Abstract
Huntington's disease (HD) is a devastating, autosomal dominant neurodegenerative disease caused by a trinucleotide repeat expansion in the huntingtin (HTT) gene. Inactivation of the mutant allele by clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 based gene editing offers a possible therapeutic approach for this disease, but permanent disruption of normal HTT function might compromise adult neuronal function. Here, we use a novel HD mouse model to examine allele-specific editing of mutant HTT (mHTT), with a BAC97 transgene expressing mHTT and a YAC18 transgene expressing normal HTT. We achieve allele-specific inactivation of HTT by targeting a protein coding sequence containing a common, heterozygous single nucleotide polymorphism (SNP). The outcome is a marked and allele-selective reduction of mHTT protein in a mouse model of HD. Expression of a single CRISPR-Cas9 nuclease in neurons generated a high frequency of mutations in the targeted HD allele that included both small insertion/deletion (InDel) mutations and viral vector insertions. Thus, allele-specific targeting of InDel and insertion mutations to heterozygous coding region SNPs provides a feasible approach to inactivate autosomal dominant mutations that cause genetic disease.
Keywords: Huntington's disease, gene editing, single nucleotide polymorphism
Introduction
Huntington's disease (HD) is due to an autosomal dominant mutation in the huntingtin gene. In the mutant allele, a tandem trinucleotide (CAG) repeat located in exon one is expanded to >36 copies.1 The resulting polyglutamine peptide in the mutant huntingtin (mHTT) protein is considered the initiating pathogenic molecule.1,2,3 A primary goal of therapy is to reduce mHTT mRNA and protein.4
Various strategies have been described to reduce HTT protein, including blocking transcription of the mutant gene,5,6 and reducing mHTT mRNA using antisense oligonucleotides (ASOs), siRNAs, and virally delivered miRNAs.6–15 These approaches have potential caveats. Oligonucleotide therapies require multiple treatments, and transcriptional inactivation requires persistent expression of an exogenous repressor. Another challenge is assessing the optimal degree of HTT reduction. Too little reduction of mHTT may be insufficient for clinical improvement, while too great a reduction in normal HTT may have negative consequences. For example, in a clinical trial with HTT lowering ASOs, the authors estimated that mHTT was reduced by 20–35% in striatum, but this intervention was insufficient to alter disease progression.16
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 based gene editing17,18 provides an alternative approach to permanently prevent expression of the toxic mHTT protein. The nuclease complex is composed of a single guide RNA (sgRNA) that pairs with DNA and the Cas9 protein that binds a short DNA sequence and cleaves the target DNA site. Repair of the induced double strand DNA breaks can disrupt protein expression by generating small frameshift mutations at single target sites or large deletions between pairs of sites.
Gene editing may be useful for HD,19,20 but important limitations must be addressed. Given the permanent nature of DNA sequence changes and the essential role of HTT in the adult mouse brain,21–25 it is prudent to retain the normal HTT. Allele-specific targeting of HTT using CRISPR-Cas9 based nucleases can distinguish between single nucleotide polymorphism (SNP) target sites.19,26,27 Previous allele-specific editing used two target sites to make deletions that remove exon 1 or make large deletions elsewhere in the gene.26,27 However, using two cut sites can limit the efficiency of mHTT disruption since insertion/deletion (InDel) mutations or viral vector insertions at the individual target sites will compete with deletions between both sites.28–30
Here, we describe an alternative gene editing approach (Fig. 1) with sgRNAs that target single SNP sites in the HTT protein coding regions.10 We describe an sgRNA that efficiently edits the mutant allele, but cannot edit the normal allele due to a single mismatch to the sgRNA at the SNP. Unlike mutations in intronic, UTR or untranscribed regions, a single frameshift mutation or vector insertion in the coding region can disrupt gene expression by evoking cellular mRNA surveillance mechanisms that block or reduce expression of the encoded protein.31 Using a mouse HD model with two human transgenes encoding mHTT and normal HTT, we demonstrate efficient, allele-specific reduction of mHTT protein in mouse striatum.
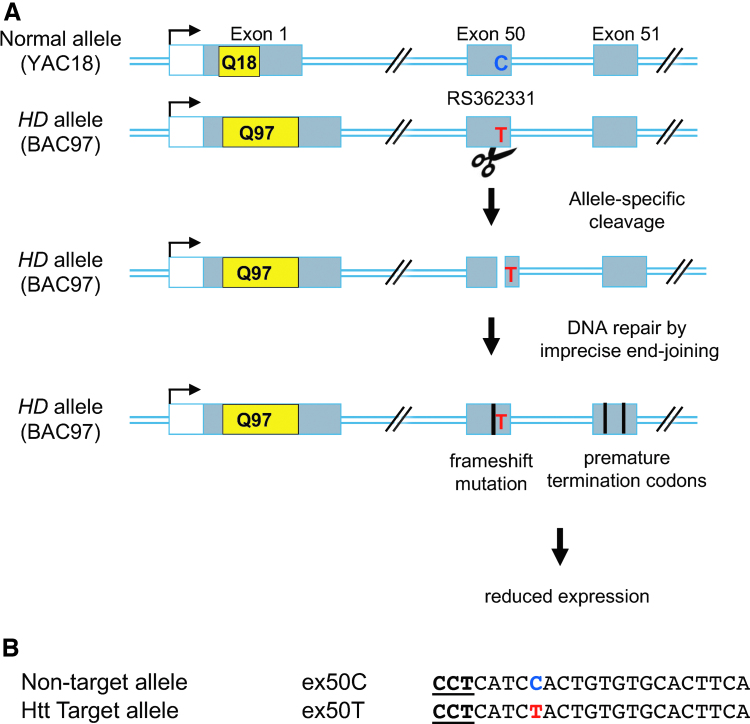
Figure 1.
Strategy for allele-specific targeting of mHTT protein by gene editing. (A) A strategy for allele-specific degradation of mHTT with SNP-specific nucleases. A schematic of the Htt gene depicts 3 of the 67 exons (not drawn to scale). Exon 1 can have a variable number of CAG repeats (indicated in yellow). HD results when a single copy of the gene has a repeat number >36. To specifically alter the disease allele, an allele-specific variable number of trinucleotide (CAG) repeats encoding glutamine (indicated in yellow) nuclease targets an SNP that is heterozygous in an HD individual. In a mouse model of HD, the YAC18 transgene carries a normal version of the human HTT locus with the C allele of SNP RS362331, and the BAC97 transgene carries a mutant human HTT locus and the T allele of SNP RS362331. DNA cleavage and imprecise repair at the mHTT gene result in frameshift mutations that disrupt the coding region of the HD allele. Premature stop codons can reduce expression of the entire HTT protein, possibly through nonsense-mediated decay of the mature mRNA. (B) CRISPR, clustered regularly interspaced short palindromic repeats-Cas9 target sequences specific for two alleles of exon 50 are shown. The heterozygous SNP is shown in blue or red. The PAM sequence is in bold. CAG; CRISPR, clustered regularly interspaced short palindromic repeats; HD, Huntington's disease; HTT, Huntingtin; mHTT, mutant HTT; PAM, protospacer adjacent motif; SNP, single nucleotide polymorphism. Color images are available online.
Methods
Detailed experimental methods are provided in Supplementary File 6.
Guide RNA design
Target sites for the CRISPR-SpCas9 nuclease were designed using the opensource Bioconductor software package CRISPRseek.32 Htt-SNP targeting sgRNAs are listed in Supplementary Table 1.
In vitro validation of guide RNAs
HEK293T cells were cotransfected with a pM427 green fluorescence protein (GFP) reporter plasmid33 with the target site and a pX330 sgRNA plasmid34 expressing sgRNA and Cas9. GFP expressing cells were measured 48 h post-transfection by flow cytometry.
Lentivirus infections
sgRNAs were cloned into lentiCRISPRv2_CMVGFP plasmid (modified from Addgene, 82416).35 Sequences are listed in Supplementary File 4. High-titer lentivirus was produced as described previously.36 Human HD fibroblasts were infected at a multiplicity of infection of 50 and analyzed after 7 days. Mouse primary neurons were infected 5 days after plating and harvested 7 days after infection. Editing efficiency, genotypes, and sgRNAs were determined for each infection by polymerase chain reaction (PCR) and Sanger sequencing.
Mice
The mouse model for allele-specific targeting of mHTT contains one transgene expressing mHTT (BAC97) and a second transgene expressing normal HTT (YAC18). The BAC97 transgenic mice (FVB/N-Tg(HTT*97Q)LXwy/ChdiJ) were obtained from William Yang at University of California, Los Angeles (UCLA).37 BAC97 mice express a neuropathogenic, full-length human mutant Huntingtin gene modified to harbor a loxP-flanked human mutant htt exon 1 sequence containing 97 mixed CAA-CAG repeats. YAC18 transgenic mice (FVB/N-Tg(HTT*18Q) were obtained from Michael Hayden at the University of British Columbia.38 YAC18 mice have a normal-length human Huntington (18 polyglutamate repeats). Both the YAC18 and BAC97 mice were crossed to Hdh−/+ (Huntingtin null) mice39 obtained from William Yang to produce YAC18 or BAC7, Hdh−/+ animals. A second cross produced YAC18 BAC97 mice with no endogenous mouse Huntington (YAC18 BAC97 Hdh−/−). Repeat lengths were checked by PCR and sequencing to ensure that there was no genetic drift.
Cas9 knock-in mice40 (Cas9: Gt(ROSA)26Sor/J) were obtained from Jackson laboratories and were crossed with BAC97 mice to produce BAC97 mice homozygous for Cas9. To generate YAC18 BAC97 mice with Cas9, BAC97 Cas9 homozygous mice were crossed with YAC18 mice (YAC18 BAC97 Hdh−/− Cas9 het). All mice used in this study were housed in the University of Massachusetts vivarium on a 12/12 light cycle with free access to food and water. All animal protocols were approved in the University of Massachusetts Chan Medical School's IACUC protocol no. A978-18.
Adeno-associated virus injections of adult mouse brains
sgRNAs were cloned into the scAAV_CB6_TurboRFP_RBG vector. Sequences are listed in Supplementary File 4. Adeno-associated virus (AAV) vectors were packaged into the AAV-AS serotype41 by the UMASS Viral Vector Core. Virus was injected into eight-week-old animals. The injection coordinates were measured from the Bregma (1.0 mm anterior, 2.0 mm lateral, and 3.0 mm from the surface of the brain). Tissue was collected as a 2 mm biopsy punch from striatal sections. Genotypes and virus were confirmed by PCR and Sanger sequencing. Editing efficiencies were surveyed by PCR and Sanger sequencing.
Illumina sequencing
Gene-specific primers with TruSeq overhang sequences were used to amplify 150 bp target sequences. Target regions were amplified using barcoded TruSeq primers. Paired-end sequences from pooled libraries were determined by the UMASS Medical School Deep Sequencing Core.
Sequence analysis
The CRISPResso software package42 quantified the frequency of each unmodified SNP allele and the frequency, size, and position of induced InDel mutation. SNPclassifier, a custom python code (Supplementary File 2), was used to assign InDel mutations to a parental SNP allele. Editing frequencies were also evaluated using Sanger sequences analyzed with the TIDE web tool (https://tide.deskgen.com/).43
Protein analysis
Lysates were prepared from frozen tissue punches from one striatal hemisphere. HTT levels were characterized using a Simple Western assay (Wes, ProteinSimple). Western blots with these lysates were performed as previously described.9
Conventional and droplet digital PCR to detect viral insertions
The primers and probes used for conventional and droplet digital PCR (ddPCR) to detect viral insertions are listed in the Key Resources Table (Supplementary File 1). For conventional PCR, ∼100 ng of template genomic DNA was amplified with Phusion High Fidelity polymerase. ddPCRs were performed using Droplet Digital PCR Supermix, and droplets were generated and analyzed with the QX200 Droplet Digital PCR System (BioRad).
Immunohistochemistry for DARPP32 and IBA1
Forty micrometers sections were prepared from treated and control animals. Every fifth section was used for immunohistochemistry staining. Sections were stained with anti-DARPP rabbit monoclonal antibody or anti-IBA1 antibody. Numbers of positively stained cells were determined for each animal using 50 images from 5 sections.
Quantification and statistical analysis
Linear regression was used to quantify and test the statistical significance between treatment conditions within genotype or treatment duration and between genotype or treatment duration within treatment groups. Analyses were conducted using STATA (v16) software's regression command to fit models for each dependent variable, then using postestimation contrasts to test each hypothesis. We considered p-values <0.05 to be statistically significant.
Results
Allele-specific nucleases can target common HTT SNP heterozygosities
We previously developed a computational tool32 to identify candidate allele-specific gRNAs. An example is diagrammed with an sgRNA that targets the “T” allele of an SNP heterozygosity in exon 50 (Fig. 1). The nontargeted “C” allele has a mismatch to the hybridizing sgRNA. We used a GFP reporter assay33 to identify gRNAs with high activity for the targeted SNP allele and low activity for the other allele (Supplementary Table 1, Supplementary Fig. 1), including allele-specific gRNAs targeting SNPs in exons 48, 50, and 57.
Allele-specific gene editing reduces mHTT in mouse primary neurons
We tested gene editing at protein coding region SNPs for allele-specific reduction of mHTT protein in neurons. We focused on the Ex50T-g1 gRNA targeting the T allele of SNP RS362331 in exon 50. RS362331 is the most frequently observed heterozygous protein coding region SNP in HD patients,10 and the T allele is more frequently associated with the mHTT haplotype.44
Primary cortical neurons were derived from a mouse model with two human HTT transgenes (Fig. 2). The BAC97 transgene37 encodes a mHTT protein with 97 glutamine codons and has the “T” allele at the exon 50 SNP. The YAC18 transgene38 expresses normal HTT and has the “C” allele. The mice were also homozygous for a null mutation of the endogenous mouse HTT gene39 and expressed transgenic Cas9.40 An sgRNA targeting the T allele at exon 50 (Ex50T) was delivered to primary neurons by lentiviral transduction 5 days after isolation, and cells were harvested one week later (Fig. 2A). An sgRNA targeting the mouse Rosa26 locus was used as a control.
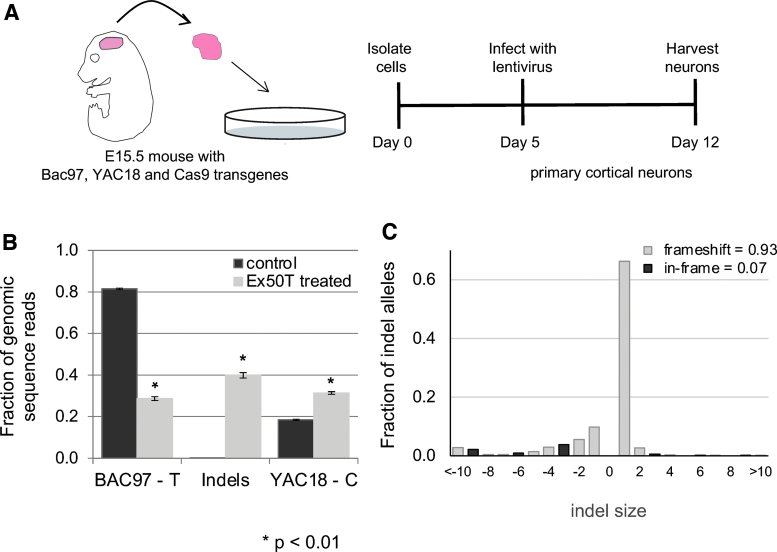
Figure 2.
Allele-specific targeting of mHTT in BAC97/YAC18 primary neurons. (A) Cartoon depicting the generation and infection of primary cortical neurons from mice heterozygous for Yac18 (wt) and Bac97 (mutant) human transgenes as well as a Cas9 transgene. (B) An allele-specific sgRNA targeting the T allele of Htt Exon 50 (Ex50T) was introduced into BAC97/YAC18/Cas9 mouse primary neurons by lentiviral transduction using the LentiCRISPRcmvGFP vector. An sgRNA targeting the Rosa26 locus was used as a control. Genomic DNA was prepared 7 days after infection. PCR amplicons containing the targeted SNPs were barcoded and analyzed by Illumina sequencing. InDel allele frequencies were determined using the CRISPResso software package. The control sample has a higher frequency of the BAC97 allele than YAC18 due to a higher copy number of the BAC97 transgene. In the treated sample, 40% of reads are induced InDel mutations. n = 3 mice and error bars represent the SEM. p-values <0.05 were considered significant. Exact p-values are reported in Supplementary File 3. (C) The distribution of InDel sizes shows a strong bias toward frameshift mutations (open bars) rather than in frame mutations (filled bars). The insertion of a single base is the dominant allele type. InDel, insertion/deletion; PCR, polymerase chain reaction; SEM, standard error of the mean; sgRNA, single guide RNA; wt, wild type. Color images are available online.
Edited genomic DNA sequences from the exon 50 SNP region were determined by Illumina sequencing of PCR amplicons. In control samples, the “T” allele present in the BAC97 transgene was present in 81% of sequences, and the C allele from the YAC18 transgene was present in 19%, reflecting a higher copy number for the BAC97 transgene (Fig. 2B). In Ex50Tg1-treated neurons, sequences with the unaltered “T” allele were reduced from 81% to 29%, while sequences harboring induced InDel mutations rose to 40%, suggesting that the InDel mutations result from inaccurate repair of DNA breaks at the targeted “T” allele. Greater than 90% of the observed InDel mutations are frameshift mutations with the most frequent allele being the addition of a single C nucleotide (Fig. 2C, Supplementary Fig. 2).
For many InDel mutations, the polymorphic base from SNP RS362331 is still present, allowing the InDel to be attributed to one of the two parental alleles. For example, the single C insertion is associated with the T allele of the SNP (Supplementary Fig. 2, Ex50Tg1 treated, third line). In contrast, the provenance of some alleles cannot be determined due to deletion of the diagnostic SNP (Supplementary Fig. 2, Ex50Tg1 treated, seventh line). A custom software tool (SNPclassifier, Supplementary File 2) was created, which assigned all alleles either to the parental T allele, or the parental C allele, or to an undetermined category. The nearly all InDel mutations in primary neurons treated with the Ex50T gRNA were either associated with the parental T allele or could not be assigned (Supplementary Table 2).
Our gene editing approach reduced full-length mHTT protein (from BAC97) with no detectable change in the nontargeted normal HTT protein (from YAC18) (Fig. 3A, B). Although the frameshift mutations are induced in exon 50, smaller HTT protein fragments were not detected by Western blot analysis (Fig. 3C). Thus, allele-specific targeting of a HTT coding region SNP reduced mutant, but not normal HTT protein in primary neurons.
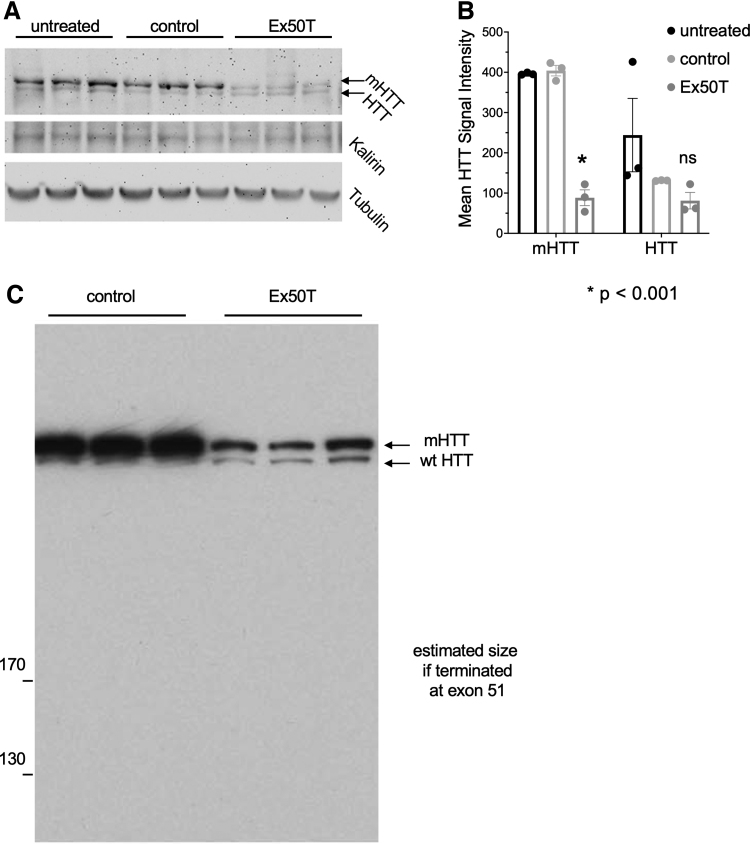
Figure 3.
Allele-specific reduction of mHTT protein in primary neurons from a HD mouse. (A) mHTT and normal HTT protein were assayed after treatment with sgRNAs targeting Exon 50 (Ex50Tg1) or Rosa26 (control). mHTT is expressed from the BAC97 transgene, and normal HTT protein is expressed from the YAC18 transgene. mHTT runs at a higher molecular weight due to the increased size of the polyQ repeat sequence (upper blot). Higher mHTT protein expression is observed because of the higher transgene copy number. Kalirin and beta-tubulin provide total protein concentration controls. The targeted mHTT protein, but not normal HTT protein, is reduced when targeted with sgRNA Ex50Tg1. (B) Quantification of HTT protein levels from digital images of the blot in (A). mHTT and HTT signals are normalized to Kalirin levels. n = 3. Error bars indicate the SEM. Exact p-values are reported in Supplementary File 3. (C) An additional protein gel was overexposed to detect accumulation of truncated protein products corresponding to translational termination after frameshift mutations in exon 50.
Gene editing at SNP heterozygosities selectively lowers mHTT in mouse striatum
We tested in vivo gene editing of mHTT in the striatum of adult BAC97/YAC18/Cas9 mice (Fig. 4). The Ex50Tg1 sgRNA was delivered using an AAV vector41 that expresses the sgRNA (Fig. 4A). AAV expressing an sgRNA targeting the Rosa26 locus was used as control. Vector was introduced by injection into the adult mouse striatum. Twenty-nine percent, 37%, and 33% InDels were observed at 2, 4, and 6 weeks, respectively, after injection. Ninety-three percent of the InDels were frameshift mutations. The increase in InDels was accompanied by a reduction in the targeted BAC97 (Fig. 4B).
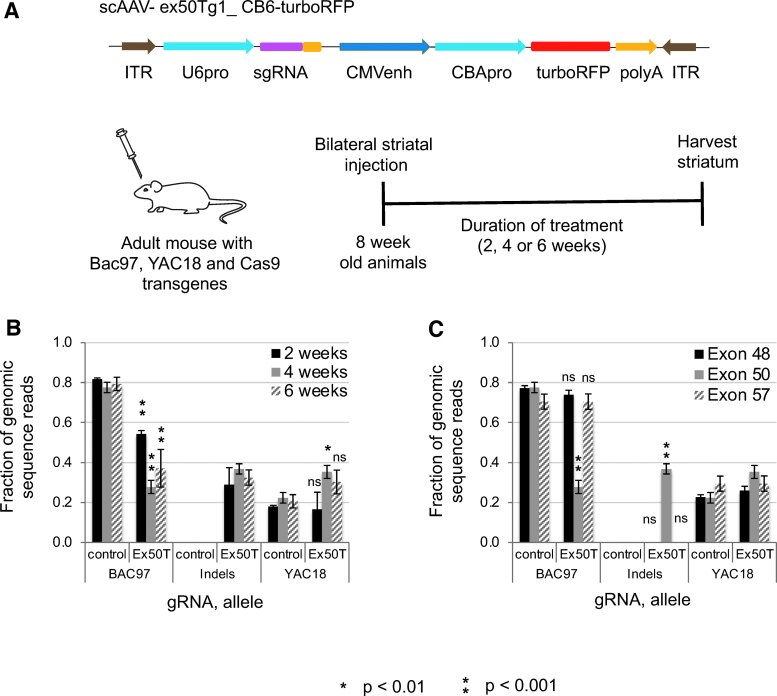
Figure 4.
Allele-specific CRISPR-Cas9 targeting of HTT SNPs in the adult striatum of a mouse model of HD. (A) Cartoon depicting the AAV vector used to deliver the Exon50Tg1 and control sgRNAs and the delivery of AAV to mouse brains. The HD model mice have two alleles at the exon50 SNP—the BAC97 transgene with the T allele and the YAC18 transgene with the C allele. The Ex50Tg1 sgRNA targets the T allele of the SNP heterozygosity. The gRNA and a turboRFP reporter gene are delivered by AAV injection into the adult striatum of 8-week-old mice. Cas9 activity is provided as a mouse transgene. (B) Analysis at multiple time points. At 2, 4 and 6 weeks after AAV treatment, the frequency of the BAC97 allele is reduced, and InDel mutations are induced by the Ex50T-programmed nuclease. (C) Analysis of flanking exons. Heterozygous SNPs were examined by Illumina sequencing at two flanking exons in the 4 weeks samples. There is no significant change in the ratio of BAC97 to YAC18 alleles in exons 48 or 57, indicating that cleavage and repair at exon 50 do not induce a high frequency of deletions large enough to remove these SNPs. n = 3 mice and error bars represent the SEM. p-values <0.05 were considered significant. Exact p-values are reported in Supplementary File 3. AAV, adeno-associated virus. Color images are available online.
To determine if InDels could be induced in the nontargeted YAC18 transgene in the absence of BAC97, animals transgenic for only the YAC18 and Cas9 transgenes were injected AAV expressing either Ex50Tg1 or Rosa26 control sgRNAs. Four weeks after injection, InDel mutation rates were determined by TIDE analysis of Sanger sequencing chromatograms43; the signal in Ex50Tg1-treated tissue (2.1%, standard error of the mean [SEM] = 1.0%) was no higher than the background signal in Rosa26-treated controls (3.5%, SEM = 2.3%). Thus, brief expression of CRISPR-Cas9 gene editing complexes in cultured neurons or extended expression in adult brain tissue induced frameshift mutations in the targeted mutant allele without altering the nontargeted allele.
Frameshift mutations in protein coding sequences can evoke RNA surveillance mechanisms that prevent translation of mRNAs with premature termination codons.31,45 We examined whether there was an allele-specific reduction in mHTT RNAs 4 weeks after editing. Reverse transcription-PCR followed by Illumina sequencing was used to determine the relative frequencies of each HTT exon 50 SNP allele and of induced InDels from AAV-treated striatum. Twenty-five percent of sequences in treated samples were InDel mutations (Supplementary Fig. 5, exon 50). The targeted BAC97 mHTT allele decreased from 68% in controls to 18% with Ex50T gRNA. Together, the frequency of all exon 50 cDNA sequences from mHTT (BAC97 sequences plus InDels) was reduced from 68% to 43%, consistent with degradation of mHTT mRNA.
The relative amounts of mHTT versus wild-type (wt) HTT RNA were also assayed by sequencing heterozygous SNPs at exons 48 and 57. In cDNA sequences, the frequency of BAC97 alleles was reduced at both flanking exons (Supplementary Fig. 5, exons 48 and 57), indicating that mHTT RNA was reduced.
We examined allele-specific reduction of mHTT protein at 4 and 6 weeks after sgRNA delivery to the striatum. The targeted mHTT protein (from BAC97) decreased approximately fourfold, while wt HTT (from YAC18) was not reduced (Fig. 5, Supplementary Fig. 3). Truncated HTT protein products were not detected (Supplementary Fig. 4).
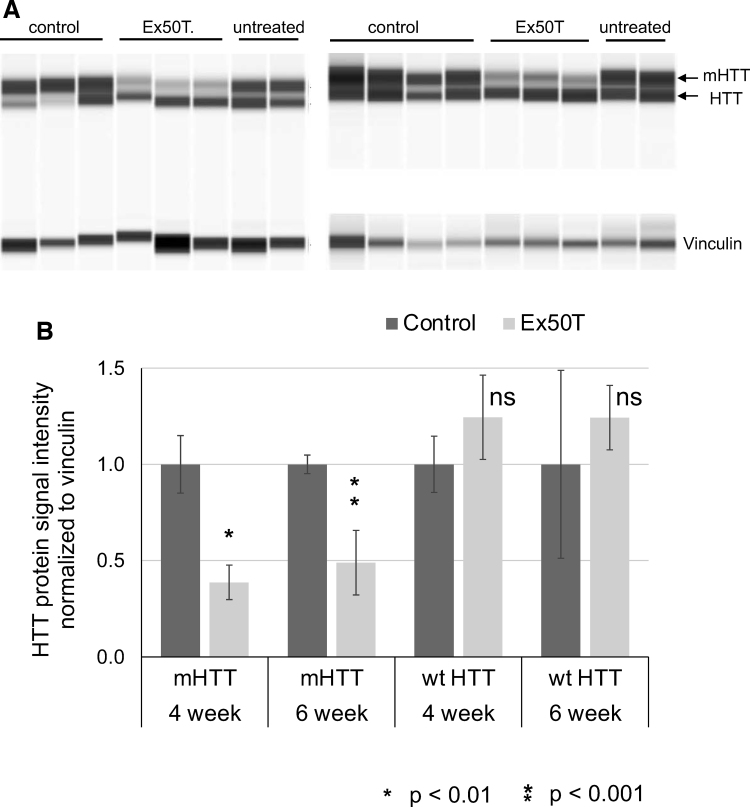
Figure 5.
Allele-specific reduction of mHTT protein in the mouse adult striatum. (A) Protein sample analysis of HTT and mHTT protein from the striatum of HD model mice with the Cas9, BAC97, and YAC18 transgenes. Each animal was bilaterally injected with AAV expressing the control (Rosa26) or Ex50Tg1 sgRNAs. Protein was isolated from striatal tissue. Vinculin serves as a total protein control. Signal is visualized as a virtual blot format. (B) Quantitative WES signals for mHTT and wt HTT were normalized to vinculin. Each pair of control and Ex50T-treated samples were further adjusted such that the control has a value of 1. At both 4- and 6-weeks after injection with AAV expressing sgRNA Ex50T, mHTT was reduced by >50% while wt HTT was not significantly changed. n = 3 mice and error bars represent the SEM. p-values <0.05 were considered significant. Exact p-values are reported in Supplementary File 3. WES, Simple Western protein analysis.
To test if either AAV delivery or CRISPR-Cas9 editing at mHTT was toxic, we examined levels of DARPP32, which is highly enriched in striatal neurons, and GFAP, which is a glial marker that increases during stress. There were no changes in either after mHTT lowering (Supplementary Fig. 7). Immunohistochemical analysis of Iba1, a marker of microglia, only revealed increased staining directly adjacent to the needle injection site (Supplementary Fig. 6C, n = 3). The total number of immunoreactive Iba1-positive microglia and DARPP32-positive neurons were not different between treatment groups (Supplementary Fig. 6D, E).
AAV- and lentiviral-mediated gene editing generates nonamplifiable alleles and viral vector insertions
Amplicon sequencing of treated primary neurons showed an increase in the percentage of YAC18 alleles (Fig. 2). This increase could occur if some editing-induced mutations in the BAC97 allele prevent PCR amplification (Fig. 6A). We quantified the fraction of HTT exon 50 sequences that could no longer be amplified using droplet digital PCR.46,47 Paired samples were obtained by treating primary neurons derived from individual animals with either control or Ex50T gRNAs. The human HTT exon 50 copy number was decreased by an average of 44% in Ex50T-treated versus control cells (Fig. 6D).
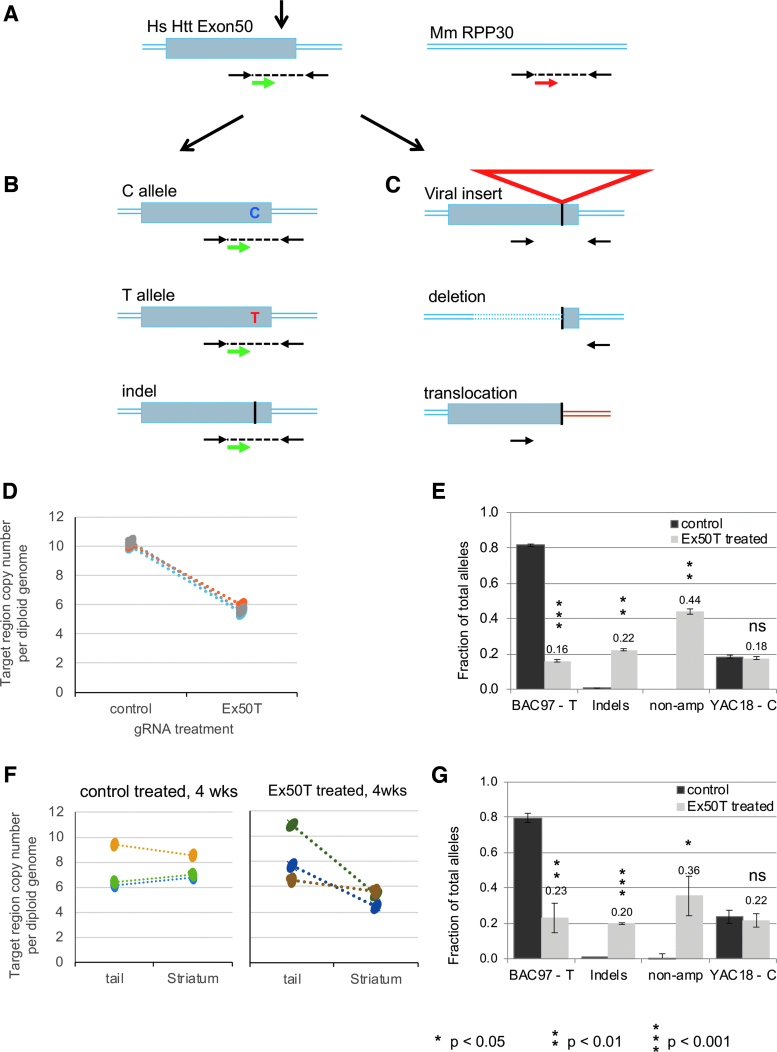
Figure 6.
Analysis of nonamplified mutations by ddPCR. (A) The relative copy number of human exon 50 was measured using primers (black arrows) and a probe (green arrow) flanking the CRISPR-Cas9 target site. A second set of primers and probe (red arrow) were used to determine the copy number of a reference locus, the mouse RPP30 gene. (B) After DNA cleavage and repair, ddPCR detects chromosomes with either of the original SNP allele sequences or small InDel alleles. (C) ddPCR does not detect chromosomes with mutations that either remove (large deletions) or separate (translations or large insertions) the primer binding sites. (D) ddPCR was used to determine the total copy number of transgenic human HTT exon 50 in the primary neurons using the mouse RPP30 as a reference for the diploid genome. Samples were treated with lentivirus expressing Ex50T gRNA or a control (Rosa26) gRNA. Exon 50 copy number was decreased from an average of 10.2 to 5.7 across three paired neuronal samples. The 44% reduction reflects alleles that can no longer be amplified using the primers for allele sequencing. (E) The fraction of each allele class is estimated by multiplying the fraction of sequenced alleles by the estimated fraction of alleles that could be amplified (0.56) based on the ddPCR results. “non-amp” indicates the estimated fraction of alleles that could not be amplified. (F) ddPCR was used to determine the total copy number of human HTT exon 50 in the 4-week striatal samples treated with AAV expressing the Ex50T or a control (Rosa26) gRNA and in untreated tail DNA from the same animals. There was no significant difference in the exon 50 copy number between tails and striatum treated with the control gRNA. In contrast, the copy number was reduced by an average of 36% in striatum treated with the Ex50T gRNA. This reduction reflects alleles that can no longer be amplified using the primers for allele sequencing. (G) The fraction of each allele class is estimated by multiplying the fraction of sequenced alleles by the estimated fraction of alleles that could be amplified (0.64) based on the ddPCR results. “non-amp” indicates the estimated fraction of alleles that could not be amplified. n = 3 mice and error bars represent the SEM. p-values <0.05 were considered significant. Exact p-values are reported in Supplementary File 3. ddPCR, droplet digital PCR. Color images are available online.
The estimated fraction of nonamplifiable alleles was added to the sequenced alleles from Fig. 2B to infer the actual fraction of each allele class (Fig. 6E). After including 44% nonamplified alleles in treated samples, the inferred frequency of unaltered BAC97 alleles was 16% in Ex50T-treated samples. The inferred frequency of induced InDel mutations was 22%. Together, the frequency of nonamplified alleles plus InDel mutations represented 66% of alleles in the treated sample. In this analysis, the nontargeted YAC18 alleles were unchanged by editing, remaining at 18%. Editing in the striatum generated a similar result (Fig. 6F, G). Thus, in both primary neurons and striatum, AAV delivery of sgRNAs induced a combination of InDel mutations, which were included in our sequenced alleles, and an additional class of mutations that were not represented in the sequenced amplicons. The relative frequency of each of these classes can be inferred by ddPCR quantification of the missing alleles.
To determine if very large deletions were removing the BAC97 allele at exon 50, we examined the frequency of SNP heterozygosities at flanking exons 48 and 57. There was no significant change in the BAC97 or YAC18 allele at either flanking SNP site (Fig. 4C), indicating that the mutations that prevent amplification were not large deletions.
Previous studies reported that AAV-mediated delivery of CRISPR-Cas9 generated a high rate of AAV insertions, including in normal mouse brain.28,29 PCR analysis revealed lentiviral and AAV insertions at the human HTT exon 50 locus or the control Rosa26 locus after gRNA delivery (Supplementary Fig. 8). Insertions were only detected at the site targeted by the corresponding gRNA, demonstrating that delivery of gene editing components by lentivirus or AAV induces viral insertion mutations at the target site.
Allele-specific editing of coding SNPs in HD patient-derived fibroblasts
To confirm allele-specific targeting in human cells, we used HD patient-derived fibroblasts heterozygous for the common HTT coding region SNPs. Unlike postmitotic neurons, fibroblasts can use the nontargeted allele to repair induced DNA breaks; this homology-dependent repair would increase the frequency of the nontargeted allele. Untreated fibroblasts had an equal frequency of each allele. Treated fibroblasts exhibited induced InDels (19%), decrease of the targeted allele (50–7%), and an increase in the nontargeted allele (50–74%). Sequencing of the individual alleles revealed that most or all InDels were derived from the targeted allele (Supplementary Fig. 9). These results confirm allele specificity in HD patient-derived cells.
Analysis of off-target sites
We characterized potential off-target sites for the Ex50Tg1 gRNA in the human genome. Loci with up to three mismatches to the target sequence were sequenced from HEK293T cells treated with ribonucleoprotein complexes programmed with either Ex50Tg1 or a control gRNA. Eighty-seven percent of HTT sequences contained InDels at the targeted sequence (Supplementary File 7). Among the predicted off-target sites, a single site had InDels above the detection limit of 0.15% in two replicates. This site, in an intronic sequence, exhibited a 0.9% and 1.5% editing rate in this experiment.
Discussion
A critical challenge for developing therapeutics for autosomal dominant disorders such as HD is to reduce the activity of the mutant protein while preserving normal protein function. We demonstrate the usefulness of CRISPR-Cas gene editing at a protein coding region SNP with high heterozygosity in the HD population. Compared with previous editing studies using mouse models of HD, our approach has some possible problems and potential advantages.
Elimination of the expanded CAG repeat region is the most straightforward way to prevent mHTT toxicity, and previous studies have demonstrated that one or two gRNAs targeting the coding region of exon 1 of a mHTT transgene can rescue organismal phenotypes.19,48 However, neither of these studies use an allele-specific approach. In another study,26 allele-specific gRNAs targeted regions flanking exon 1, but only an estimated 20% of HD patients are estimated to be heterozygous at both sites. In addition, these sgRNAs reduced expression of the endogenous mouse htt gene,26 so whether they would be allele specific in a model such as the BAC97/YAC18 used in our study is unclear. Our study is the first demonstration of allele-specific knockdown of mHTT in a mouse model with two human transgenes expressing both mHTT and normal HTT. Furthermore, a significant percentage of HD patients are candidates for this approach. SNP RS362331 is heterozygous in 40–46% of HD patients.10,49 Targeting three coding region SNPs using our strategy should enable treatments for 70–80% of patients.10,32
A second advantage of allele-specific targeting of HTT at single coding region sites is that both InDels and vector insertions can disrupt gene function. A possible limitation of creating deletions between two target sites is that the frequent induction of viral insertions or InDel mutations at one or both sites will prevent the formation of deletions between the two sites.28 The in vivo frequencies of these different outcomes can be challenging to measure and were not tested in the previous mouse study of allele-specific editing.26 In contrast, when targeting single SNP heterozygosities that occur within the protein coding region, either individual InDel mutations or other mutations such as large insertions or deletions are sufficient to reduce protein levels.
Our results targeting a single coding region SNP heterozygosity demonstrate a high efficiency of allele-specific reduction of the targeted gene. The 40–50% reduction of mHTT protein we observed when targeting an SNP in exon 50 is similar to that described using one or two sgRNAs to target the nonpolymorphic regions of exon1.19,48
An unexpected outcome of our analysis was the increase in the percentage of sequence reads with the nontargeted allele in treated neurons (the YAC18 allele in Figs. 2B and 4B). In dividing cells, this increase could reflect homology-dependent repair of the targeted allele using the nontargeted allele as a donor. However, homology-dependent repair is low or absent in most postmitotic cells such as neurons. We found that most induced mutations were excluded from our sequence analysis (Fig. 6). If these “unamplified” alleles were added to the sequenced alleles, the nontargeted alleles were unchanged after gene editing (Fig. 6E, G).
Recent studies suggest that a combination of AAV vector insertions, large deletions, or other rearrangements can comprise a major fraction of edited alleles when AAV is used to deliver gene editing components.28–30 In our experiments, most mutations induced by AAV or by lentivirus delivery of sgRNAs were not amplified using primers that flank the targeted site. With both AAV and lentivirus, the viral vector sequences were found at the CRISPR-Cas9 targeted site. These results suggest that many gene editing experiments using either lentivirus or AAV have the potential for frequent viral insertions at the targeted site. While AAV and integrase-defective lentivirus were known to insert at sites of induced DNA breaks, this is, to the best of our knowledge, the first demonstration that integration-competent lentivirus, which typically integrates at random genomic sites, was directed to integrate at DNA breaks induced by CRISPR-Cas9 gene editing. Depending on the types of sequences carried by the viral vector, such insertions might result in a different cellular outcome than would be expected with a simple InDel or deletion allele.
The terminal regions of both viral vectors used in this study have stop codons in all three reading frames; when these sequences insert at the exon 50 editing site, they are predicted to result in a truncated mHTT protein coding region. For the HTT locus, we demonstrated that while there are both the expected InDel mutations and unanticipated viral insertions, the outcome of editing at the exon 50 SNP is a significant reduction in full-length HTT protein and no detectable truncated products. These results suggest that either InDel or insertion mutations result in altered RNA and/or protein products with reduced expression.
For clinical applications of the approach described here, improvements in specificity and delivery are needed. We detected low rates of editing at one potential off-target site; one possible way to reduce off-target editing is use of higher fidelity editing enzymes.50 In addition, alternative methods for gRNA and Cas9 delivery are required. For in vivo expression of Cas9 in mice, we have taken advantage of a Cas9 transgene; however, previous HTT studies have confirmed that it is possible to deliver both sgRNAs and SpCas9 using two AAV viruses or sgRNAs with SaCas9 using a single AAV virus.19,48 In addition to viral delivery, future therapeutic implementation of allele-specific editing may deliver transient editing complexes containing gRNAs with Cas9 mRNA or gRNAs with Cas9 protein, which could lower off-target activity and improve in vivo safety.51
Conclusion
We conclude that using viral delivery to target coding region SNPs for gene editing is a viable approach to selectively reduce mHTT protein expression.
Supplementary Material
Acknowledgments
The authors thank David Howland and Richard Chen of the CHDI Foundation for their support and scientific insights. They also thank Wendy Cullinane and Rina Paladino for their administrative support. Illumina sequencing was performed at the University of Massachusetts Chan Medical School Deep Sequencing Core.
Authors' Contributions
S.R.O., E.L.P., M.S.E., S.A.W., M.D., N.A., and M.H.B. conceived and planned experiments. S.R.O., E.L.P., E.S., K.O.C., L.A.K., E.H., R.M., and A.C. carried out the experiments. S.R.O., L.J.Z., E.O.M., M.D., and M.H.B. analyzed the data. S.R.O., N.A., and M.H.B. wrote the article, with feedback from all authors.
Author Disclosure
No competing financial interests exist.
Funding Information
This work was supported by NIH R01 NS106245 to M.H.B. and N.A., CHDI Foundation Research Agreement A-10199 to M.H.B., CHDI Foundation Research Agreement A-5038 to N.A., CHDI Research Agreement A-6367 to M.D., the Dake Family Fund to M.D., and NIH UL1 TR000161-05 to the University of Massachusetts Center for Clinical and Translation Science.
Supplementary Material
References
- 1. Huntingtons Disease Collaborative. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 1993;72:971–983. [DOI] [PubMed] [Google Scholar]
- 2. Aronin N, Chase K, Young C, et al. . CAG expansion affects the expression of mutant huntingtin in the Huntington's disease brain. Neuron 1995;15:1193–1201. [DOI] [PubMed] [Google Scholar]
- 3. DiFiglia M, Sapp E, Chase K, et al. . Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 1995;14:1075–1081. [DOI] [PubMed] [Google Scholar]
- 4. Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest 2011;121:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garriga-Canut M, Agustin-Pavon C, Herrmann F, et al. . Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc Natl Acad Sci U S A 2012;109:E3136–E3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zeitler B, Froelich S, Marlen K, et al. . Allele-selective transcriptional repression of mutant HTT for the treatment of Huntington's disease. Nat Med 2019;25:1131–1142. [DOI] [PubMed] [Google Scholar]
- 7. Kordasiewicz HB, Stanek LM, Wancewicz EV, et al. . Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron 2012;74:1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pfister EL, DiNardo N, Mondo E, et al. . Artificial miRNAs reduce human mutant huntingtin throughout the striatum in a transgenic sheep model of Huntington's disease. Hum Gene Ther 2018;29:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keeler AM, Sapp E, Chase K, et al. . Cellular analysis of silencing the Huntington's disease gene using AAV9 mediated delivery of artificial micro RNA into the striatum of Q140/Q140 mice. J Huntingtons Dis 2016;5:239–248. [DOI] [PubMed] [Google Scholar]
- 10. Pfister EL, Kennington L, Straubhaar J, et al. . Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol 2009;19:774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harper SQ, Staber PD, He X, et al. . RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci U S A 2005;102:5820–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alterman JF, Godinho B, Hassler MR, et al. . A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat Biotechnol 2019;37:884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evers MM, Miniarikova J, Juhas S, et al. . AAV5-miHTT gene therapy demonstrates broad distribution and strong human mutant huntingtin lowering in a Huntington's disease minipig model. Mol Ther 2018;26:2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miniarikova J, Zimmer V, Martier R, et al. . AAV5-miHTT gene therapy demonstrates suppression of mutant huntingtin aggregation and neuronal dysfunction in a rat model of Huntington's disease. Gene Ther 2017;24:630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stanek LM, Sardi SP, Mastis B, et al. . Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington's disease. Hum Gene Ther 2014;25:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith AV, Tabrizi SJ. Therapeutic antisense targeting of huntingtin. DNA Cell Biol 2020;39:154–158. [DOI] [PubMed] [Google Scholar]
- 17. Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci 2016;17:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang S, Chang R, Yang H, et al. . CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington's disease. J Clin Invest 2017;127:2719–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Merienne N, Vachey G, de Longprez L, et al. . The self-inactivating KamiCas9 system for the editing of CNS disease genes. Cell Rep 2017;20:2980–2991. [DOI] [PubMed] [Google Scholar]
- 21. Liu JP, Zeitlin SO. Is huntingtin dispensable in the adult brain? J Huntingtons Dis 2017;6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burrus CJ, McKinstry SU, Kim N, et al. . Striatal projection neurons require huntingtin for synaptic connectivity and survival. Cell Rep 2020;30:642.e646–657.e646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet 2000;26:300–306. [DOI] [PubMed] [Google Scholar]
- 24. Mehler MF, Petronglo JR, Arteaga-Bracho EE, et al. . Loss-of-huntingtin in medial and lateral ganglionic lineages differentially disrupts regional interneuron and projection neuron subtypes and promotes Huntington's disease-associated behavioral, cellular, and pathological hallmarks. J Neurosci 2019;39:1892–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKinstry SU, Karadeniz YB, Worthington AK, et al. . Huntingtin is required for normal excitatory synapse development in cortical and striatal circuits. J Neurosci 2014;34:9455–9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Monteys AM, Ebanks SA, Keiser MS, et al. . CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther 2017;25:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shin JW, Kim KH, Chao MJ, et al. . Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet 2016;25:4566–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nelson CE, Wu Y, Gemberling MP, et al. . Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med 2019;25:427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanlon KS, Kleinstiver BP, Garcia SP, et al. . High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun 2019;10:4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCullough KT, Boye SL, Fajardo D, et al. . Somatic gene editing of GUCY2D by AAV-CRISPR/Cas9 alters retinal structure and function in mouse and macaque. Hum Gene Ther 2019;30:571–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Ann Rev Genet 2013;47:139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu LJ, Holmes BR, Aronin N, et al. . CRISPRseek: a bioconductor package to identify target-specific guide RNAs for CRISPR-Cas9 genome-editing systems. PLoS One 2014;9:e108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilson KA, Chateau ML, Porteus MH. Design and development of artificial zinc finger transcription factors and zinc finger nucleases to the hTERT locus. Mol Ther Nucleic Acids 2013;2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walter DM, Venancio OS, Buza EL, et al. . Systematic in vivo inactivation of chromatin-regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res 2017;77:1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sena-Esteves M, Tebbets JC, Steffens S, et al. . Optimized large-scale production of high titer lentivirus vector pseudotypes. J Virol Methods 2004;122:131–139. [DOI] [PubMed] [Google Scholar]
- 37. Gray M, Shirasaki DI, Cepeda C, et al. . Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci 2008;28:6182–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hodgson JG, Agopyan N, Gutekunst CA, et al. . A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 1999;23:181–192. [DOI] [PubMed] [Google Scholar]
- 39. Zeitlin S, Liu JP, Chapman DL, et al. . Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet 1995;11:155–163. [DOI] [PubMed] [Google Scholar]
- 40. Platt RJ, Chen S, Zhou Y, et al. . CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014;159:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choudhury SR, Harris AF, Cabral DJ, et al. . Widespread central nervous system gene transfer and silencing after systemic delivery of novel AAV-AS vector. Mol Ther 2016;24:726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pinello L, Canver MC, Hoban MD, et al. . Analyzing CRISPR genome-editing experiments with CRISPResso. Nat Biotechnol 2016;34:695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brinkman EK, van Steensel B. Rapid quantitative evaluation of CRISPR genome editing by TIDE and TIDER. Methods Mol Biol 2019;1961:29–44. [DOI] [PubMed] [Google Scholar]
- 44. Chao MJ, Gillis T, Atwal RS, et al. . Haplotype-based stratification of Huntington's disease. Eur J Hum Genet 2017;25:1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palacios IM. Nonsense-mediated mRNA decay: from mechanistic insights to impacts on human health. Brief Funct Genom 2013;12:25–36. [DOI] [PubMed] [Google Scholar]
- 46. Hatch AC, Fisher JS, Tovar AR, et al. . 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab Chip 2011;11:3838–3845. [DOI] [PubMed] [Google Scholar]
- 47. Hindson BJ, Ness KD, Masquelier DA, et al. . High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ekman FK, Ojala DS, Adil MM, et al. . CRISPR-Cas9-mediated genome editing increases lifespan and improves motor deficits in a Huntington's disease mouse model. Mol Ther Nucleic Acids 2019;17:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lombardi MS, Jaspers L, Spronkmans C, et al. . A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol 2009;217:312–319. [DOI] [PubMed] [Google Scholar]
- 50. Vakulskas CA, Dever DP, Rettig GR, et al. . A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med 2018;24:1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Haasteren J, Li J, Scheideler OJ, et al. . The delivery challenge: fulfilling the promise of therapeutic genome editing. Nat Biotechnol 2020;38:845–855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.