Abstract
We developed a rapid thermocycling, real-time detection (also known as real-time PCR) method for the detection of Legionella species directly from clinical specimens. This method uses the LightCycler (Roche Molecular Biochemicals, Indianapolis, Ind.) and requires approximately 1 to 2 h to perform. Both a Legionella genus PCR assay and Legionella pneumophila species-specific PCR assay were designed. A total of 43 archived specimens from 35 patients were evaluated, including 19 bronchoalveolar lavage (BAL) specimens and 24 formalin-fixed, paraffin-embedded open lung biopsy specimens. Twenty-five of the specimens were culture-positive for Legionella (9 BAL specimens and 16 tissue specimens). BAL specimens were tested by LightCycler PCR (LC-PCR) methods and by a direct fluorescent antibody (DFA) assay, which detects L. pneumophila serogroups 1 to 6 and several other Legionella species. Tissue sections were tested by the two LC-PCR methods, by DFA, by an in situ hybridization (ISH) assay, specifically designed to detect L. pneumophila, and by Warthin-Starry (WS) staining. The results were compared to the “gold standard” method of bacterial culture. With BAL specimens the following assays yielded the indicated sensitivities and specificities, respectively: Legionella genus detection by Legionella genus LC-PCR, 100 and 100%; Legionella genus detection by DFA assay, 33 and 100%; and L. pneumophila detection by L. pneumophila species-specific LC-PCR, 100 and 100%. With open lung biopsy specimens the following assays yielded the indicated sensitivities and specificities, respectively: Legionella genus detection by LC-PCR 68.8 and 100%; Legionella genus detection by DFA assay, 44 and 100%; Legionella genus detection by WS staining, 63 and 100%; L. pneumophila species-specific detection by LC-PCR, 17 and 100%; and L. pneumophila species-specific detection by ISH, 100 and 100%. The analytical sensitivity of both LC-PCR assays was <10 CFU/reaction. LC-PCR is a reliable method for the direct detection of Legionella species from BAL specimens. The Legionella genus LC-PCR assay could be performed initially; if positive, L. pneumophila species-specific LC-PCR could then be performed (if species differentiation is desired). The speed with which the LC-PCR procedure can be performed offers significant advantages over both culture-based methods and conventional PCR techniques. In contrast, for the methods evaluated, culture was the best for detecting multiple Legionella species in lung tissue. WS staining, Legionella genus LC-PCR, and L. pneumophila species-specific ISH were useful as rapid tests with lung tissue.
The genus Legionella, family Legionellaceae, includes over 40 different species of fastidious gram-negative bacilli, with over 60 described serogroups (2, 21, 41, 60). While these organisms represent normal environmental flora, many have been shown to cause human disease, most commonly opportunistic pneumonia in immunocompromised patients. The vast majority of such cases (approximately 85%) are due to L. pneumophila, with a substantial minority due to other species, most commonly L. micdadei, L. bozemanii, L. dumoffii, and L. longbeachae (17, 41, 47). Legionella pneumonia can be community acquired or nosocomial and sporadic or epidemic in nature. Pulmonary infection may be subclinical or severe and life threatening. The fatality rate can approach 50% in immunocompromised patients (60). The organism often responds to antimicrobial therapy, usually with macrolides, and clinical responses usually occur within 3 to 5 days. The latter fact, combined with clinical and radiographic features that are often nonspecific, serves to underscore the value of a prompt and accurate laboratory diagnosis.
The diagnosis of Legionella infection can be made from a number of specimen types and by a number of testing modalities. Bacterial culture of bronchoscopy or lung biopsy specimens remains the most sensitive means of detection (7, 11, 61). Specialized growth medium, such as buffered charcoal-yeast extract agar (BCYEα), is required, with up to 2 weeks of incubation recommended to ensure maximal recovery (11). Isolates are typically identified by a combination of colony and Gram stain morphology, with serologic confirmation and species identification, using specific fluorescein-labeled antibodies. Direct detection of organisms in uncultured clinical specimens, usually performed with immunofluorescent methods, is much more rapid than culture, but the sensitivity of these methods has been reported to be poor (11, 12, 56). A variety of means, including radioimmunoassay, enzyme immunoassay, and latex agglutination, can be used to detect a soluble polysaccharide antigen of L. pneumophila (serogroup 1 only), in urine, with a reported sensitivity of 55 to 90% (8–10, 25, 48, 56). Serologic methods are highly sensitive (56, 60), but their utility is generally limited to epidemiologic studies, due to the time lag needed to detect seroconversion. A number of methods have been used in attempt to identify these organisms in paraffin-embedded tissue sections, including various histochemical and immunohistochemical techniques (3–5, 50, 52). Silver impregnation stains, such as the Warthin-Starry (WS) stain (39), serve as the current mainstay of detection in such cases.
Assays based on molecular diagnostic techniques have included DNA probes for in situ hybridization (ISH), as well as PCR-based methods. Probes have largely been directed against rRNA sequences, with sensitivities of approximately 30 to 75% in both bronchoalveolar lavage (BAL) and fixed tissue specimens (11, 13, 14, 16, 18, 19, 45, 55). PCR methodology has been used primarily against the 5S and 16S rRNA genes and against the macrophage infectivity potentiator (mip) gene of L. pneumophila. The latter amplification assays have been utilized for detection of Legionella species in environmental specimens, serum, urine, throat swabs, and BAL specimens (1, 6, 22, 24, 27, 28, 32, 33, 35–38, 40, 42, 46, 49, 53). Several studies have shown 100% sensitivity when such methods are used on BAL specimens. A few investigators have suggested that PCR may exceed culture in its ability to detect Legionella in these specimens (6, 22, 24, 33). To our knowledge, no studies exist that examine the sensitivity of these techniques in tissue specimens.
Conventional molecular methods, used in the above-noted studies, require PCR-based amplification, followed by probe hybridization detection (24). These methods are labor intensive and frequently require at least 1 day to perform. Additionally, the required manipulation of postamplification products increases the risk of carryover contamination and resultant false positivity.
By using commercially available, rapid cycle, real-time PCR instrumentation (LightCycler; Roche Molecular Biochemicals, Indianapolis, Ind.), PCR amplification and detection can be combined in a single closed cuvette, with dramatically reduced cycling time (58). This method obviates the need for further manipulation of the specimen, greatly reduces turnaround time, and diminishes the risk of cross-contamination between samples (54, 57). As recently demonstrated (15, 26, 31, 44, 59), this and other real-time PCR methods are attractive alternatives to conventional PCR techniques in the clinical laboratory.
We describe the development of a real-time LightCycler PCR (LC-PCR) method, for the detection of Legionella species directly from clinical specimens. Both a Legionella genus LC-PCR assay and an L. pneumophila species-specific LC-PCR assay were designed. With conventional culture serving as the “gold standard,” the results of LC-PCR were compared to direct fluorescent antibody (DFA) assay for the detection of Legionella species in BAL specimens, and to DFA assay, ISH, and WS staining for the detection of Legionella species in open lung biopsy specimens.
MATERIALS AND METHODS
Control organisms.
All experiments to optimize PCR conditions, as well as dilution studies to evaluate sensitivity and plasmid construction, were performed using L. pneumophila serogroup 1 (ATCC 33152). Other strains of Legionella, used for validation of the assay, are listed in Table 1, and included L. pneumophila serogroups 1 to 6, as well as several other strains of Legionella, representing the most commonly isolated non-L. pneumophila species. The specificity of the assay was assessed using a panel of control strains of bacteria (Table 2), representing both commonly isolated respiratory pathogens and nonpathogens that might be detected in respiratory specimens.
TABLE 1.
Control strains of Legionella
| Bacterial strain | Sourcea | Result of PCR with primer-probe set
|
|
|---|---|---|---|
| 5S rDNA | mip | ||
| L. pneumophila | |||
| Serogroup 1 | ATCC 33152 | + | + |
| Serogroup 1 | CDC Philadelphia strain | + | + |
| Serogroup 1 | + | + | |
| Serogroup 2 | + | + | |
| Serogroup 3 | + | + | |
| Serogroup 4 | + | + | |
| Serogroup 5 | + | + | |
| Serogroup 6 | + | + | |
| L. longbeachae | + | − | |
| Serogroup 1 | + | − | |
| Serogroup 2 | + | − | |
| L. dumoffii | + | − | |
| L. dumoffii | + | − | |
| L. bozemanii | ATCC 33204 | + | − |
| L. micdadei | + | − | |
| L. micdadei | ATCC 33623 | + | − |
| L. jordanis | + | − | |
If not otherwise specified, strains are Mayo Clinic isolates.
TABLE 2.
Specificity panel for Legionella LCR-PCR assays
| Bacterial strain | Sourcea | Result of PCR with primer-probe set
|
|
|---|---|---|---|
| 5S rDNA | mip | ||
| Stenotrophomonas maltophilia | CDC AB9-D19-80 | − | − |
| Morganella morganii | − | − | |
| Bordetella bronchoseptica | − | − | |
| Bordetella pertussis | ATCC VR1310 | − | − |
| Bordetella parapertussis | ATCC 27853 | − | − |
| Chlamydia pneumoniae | ATCC 25922 | − | − |
| Pseudomonas aeruginosa | − | − | |
| Escherichia coli | CDC B2-003-72 | − | − |
| Klebsiella pneumoniae | − | − | |
| Streptococcus pneumoniae | − | − | |
| Mycoplasma pneumoniae | − | − | |
| Mycobacterium tuberculosis | − | − | |
| Mycobacterium avium complex | CDC ABD-D20-82 | − | − |
| Streptococcus sp. viridans group | − | − | |
| Listeria monocytogenes | − | − | |
| Staphylococcus epidermidis | CDC AB4-BID-84 | − | − |
| Aeromonas hydrophila | − | − | |
| Pseudomonas fluorescens | − | − | |
| Moraxella catarrhalis | − | − | |
| Mycoplasma pneumoniae | − | − | |
| Pseudomonas cepacia | − | − | |
| Acinetobacter sp. | ATCC 25923 | − | − |
| Klebsiella oxytoca | − | − | |
| Staphylococcus aureus | CDC AB4-B08-84 | − | − |
| Proteus mirabilis | − | − | |
| Streptococcus pyogenes | ATCC 10211 | − | − |
| Proteus vulgaris | − | − | |
| Haemophilus influenzae | − | − | |
| Bacteroides fragilis | CDC AB2-C15-82 | − | − |
| Citrobacter freundii | − | − | |
| Campylobacter jejuni | − | − | |
If not otherwise specified, strains are Mayo Clinic isolates.
Clinical specimens.
A retrospective review of positive Legionella culture results at the Mayo Clinic, from 1979 to 1999, revealed nine BAL specimens for which frozen, archived material was available. In seven of these cases, frozen cell suspensions were available; in two cases, only supernatant was available for analysis. In addition, cell suspensions from 10 BAL specimens which were culture-negative for Legionella, were randomly selected from similarly archived material. These cell suspensions were originally prepared from BAL specimens using a cytospin method. Only cell suspensions having >2 × 106 cells counted by microscopy on a 4-mm2 grid were cultured for Legionella spp. and archived. Cultures of BAL or lung biopsy specimens were performed at the time the specimens were collected using standard techniques (43); cultures were not repeated on archived portions of these specimens during the present study. In all cases, archived results of DFA assays for Legionella, performed on fresh specimens, were also available. A retrospective review, also from 1979 to 1999, revealed nine open lung biopsy specimens culture positive for Legionella species, for which a total of 16 formalin-fixed, paraffin-embedded tissue blocks were available for evaluation. Eight open lung biopsy specimens, from the same time period, all showing nonspecific histologic findings of pneumonia or pneumonitis and all culture-negative for Legionella species, were also selected for evaluation. A single tissue block was used from each of these culture-negative cases.
BAL processing and culture methodology.
Prior to culture, BAL specimens were centrifuged for 15 min at 1,200 × g and 2,500 rpm, and the top 7.5 ml of the resulting suspension was removed. The remaining cell concentrate was mixed and used for culture. Fresh tissue from open lung biopsy specimens was homogenized in enriched brain heart infusion broth (Difco formulation; Becton Dickinson, Sparks, Md.) prior to plating. Culture for Legionella species was performed on BCYEα and BCYEα with polymyxin B, anisomycin, and vancomycin (Becton Dickinson), and plates were incubated at 35°C, in room air, for up to 14 days. Organisms from characteristic colonies were Gram stained and identified to the species level using a commercially available panel of fluorescein isothiacyanate (FITC)-labeled antibodies (SciMedX, Denville, N.J.).
Histopathologic examination.
Tissues stained with hematoxylin and eosin and WS stain were cut and stained concurrent with and consecutive to sections taken for ISH, DFA assay, and PCR. All tissue sections were cut at 4 μm. Hematoxylin and eosin staining and WS staining were carried out using standard histologic laboratory methods (39).
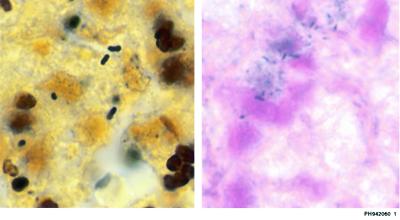
The WS-stained slides were evaluated in a blinded manner by one of the authors (M.C.A.) Slides positive for Legionella-like organisms showed dark-brown-staining bacillary structures (Fig. 1).
FIG. 1.
Legionella bacilli, demonstrated by WS staining (left) and by ISH (right).
DFA detection.
Prior to direct examination, fresh BAL specimen was centrifuged for 5 min, at 2,500 rpm, and the resultant supernatant was removed (in some cases this supernatant was later used in the PCR assay; see below, under “Real-time PCR”). The cell pellet was resuspended in normal saline, with lysis agent and/or mucolytic agents added as necessary. This cell suspension was used both for DFA assay and PCR (see below). For each smear to be examined by DFA, 200 μl was cytocentrifuged at 700 rpm, for 7 min, onto a clean glass slide and allowed to air dry. Histologic sections were deparaffinized through xylene and graded ethanol dilutions and allowed to air dry prior to examination. DFA assay was performed per the manufacturer's instructions (SciMedX). Two polyvalent FITC-labeled rabbit anti-Legionella conjugate pools were applied to a separate replicate smear or tissue section. FITC-labeled negative rabbit globulin was applied to a third replicate of the specimen, to serve as a negative control. Antibody pools were also applied to a positive control slide prepared from a known Legionella-positive lung tissue specimen in formalin. Positives were interpreted based on the presence of fluorescent bacillary structures in a given smear or section.
ISH.
ISH was performed by a procedure, as previously described with some modifications (34).
(i) Oligonucleotide probes.
Two oligonucleotide probes (Table 3), both directed against the 16S rRNA sequence of L. pneumophila, were used. One probe was previously described (19). The other probe was designed based on the analysis of sequence matches and mismatches (GenBank). The specificity of probes was checked against the sequences of other bacteria, fungi, parasites, and animals, using Genetics Computer Group software (Madison, Wis.). Probes were 3′ tailed with digoxigenin-11-dUTP (Enzo Diagnostic, Inc.) and then diluted to a final concentration of 2.0 ng/μl in hybridization buffer.
TABLE 3.
Nucleic acid targets for primers and probes
| Assay | Purpose | Targetb | Product size | Primer name | Sequence and label(s) (5′→3′)d |
|---|---|---|---|---|---|
| ISH | Probe | L. pneumophila 16S M36026a | P1a | ATC TGA CCG TCC CAG GTT | |
| ISH | Probe | L. pneumophila 16S M36026 | P2 | AGC TTT CAT CCA AAG ATA | |
| PCR | Primer | L. pneumophila mipS72442 | 24 | LPmipAf | ACC GAA CAG CAA ATG AAA GA |
| PCR | Primer | L. pneumophila mipS72442 | 124 | LPmipAr | AAC GCC TGG CTT GTT TTT GT |
| PCR | Primer | Legionella species 5S X05081 | 105 | L5Fc | ACT ATA GCG ATT TGG AAC C |
| PCR | Probe | Legionella species 5S X05081 | L5Pc | RCAT GAG GAA GCC TCA CAC TAT CAP | |
| PCR | Primer | Legionella species 5S X05081 | 105 | L5RBc | GGC GAT GAC CTA CTFT TC |
| PCR | Probe | L. pneumophila mipS72442 | LPP1 | AAC AAG TTT CAG AAA GAT TTG ATG GCA AAGF | |
| PCR | Probe | L. pneumophila mipS72442 | LPP2 | RGTA CTG CTG AAT TCA ATA AGT AAG CGG ATGP |
(ii) Pretreatment of sections for ISH.
Paraffin sections, after deparaffinizing and rehydration, were rinsed twice in diethyl pyrocarbonate-treated H2O for 2 min each. Endogenous alkaline phosphatase activity was quenched with 0.2 M HCl for 20 min at room temperature, and slides were microwaved for 10 min in 10 mM citric acid, pH 6.0, and cooled to room temperature. Sections were then digested with proteinase K (25 μg/ml; Sigma, St. Louis, Mo.) in 10 mM phosphate-buffered saline, pH 7.2, for 10 min at room temperature, followed by acetylation for 15 min with freshly prepared 0.6% acetic anhydride in 0.1 M triethanolamine (pH 8.0). Prehybridization was performed for 30 min, at room temperature, with a mixture containing 50% deionized formamide (Sigma), 10% dextran sulfate (Sigma), 1× Denhardt's solution (Sigma), 3× standard saline citrate (SSC), salmon sperm DNA (100 μg/ml, Sigma), yeast tRNA (125 μg/ml), polyadenylic-cytidylic acid (10 μg/ml), Tris (0.05 M), EDTA (5 mM), 600 mM NaCl, and 0.1% sodium pyrophosphate (inorganic).
(iii) Hybridization and post hybridization washes.
Following prehybridization, residual prehybridization buffer was thoroughly removed from around the tissue section. An oligonucleotide probe cocktail specific for L. pneumophila (2 ng/μl in prehybridization buffer) was applied to sections. A Sigmacote (Sigma) coverglass was placed on each slide, and the slides were heat treated at 95°C for 5 min and hybridized in a humid environment for 3 h at 50°C. Sections were rinsed twice in 2× SSC for 10 min at room temperature, washed in 0.5× SSC at 37°C for 20 min (to remove excess probe), and rinsed twice in buffer A (1% normal sheep serum in 0.3% Triton X-100) for 2 min at room temperature.
(iv) Immunochemical detection.
After posthybridization washing, digoxigenin-labeled probes were detected according to the manufacturer's instructions (digoxigenin detection kit; Boehringer Mannheim). Briefly, after preincubation of sections for 30 min in blocking buffer A (1% normal swine serum and 0.3% Triton X-100), the sections were incubated in a 1:200 dilution of alkaline phosphatase-conjugated antidigoxigenin Fab fragment in blocking buffer A for 1 h at room temperature. Rinsing with buffer A and buffer C (Tris-HCl and MgCl, pH 9.5) was performed, and sections were subsequently reacted with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyphosphate (BCIP), forming an insoluble blue precipitate at the site of reaction. Sections were then rinsed in buffer C, counterstained with 0.1% nuclear fast red, rinsed again in buffer C, dehydrated in graded ethanols, and cleared in xylene, and a coverslip was placed on each section with a xylene-based synthetic mounting medium. Positive interpretation of a slide was based on the presence of blue-staining bacillary structures, against a pink-red background (Fig. 1).
(v) ISH negative controls and probe specificity tests.
Negative controls used for ISH consisted of (i) omission of the probes from the hybridization reaction; (ii) slides hybridized with nonlabeled probe; (iii) cross-reactivity testing for target specificity, using an ISH probe for albumin, hybridized to Legionella-positive tissue sections; and (iv) cross-reactivity testing for probe specificity, using five additional specimens with tissue involvement by other gram-positive and gram-negative bacteria.
Real-time PCR.
PCR and product detection were performed simultaneously on LightCycler instrumentation (Roche Molecular Biochemicals). The LightCycler is a combined thermocycler and fluorimeter that offers rapid PCR thermocycling (20 to 40 min). The temperature is controlled with circulated heated and ambient air. Samples and PCR master mix are contained in 30-μl glass cuvettes (58). Sample detection is based on the principle of fluorescence resonance energy transfer (57, 58), with adjacent hybridization probes directed against the intended PCR product. With fluorescein serving as the donor fluorophore and LC-Red 640 (Roche Molecular Biochemicals) serving as the acceptor fluorophore, the presence of PCR amplicons can be assessed by detection of LC-Red 640 fluorescence. Samples can be assayed for the presence of this signal during each PCR cycle, and the cycle number at which the signal is first detected can be correlated to the original concentration of target. The specificity of amplification can be confirmed by melting curve analysis. Single melting peaks can be generated by depicting the negative derivative of fluorescence versus temperature (−dF/dT) over the course of a gradual PCR product melt (see below, “PCR cycling and melting curve conditions”).
(i) Extraction of control bacterial strains.
Pure culture isolates, used as control organisms, were extracted by two different methods. (i) In the case of gram-negative isolates, including Legionella species, bacterial colonies were suspended in sterile H2O, to a turbidity of approximately 1 McFarland standard, lysed in a 100°C heat block for 10 min, and centrifuged for 1 min at 20,000 × g. The resulting supernatant was used for analysis. (ii) In the case of gram-positive isolates, bacterial colonies were suspended in 1.0 ml of 1 N NaOH, to a turbidity of approximately 1 McFarland standard and incubated at room temperature for 5 min. The cells were pelleted, washed in an equal volume of 0.5 M Tris-HCl (pH 8.0), pelleted again, and resuspended in 100 μl of H2O. After heating in a 100°C heat block for 10 min, the suspension was centrifuged for 1 min at 20,000 × g, and the supernatant was used for analysis. Control extracts were used at a final dilution of 1:100 in sterile H2O. The presence of amplifiable DNA in specificity controls was verified by utilizing broad-range 16S ribosomal (rDNA) amplification by standard methods (23). Specific PCR assays were used to verify the presence of nucleic acid in extracts of mycobacteria and chlamydiae.
(ii) Extraction of BAL specimens.
BAL specimens were extracted using Chelex 100 (InstaGene Matrix; Bio-Rad Laboratories, Hercules, Calif.). Briefly, the specimen was mixed thoroughly, and 20 μl was added to 200 μl of InstaGene matrix. The resulting solution was mixed and placed in a 100°C heat block for 10 min. After mixing again, the sample was centrifuged for 2 min at 12,000 rpm. The resulting supernatant was used for analysis.
(iii) Extraction of tissue specimens.
Tissue sections (25 μm thick) were cut consecutively with other sections that were used for DFA and ISH assays. The sections were each cut with a clean blade and placed in a sterile glass tube. Sections were deparaffinized in xylene, washed twice in absolute ethanol, subjected to proteinase K digestion overnight at 55°C, and then placed in a 100°C heat block for 15 min. Extractions were carried out using QIAmp DNA spin columns (Qiagen Inc., Valencia, Calif.), following a standard protocol. DNA was eluted from the spin columns twice, each using 50 μl of elution buffer AE (from the QIAmp DNA mini kit). Elution buffer was incubated in the column for 1 min at room temperature, prior to the first elution, and for 5 min at RT prior to the second elution. The first and second eluants were analyzed separately by PCR.
(iv) Primers and probes.
All nucleic acid targets for primers and probes used in the study are listed in Table 3. Amplicons were kept to a minimum length (105 bp for the 5S gene and 124 bp for mip) in order to enhance the utility of these assays in formalin-fixed tissue. Probes were constructed in order to juxtapose donor (fluorescein) and acceptor (LC-Red 640) fluorophore dyes, when probes were annealed to amplicon. The mip primer-probe set was constructed for specific detection of L. pneumophila (Legionella pneumophila species-specific LC-PCR). The 5S rRNA primer-probe set was constructed to allow detection of all common Legionella species (Legionella genus LC-PCR). Primers for the latter assay were slightly truncated versions of the 20-mer sequences, L5SL9 and L5SR93, previously reported by Mahbubani et al. (35). Due to constraints imposed by the size and sequence of the 5S amplicon, only a single probe, a 23-bp portion of the 50-mer probe, also reported by Mahbubani et al. (35) was used. This probe was labeled at its 5′ end with LC-Red 640, while the fluorescein label was placed near the 3′ end of the reverse primer, for fluorescence resonance energy transfer signal production.
(v) PCR master mix (5S).
A 5-μl aliquot of sample (5 μl of H2O was used as a negative control for each run) was added to 15 μl of PCR mix in each sample cuvette. Optimized PCR master mix consisted of 50 mM KCl–20 mM Tris-HCl (pH 8.4) with a 0.1 mM concentration of each of the deoxyribonucleoside triphosphates, 6 mM MgCl2, a 0.5 μM concentration of both 5S primers, a 0.1 μM concentration of the single 5S probe, 0.05% IGEPAL CA-630 (Sigma), 0.025% bovine serum albumin, and 0.025 U of PLATINUM Taq DNA Polymerase (Life Technologies, Rockville, Md.) per μl.
(vi) PCR Master Mix(mip).
A 5-μl aliquot of sample (5 μl of H2O was used as a negative control for each run) was added to 15 μl of PCR mix in each sample cuvette. Optimized master mix consisted of 50 mM KCl–20 mM Tris-HCl (pH 8.4) with a 0.2 mM concentration of each of the deoxyribonucleoside triphosphates, 6 mM MgCl2, a 0.5 μM concentration of both mip primers, a 0.2 μM concentration of the fluorescein mip probe, a 0.4 μM concentration of the LC-Red 640 mip probe, 0.05% IGEPAL CA-630 (Sigma), 0.025% bovine serum albumin, and 0.025 U of PLATINUM Taq DNA Polymerase (Life Technologies) per μl.
(vii) PCR cycling and melting curve conditions.
PCR reagents and specimen extracts were sealed in glass capillary cuvettes with plastic plugs, centrifuged to allow mixing and to drive the mix into the distal end of each tube, and then placed on the LightCycler instrument. The cycling protocol was identical for both the mip and the 5S amplification reactions: one cycle of 95°C for 2 min followed by 50 cycles of denaturation at 95°C, annealing for 10 s at 57°C, and extension for 5 s at 72°C. Melting curves were generated as follows: starting at 55°C, the thermal chamber temperature was slowly raised to 85°C, during which time fluorescence was measured at frequent intervals. Analysis of PCR amplification and melting curves was carried out using LightCycler software.
(viii) Sensitivity and inhibition studies.
The sensitivities of both PCR assays were assessed by testing serial dilutions of a known number of CFU of L. pneumophila. PCR inhibition was assessed by spiking all culture- and PCR-negative eluants of both BAL and tissue specimens with low concentrations of L. pneumophila. Concentrations of organism used were within 1 log unit of each assay's limit of sensitivity. Inhibition was demonstrated by loss of amplification signal and/or by appearance of signal at a later cycle number than that seen in similar concentrations of organisms, diluted in sterile water as a control.
Analysis of results.
For direct (WS, DFA, ISH) assays, positives were defined by the presence of five or more identifiable bacilli, with the proper staining characteristics for the given assay (as defined above). For PCR, positives were defined by a fluorescent signal from the reporter dye (either during PCR amplification or during melting curve analysis) of three times the baseline level of fluorescence (in turn defined by the signal from the negative control cuvette for each run). The results of culture were considered to be the gold standard against which all other assays were compared. Several tissue isolates were originally designated “Legionnaires' disease bacillus” (LDB), as they were recovered before methods were in use for species determination. These were all retrospectively classified as L. pneumophila, based on the results of ISH, for the current study. Analysis of results was based on the number of specimens, rather than on the number of cases or patients tested. For BAL fluid samples, cells and supernatants from a single lavage procedure were classified as a single specimen. A breakdown of results, by case, as well as by specimen, is given in Tables 4 and 5.
TABLE 4.
Comparison of results for BAL specimens
| Case no. | Sample type | Culture result | DFA assay result | Result of PCR with primer-probe set
|
|
|---|---|---|---|---|---|
| 5S rDNA | mip | ||||
| 1 | Cells | L. pneumophila serogroup 3 | − | + | + |
| 2 | Cells | L. pneumophila serogroup 1 | − | + | + |
| 3 | Cells | L. pneumophila serogroup 1 | − | + | + |
| 4 | Cells | L. bozemanii | − | + | − |
| 5 | Supernatant | L. pneumophila serogroup 1 | + | + | + |
| 6 | Supernatant | L. micdadei | + | + | − |
| 7 | Supernatant and cells | L. pneumophila serogroup 1 | − | −/+a | −/+a |
| 8 | Supernatant and cells | L. pneumophila serogroup 1 | + | +/+a | +/+a |
| 9 | Cells | L. pneumophila serogroup 1 | − | + | + |
| 10 | Cells | − | − | − | − |
| 11 | Cells | − | − | − | − |
| 12 | Cells | − | − | − | − |
| 13 | Cells | − | − | − | − |
| 14 | Cells | − | − | − | − |
| 15 | Cells | − | − | − | − |
| 16 | Cells | − | − | − | − |
| 17 | Cells | − | − | − | − |
| 18 | Cells | − | − | − | − |
| 19 | Cells | − | − | − | − |
| Total no. of positive specimens | 9 | 3 | 9 | ||
| Total no. of cases of L. pneumophila | 7 | 7 | |||
Supernatant result/cell concentrate result.
TABLE 5.
Comparison of results for open lung biopsy specimens
| Case no. | Slide | Result of:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cultureb | 5S rDNA PCR with
|
mipa PCR with:
|
WS staining | DFA assay | ISHa | ||||
| 1st eluant | 2nd eluant | 1st eluant | 2nd eluant | ||||||
| 1 | O | Lp 1 | + | − | − | − | − | − | + |
| 1 | G | Lp 1 | − | − | − | − | + | − | + |
| 2 | E | Lp 1 | − | − | − | − | − | − | + |
| 2 | H | Lp 1 | − | + | − | − | + | − | + |
| 3 | B | L. bozemanii | − | + | −c | −c | − | + | −c |
| 3 | N | L. bozemanii | − | + | −c | −c | + | − | −c |
| 4 | M | LDB | − | − | − | − | − | + | + |
| 5 | P | LDB | − | − | − | − | − | − | + |
| 5 | C | LDB | − | − | − | − | − | − | + |
| 6 | D | LDB | + | + | + | − | + | + | + |
| 6 | L | LDB | − | + | − | + | + | + | + |
| 7 | I | LDB | + | + | − | − | + | + | + |
| 7 | K | LDB | + | + | − | − | + | + | + |
| 8 | A | Lp 1 | + | + | − | − | + | + | + |
| 9 | F | Lp 1 | + | − | − | − | + | − | + |
| 9 | J | LDB | − | + | − | − | + | − | + |
| Total no. of positive specimensd | 6 | 9 | 1 | 1 | 10 | 7 | 14 | ||
| 10 | Q | − | − | − | − | − | − | − | − |
| 11 | R | − | − | − | − | − | − | − | − |
| 12 | S | − | − | − | − | − | − | − | − |
| 13 | T | − | − | − | − | − | − | − | − |
| 14 | U | − | − | − | − | − | − | − | − |
| 15 | V | − | − | − | − | − | − | − | − |
| 16 | W | − | − | − | − | − | − | − | − |
| 17 | X | − | − | − | − | − | − | − | − |
mip PCR and ISH assays were both directed only at L. pneumophila; non-L. pneumophila cases were not included in evaluation of these tests (therefore, only 14 specimens were considered).
Lp 1, L. pneumophila serogroup 1. For LDB cultures when these tissue specimens were collected, they were not identified to the species level because fluorescent antibody conjugates for species identification of isolated colonies were not available. LDB organisms were identified as L. pneumophila from the ISH assay performed in the present study. As these specimens were fixed in formalin it was not possible to culture them at the time the current study was performed).
Result negative, but this assay was not designed for detection of L. bozemanii.
For cases 1 to 9, 11 of 16 (5S target) were positive when the results from both the first and second eluants were combined, and 2 of 14 (mip target) were positive when the results from the first and second eluants were combined.
RESULTS
Validation of PCR assay on culture isolates.
Both Legionella primer-probe sets were tested against a total of 17 different known strains of Legionella (Table 1). All Legionella species and strains were detected by the Legionella genus (5S rDNA) primers and probes, with all L. pneumophila serotypes detected by the L. pneumophila species-specific (mip) primers and probes. Both PCR assays showed 100% specificity (Table 2), with no evidence of cross-reactivity against any of the non-Legionella isolates. Both tests showed an analytic sensitivity of <10 CFU when used with serial dilutions of a known concentration of control organism.
BAL specimens.
As shown in Tables 4 and 6, the 5S rDNA Legionella genus LC-PCR assay detected nine of nine culture-positive specimens (100% clinical sensitivity), including two non-L. pneumophila species (L. bozemanii and L. micdadei). All 10 culture-negative specimens were negative by this LC-PCR assay (10 of 10; 100% specificity). Similarly, the mip L. pneumophila species-specific PCR assay detected seven of seven L. pneumophila culture-positive specimens and 12 of 12 specimens were correctly identified as negative for L. pneumophila (100% clinical sensitivity and specificity). Inhibition studies showed minimal evidence of inhibitory effect for each of the 10 Legionella-negative extracts, based on the ability to detect low concentrations of spiked organisms. Results of inhibition assays were similar for both the 5S rRNA Legionella genus and the L. pneumophila species-specific tests. Only three of nine Legionella-culture-positive specimens were detected by DFA assay (33% sensitivity and 100% specificity).
TABLE 6.
Sensitivities and specificities of all assays evaluated
| Specimen type | Assay evaluated and result | No. of specimens
|
Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|
| Culture positive | Culture negative | ||||
| BAL | Legionella genus LC-PCRa | ||||
| Positive | 9 | 0 | 100 | 100 | |
| Negative | 0 | 10 | |||
| DFA (genus pool) | |||||
| Positive | 3 | 0 | 33 | 100 | |
| Negative | 6 | 10 | |||
| L. pneumophila LC-PCRb | |||||
| Positive | 7 | 0 | 100 | 100 | |
| Negative | 0 | 12 | |||
| Open lung biopsy | Legionella genus LC-PCRa | ||||
| Positive | 11 | 0 | 69 | 100 | |
| Negative | 5 | 8 | |||
| DFA (genus pool) | |||||
| Positive | 7 | 0 | 44 | 100 | |
| Negative | 9 | 8 | |||
| WS staining | |||||
| Positive | 10 | 0 | 63 | 100 | |
| Negative | 6 | 8 | |||
| L. pneumophila LC-PCRb | |||||
| Positive | 2 | 0 | 17 | 100 | |
| Negative | 12 | 8 | |||
| L. pneumophila ISHc | |||||
| Positive | 14 | 0 | 100 | 100 | |
| Negative | 0 | 10 | |||
Assay target is 5S rDNA and is genus specific only.
Assay target is mip gene and is L. pneumophila species specific.
Assay target is 16S rDNA and is L. pneumophila species specific.
Open lung biopsy specimens.
Tables 5 and 6 show the results obtained from open lung biopsy specimens. The method used most commonly by surgical pathologists, examination of WS-stained slides, showed positive results in 10 of 16 specimens which were culture-positive for Legionella species (63% sensitivity); there were no false-positive stains (100% specificity). The sensitivity and specificity for the DFA method were 44% (7 of 16 culture-positive specimens detected) and 100%, respectively. ISH detected all 14 L. pneumophila culture-positive specimens (100% sensitivity and 100% specificity). As noted previously, the ISH assay was not designed for detection of non-L. pneumophila species; therefore, the slides with L. bozemanii were counted as culture negative for the purposes of this analysis. Specificity controls (see Materials and Methods)—including specimens which were culture-negative for Legionella, probe-negative assays, assays performed with nonlabeled probe, and both target and probe cross-reactivity assays—were all negative.
PCR was performed separately on the first and second 50-μl eluants from the extraction columns (see Materials and Methods). Table 4 shows the results of these two eluants separately and in aggregate for each case. In all, 11 of 16 tissue specimens tested positive for Legionella (69% clinical sensitivity), using the Legionella genus (5S rDNA) primer-probe set. Although not uniformly advantageous, the second eluant showed a higher rate of positivity (nine slides positive) than the first eluant (six slides positive). The L. pneumophila species-specific assay (mip) was much less sensitive, being positive in only 2 of 14 cases (17% clinical sensitivity). Inhibition assays, performed with the extracts of culture-negative specimens, showed similar results for both the Legionella genus and the L. pneumophila species-specific assays. There was minimal inhibition by the second eluants of all extracts. In contrast, the first eluants showed variable, but in some cases marked, inhibition.
Turnaround time.
Legionella culture, while usually positive in 3 to 5 days, is not reported as negative by our laboratory until a full 2-week incubation period has elapsed. DFA assay of BAL specimens, including BAL prep and cytospin, in our hands requires 1 to 2 h. Similarly, real-time PCR of BAL specimens requires approximately 1 to 2 h, including sample preparation, cycling, and detection. Studies performed on fixed tissue sections require more time, due to the need for overnight tissue processing and paraffin embedding. Including that processing time, WS, DFA, and ISH assays of tissue all require ∼24 h for assay turnaround. LC-PCR of tissue required an additional overnight digestion, bringing its reporting time to 2 days.
DISCUSSION
For many years the gold standard test for diagnosing Legionella infection has been culture. A drawback of culture is that results may not be available for several days. Rapid testing methods, which include direct histochemical staining of tissue, fluorescent antibody staining of tissue or pulmonary secretions, urine antigen detection, and serology, have been useful but often lack sensitivity and/or specificity. Recently, real-time PCR assays have become available which are highly sensitive and can be performed in as little as 1 to 2 h. In the present study, we evaluated the utility of the LC-PCR method for detecting Legionella species in pulmonary tissues and in BAL fluid. We compared this method, along with DFA assay, ISH, and WS staining, to the gold standard culture method.
With BAL specimens, LC-PCR showed a high degree of sensitivity and specificity. These results agree with previous publications, which used conventional nucleic acid amplification methods (22, 24, 28, 29, 53). By using the combination of both 5S rDNA- and mip-directed primer-probe sets, we demonstrated detection of all commonly reported pathogenic species of Legionella and were able to differentiate L. pneumophila. Specificity was demonstrated by testing both culture-negative BAL specimens, and an extensive panel of non-Legionella bacterial isolates. Sensitivity was 100% in clinical specimens, and additional spiking studies showed detection to <10 CFU of Legionella organisms. Inhibition of LC-PCR was minimal, as demonstrated with artificially spiked extracts of Legionella culture-negative BAL specimens. The entire LightCycler assay, including preparation of the BAL specimen, can be easily performed in 1 to 2 h. This compares to the extended incubation periods needed for Legionella cultures. The DFA assay, while requiring a similar time frame for testing, was shown to have a relatively low sensitivity (33%). While a wide range of sensitivities for this assay has been reported (20), the lower number is consistent with the findings of some investigators (11, 12). The DFA assay performed better in paraffin-embedded tissue sections than in BAL specimens. Our results for sensitivity (44%) are slightly lower than those reported by Theaker et al. (51) (six of nine positives detected) and higher that those noted by Koide et al. (29) (two of eight positives detected). In comparison, for the current study examination of WS-stained slides demonstrated a sensitivity of 63%, which is similar to the results obtained in an earlier study using the Dieterle silver impregnation method (4).
We are aware of a single report evaluating the ISH method (16). This investigation showed ISH to have a sensitivity of 69%, compared to other methods (culture, DFA assays, and serology), for the identification of Legionella in fixed tissue specimens (16). The present study demonstrated that both real-time PCR and ISH were able to detect a majority of cases of legionellosis. ISH, in this case limited to the detection of L. pneumophila, was positive in all specimens infected by that species (100% sensitivity). This method has a rapid turnaround time (roughly 24 h, including tissue processing—same day, excluding tissue processing). The LC-PCR Legionella genus assay had a sensitivity of 69% and a specificity 100%; the assay was able to detect the single patient infected by L. bozemanii. In contrast, the mip L. pneumophila species-specific PCR assay performed poorly with tissue sections, with a sensitivity of only 17%.
The lower sensitivity of LC-PCR with tissue specimens, compared to that with BAL specimens, may be explained by several factors. Inhibition of the PCR, either by components of the extraction process, or by substances in tissue specimens, may have played a role. We noted that in the initial 50 μl of eluant from the extraction column, there was significant inhibition of PCR. Inhibition was minimal in the second eluant, suggesting that an inhibitory compound(s) was either diluted or washed through by that point in the procedure. Consistent with this is the fact that more specimens were positive in the second eluant (nine samples positive) than in the first eluant (six samples positive). Unfortunately, while the inhibitory effect may have been reduced, the second eluant also would have a markedly reduced quantity of target DNA. Overall efficiency of the extraction protocol may also be a factor. Additionally, amplification of DNA from fresh tissue samples versus formalin-fixed samples may have resulted in higher sensitivities. No fresh tissue was available for the current study. Formalin fixation may fragment DNA, making fewer targets of acceptable size available for PCR. Moreover, the higher sensitivity of the Legionella 5S rDNA genus assay versus the L. pneumophila mip species-specific assays may also relate to this effect; smaller amplicons were produced with the Legionella 5S rDNA genus assays (105 bp) than with the L. pneumophila mip species-specific assay (124 bp). These factors aside, decreased sensitivity of the mip gene assay compared with the 5S rDNA assay may also be explained by the fact that the mip gene is present as a single copy per genome, as opposed to the 5S rDNA sequence, which may present in multiple copies.
Despite some limitations in sensitivity for detecting Legionella species in tissue specimens, LC-PCR appears to be as sensitive as culture for detecting Legionella species in BAL specimens. While the total number of cases in our study was relatively small, the findings are corroborated by those of several other authors, using conventional PCR methods (22, 24, 28, 29, 53); several of these authors showed PCR to have a higher rate of detection than culture-based methods. The use of real-time PCR increases the ease and practicality with which PCR can be introduced into the clinical laboratory. It also offers a dramatic decrease in turnaround time for results and a marked reduction in the risk of carryover contamination (inherent in single-tube amplification assays).
Molecular detection methodologies are particularly useful in the case of slow-growing organisms, such as Legionella species. If these molecular methods are demonstrated to be as sensitive as culture, then culture should not be required to confirm negative molecular test results. If an organism has a predictable antimicrobial susceptibility profile, the need to cultivate the organism may be also obviated. However, if DNA fingerprinting by pulsed-field gel electrophoresis of genomic DNA is required for epidemiological purposes, then the organism must be cultured. As a precaution, one may attempt to culture all specimens which are positive by LC-PCR, with the cultures to be used in the event that further studies (antimicrobial susceptibility or DNA fingerprinting) are needed.
With respect to detection of Legionella in tissue, it is probable that further optimization of extraction methods would increase the sensitivity of LC-PCR. Based on the limited number of specimens included in this study, the ISH method we used was a more accurate means than LC-PCR for detection of L. pneumophila in tissue. ISH offers a faster turnaround time for results than culture. Also of benefit, compared both to culture and to PCR, ISH preserves tissue morphology and requires only a 4-μm-thick tissue section. This might be of particular benefit in the instances in which limited biopsy material is available for culture or for PCR assays. The primary drawback to the ISH assay that we used is that its detection is limited to L. pneumophila; however, the development of additional ISH probes could remedy this problem. Both ISH and PCR offer the advantage over culture of using formalin-fixed tissue. This allows retrospective study when culture results are not available or when fresh tissue is unavailable for culture, either due to quantitative limitations or because Legionella may not have been a diagnostic consideration at the time of biopsy.
The reported sensitivity of L. pneumophila serogroup 1 polysaccharide urinary antigen determination in active cases of disease has ranged from 55 to 90% (8–10, 25, 48, 56). For the present study, the sensitivity of L. pneumophila species-specific LC-PCR was 100%; with open lung biopsies, the sensitivity of L. pneumophila species-specific ISH was 100%. As we did not perform L. pneumophila urinary antigen assays of samples from these patients, we cannot effectively deduce what the sensitivity of the antigen assay would be. Therefore, the utility of urinary antigen assays in our patients is undetermined.
The availability of real-time PCR offers the potential for dramatically increasing the speed with which legionellosis can be diagnosed. PCR appears equal to culture in sensitivity and specificity for BAL specimens. The use of real-time PCR offers advantages over conventional amplification methods, including an easily performed test, procedure, a marked decrease in turnaround time for results, and a reduction in the risk of cross-contamination. These factors should make this technology adaptable for routine use in the clinical laboratory.
While a prospective study is needed to prove the clinical value of the LC-PCR assay described herein, it appears suitable as a first-line assay for the detection of Legionella species in BAL specimens. Samples positive by PCR could be cultured in order to allow for supplemental studies (i.e., epidemiologic investigations or antimicrobial susceptibility testing). In contrast, considering the assays we evaluated, culture was the best method for detecting Legionella species in lung tissue. WS staining, Legionella genus LC-PCR, and L. pneumophila-specific ISH were useful as rapid tests.
ACKNOWLEDGMENT
Roberta Kondert is thanked for her effort in preparing the manuscript.
REFERENCES
- 1.Bej A K, Mahbubani M H, Miller R, DiCesare J L, Haff L, Atlas R M. Multiplex PCR amplification and immobilized capture probes for detection of bacterial pathogens and indicators in water. Mol Cell Probes. 1990;4:353–365. doi: 10.1016/0890-8508(90)90026-v. [DOI] [PubMed] [Google Scholar]
- 2.Benson R F, Fields B S. Classification of the genus Legionella. Semin Respir Infect. 1998;13:90–99. [PubMed] [Google Scholar]
- 3.Blackmon J A, Chandler F W, Hicklin M D. Legionnaires' disease: a review for pathologists. Pathol Annu. 1979;14:383–404. [PubMed] [Google Scholar]
- 4.Chandler F W, Hicklin M D, Blackmon J A. Demonstration of the agent of Legionnaires' disease in tissue. N Engl J Med. 1977;297:1218–1220. doi: 10.1056/NEJM197712012972206. [DOI] [PubMed] [Google Scholar]
- 5.Cherry W B, Pittman B, Harris P P, Hebert G A, Thomason B M, Thacker L, Weaver R E. Detection of Legionnaires disease bacteria by direct immunofluorescent staining. J Clin Microbiol. 1978;8:329–338. doi: 10.1128/jcm.8.3.329-338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloud J L, Carroll K C, Pixton P, Erali M, Hillyard D R. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J Clin Microbiol. 2000;38:1709–1712. doi: 10.1128/jcm.38.5.1709-1712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doern G V. Detection of selected fastidious bacteria. Clin Infect Dis. 2000;30:166–173. doi: 10.1086/313586. [DOI] [PubMed] [Google Scholar]
- 8.Domínguez J A, Galí N, Pedroso P, Fargas A, Padilla E, Manterola J M, Matas L. Comparison of the Binax Legionella urinary antigen enzyme immunoassay (EIA) with the Biotest Legionella Urin Antigen EIA for detection of Legionella antigen in both concentrated and nonconcentrated urine samples. J Clin Microbiol. 1998;36:2718–2722. doi: 10.1128/jcm.36.9.2718-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominguez J A, Manterola J M, Blavia R, Sopena N, Belda F J, Padilla E, Gimenez M, Sabria M, Morera J, Ausina V. Detection of Legionella pneumophila serogroup 1 antigen in nonconcentrated urine and urine concentrated by selective ultrafiltration. J Clin Microbiol. 1996;34:2334–2336. doi: 10.1128/jcm.34.9.2334-2336.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez J A, Matas L, Manterola J M, Blavia R, Sopena N, Belda F J, Padilla E, Gimenez M, Sabria M, Morera J, Ausina V. Comparison of radioimmunoassay and enzyme immunoassay kits for detection of Legionella pneumophila serogroup 1 antigen in both concentrated and nonconcentrated urine samples. J Clin Microbiol. 1997;35:1627–1629. doi: 10.1128/jcm.35.6.1627-1629.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelstein P H. The laboratory diagnosis of Legionnaires' disease. Semin Respir Infect. 1987;2:235–241. [PubMed] [Google Scholar]
- 12.Edelstein P H, Beer K B, Sturge J C, Watson A J, Goldstein L C. Clinical utility of a monoclonal direct fluorescent reagent specific for Legionella pneumophila: comparative study with other reagents. J Clin Microbiol. 1985;22:419–421. doi: 10.1128/jcm.22.3.419-421.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelstein P H, Bryan R N, Enns R K, Kohne D E, Kacian D L. Retrospective study of Gen-Probe rapid diagnostic system for detection of legionellae in frozen clinical respiratory tract samples. J Clin Microbiol. 1987;25:1022–1026. doi: 10.1128/jcm.25.6.1022-1026.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engleberg N C, Carter C, Demarsh P, Drutz D J, Eisenstein B I. A Legionella-specific DNA probe detects organisms in lung tissue homogenates from intranasally inoculated mice. Isr J Med Sci. 1986;22:703–705. [PubMed] [Google Scholar]
- 15.Espy M J, Uhl J R, Mitchell P S, Thorvilson J N, Svien K A, Wold A D, Smith T F. Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J Clin Microbiol. 2000;38:795–799. doi: 10.1128/jcm.38.2.795-799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fain J S, Bryan R N, Cheng L, Lewin K J, Porter D D, Grody W W. Rapid diagnosis of Legionella infection by a nonisotopic in situ hybridization method. Am J Clin Pathol. 1991;95:719–724. doi: 10.1093/ajcp/95.5.719. [DOI] [PubMed] [Google Scholar]
- 17.Fang G D, Yu V L, Vickers R M. Disease due to the Legionellaceae (other than Legionella pneumophila). Historical, microbiological, clinical, and epidemiological review. Medicine (Baltimore) 1989;68:116–132. doi: 10.1097/00005792-198903000-00005. . (Erratum, 68:209.) [DOI] [PubMed] [Google Scholar]
- 18.Finkelstein R, Brown P, Palutke W A, Wentworth B B, Geiger J G, Bostic G D, Sobel J D. Diagnostic efficacy of a DNA probe in pneumonia caused by Legionella species. J Med Microbiol. 1993;38:183–186. doi: 10.1099/00222615-38-3-183. [DOI] [PubMed] [Google Scholar]
- 19.Grimm D, Merkert H, Ludwig W, Schleifer K H, Hacker J, Brand B C. Specific detection of Legionella pneumophila: construction of a new 16S rRNA-targeted oligonucleotide probe. Appl Environ Microbiol. 1998;64:2686–2690. doi: 10.1128/aem.64.7.2686-2690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimont P A. Rapid methods for identification of Legionella—a review. Isr J Med Sci. 1986;22:697–702. [PubMed] [Google Scholar]
- 21.Helbig J H, Kurtz J B, Pastoris M C, Pelaz C, Luck P C. Antigenic lipopolysaccharide components of Legionella pneumophila recognized by monoclonal antibodies: possibilities and limitations for division of the species into serogroups. J Clin Microbiol. 1997;35:2841–2845. doi: 10.1128/jcm.35.11.2841-2845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaulhac B, Nowicki M, Bornstein N, Meunier O, Prevost G, Piemont Y, Fleurette J, Monteil H. Detection of Legionella spp. in bronchoalveolar lavage fluids by DNA amplification. J Clin Microbiol. 1992;30:920–924. doi: 10.1128/jcm.30.4.920-924.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson J L. Similarity analysis of rRNAs. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 683–700. [Google Scholar]
- 24.Jonas D, Rosenbaum A, Weyrich S, Bhakdi S. Enzyme-linked immunoassay for detection of PCR-amplified DNA of legionellae in bronchoalveolar fluid. J Clin Microbiol. 1995;33:1247–1252. doi: 10.1128/jcm.33.5.1247-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashuba A D, Ballow C H. Legionella urinary antigen testing: potential impact on diagnosis and antibiotic therapy. Diagn Microbiol Infect Dis. 1996;24:129–139. doi: 10.1016/0732-8893(96)00010-7. [DOI] [PubMed] [Google Scholar]
- 26.Ke D, Menard C, Picard F J, Boissinot M, Ouellette M, Roy P H, Bergeron M G. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin Chem. 2000;46:324–331. [PubMed] [Google Scholar]
- 27.Kessler H H, Reinthaler F F, Pschaid A, Pierer K, Kleinhappl B, Eber E, Marth E. Rapid detection of Legionella species in bronchoalveolar lavage fluids with the EnviroAmp Legionella PCR amplification and detection kit. J Clin Microbiol. 1993;31:3325–3328. doi: 10.1128/jcm.31.12.3325-3328.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koide M, Saito A. Diagnosis of Legionella pneumophila infection by polymerase chain reaction. Clin Infect Dis. 1995;21:199–201. doi: 10.1093/clinids/21.1.199. [DOI] [PubMed] [Google Scholar]
- 29.Koide M, Saito A, Kusano N, Tateyama M, Inadome J, Kyan Y, Kishaba T, Miyagi S. Relation between the polymerase chain reaction and the indirect fluorescent antibody method in the diagnosis of Legionella infection. Clin Infect Dis. 1996;23:656–657. doi: 10.1093/clinids/23.3.656. [DOI] [PubMed] [Google Scholar]
- 30.Koneman, E. W., S. D. Allen, W. M. Janda, P. C. Schreckenberger, and W. C. Winn (ed.). Color atlas and textbook of diagnostic microbiology. 5th ed., p. 473–489. Lippincott-Raven Publishers, Philadelphia, Pa.
- 31.Lee M A, Brightwell G, Leslie D, Bird H, Hamilton A. Fluorescent detection techniques for real-time multiplex strand specific detection of Bacillus anthracis using rapid PCR. J Appl Microbiol. 1999;87:218–223. doi: 10.1046/j.1365-2672.1999.00908.x. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay D S, Abraham W H, Fallon R J. Detection of mip gene by PCR for diagnosis of Legionnaires' disease. J Clin Microbiol. 1994;32:3068–3069. doi: 10.1128/jcm.32.12.3068-3069.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lisby G, Dessau R. Construction of a DNA amplification assay for detection of Legionella species in clinical samples. Eur J Clin Microbiol Infect Dis. 1994;13:225–231. doi: 10.1007/BF01974541. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd R V, Jin L. In situ hybridization analysis of chromogranin A and B mRNAs in neuroendocrine tumors with digoxigenin-labeled oligonucleotide probe cocktails. Diagn Mol Pathol. 1995;4:143–151. doi: 10.1097/00019606-199506000-00011. [DOI] [PubMed] [Google Scholar]
- 35.Mahbubani M H, Bej A K, Miller R, Haff L, DiCesare J, Atlas R M. Detection of Legionella with polymerase chain reaction and gene probe methods. Mol Cell Probes. 1990;4:175–187. doi: 10.1016/0890-8508(90)90051-z. [DOI] [PubMed] [Google Scholar]
- 36.Maiwald M, Schill M, Stockinger C, Helbig J H, Luck P C, Witzleb W, Sonntag H G. Detection of Legionella DNA in human and guinea pig urine samples by the polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1995;14:25–33. doi: 10.1007/BF02112614. [DOI] [PubMed] [Google Scholar]
- 37.Matsiota-Bernard P, Pitsouni E, Legakis N, Nauciel C. Evaluation of commercial amplification kit for detection of Legionella pneumophila in clinical specimens. J Clin Microbiol. 1994;32:1503–1505. doi: 10.1128/jcm.32.6.1503-1505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menendez R, Cordoba J, de La Cuadra P, Cremades M J, Lopez-Hontagas J L, Salavert M, Gobernado M. Value of the polymerase chain reaction assay in noninvasive respiratory samples for diagnosis of community-acquired pneumonia. Am J Respir Crit Care Med. 1999;159:1868–1873. doi: 10.1164/ajrccm.159.6.9807070. [DOI] [PubMed] [Google Scholar]
- 39.Mikel U V. Advanced laboratory methods in histology and pathology. Washington, D.C.: American Registry of Pathology; 1994. [Google Scholar]
- 40.Miyamoto H, Yamamoto H, Arima K, Fujii J, Maruta K, Izu K, Shiomori T, Yoshida S. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl Environ Microbiol. 1997;63:2489–2494. doi: 10.1128/aem.63.7.2489-2494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munder R U. Other Legionella Species. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 5th ed. Vol. 2. Philadelphia, Pa: Churchill Livingstone; 2000. pp. 2435–2441. [Google Scholar]
- 42.Murdoch D R, Walford E J, Jennings L C, Light G J, Schousboe M I, Chereshsky A Y, Chambers S T, Town G I. Use of the polymerase chain reaction to detect Legionella DNA in urine and serum samples from patients with pneumonia. Clin Infect Dis. 1996;23:475–480. doi: 10.1093/clinids/23.3.475. [DOI] [PubMed] [Google Scholar]
- 43.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: ASM Press; 1999. [Google Scholar]
- 44.Nitsche A, Steuer N, Schmidt C A, Landt O, Siegert W. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin Chem. 1999;45:1932–1937. [PubMed] [Google Scholar]
- 45.Pasculle A W, Veto G E, Krystofiak S, McKelvey K, Vrsalovic K. Laboratory and clinical evaluation of a commercial DNA probe for the detection of Legionella spp. J Clin Microbiol. 1989;27:2350–2358. doi: 10.1128/jcm.27.10.2350-2358.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez J A, Ahkee S, Tolentino A, Miller R D, Summersgill J T. Diagnosis of Legionella pneumophila, Mycoplasma pneumoniae, or Chlamydia pneumoniae lower respiratory infection using the polymerase chain reaction on a single throat swab specimen. Diagn Microbiol Infect Dis. 1996;24:7–14. doi: 10.1016/0732-8893(95)00254-5. [DOI] [PubMed] [Google Scholar]
- 47.Reingold A L, Thomason B M, Brake B J, Thacker L, Wilkinson H W, Kuritsky J N. Legionella pneumonia in the United States: the distribution of serogroups and species causing human illness. J Infect Dis. 1984;149:819. doi: 10.1093/infdis/149.5.819. [DOI] [PubMed] [Google Scholar]
- 48.Sathapatayavongs B, Kohler R B, Wheat L J, White A, Winn W C., Jr Rapid diagnosis of Legionnaires' disease by latex agglutination. Am Rev Respir Dis. 1983;127:559–562. doi: 10.1164/arrd.1983.127.5.559. [DOI] [PubMed] [Google Scholar]
- 49.Starnbach M N, Falkow S, Tompkins L S. Species-specific detection of Legionella pneumophila in water by DNA amplification and hybridization. J Clin Microbiol. 1989;27:1257–1261. doi: 10.1128/jcm.27.6.1257-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suffin S C, Kaufmann A F, Whitaker B, Muck K B, Prince G A, Porter D D. Legionella pneumophila: identification in tissue sections by a new immunoenzymatic procedure. Arch Pathol Lab Med. 1980;104:283–286. [PubMed] [Google Scholar]
- 51.Theaker J M, Tobin J O, Jones S E, Kirkpatrick P, Vina M I, Fleming K A. Immunohistological detection of Legionella pneumophila in lung sections. J Clin Pathol. 1987;40:143–146. doi: 10.1136/jcp.40.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomason B M, Van Orden A, Chandler F W, Hicklin M D. Effect of various histological fixatives on fluorescent antibody detection of Legionnaires disease bacteria. J Clin Microbiol. 1979;10:106–108. doi: 10.1128/jcm.10.1.106-108.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weir S C, Fischer S H, Stock F, Gill V J. Detection of Legionella by PCR in respiratory specimens using a commercially available kit. Am J Clin Pathol. 1998;110:295–300. doi: 10.1093/ajcp/110.3.295. [DOI] [PubMed] [Google Scholar]
- 54.Whitcombe D, Newton C R, Little S. Advances in approaches to DNA-based diagnostics. Curr Opin Biotechnol. 1998;9:602–608. doi: 10.1016/s0958-1669(98)80137-7. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson H W, Sampson J S, Plikaytis B B. Evaluation of a commercial gene probe for identification of Legionella cultures. J Clin Microbiol. 1986;23:217–220. doi: 10.1128/jcm.23.2.217-220.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winn W C. Legionella. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C.: ASM Press; 1999. pp. 572–585. [Google Scholar]
- 57.Wittwer C T, Herrmann M G, Moss A A, Rasmussen R P. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques. 1997;22:130–131. doi: 10.2144/97221bi01. , 134–138. [DOI] [PubMed] [Google Scholar]
- 58.Wittwer C T, Ririe K M, Andrew R V, David D A, Gundry R A, Balis U J. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 59.Woo T H, Patel B K, Smythe L D, Symonds M L, Norris M A, Weyant R S, Dohnt M F. Identification of Leptospira inadai by continuous monitoring of fluorescence during rapid cycle PCR. Syst Appl Microbiol. 1998;21:89–96. doi: 10.1016/S0723-2020(98)80011-8. [DOI] [PubMed] [Google Scholar]
- 60.Yu V L. Legionella pneumophila. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 5th ed. Vol. 2. Philadelphia, Pa: Churchill Livingstone; 2000. pp. 2424–2435. [Google Scholar]
- 61.Zuravleff J J, Yu V L, Shonnard J W, Davis B K, Rihs J D. Diagnosis of Legionnaires' disease. An update of laboratory methods with new emphasis on isolation by culture. JAMA. 1983;250:1981–1985. doi: 10.1001/jama.250.15.1981. [DOI] [PubMed] [Google Scholar]