natl. Acad. Sci. U S A. 2017;114:4591–4596. doi: 10.1073/pnas.1700773114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari Belinda C., Oregaard G., Sørensen S.J. Recovery of GFP-labeled bacteria for culturing and molecular analysis after cell sorting using a benchtop flow cytometer. Microb. Ecol. 2004;48:239–245. doi: 10.1007/s00248-003-1069-9. [DOI] [PubMed] [Google Scholar]
- Gascoyne P.R.C., Shim S. Isolation of circulating tumor cells by dielectrophoresis. Cancers. 2014;6:545–579. doi: 10.3390/cancers6010545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Jafferji I., Garza M., Melnikova V.O., Hasegawa D.K., Pethig R., Davis D.W. ApoStream™, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood Dielectrophoresis of chloroplasts. Biomicrofluidics. 2012;6:024133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M.P. Fifty years of dielectrophoretic cell separation technology. Biomicrofluidics. 2016;10:032801. doi: 10.1063/1.4954841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang N.S., Zhang C., Hwang Y.S., Varghese S. Mesenchymal stem cell differentiation and roles in regenerative medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:97–106. doi: 10.1002/wsbm.26. [DOI] [PubMed] [Google Scholar]
- Ikejima H., Friedman H., Leparc G.F., Yamamoto Y. Depletion of resident Chlamydia pneumoniae through leukoreduction by filtration of blood for transfusion. J. Clin. Microbiol. 2005;43:4580–4589. doi: 10.1128/JCM.43.9.4580–4584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Hwang B., Kim B. The potential of a dielectrophoresis activated cell sorter (DACS) as a next generation cell sorter. Micro Syst. Lett. 2016;4 doi: 10.1186/s40486-016-0028-4. [DOI] [Google Scholar]
- Markx G.H., Dyda P.A., Pethig R. Dielectrophoretic separation of bacteria using a conductivity gradient. J. Biotechnol. 1996;51:175–180. doi: 10.1016/0168-1656(96)01617-3. [DOI] [PubMed] [Google Scholar]
- Miltenyi S., Müller W., Weichel W., Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- Moon H.-S., Kwon K., Kim S.-I., Han H., Sohn J., Lee S., Jung H.-I. Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP) Lab Chip. 2011;11:1118–1125. doi: 10.1039/c0lc00345j. [DOI] [PubMed] [Google Scholar]
- Morgan H., Hughes M.P., Green N.G. Separation of submicron bioparticles by dielectrophoresis. Biophys. J. 1999;77:516–525. doi: 10.1016/S0006-3495(99)76908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti I., Migliorati G., Pagliacci M.C., Grignani F., Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods. 1991;139:271–279. doi: 10.1016/0022-1759(91)90198-O. [DOI] [PubMed] [Google Scholar]
- Park J., Komori T., Uda T., Miyajima K., Fujii T., Kim S.H. Sequential cell-processing system by integrating hydrodynamic purification and dielectrophoretic trapping for analyses of suspended cancer cells. Micromachines (Basel) 2019;11:47. doi: 10.3390/mi11010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pethig R. Review article-dielectrophoresis: status of the theory, technology, and applications. Biomicrofluidics. 2010;4:022811. doi: 10.1063/1.3456626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl H.A., Crane J.S. Dielectrophoretic force. J. Theor. Biol. 1972;37:1–13. doi: 10.1016/0022-5193(72)90112-9. [DOI] [PubMed] [Google Scholar]
- Pohl H.A., Hawk I. Separation of living and dead cells by dielectrophoresis. Science. 1966;152:647–649. doi: 10.1126/science.152.3722.647-a. [DOI] [PubMed] [Google Scholar]
- Rosenberg R., Gertler R., Friederichs J., Fuehrer K., Dahm M., Phelps R., Thorban S., Nekarda H., Siewert J.R. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry. 2002;49:150–158. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- Rosenbluth M.J., Lam W.A., Fletcher D.A. Force microscopy of nonadherent cells: a comparison of leukemia cell deformability. Biophys. J. 2006;90:2994–3003. doi: 10.1529/biophysj.105.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto C., Yamaguchi N., Nasu M. Rapid and simple quantification of bacterial cells by using a microfluidic device. Appl. Environ. Microbiol. 2005;71:1117–1121. doi: 10.1128/aem.71.2.1117-1121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Stemke-Hale K., Noshari J., Becker F.F., Gascoyne P.R.C. Dielectrophoresis has broad applicability to marker-free isolation of tumor cells from blood by microfluidic systems. Biomicrofluidics. 2013;7:011808. doi: 10.1063/1.4774307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeth E., Röder C., Juhl H., Krüger U., Kremer B., Kalthoffet H. The detection of disseminated tumor cells in bone marrow from colorectal-cancer patients by a cytokeratin-20-specific nested reverse-transcriptase-polymerase-chain reaction is related to the stage of disease. Int. J. Cancer. 1996;69:278–282. doi: 10.1002/(SICI)1097-0215(19960822)69:4<278::AID-IJC7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Song H., Rosano J.M., Wang Y., Garson C.J., Prabhakarpandian B., Pant K., Klarmann G.J., Perantoni A., Alvarez L.M., Lai E. Continuous-flow sorting of stem cells and differentiation products based on dielectrophoresis. Lab Chip. 2015;15:1320–1328. doi: 10.1039/c4lc01253d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Talary M.S., Pethig R., Burnett A.K., Mills K.I. The dielectrophoresis enrichment of CD34+ cells from peripheral blood stem cell harvests. Bone Marrow Transplant. 1996;18:777–782. [PubMed] [Google Scholar]
- Tada S., Hayashi M., Eguchi M., Tsukamoto A. High-throughput separation of cells by dielectrophoresis enhanced with 3D gradient AC electric field. Biomicrofluidics. 2017;11:064110. doi: 10.1063/1.5007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting I.P., Jolley K., Beasley C.A., Pohl H.A. Dielectrophoresis of chloroplasts. Biochim. Biophys. Acta. 1971;234:324–329. doi: 10.1016/0005-2728(71)90198-8. [DOI] [PubMed] [Google Scholar]
- Turnbull P.C. Bacillus cereus toxins. Pharmacol. Ther. 1981;13:453–505. doi: 10.1016/0163-7258(81)90026-7. [DOI] [PubMed] [Google Scholar]
- Vahey M.D., Voldman J. An equilibrium method for continuous-flow cell sorting using dielectrophoresis. Anal. Chem. 2008;80:3135–3143. doi: 10.1021/ac7020568. [DOI] [PubMed] [Google Scholar]
- Wolff A., Perch-Nielsen R., Larsen U.D., Friis P., Goranovic G., Poulsen C.R., Kutter J.P., Telleman P. Integrating advanced functionality in a microfabricated high-throughput fluorescent-activated cell sorter. Lab Chip. 2003;3:22–27. doi: 10.1039/b209333b. [DOI] [PubMed] [Google Scholar]
- Yamada M., Seki M. Hydrodynamic filtration for on-chip particle concentration and classification utilizing microfluidics. Lab Chip. 2005;5:1233–1239. doi: 10.1039/b509386d. [DOI] [PubMed] [Google Scholar]
- Yamada M., Seki M. Microfluidic particle sorter employing flow splitting and recombining. Anal. Chem. 2006;78:1357–1362. doi: 10.1021/ac0520083. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N., Torii M., Uebayashi Y., Nasu M. Rapid, semiautomated quantification of bacterial cells in freshwater by using a microfluidic device for on-chip staining and counting. Appl. Environ. Microbiol. 2011;77:1536–1539. doi: 10.1128/aem.01765-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka J., Ohsugi Y., Yoshitomi T., Yasukawa T., Sasaki N., Yoshimoto K. Label-free rapid separation and enrichment of bone marrow-derived mesenchymal stem cells from a heterogeneous cell mixture using a dielectrophoresis device. Sensors. 2018;18:3007. doi: 10.3390/s18093007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data produced in this study are included in the published article and its supplemental information, or are available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.