Abstract
Cereblon (CRBN) is a substrate recruiter element of the E3 cullin 4-RING ubiquitin ligase complex, and a binding target of immunomodulatory agents (IMiDs). CRBN is responsible for the pleiotropic effects of IMiDs, yet its function in angiogenesis and in mediating the antiangiogenic effects of IMiDs remains unclear. We investigated the role of CRBN in the angiogenic process and in propagating the antiangiogenic effects of IMiDs in vitro. siRNA-mediated CRBN knock down in human endothelial cells (HUVEC and HMVEC-L), did not affect endothelial cell proliferation, migration, or tube formation. Using CRBN-deficient mice, we further demonstrated that microvessal formation can occur independently of cereblon in the ex vivo mouse aortic ring model. The cereblon E3 ubiquitin ligase complex can recruit endothelial cell-specific factors, AGO2 (associated with angiogenesis), and SALL4 (associated with embryogenesis/angiogenesis), for ubiquitin-mediated degradation. Knockdown of CRBN caused a corresponding increase in AGO2 and SALL4 protein expression and IMiD treatment was able to rescue the siCRBN effect to increase the CRBN expression. These findings suggest one potential mechanism of action that likely involves a tightly coordinated regulation of CRBN with endothelial cell targets and highlight the need to further elucidate the mechanism(s), which could include cereblon-independent pathways, through which IMiDs exert their antiangiogenic effects.
Keywords: angiogenesis, argonaute-2, cereblon, endothelial cell, immunomodulatory drugs, SALL4
1 |. INTRODUCTION
Cereblon (CRBN), an evolutionarily conserved and ubiquitously expressed protein, is a component of the E3 cullin 4-RING ubiquitin ligase complex (known as CRL4-CRBN), and recently established binding target of the immunomodulatory drugs (IMiDs) thalidomide, lenalidomide, and pomalidomide.1–4 CRBN functions as a substrate receptor of the E3 ubiquitin ligase complex to target proteins for proteolysis via the ubiquitin-proteasomal pathway. Under normal conditions, cellular CRBN undergoes auto-ubiquitination; but in the presence of thalidomide, the auto-ubiquitination process is inhibited, resulting in the accumulation of cereblon and leading to increased cullin-4 RING E3 ligase-mediated degradation of target proteins.1,5,6
CRL4-CRBN can mediate the ubiquitination and degradation of various endogenous substrates to elicit differential cellular functions. IMiD binding to CRBN may interrupt the binding of CRBN to its endogenous substrates, resulting in intracellular accumulation of substrates, and may simultaneously recruit altered substrates to promote their degradation by E3 ligase ubiquitination.5 Evidence for the necessity of cereblon in thalidomide-mediated immunomodulatory activity can be seen in both in vitro and in vivo models of multiple myeloma (MM), and involves the recruitment of downstream substrates, such as MEIS2, IKZF1, and IKZF3, and CD147 and MCT1, and their subsequent effects on cell proliferation and apoptosis.4,7–9 Silencing of cereblon in MM cells by transfection with a shRNA lentiviral expression system results in a significant reduction of cell viability (65%−78% reduction), and surviving cells acquire resistance to thalidomide treatment.2 Thalidomide-resistant MM cell lines (eg, OCI-My5) express lower levels of cereblon (both mRNA and protein) than sensitive cell lines (eg, MM1.S).2 Drug resistance has been reported to be reversed with introduction of wild-type cereblon to non-sensitive cell lines.10
Argonaute-2 (AGO2), a cancer microRNA regulator, was recently discovered as a binding partner for cereblon in lenalidomide-treated MM cells.11 AGO2 plays an important role as a mediator of small RNA-guided gene silencing.12 In endothelial cells, AGO2 is essential for endothelial cell survival and tube formation in vitro, suggesting that AGO2 modulates angiogenesis.13 Loss of early forming blood vessels has been proposed to be a cause of thalidomide-induced embryopathy.14 Misregulation of AGO2 may also impair vessel development during early angiogenesis, or promote the anticancer effects of thalidomide by miRNA misregulation, but this remains to be studied. SALL4, a spalt-like developmental transcription factor important for limb development, is another IMiD-dependent ubiquitination target of CRBN that has recently been implicated in thalidomide pathogenesis, with dose-dependent decreases in SALL4 protein expression observed following treatment of human embryonic stem cells with thalidomide, lenalidomide, and pomalidomide.15,16 SALL4 was recently shown to play a role in activating vascular endothelial growth factor A signaling, thereby promoting angiogenesis.17
CRBN plays important roles in brain development, inflammatory response, ion transport, modulating the AMP-activated protein kinase signaling pathway and cell metabolism as well as teratogenesis (reviewed in 18). In recent years, CRBN has been extensively studied because it is involved in many biological processes and may be responsible for the pleiotropic effects of IMiDs. However, its effect on embryonic development remains unclear, especially since CRBN knock out (KO) mice appear to be normal.19,20 In addition, its function in angiogenesis and in mediating the antiangiogenic effects of IMiDs is unknown. In this study, we investigated the role of CRBN in endothelial cells and its effect on angiogenesis, and in propagating the antiangiogenic effects of IMiDs.
2 |. MATERIALS AND METHODS
2.1 |. Cell culture
Human umbilical vein endothelial cells (HUVECs) and human lung microvascular endothelial cells (HMVEC-Ls) (Lonza, Walkersville, MD) were cultured in EGM-Plus and EGM-II MV, respectively (Lonza, Walkersville, MD). Cells were split by washing with HBSS, detaching with TryplE Express (Thermo Fisher Scientific, Waltham, MA), and spinning in a centrifuge (300G, 5 minutes). Cells were split at a density of 1:2–1:4 and used below passage 12.
2.2 |. Small interfering RNA transfection experiments
We optimized the siRNA transfection for each cellular subtype using Qiagen flexitube siRNA for CRBN (Stock 5μM). Specifically, HMVEC-L cells were transfected for 24 hours using Qiagen AllStars siCRBN #7 (25 nM) (Qiagen, Valencia, CA) and HUVEC cells were transfected for 24 hours using Qiagen AllStars siCRBN #3 (25 nM) (Qiagen, Valencia, CA). Both siRNA #3 and siRNA #7 were transfected with DharmaFect (Thermo Fisher Scientific, Walkersville, MD), using a method optimized for endothelial cells and small volume procedures.21 For all western blot experiments, and a subset of complimentary qPCR experiments, cells were transfected for 24 or 48 hours using CRBN siRNA-STEALTH-1 (Invitrogen) (5 μM) diluted in Lipofectamine 3000 transfection reagent (final concentration, 25 nM) according to the manufacturer’s specifications. Unless otherwise stated, experiments are performed in HUVEC cells, transfected with 25 nM Qiagen siCRBN #3, for 24 hours. Nonsilencing small interfering RNA (AllStars Negative Control, Qiagen, Valencia, CA) with no homology to any known mammalian gene was used as a negative control according to the manufacturer’s instructions. In some subsets of experiments, cells were incubated with drugs for 24 hours after the 24-hour transfection period. RNA was extracted using RNEasy kits (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
2.3 |. Quantitative RT-PCR
RNA was extracted from HUVEC and HMVEC-L cells using RNEasy kits (Qiagen, Valencia, CA) and concentrations were quantified using a Nanodrop. Stock solutions of 80 ng/μL were made for cDNA synthesis. cDNA was synthesized using the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). To quantify the gene expression levels, cDNA samples were amplified by combining 2 μL with 10 μL of gene expression master mix solution (Applied Biosystems, Foster City, CA), 7 μL sterile water, and 1 μL of the primer of interest. Primers were purchased as Taqman gene expression assays with 20× dye preloaded (Thermo Fisher Scientific, Waltham, MA). Samples were added to a clear 96-well plate at a final cDNA concentration of 64 ng/well. qPCR was run on a StepOnePlus RT-PCR system and optimized at 40 cycles. All qPCR reactions were run in triplicate and the expression levels were normalized to β-actin. The Taqman assay ID for cereblon was Hs00372271_m1.
2.4 |. Migration assay
Upon reaching 80% confluency the HUVECs were harvested, treated with negative or cereblon siRNA using DharmFECT Reagent 3 and plated at a density of 60 000 cells/mL (500 μL/well) to a 24-well plate. They were left to incubate at 37°C for 24 hours. A 1 mL pipette tip was used to scratch the monolayer across the center of the well. After scratching, the wells were washed twice with medium to remove the detached cells. The cells were imaged after 24 hours. Wells were imaged using an EVOS scope, and cell migration was quantified using ImageJ Version 1.51s (National Institutes of Health, US).
2.5 |. Endothelial cell tube formation assay
ECMatrix gel solution and 10× diluent buffer (Millipore, Darmstadt, Germany) were thawed slowly overnight on ice. One part diluent was added to nine parts gel using frozen pipette tips. The gel (50 μL/well) was plated in wells of a 96-well plate and left to set for 1 hour. HUVECs were then plated atop the gel and treated with 0.1% DMSO, 10 μg/mL (30 μM) to 60 μg/mL (180 μM) CPS49 as the positive control, 25 μg/mL (100 μM) to 100 μg/mL (380 μM) thalidomide, or 10 μM lenalidomide. The plates were returned to the incubator and imaged after 18 hours of incubation. Tubule formation was quantified using ImageJ.
2.6 |. Cell proliferation assay
HUVECs and HMVEC-Ls were seeded in 96-well plates in their respective medium. Following overnight incubation at 37°C, cells were transfected with siCRBN (AllStars #3 (optimized for HUVEC)/#7 (optimized for HMVEC-L) Qiagen, Valencia, CA) or a negative control (AllStars Negative Control, Qiagen, Valencia, CA) using DharmaFECT Reagent 3 (Thermo Fisher Scientific, Walkersville, MD). Twenty-four hours after siRNA transfection, cells were then treated with 100 μM thalidomide or 10 μM lenalidomide for 24 hours. Cell proliferation was measured using the Cell Counting Kit-8 assay according to the manufacturer’s instructions (Dojindo, Rockville, MD), and absorbance was read at 450 nm using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA).
2.7 |. Western blotting
Cells were transfected for 24 hours, and if required, treated with drugs (thalidomide, 100 μM/lenalidomide, 10 μM) for 24 hours. In experiments with drug treatments, a time 0 (0.1% DMSO) control was included. Cells were washed in ice-cold PBS, then lysed with RIPA lysis buffer (Sigma) and proteasomal inhibitors. Samples were left on ice for 15 minutes, briefly vortexed, and returned to the ice for 15 minutes. Lysed cells were centrifuged for 15 minutes at 16,000 × g at 4°C. The supernatant containing protein was carefully removed and quantification of total protein concentration was assessed using the Pierce bicinchoninic acid (BCA) assay (Thermo Fisher Scientific, Waltham, MA). Samples were run on 4%−12% Nupage Bis-Tris gels (Thermo Fisher Scientific, Waltham, MA) and nitrocellulose membranes were probed with antibodies against GAPDH (1:2000) (Sigma, St. Louis, MO), CRBN (1:500) (HPA045910, Sigma Prestige, St. Louis, MO), AGO2 (1:500) (Sigma Prestige, St. Louis, MO), or SALL4 (1:50) (abcam, Cambridge, MA). Blotting was detected by incubating with IR-secondary antibodies and scanning the membrane at the appropriate wavelengths (Licor, Lincoln, NE).
2.8 |. Animals
Germline cereblon-deficient mice (Crbn−/−) were a generous gift from Dr Pearlie Epling-Burnett (Moffitt Cancer Centre, FL) and Dr Anjali Rajadhyaksha (Weill Cornell Medical College, NY). C57BL/6 (Crbn+/+) mice were purchased from Charles River Laboratories (Wilmington, MA). Generation of Crbn−/− mice was described previously20,22,23 with germ-line CRBN deletion in all tissues. Gene deletion was confirmed using wild-type and CRBN-knockout specific primers per published protocol.20,22 The National Cancer Institute (NCI) is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International and follows the Public Health Service (PHS) Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the Guide for the Care and Use of Laboratory Animals. The study protocol was approved by the NCI Animal Care and Use Committee.
2.9 |. Aortic ring angiogenesis assay
Mouse aortas were extracted from Crbn+/+ and Crbn−/− mice and cleaned in EBM media. The aortas were then sliced to approximately 1 mm sections and embedded in Matrigel (Corning, Corning, NY) for 1 hour. EGM-II media was then added overnight. Mouse aortic rings were imaged 5 days later. Images of microvessel outgrowths were quantified using ImageJ Version 1.51s (National Institutes of Health, US) and Adobe Photoshop CC (Adobe Systems Incorporated, San Jose, CA). This was replicated three times using aortas from different mice.
2.10 |. Statistical analysis
Analysis was conducted using GraphPad Prism Version 7.0 (GraphPad Software, La Jolla, CA) and statistical significance was assessed using two-tailed Student’s t tests or ANOVA analyses. Error bars represent standard error of the mean.
3 |. RESULTS
To investigate the function of cereblon in vitro angiogenesis, we used small interfering RNAs (siRNAs) to knock down CRBN expression in endothelial cells. Endothelial cell lines are notoriously difficult to transfect.21 In order to confirm knockdown could be achieved robustly, we assessed previously used siRNAs for CRBN (Stealth, Invitrogen) in two endothelial cell lines (HUVEC and HMVEC-L), and optimized a commercial (Qiagen flexitube) siRNA for each cell line. CRBN siRNA (Qiagen si3 and Invitrogen Stealth siRNA) reduced CRBN mRNA expression in HUVECs to 14.39% and 12.96% (>85% knockdown efficiency), respectively (P < .0001). In HMVEC-Ls, a similar knockdown efficiency (~77%) was observed with Qiagen si7 (23.18% expression) and Invitrogen stealth siRNA (22.83% expression) for CRBN (Figure 1A). Knockdown was confirmed at the protein level by western blot analysis demonstrating an increase in knockdown efficiency that is time-dependent. (Figure 1B). In addition, transfection was confirmed by observing GFP-tagged siRNA. The fluorescently tagged siRNA was seen to localize in the cytoplasm of HUVECs (Figure 1C). Surprisingly, no significant difference in cell proliferation was observed in either HUVEC or HMVEC-L cells following loss of cereblon after 24-hour transfection (Figure 1D). This suggests that silencing of cereblon expression does not alter the endothelial cell viability in vitro.
FIGURE 1.
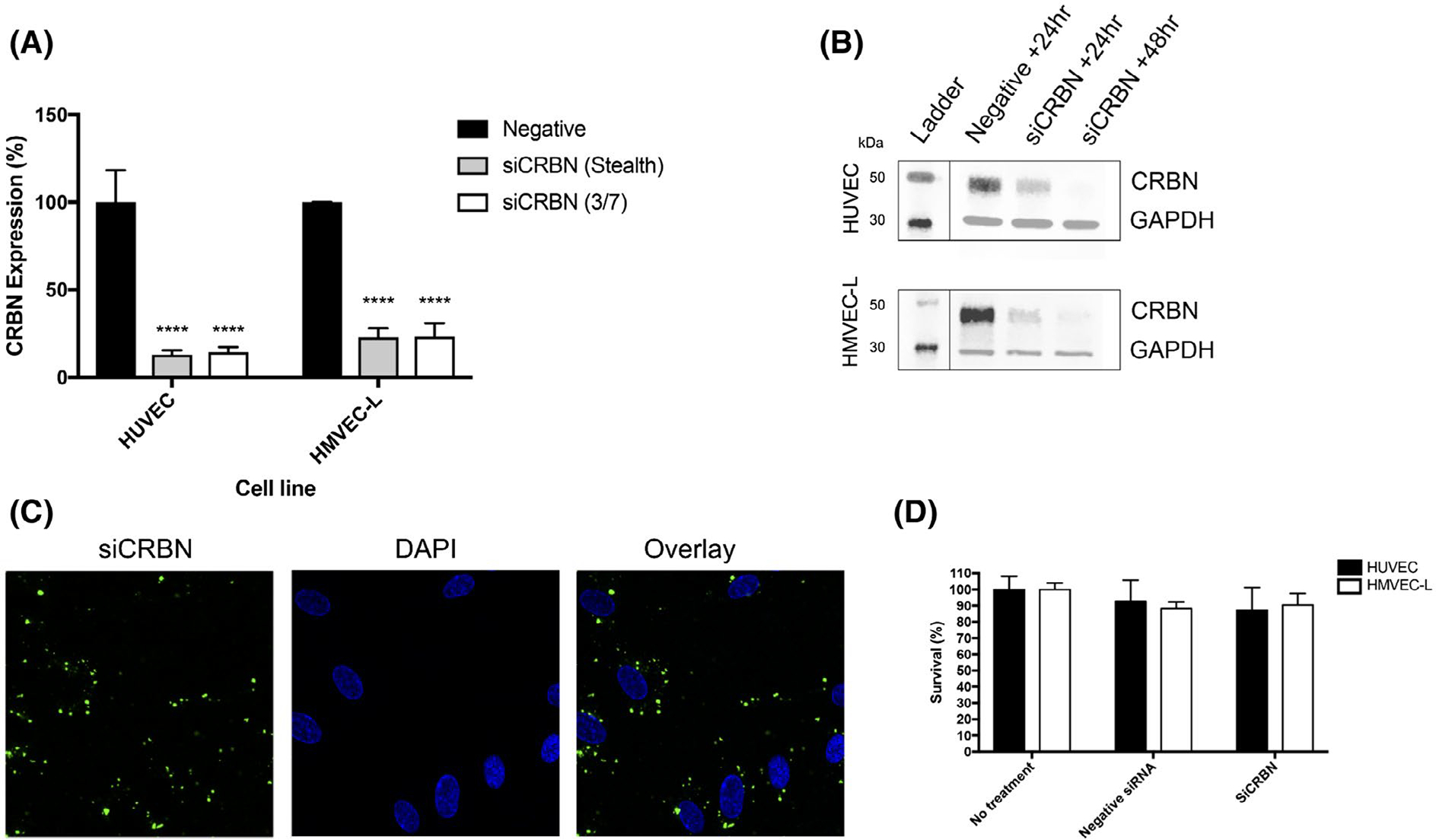
Loss of cereblon can be achieved in both HUVEC and HMVEC-L endothelial cells. A, Gene expression of cereblon is reduced following 24-hour transfection with siRNA for CRBN from Invitrogen (Stealth) and Qiagen (CRBN 3 HUVEC, CRBN 7 HMVEC-L) (P < .0001). B, Protein expression is reduced between 24 and 48 hours following transfection with Lipofectamine 3000. C, Representative images of fluorescently tagged siCRBN can be visualized in the cells. D, 24-hour transfection with siCRBN using Dharmafect Reagent 3 does not reduce cell viability in either cell line. Results shown are representative of at least three independent experiments.
We evaluated for any phenotypic or functional changes to the endothelial cells as a result of the siRNA-mediated knockdown of CRBN. We observed that there were no differences in the phenotype of HUVECs after transfection (Figure 2A). Next, we conducted an in vitro angiogenesis assay to determine the effect of CRBN knockdown on endothelial cell tube formation. Transfected HUVECs were cultured on ECM gel and left to form tubule structures. We observed no difference in the tubules formed by either negative siRNA transfected cells or those transfected with siCRBN (Figure 2B). Finally, we used the scratch assay to monitor migration and proliferation of the cells. By the 24-hour time point, both treatment groups were able to heal the wound area, suggesting migration and proliferation is not affected by loss of cereblon (Figure 2C).
FIGURE 2.
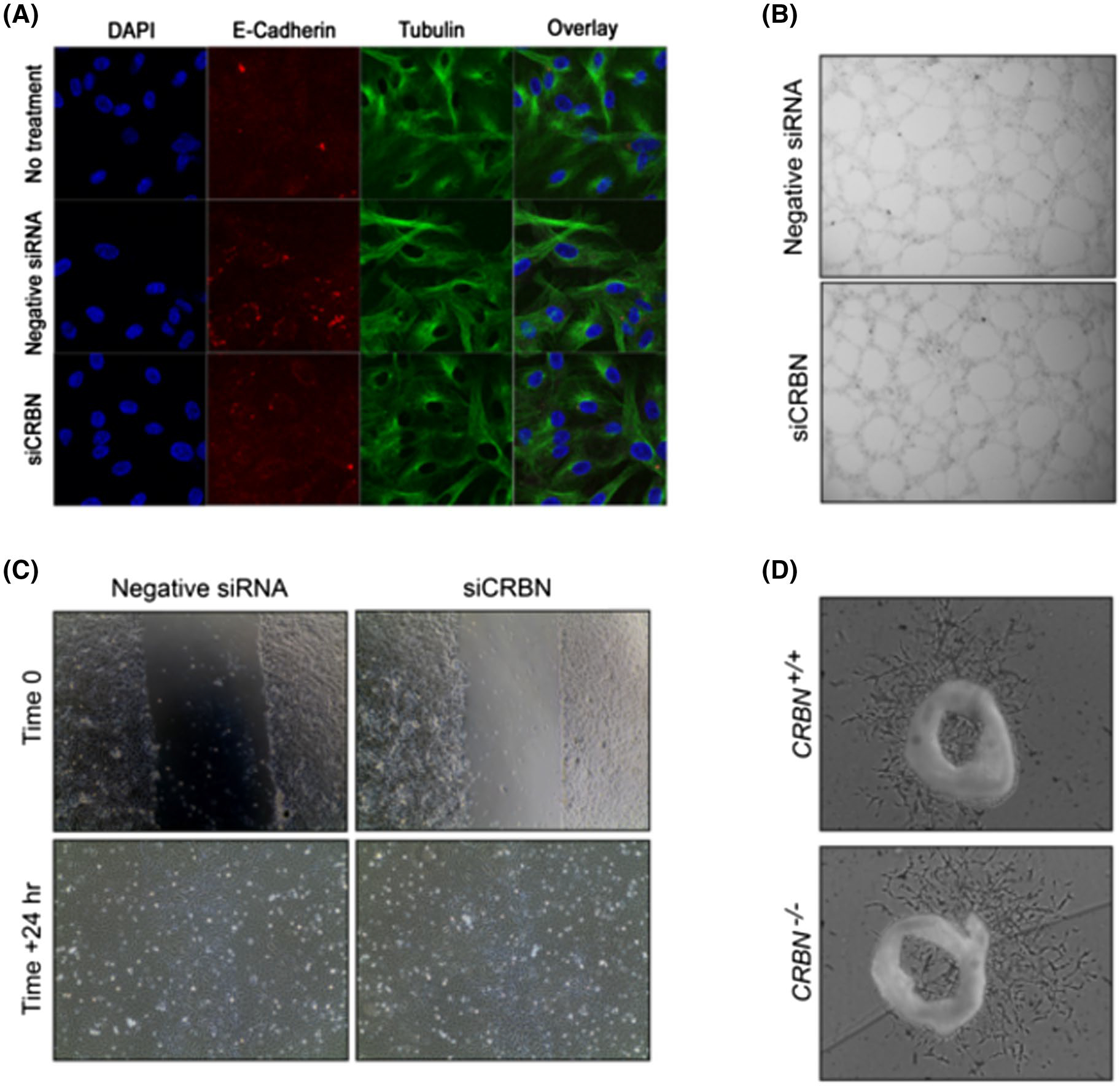
Loss of cereblon does not affect endothelial cell phenotype and does not change functional properties in select in vitro and ex vivo angiogenesis assays. A, Antibody staining shows HUVECs are intact after transfection. Red is E-Cadherin, green is tubulin, blue is DAPI DNA stain. B, HUVECs can form tubules regardless of cereblon expression. C, Loss of cereblon does not affect migration and proliferation of HUVECs. D, Loss of cereblon in CRBN−/− mice does not inhibit mouse aortic ring outgrowth.
We then examined whether loss of cereblon affects microvessel formation using the three-dimensional ex vivo mouse aortic ring model,24 which recapitulates the complexities of angiogenesis and combines the advantages of in vitro and in vivo models. We sectioned the mouse aorta obtained from germ-line CRBN knockout (Crbn−/−) mice. Aortic rings were cultured in Matrigel and the extent of microvessel outgrowth was quantified. After a 5-day incubation, no differences were observed in the extent of microvessel outgrowths from Crbn+/+ or Crbn−/− tissues (Figure 2D). Together, these data suggest that CRBN is dispensable for in vitro angiogenesis.
We next asked whether CRBN is required for the antiangiogenic activity of IMiDs in endothelial cells. We knocked down CRBN followed by drug treatment and assessed for the effect on endothelial cell tube formation. Following gene knockdown and IMiD treatment, tube formation was inhibited by drug treatments in both siCRBN and mock-transfected (wildtype) cells (Figure 3A). Notably, lenalidomide treatment of siCRBN-mediated knockdown cells showed less inhibition in tubule formation (as compared to lenalidomide-treated wild-type cells) following treatment for 18 hours (Figure 3B). The positive control, CPS49, a well-characterized potent antiangiogenic analog of thalidomide25–28 also inhibited lattice formation regardless of CRBN knockdown.
FIGURE 3.
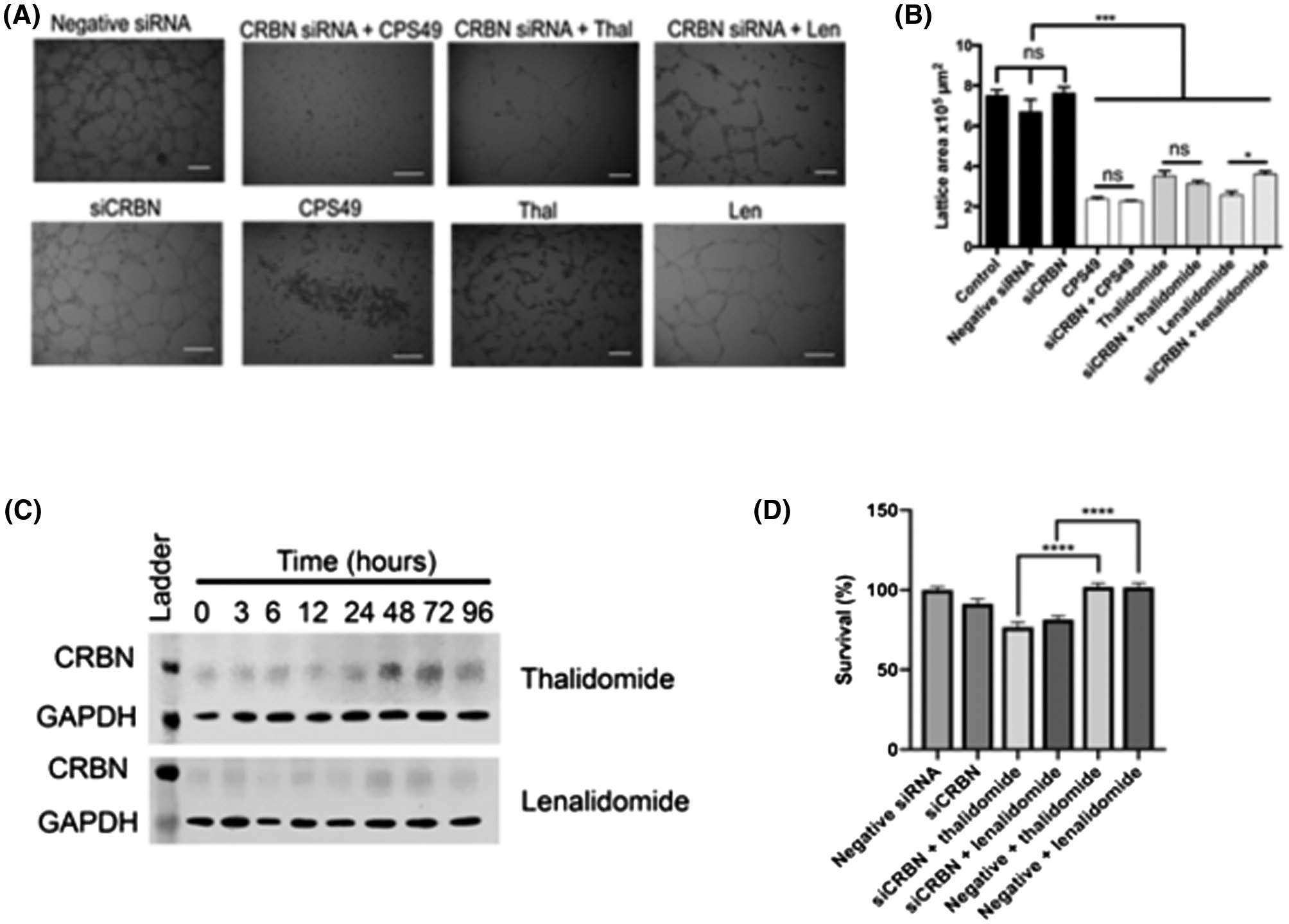
Loss of cereblon partially mediates IMiD-induced antiangiogenic effects in vitro. A, Tubule formation in HUVECs is inhibited following treatment with CPS49, and reduced with thalidomide and lenalidomide treatment. This occurs in both siCRBN-transfected and mock-transfected cells. B, Tubule formation area quantified (****P < .001, *P = .01). C, Cereblon protein appears to increase following treatment with thalidomide and lenalidomide. D, HUVEC cell survival following 24-hour drug treatment (thalidomide or lenalidomide) is significantly reduced in siCRBN-transfected cells, compared to WT (mock-transfected) cells. Results shown are representative of at least three independent experiments with representative images shown.
Since thalidomide has been shown to inhibit CRBN autoubiquitination, resulting in accumulation of CRBN,5 we assessed for cereblon protein expression following treatment with IMiDS. Consistent with previous studies in myeloma cells,5 we found there was an increase in cereblon protein band intensity following thalidomide and lenalidomide treatment in endothelial cells in a time-dependent manner (Figure 3C). This suggests that cereblon may be partially responsible for mediating the effects of IMiDs on endothelial cells. In addition, while HUVEC proliferation was not significantly affected by CRBN knockdown or drug treatment (thalidomide or lenalidomide) alone, cell proliferation was decreased following drug treatment in siCRBN-transfected cells compared to mock-transfected cells (Figure 3D). This raises the question of whether other targets besides cereblon may be involved in mediating the effect of the drug.
We further explored possible endogenous substrates for CRL4-CRBN that may be involved in regulating angiogenesis. AGO2 is the first identified binding partner recruited by cereblon with a known role in endothelial cell viability and in vitro angiogenesis.13,29 Treatment of MM cells with lenalidomide caused an increase in CRBN expression, with a subsequent decrease in AGO2 expression11; the reduction in cellular AGO2 levels led to a decrease in cell survival, regardless of drug sensitivity. In addition, treatment of human embryonic stem cells with thalidomide, lenalidomide, and pomalidomide has been shown to induce the degradation of SALL4, which has been causatively linked to limb and inner organ birth defects.15,16 Thus, we aimed to determine whether cereblon can also modulate AGO2 and SALL4 regulation in endothelial cells. Protein expression was assessed following 24-hour and 48-hour transfection with non-coding siRNA or siCRBN. Knockdown of cereblon in siCRBN-transfected cells caused a significant increase in both AGO2 and SALL4 protein levels (Figure 4A), suggesting that CRBN may modulate levels of AGO2 and SALL4 in endothelial cells.
FIGURE 4.
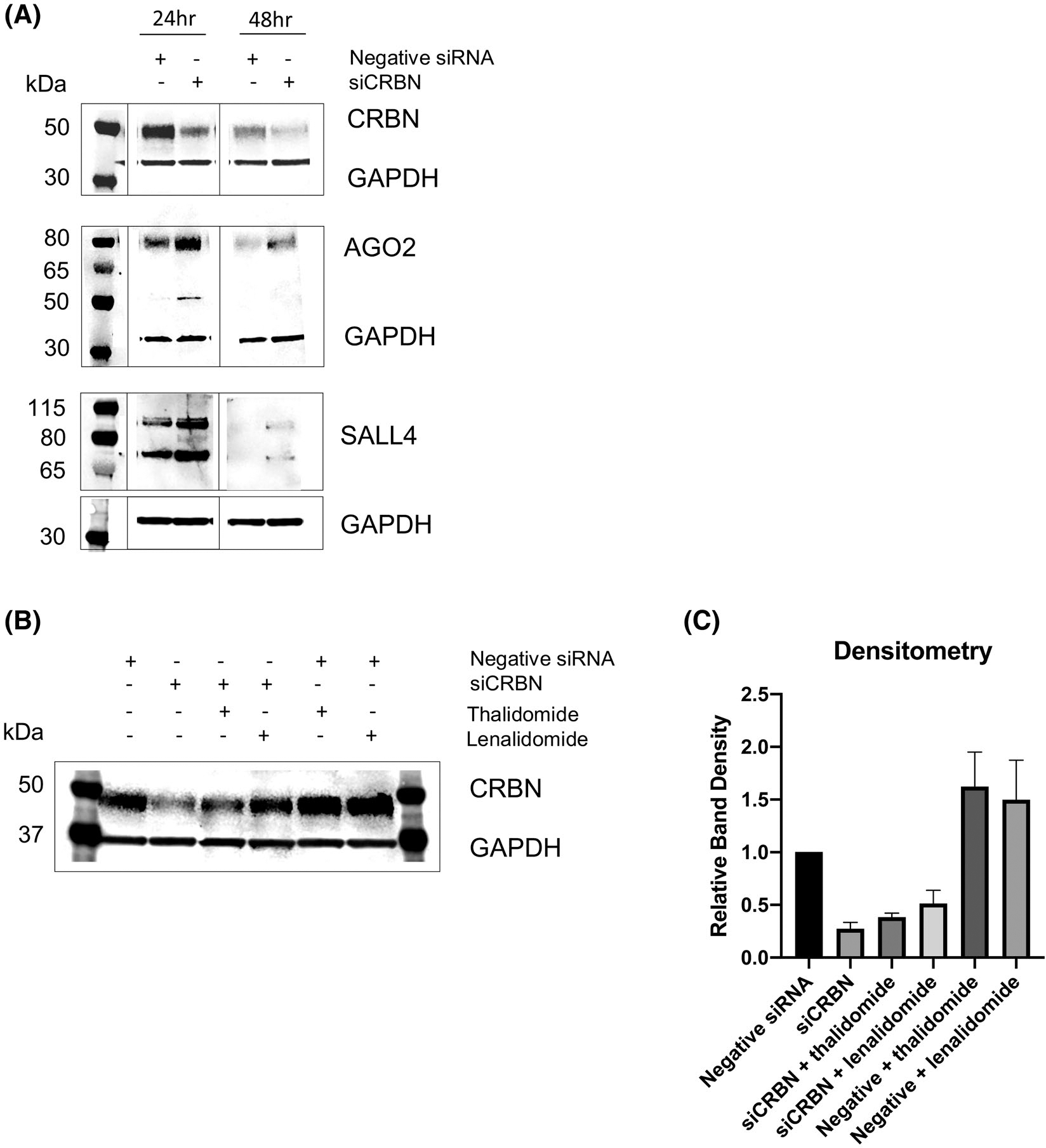
Protein expression of cereblon binding partner Argonaut-2 (AGO2) and spalt-like transcription factor (SALL4) following siCRBN knockdown and IMiD drug treatment in HUVECs. A, Loss of cereblon following 24-hour and 48-hour transfection using Stealth siRNA and Lipofectamine 3000 increases both AGO2 and SALL4 protein levels. B, CRBN protein expression levels returns to baseline following thalidomide treatment and recovers above baseline following thalidomide/lenalidomide treatment in siCRBN knockdown HUVECs. C, Corresponding densitometry analysis of blot shown in (B). Results shown are representative of at least three independent experiments with one representative blot shown.
We conducted Western analysis for cereblon expression in siCRBN knockdown HUVECs treated with IMiDs and discovered that CRBN protein expression returned to baseline levels for thalidomide and recovered above baseline levels for lenalidomide (Figure 4B). These results demonstrated that treatment with IMiDs following siCRBN knockdown rescued the siRNA effect to increase cereblon expression back to at least baseline levels (ie, they are no longer cereblon knockdowns). Cereblon expression was also significantly increased in mock-transfected HUVECs when treated with IMiDs, consistent with our protein data in Figure 3C.
4 |. DISCUSSION
Angiogenesis is essential for proper development of the embryo, and evidence exists indicating that the antiangiogenic activity of thalidomide can cause teratogenic effects.27,30 However, the molecular mechanism of IMiD-induced antiangiogenic effects is not well documented. Cereblon (in complex with DDB1 as part of the E3 cullin 4-RING ubiquitin ligase complex) is a recently identified binding target of thalidomide, and is thought to be the instigator of thalidomide embryopathy1 and mediator of the immunodulatory effects of the drug. Zebrafish embryos expressing zCRBNYW/AA produce a mutant cereblon protein that cannot bind to thalidomide, and do not develop any discernible defects following thalidomide treatment.1 From this observation, the authors concluded that thalidomide might exert teratogenic effects by binding to, and inhibiting the function of, cereblon. They also hypothesized that the teratogenic effects of thalidomide-cereblon binding may be due to antiangiogenic actions.
Our study aimed to (i) elucidate the role of cereblon in in vitro angiogenesis and (ii) examine the role of cereblon in mediating the antiangiogenic effects of IMiDs. We first evaluated how to best detect cereblon in endothelial cells. An evaluation of commercial cereblon antibodies by Gandhi et al found that only 3 of 7 tested detected a putative cereblon band, and often a large number of bands of varying molecular weights were also detected.31 We have found that the Sigma Prestige antibody appears suitable for in vitro purposes and gives similar results to qPCR assays (Figure 1A). We then showed that cereblon is expressed in endothelial cells, and investigated the role of cereblon in the angiogenic process using in vitro and ex vivo angiogenesis assays in human and mouse systems. We found that silencing of cereblon did not affect human endothelial cell proliferation, migration, or tube formation. Using CRBN-deficient mice, we further demonstrated that microvessel formation can occur independently of cereblon in the ex vivo mouse aortic ring model. This suggests that cereblon is not essential for angiogenesis in both human and mouse in vitro systems, and is not surprising since CRBN knockout mice appear morphologically normal,19,20 although the genetic background of a knockout mouse can have effects on disease phenotype.32 Interestingly, cereblon knockdown had no effect on the survival of primary effusion lymphoma (PEL) cells but is essential for the antiproliferative activity of IMiDs in PEL cells.33
We next determined whether CRBN is involved in mediating the antiangiogenic activity of IMiDs. We found that IMiD treatment of wild-type or siCRBN-transfected endothelial cells resulted in inhibition of tube formation, and showed that CRBN can be increased upon treatment with IMiDs in endothelial cells, such that the siCRBN knockdown effect can be rescued. This implies that cereblon may be partly involved in mediating the activity of IMiDs in endothelial cells. Given that CRL4-CRBN binds a plethora of endogenous substrates, we hypothesized that AGO2, which is essential for endothelial cell function, may be a possible target substrate that cooperates with CRBN to mediate the antiangiogenic effects of IMiDs. Our data showed that loss of CRBN caused a corresponding increase in AGO2 (and SALL4) protein expression, suggesting that thalidomide and lenalidomide may interact with other targets besides CRBN in endothelial cells to exert its antiangiogenic effect. The interplay of AGO2 (and SALL4) expression is tightly coordinated and closely regulated by its interaction with CRBN. Indeed, previous studies have identified AGO2 as a cereblon binding partner and shown that steady-state levels of AGO2 were regulated by CRBN in MM cells.11 Recent research has demonstated that ubiquitously expressed genes can interact with cell type-specific factors to establish cell type-specific features or function.34 Similarly, our findings established the link that the ubiquitously expressed cereblon may cooperate with AGO2, a known modulator of endothelial cell function, to mediate the antiangiogenic effects of IMiDs like lenalidomide. Further studies are required to elucidate the precise relationship between cereblon and AGO2 regulation.
Given the multitude of biological effects of thalidomide, it is likely there are multiple mechanisms causing teratogenesis. Indeed, the original paper identifying cereblon as a binding target discusses the possibility of multiple targets.1 More recent structural studies also discuss how teratogenicity and other physiological effects may occur through distinct mechanisms, including highlighting the likelihood of multiple endogenous substrates, as well as ubiquitin-independent roles for CRBN or CRBN-independent mechanisms.35 This hypothesis is entirely conceivable based upon the vast number of actions and the species specificity of thalidomide alone. Indeed data have supported the increased activity of thalidomide analogs with minor functional group changes (lenalidomide, pomalidomide) and also those without the glutarimide ring, such as apremilast, or those with large functional groups added to the phthalimide group, such as CC-885.36 In addition, a study using mice with humanized livers (CYP3A-HAC) to identify target binding proteins of thalidomide did not pull down cereblon.37 Although the study used a modified hydroxy-thalidomide compound, it emphasized that multiple binding targets are likely to exist.
The discovery of new cerebon binding targets is on the rise, with the most recent studies identifying p63 as a cereblon substrate involved in thalidomide teratogenicity38 and several C2H2 zinc finger transcription factors, including SALL4, a member of the spalt-like family of developmental transcription factors.15,16 While SALL4 was identified as a potential binding target and mutations in SALL4 lead to congenital defects in humans,15,16 the mechanism of how thalidomide-induced degradation of SALL4 contributes to these defects remains to be determined (reviewed in 39). The expression of SALL4 has recently been identified in the endothelium of venous malformations.40 SALL4 was also recently shown to promote endothelial cell growth, migration, and tube formation via targeting VEGFA4; however, further work is needed to address the mechanistic role of SALL4 in angiogenesis and thalidomide embryopathy. Moreover, Fink et al demonstrated in the CrbnI391V knock-in mouse that thalidomide induced fetal loss but phocomelia and amelia were absent in this model, perhaps due to additional species-specific differences in the sequence of downstream targets, the phenotypic consequences of their degradation, or their dependence on Crbn residues other than I391V.40
Angiogenesis is a highly complex multistep process indispensable for developmental growth coupled with endothelial cell behavior that is tightly controlled by various transcription factors for gene expression throughout the development (and adulthood). Our findings suggest that the antiangiogenic properties of IMiDs are essential to thalidomide embryopathy syndrome and likely involve a tightly regulated association of CRBN with developmental-specific and/or endothelial cell-specific factors such as AGO2 and SALL4. Since CRBN is dispensable for angiogenesis and knockdown of CRBN caused an increase in endothelial cell-specific factors, this suggests the existence of alternative targets or a cereblon-independent mechanism of IMiD action. Therefore, future studies are warranted to elucidate whether additional co-factor(s)/conditions or a multi-gene coordinated effort is needed for CRBN as well as CRBN-independent pathways in fully mediating the antiangiogenic effects of IMiDs. Whether these targets or other new undiscovered targets can compensate for cereblon’s interaction with IMiDs to exert their anti-angiogenic activity remains to be determined. Understanding the precise role of CRBN in mediating IMiDs’ antiangiogenic action may help to identify more novel and safer therapeutics with increased specificity or unwanted side effects.
ACKNOWLEDGMENTS
The authors thank Prof. Lynda Erskine and Dr Lucas Rosa Fraga for discussions on cereblon and Dr Tristan Sissung for advice on primers and genotyping, and to Afua Akuffo for helpful discussion. We are especially grateful to Drs. Pearlie Burnette (Moffitt Cancer Institute, FL) and Anjali Rajadhyaksha (Weill Cornell Medical College, NY) for providing the cereblon knockout mice. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the US Government. This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health (ZIA SC006538) and a Wellcome Trust-NIH PhD Studentship to SB, WDF, and NV (Grant number 098252/Z/12/Z).
Funding information
National Cancer Institut; National Institutes of Health, Grant/Award Number: ZIA SC006538; Wellcome Trust-NIH, Grant/Award Number: 098252/Z/12/Z
Abbreviations:
- AGO2
argonaute-2
- CRBN
cereblon
- CRL4-CRBN
E3 cullin 4-RING ubiquitin ligase complex
- HUVECs
human umbilical vein endothelial cells
- HMVEC-Ls
human lung microvascular endothelial cells
- IMiDs
immunomodulatory agents
- SALL4
spalt like transcription factor 4
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interest.
REFERENCES
- 1.Ito T, Ando H, Suzuki T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. [DOI] [PubMed] [Google Scholar]
- 2.Zhu YX, Braggio E, Shi CX, et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118:4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chamberlain PP, Lopez-Girona A, Miller K, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol. 2014;21:803–809. [DOI] [PubMed] [Google Scholar]
- 4.Kronke J, Udeshi ND, Narla A, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Huang X, He X, et al. A novel effect of thalidomide and its analogs: suppression of cereblon ubiquitination enhances ubiquitin ligase function. FASEB J. 2015;29:4829–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Girona A, Mendy D, Ito T, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjorklund CC, Lu L, Kang J, et al. Rate of CRL4(CRBN) substrate Ikaros and Aiolos degradation underlies differential activity of lenalidomide and pomalidomide in multiple myeloma cells by regulation of c-Myc and IRF4. Blood Cancer J. 2015;5:e354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu G, Middleton RE, Sun H, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichner R, Heider M, Fernandez-Saiz V, et al. Immunomodulatory drugs disrupt the cereblon-CD147-MCT1 axis to exert antitumor activity and teratogenicity. Nat Med. 2016;22:735–743. [DOI] [PubMed] [Google Scholar]
- 10.Zhu YX, Kortuem KM, Stewart AK. Molecular mechanism of action of immune-modulatory drugs thalidomide, lenalidomide and pomalidomide in multiple myeloma. Leuk Lymphoma. 2013;54:683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Wang X, Cheng Z, Zhu Q. AGO2 and its partners: a silencing complex, a chromatin modulator, and new features. Crit Rev Biochem Mol Biol. 2020;55:33–53. [DOI] [PubMed] [Google Scholar]
- 12.Asai T, Suzuki Y, Matsushita S, et al. Disappearance of the angiogenic potential of endothelial cells caused by Argonaute2 knockdown. Biochem Biophys Res Commun. 2008;368:243–248. [DOI] [PubMed] [Google Scholar]
- 13.Vargesson N Thalidomide-induced limb defects: resolving a 50-year-old puzzle. BioEssays. 2009;31:1327–1336. [DOI] [PubMed] [Google Scholar]
- 14.Donovan KA, An J, Nowak RP, et al. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. eLife. 2018; 7: pii: e38430. 10.7554/elife.38430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matyskiela ME, Couto S, Zheng X, et al. SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat Chem Biol. 2018;14:981–987. [DOI] [PubMed] [Google Scholar]
- 16.Sun J, Zhao Z, Zhang W, et al. Spalt-like protein 4 (SALL4) promotes angiogenesis by activating vascular endothelial growth factor A (VEGFA) signaling. Med Sci Monit. 2020;26:e920851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HK, Ko TH, Nyamaa B, et al. Cereblon in health and disease. Pflugers Arch. 2016;468:1299–1309. [DOI] [PubMed] [Google Scholar]
- 18.Lee KM, Yang SJ, Kim YD, et al. Disruption of the cereblon gene enhances hepatic AMPK activity and prevents high-fat diet-induced obesity and insulin resistance in mice. Diabetes. 2013;62:1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajadhyaksha AM, Ra S, Kishinevsky S, et al. Behavioral characterization of cereblon forebrain-specific conditional null mice: a model for human non-syndromic intellectual disability. Behav Brain Res. 2012;226:428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennstedt E, Bryan B. siRNA knockdown of gene expression in endothelial cells. Methods Mol Biol. 2011;764:215–222. [DOI] [PubMed] [Google Scholar]
- 21.Akuffo AA, Alontaga AY, Metcalf R, et al. Ligand-mediated protein degradation reveals functional conservation among sequence variants of the CUL4-type E3 ligase substrate receptor cereblon. J Biol Chem. 2018;293:6187–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bavley CC, Rice RC, Fischer DK, et al. Rescue of learning and memory deficits in the human nonsyndromic intellectual disability cereblon knock-out mouse model by targeting the AMP-activated protein kinase-mTORC1 translational pathway. J Neurosci. 2018;38:2780–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruger EA, Duray PH, Tsokos MG, et al. Endostatin inhibits microvessel formation in the ex vivo rat aortic ring angiogenesis assay. Biochem Biophys Res Commun. 2000;268:183–191. [DOI] [PubMed] [Google Scholar]
- 24.Mahony C, McMenemy S, Rafipay AJ, et al. CPS49-induced neurotoxicity does not cause limb patterning anomalies in developing chicken embryos. J Anat. 2018;232:568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahony C, Erskine L, Niven J, Greig NH, Figg WD, Vargesson N. Pomalidomide is nonteratogenic in chicken and zebrafish embryos and nonneurotoxic in vitro. Proc Natl Acad Sci USA. 2013;110:12703–12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Therapontos C, Erskine L, Gardner ER, Figg WD, Vargesson N. Thalidomide induces limb defects by preventing angiogenic outgrowth during early limb formation. Proc Natl Acad Sci U S A. 2009;106:8573–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng SS, Gutschow M, Weiss M, et al. Antiangiogenic activity of N-substituted and tetrafluorinated thalidomide analogues. Cancer Res. 2003;63:3189–3194. [PubMed] [Google Scholar]
- 28.Wu S, Yu W, Qu X, et al. Argonaute 2 promotes myeloma angiogenesis via microRNA dysregulation, Journal of Hematology & Oncology. 2014; 7: 40. 10.1186/1756-8722-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vargesson N Thalidomide-induced teratogenesis: history and mechanisms. Birth Defects Res C Embryo Today. 2015;105:140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gandhi AK, Mendy D, Waldman M, et al. Measuring cereblon as a biomarker of response or resistance to lenalidomide and pomalidomide requires use of standardized reagents and understanding of gene complexity. Brit J Haematol. 2014;164:233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisener-Dorman AF, Lawrence DA, Bolivar VJ. Cautionary insights on knockout mouse studies: the gene or not the gene? Brain Behav Immun. 2009;23:318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gopalakrishnan R, Matta H, Tolani B, Triche T Jr, Chaudhary PM. Immunomodulatory drugs target IKZF1-IRF4-MYC axis in primary effusion lymphoma in a cereblon-dependent manner and display synergistic cytotoxicity with BRD4 inhibitors. Oncogene. 2016;35:1797–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng G, Tong M, Xia B, et al. Ubiquitously expressed genes participate in cell-specific functions via alternative promoter usage. EMBO Rep. 2016;17:1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akuffo A A, Alontaga A Y, Lawrence H R, Lawrence N J, Epling-Burnette P K. Controversy regarding the functional conservation of cereblon CUL4-type E3 ligase substrate receptor. Integr Cancer Sci Therap. 2018;5:1–3. [Google Scholar]
- 35.Matyskiela ME, Lu G, Ito T, et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature. 2016;535:252–257. [DOI] [PubMed] [Google Scholar]
- 36.Yamazaki H, Suemizu H, Kazuki Y, et al. Assessment of protein binding of 5-hydroxythalidomide bioactivated in humanized mice with human P450 3A-chromosome or hepatocytes by two-dimensional electrophoresis/accelerator mass spectrometry. Chem Res Toxicol. 2016;29:1279–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asatsuma-Okumura T, Ando H, De Simone M, et al. p63 is a cereblon substrate involved in thalidomide teratogenicity. Nat Chem Biol. 2019;15:1077–1084. [DOI] [PubMed] [Google Scholar]
- 38.Vargesson N The teratogenic effects of thalidomide on limbs. J Hand Surg Eur. 2019;44:88–95. [DOI] [PubMed] [Google Scholar]
- 39.Tan EMS, Siljee SD, Brasch HD, Enriquez S, Tan ST, Itinteang TEmbryonic Stem Cell-Like Subpopulations in Venous Malformation, Front. Med. 2017; 4: 162. 10.3389/fmed.2017.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fink EC, McConkey M, Adams DN, et al. Crbn(I391V) is sufficient to confer in vivo sensitivity to thalidomide and its derivatives in mice. Blood. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]


