Abstract
A Pasteurella multocida species-specific oligonucleotide probe, pmhyb449, targeting 16S rRNA was designed and evaluated by whole-cell hybridization against 22 selected reference strains in animal tissues. It differentiated P. multocida from other bacterial species of the families Pasteurellaceae and Enterobacteriaceae and also from divergent species of the order Cytophagales (except biovar 2 strains of Pasteurella avium and Pasteurella canis, which have high 16S rRNA similarity to P. multocida). The potential of the probe for specific identification and differentiation of P. multocida was further detected in formalin-fixed paraffin-embedded lung tissues from experimental fowl cholera in chickens and infections in pigs. In chicken lung tissues P. multocida cells were detected singly, in pairs, as microcolonies, and as massive colonies within air capillaries (septa and lumen), parabronchial septa, and blood vessels (wall and lumen). In pig lung, postmortem-injected P. multocida was detected in the alveoli (lumen and wall), and in both animals the bacterial cells were seen in the bronchi. The results showed that with the oligonucleotide probe pmhyb449, fluorescent in situ hybridization is a suitable and fast method for specific detection of P. multocida in histological formalin-fixed tissues. The test was replicable and reproducible and is recommended as a supplementary test for diagnosis and as a tool in pathogenesis studies of fowl cholera and respiratory tract infections in pigs due to P. multocida.
Pasteurella multocida is an important pathogen that infects many production animals and is an opportunistic human pathogen (7, 21). In poultry, infection with P. multocida may result in fowl cholera, a disease of economic importance in commercial production that may occur in different forms, such as peracute, acute, and chronic infections (16, 43). In pigs, P. multocida is commonly associated with atrophic rhinitis, pneumonia, and septicemic pasteurellosis. Pneumonic pasteurellosis due to P. multocida is common and is of major economic importance for industrial pig production (41). The histology, clinical signs, and macroscopic lesions associated with P. multocida infections in poultry and pigs are not pathognomonic and can be mixed up with other respiratory system infections characterized by upper respiratory tract inflammations, pneumonia, airsacculitis, polyserositis, and septicemia (14, 33, 41, 43), and hence the diagnosis depends on specific detection of the causative organism.
Detection and characterization of P. multocida by phenotypic characteristics including serotype have been dependent on the ability to cultivate and purify the bacteria in the laboratory (16). Cultivation and identification by standard bacteriological methods can be ambiguous because of V-factor requirements or nontypeable strains, including cross-reaction in serotyping and viable but nonculturable cells (30).
In situ hybridization (ISH) allows precise localization of a specific segment of nucleic acid within a histologic section (13) or detection of specific rRNA in morphologically intact bacteria cells (5). ISH combines basic molecular biological techniques and the ability to appreciate subtle histomorphologic changes (13). The key feature distinguishing ISH from other molecular methodologies (filter hybridization and PCR) is that the sample DNA or RNA is detected directly in the intact cell rather than being extracted from the cell before testing (40). The cell morphology and its abundance and spatial distribution can be analyzed in situ (5, 42).
With fluorescent-labeled probes, in situ hybridization (FISH) has excellent spatial resolution (42). FISH has been used to visualize the spatial distribution of Escherichia coli in intestinal mucosa (42); to evaluate colonization of mouse intestines by Salmonella enterica serovar Typhimurium (32); and to specifically detect Salmonella serovars in pig intestines and mouse lungs (40), Actinobacillus pleuropneumoniae strains in diseased porcine tissues (23, 26), Brachyspira (Serpulina) pilosicoli infections in the intestines of growing pigs (11, 27), Lawsonia intracellularis in porcine proliferative enteropathy (10), Streptococcus suis infection in pigs (12), and bacteria in blood cultures (25, 29).
The aim of the present study was to develop a culture-independent FISH test for P. multocida based upon hybridization of tagged oligonucleotide probes to bacterial rRNA. The diagnostic potential of this test was evaluated using lungs from chickens infected with P. multocida that developed clinical fowl cholera and with pig lung tissues injected with P. multocida.
MATERIALS AND METHODS
Cultivation of bacteria.
Twenty-two strains of P. multocida and other bacterial species isolated from normal and diseased lungs of poultry and other animals were obtained from publicly accessible service collections and a local culture collection (Table 1).
TABLE 1.
Specificity test of oligonucleotide probe pmhyb449 and its complementary non-pmhyb449 probe labeled by Cy3 with bacteria grown in pure culture
| Bacterial straina | Bacterial species | Animal source | Hybridization signal intensity withb:
|
|
|---|---|---|---|---|
| pmhyb449 | Non-pmhyb449 | |||
| NCTC 10322T | P. multocida subsp. multocida | Pig | +++ | − |
| 214 | P. multocida subsp. multocida | Calf | +++ | − |
| 40605-1 | P. multocida subsp. multocida | Eider | +++ | − |
| RA 12/2 | P. multocida subsp. multocida | Calf | +++ | − |
| NCTC 10204T | P. multocida subsp. gallicida | Cow | +++ | − |
| 77179 | P. multocida subsp. gallicida | Chicken | +++ | − |
| NCTC 11619T | P. multocida subsp. septica | Humanc | +++ | − |
| 5 | P. avium biovar 2 | Calf | +++ | − |
| 25 | P. canis biovar 2 | Calf | +++ | − |
| NCTC 11188T | P. gallinarum | Chicken | − | − |
| ATCC 43326T | P. canis biovar 1 | Dog | − | − |
| F 149T | P. anatis | Duck | − | − |
| CCM 5974T | Actinobacillus salpingitidis | Chicken | − | − |
| NCTC 4189T | A. lignieresii | Cow | − | − |
| P737 | Mannheimia glucosida | Sheep | − | − |
| 14R525 | E. coli | Human | − | − |
| SA 4461 | Haemophilus paragallinarum | Chicken | − | − |
| SA 7191 | H. paragallinarum | Chicken | − | − |
| 4237/2sv | Riemerella anatipestifer | Duck | − | − |
| 4280/2sv | R. anatipestifer | Duck | − | − |
| 726-82T | Coenonia anatina | Duck | − | − |
| 1912+pSD | S. enterica serotype Gallinarum | Chicken | − | − |
| 19110+pSD | S. enterica serotype Gallinarum | Chicken | − | − |
A superscript T indicates the type strain.
+++, high signal intensity; −, no signal observed.
Infection was by means of a cat bite.
Bacteria were stored at −80°C and cultivated overnight on blood agar base (CM55; Oxoid Ltd., Basingstoke, Hampshire, England) containing 5% citrated calf blood. Single colonies were cultured in brain heart infusion broth (Oxoid) at 37°C for 3 to 4 h in a shaker incubator, and optical density (A600) was measured before fixation.
Fixation of bacterial cells.
The bacterial cell cultures were fixed in 4% paraformaldehyde (Sigma Chemical Co., St. Louis, Mo.) as previously described (2) and were collected by centrifugation at 10,000 × g for 5 min. The supernatant was discarded, and fixed cells were washed in phosphate-buffered saline with 0.1% Nonidet P-40 (Sigma) and resuspended in 200 μl of 2× storage buffer (40 mM Tris [pH 7.5], 0.2% [vol/vol] Nonidet P-40) and 200 μl of 96% alcohol and were stored at −20°C until use.
Tissue section processing.
Lung tissue from chickens infected with P. multocida subsp. multocida strain 40605-1 (17) was used. Chickens were infected intratracheally with 104 CFU per 0.5 ml and sacrificed 12, 24, and 48 h postinoculation. These birds developed typical clinical signs and gross lesions of fowl cholera, and P. multocida subsp. multocida was reisolated from their spleens after postmortem examination. For comparison to other animal species, lung tissue from a healthy sacrificed pig injected postmortem with P. multocida subsp. septica strain HIM 746-6T was kindly provided by Tim K. Jensen, Danish Veterinary Laboratory. All infected chicken and pig tissues as well as noninfected tissues (controls) were immediately (within 1 to 2 h) fixed in 10% neutral buffered formalin for at least 24 h, processed for histology, embedded in paraffin wax, sectioned 3 to 4 μm thick, mounted on adhesive slides (Super Frost/plus slides; Menzel-Gläser, Braunschweig, Germany), and kept at 4°C until use.
Oligonucleotide probes.
Unpublished sequences of Pasteurella canis biovar 2 and 16S rRNA sequences obtained from GenBank were aligned with Pileup (Wisconsin Sequence Analysis Package; GCG, Madison, Wis.) (Table 2). A region was found where at least four mismatches separated P. multocida and biovar 2 of Pasteurella avium and P. canis from other members of Pasteurellaceae as well as from other bacterial species (Table 2). Comparison with similar 16S rRNA secondary structures of E. coli predicted this region to give strong detection (22).
TABLE 2.
Oligonucleotide probe DNA sequence of the P. multocida-specific probe pmhyb449 compared with selected 16S rRNA sequences of strains of bacteria within the genera Pasteurella, Actinobacillus, and Mannheimia used for ISH
| Probe or organism | 16S rRNA gene sequencea | Accession no. | Reference |
|---|---|---|---|
| Probe | |||
| pmhyb449 | 3′ CTTCCCTACAACAATTTATC | ||
| 5′ GAAGGGATGTTGTTAAATAG | |||
| Organisms | |||
| P. multocida subsp. multocida | .................... | M35018 | Unpublished |
| P. multocida subsp. gallicida | .................... | AF294412 | Unpublished |
| P. multocida subsp. septica | .................... | AF294423 | Unpublished |
| P. avium biovar 2 | .................... | L06085 | 19 |
| P. canis biovar 2 | .................... | Unpublished | Unpublished |
| P. canis biovar 1 | ......G.A..A..G..... | M75049 | 19 |
| P. gallinarum | .....CGGTAGTG.TN.... | M75059 | 19 |
| P. anatis | .....CGGTAGTG.T..... | M75054 | 19 |
| P. avium biovar 1 | .....TTG.AGTG.T..... | M75058 | 19 |
| A. lignieresii | .....TA.CAAA..T..... | M75068 | 19 |
| A. salpingitidis | .....TTNA.ATG.T..... | L06077 | 19 |
| M. glucosida | .....CGAT.GT..T..... | AF053889 | 6 |
A dot indicates that the nucleotide is complementary to that of the probe at the same position.
A specific oligonucleotide probe, pmhyb449, 5′-CTATTTAACAACATCCCTTC-3′ (S-S-Pmul-0449-a-A-20 [1]) (Tags, Copenhagen, Denmark), for specific detection of P. multocida was selected based on 16S rRNA sequence comparison. The probe and its complementary probe, non-pmhyb449 (S-*-Npmol-0449-a-S-20 [1]), were labeled by fluorescein or Cy3 and purified by high-performance liquid chromatography. The Cy3 label was chosen for further experiments because it gave a stronger signal than fluorescein or rhodamine (data not shown) when tested with strains of P. multocida. Probe EUB338 (S-D-Bact-0338-a-A-18 [1, 3]), which is specific for the bacterial domain, and its complementary probe, non-EUB338 (S-*-non-0338-a-S-18 [1]), labeled with rhodamine or Cy3, were used as controls.
ISH of cultured bacterial cells.
Prior to hybridization, fixed bacterial cells were acclimatized at ambient (room) temperature, mixed by vortexing, and bound to 10-well Teflon-coated slides (Novakemi; AB, Enskede, Sweden) which had been coated by immersion in a 1:10, 0.1% (wt/vol) aqueous solution of poly-l-lysine (Sigma) for 2 min and air dried. One microliter of bacterial cells was applied in each well. Slides were air dried, dehydrated in serial concentrations of 50, 80, and 96% ethanol for 3 min each, and air dried again. Ten microliters of hybridization buffer (15% formamide, 100 mM Tris [pH 7.2], 0.9 NaCl, 0.1% sodium dodecyl sulfate) containing 5 ng of probe was applied per well, and hybridization was performed in a humidified moist chamber overnight at 37°C. Slides were washed in 100 ml of hybridization buffer prewarmed to 37°C in a coplin jar for 10 min and thereafter changed to 100 ml of prewarmed (37°C) hybridization buffer containing 20% formamide for 10 min. Finally, the slides were rinsed in 100 ml of Milli-Q water prewarmed to 37°C and air dried in the dark.
ISH of tissue sections.
Tissue sections were deparaffinized in coplin jars by two changes of xylene and were dehydrated twice with 99.9% ethanol for 3 min at each step. Slides were air dried, and a circle was drawn around the tissue section using a hydrophobic pen (Dako PAP pen; Glostrup, Denmark). The hybridization and wash steps were done as described for the cultured bacterial cells but with 50 μl of hybridization buffer and 30 ng of probe per tissue section. The PAP pen circle was removed from the slides with tissue sections by using xylene solvent.
DAPI staining.
To evaluate the proper morphology and the presence of the bacterial cells where no signal was obtained with probe pmhyb449, DAPI (4′, 6–diamidino-2-phenylindole) (Sigma) staining was performed for selected bacteria on hybridized slides. Prior to DAPI staining, coverslips were removed and slides were immersed in 99.9% ethanol for 10 min to remove mountant and then were air dried.
Bacteria were stained with 40 μl of DAPI solution (6 ng/ml)/well for 10 min. Slides were washed with 100 ml of Milli-Q water in a coplin jar for 10 min and were air dried prior to epifluorescence microscopy.
Epifluorescence microscopy.
A coverslip was mounted on hybridized slides with the application of a small amount (2 to 3 drops) of paraffin oil (cultured bacterial cells) or Vectashield oil (Vector Laboratories, Inc., Burlingame, Calif.) (tissue sections) on the dry slide. Nonfluorescent oil was applied, and slides were examined using Zeiss filter sets 05, 09, and 15 for visualization of DAPI, fluorescein, and rhodamine (tetramethyl rhodamine isothiocyanate) or Cy3, respectively, using a Zeiss Axioplan II microscope (Oberkochen, Germany) (magnification, ×1,000) with a 100-W mercury lamp. Filter set XF53 (Omega Optical, Brattleboro, Vt.) was applied during micrograph photography with an MC 200 Zeiss camera and exposed on Kodak Ektachrome Elite 400 film.
RESULTS
Specificity of the probe sequence.
A suitable oligonucleotide probe, pmhyb449, and its complementary oligonucleotide probe, non-pmhyb449 (Table 2), were selected based on comparison of 16S rRNA sequences specific for P. multocida. The probe and its complementary probe were initially tested with fixed bacterial strains. By use of the universal bacterial probe EUB338, all strains tested were found positive, which showed that the rRNA in the fixed bacterial cells was accessible for the probe to bind and give a strong fluorescence signal. Its complementary probe, non-EUB338, did not give any signal. Probe pmhyb449 was tested on strains of P. multocida and gave good signals (Table 1 and Fig. 1A) comparable to those of EUB338 labeled with Cy3, while its complementary probe, non-pmhyb449, did not give any signals with the same bacterial strains. All other bacteria tested, except P. avium biovar 2 and P. canis biovar 2, were negative with probe pmhyb449 (Table 1). No signal was detected with controls, including empty wells, bacterial cells only, probe only, cells plus buffer (no probe), and buffer plus probe (no cells), showing that there was no nonspecific staining.
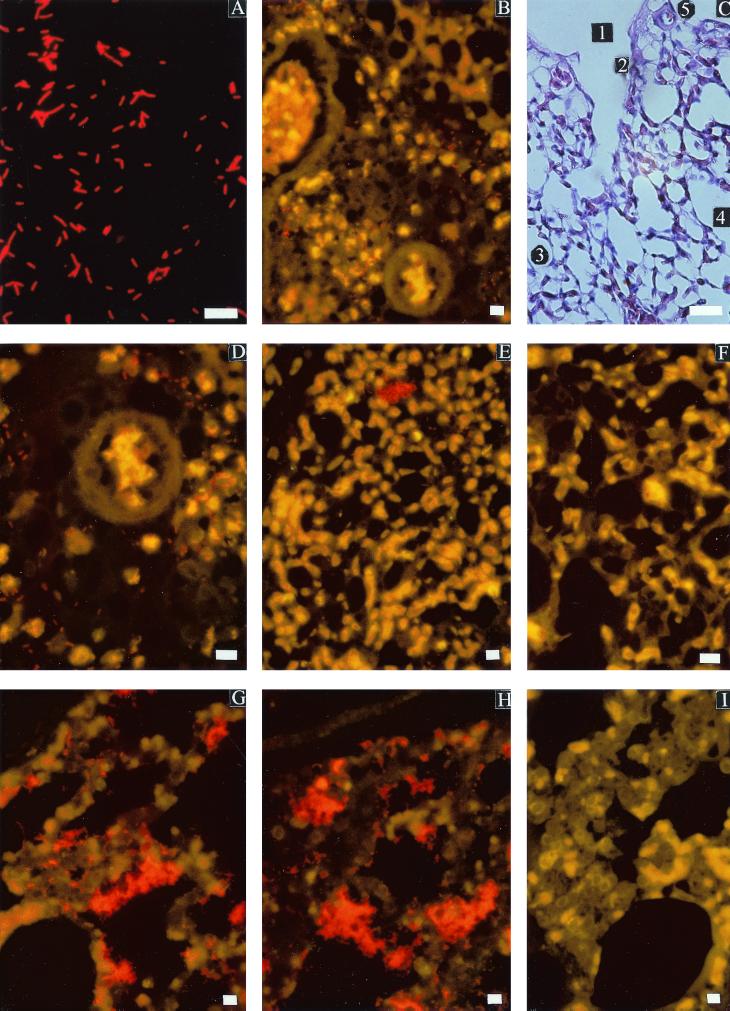
FIG. 1.
Photomicrographs demonstrating specific detection of P. multocida in pure culture and formalin-fixed lung tissues by use of the FISH technique. Shown are a pure culture of P. multocida subsp. multocida strain 40605-1 (A) and lung tissue sections of chicken (B, C, D, E, and F) or pig (G, H, and I) following light microscopy (C) or epifluorescence microscopy (A, B, and D through I) and ISH with the P. multocida-specific Cy3-labeled probe pmhyb449 (A, B, D, E, G, and H) and a nonsense Cy3-labeled probe, non-pmhyb449 (F and I), used as a control. A normal chicken lung (C) shows the location of the bacteria detected in cases of fowl cholera with an indication of the parabronchi (1), infundibulum (2), air capillary (3), pleura (4), and arterioles (5) (hematoxylin and eosin stain). Also shown are chicken lung infected with fowl cholera (experimentally infected with P. multocida subsp. multocida strain 40605-1), with the location of P. multocida cells near two blood vessels (artery and arteriole) (B), with P. multocida cells within the lumen and the walls of the air capillaries and perivascular space (D), and with P. multocida cells as individual organisms or as microcolonies (E); a control tissue section hybridized with probe non-pmhyb449 is also shown (F). Also shown is lung of a pig infected postmortem with P. multocida subsp. septica strain HIM 746-6T showing P. multocida bacteria in the lumen and wall of the alveoli (G), the area close to the pleura (H), and a control tissue section hybridized with non-pmhyb449 (I). Bars = 10 μm (A, B, D, E, F, G, H, and I) and 100 μm (C).
Optimization of hybridization conditions with fixed bacterial strains.
Posthybridization washing with formamide was evaluated using different formamide concentrations (15, 20, 25, and 30%) in two steps, where hybridization time was varied between 5 and 30 min at each step. Ten minutes of washing time at each step gave the best results without background noise. By use of 30 or 25% formamide in the second washing buffer, probe pmhyb449 resulted in weaker signal compared with that derived with 20% formamide, while under stringency conditions of hybridization the washing buffer containing 15% formamide gave a strong signal with P. multocida but weak signals with other Pasteurella spp. and non-Pasteurella bacteria at the hybridization temperature of 37°C (data not shown).
The optimum formamide concentration during posthybridization washing for probe pmhyb449 was found to be 15% in the first wash and 20% in the second wash using a 10-min washing period at each step.
Tissue hybridization with probe pmhyb449.
The ability of the pmhyb449 to detect P. multocida in tissues was tested using chicken lungs from experimentally induced fowl cholera and normal pig lungs injected with P. multocida.
The specific and narrow fluorescence of probe pmhyb449 distinguished P. multocida bacterial cells by a red fluorescence, while lung tissue cells fluoresced greenish to brownish and red blood cells were yellow on examination after ISH (Fig. 1A, B, and D through I).
In fowl cholera cases, P. multocida was clearly seen within the lung tissues. However, differences in distribution were observed (Table 3). In early cases (12 h postinfection), most bacteria were seen in the air capillaries (lumen and wall), bronchi, parabronchial areas (lumen and interparabronchial septa), perivascular areas (arterioles), and as masses in anatomically necrotic lung tissues. A few bacteria were also seen in the infundibulum space linings, pleura, and vascular lumen and their walls (Table 3). The bacteria appeared small in size (compared to pure culture cells) and were shaped as coccobacilli and rods and occurred singly, in pairs, in fours, in clusters of microcolonies (Fig. 1B, D, and E), and as massive colonies. Twenty-four hours postinfection the bacteria were commonly present in air capillaries and parabronchial lumen and as masses of bacteria in necrotic lung tissues and air capillaries. Low numbers of bacteria were seen in air capillary walls, blood vessels (lumen and perivascular), infundibulum spaces, and respiratory atria. Some bacteria were seen in pairs or in fours in a ring formation in phagocytic cells (macrophages or heterophils) within the parabronchial spaces and blood vessels and in aggregation of lymphoid tissues within the lung tissues. At 48 h most bacteria were detected in the air capillaries, air capillary walls, and infundibulum spaces, and a few were detected on the spiral smooth muscles of the parabronchi (Table 3). Some bacteria were found in a ring formation in phagocytes (in bronchi and blood vessels) and in lymphoid aggregation areas of the lung tissue, indicating either phagocytosis or the presence of circulating phagocytes with bacteria. The role of these cells in pathogenesis of P. multocida needs further investigation.
TABLE 3.
Results of ISH lung tissues showing the anatomical location where P. multocida was detected by probe pmhyb449
| Tissue source | Bacteria detection ina
|
|||
|---|---|---|---|---|
| Chicken after an infection period (h) of:
|
Pigb | |||
| 12 | 24 | 48 | ||
| Bronchi | ++ | − | − | +c |
| Parabronchi | ++ | ++ | − | ++ |
| Interstitium spaces | + | − | − | + |
| Smooth muscles | − | − | + | + |
| Infundibulum spaces | + | + | + | NAd |
| Air capillaries or alveoli spaces | +++ | ++ | + | +++ |
| Air capillaries or alveolar septa | +++ | ++ | + | +++ |
| Pleura | + | − | + | − |
| Vascular areas | + | + | − | − |
−, no bacteria detected;+, few bacteria detected; ++, moderate number of bacteria detected; +++, many bacteria detected.
Bacteria were injected into pig lung postmortem.
From bronchiole lumen.
NA, not applicable.
In the pig lung, ISH with probe pmhyb449 clearly detected P. multocida among tissues and cells of the lung. The bacteria were found mainly in the alveoli, terminal bronchioles, and interstitial areas, and a few bacteria were observed in the bronchi, bronchioles, and terminal bronchioles (Table 3 and Fig. 1G and H).
Infected as well as noninfected tissues were negative when hybridized with the non-pmhyb449 probe (Fig. 1F and I). The inflammatory reaction of the lung tissues in response to the bacteria did not hinder the binding of the probe in fowl cholera. The results of this study indicate that probe pmhyb449 is suitable for detection and determination of the in vivo localization of P. multocida in tissues.
DISCUSSION
FISH of whole cells with rRNA-targeted oligonucleotide probes has been extensively used as a tool for specific detection of bacteria (4, 5, 12, 15, 18, 23, 25, 39). To our knowledge no FISH procedure has been developed and used for detection or diagnosis of P. multocida. To aid in diagnosis of infections caused by P. multocida, a species-specific probe, pmhyb449, targeting 16S rRNA was designed and used for FISH. The specificity of this oligonucleotide probe was examined by whole-cell hybridization against selected species representing both 16S rRNA clusters described by Dewhirst et al. (19) outlined within genus Pasteurella sensu stricto as defined by Mutters et al. (38) and other bacterial species commonly associated with respiratory tract infections and septicemia in poultry. The probe was able to differentiate bacteria by at least four base pair mismatches. Exceptions were P. avium biovar 2 and P. canis biovar 2. These taxa were originally described by Madsen et al. (34) and subsequently were named by Mutters et al. (36, 37). Both taxa have been reported from pneumonia in calves in several countries (8, 9). 16S rRNA sequence studies of members of the family Pasteurellaceae Pohl 1981, however, indicated that taxon 13 of Bisgaard should be reinvestigated, since CCUG 16497 (P. avium biovar 2) clustered with the type strain of P. multocida and not with P. avium (19). Recently it was found that the 16S rRNA similarities between P. multocida and biovar 2 of P. avium and P. canis are greater than 98.6%, while similarities between P. multocida and biovar 1 of P. avium and P. canis are 94 and 96%, respectively (data not shown). However, similarities of organisms based on 16S rRNA sequence comparison are insufficient per se for species separation (20). Just as the outlining of species based upon a few selected DNA-DNA hybridizations might result in uncertain species definitions (6), on this background it appears that biovar 2 of P. avium and P. canis are misclassified and that future reclassification will place them with P. multocida. With this view, probe pmhyb449 is concluded to be specific for P. multocida.
The ISH assay was used for detection of P. multocida in formalin-fixed, paraffin-embedded tissues of a pig lung injected with a pure culture of P. multocida subsp. septica and in lungs from chickens that developed clinical fowl cholera infection with P. multocida subsp. multocida. The pmhyb449 probe was able to detect single cells of P. multocida in situ in the respective lung tissues, whereas no signal was observed for control lung tissue sections from noninfected birds that contained no organisms or infected lung tissues that were hybridized with the complementary non-pmhyb449 probe.
Methods based on 16S rRNA are advantageous in the detection and identification of microorganisms due to the fact that each bacterial cell contains multiple copies of the 16S rRNA that eases its detection, with evolutionarily highly conserved 16S rRNA regions common to bacteria and other regions which might be species specific (18, 31). 16S rRNA-based methods allow identification of microorganisms independently of bacterial growth rates and metabolic activities (31, 44), although cellular ribosomes are more abundant in rapidly growing bacteria (18). The amount of bound probe is directly correlated to the cellular rRNA content, which is dependent on physiological activity at the time of fixation (4, 24). In cases of fowl cholera the P. multocida cells could have been highly active within the lung tissue, as they gave strong signals with probe pmhyb449. In fast-growing bacteria, the rRNA content correlates directly with the growth rate (18), although it may not be valid in the case of slow-growing or starved organisms (28, 44). Starved and dormant bacteria can, however, be detected with FISH as active bacteria if they still possess relatively high 16S rRNA levels, as reported for S. enterica serovar Typhimurium (44).
Probe pmhyb449 clearly detected short rods of bacteria in various parts of the chicken lungs under mild or severe lung inflammation. In chickens, P. multocida was shown to occur singly, in aggregates probably representing microcolonies, or in masses occupying large areas of the lungs. Their individual morphologies were clearly detected by the pmhyb449 probe, as reported in similar FISH procedures with other bacteria (4, 18, 24).
There are limitations to the ISH assay, as the test depends on the number of copies of rRNA in the cell (5) and hence on the physiological activity of the microorganisms prior to fixation of samples and as it is restricted to eutrophic environments such as the lungs of the chickens. Another limitation is the autofluorescence background of the eucaryotic tissue. However, in contrast to immunological methods that rely on the expression of specific antigenic markers which may not be constant, phenotypic variation does not pose a problem when rRNA is used as a target (11, 12). Furthermore, the inflammatory reactions observed with P. multocida infections during this study did not hinder the detection of bacteria by FISH. The stability of the ribosome target allowed for the detection of single cells even in clinical material as well as identification in smears of pure culture (39), which correlates with our observations here.
In their study of P. multocida carriers in commercial poultry in Denmark, Muhairwa et al. (35) found mouse inoculation to be more effective in the recovery of this organism than the commonly used selective media. Yet mouse passage may only select strains pathogenic for mice. The developed test can offer a complementary role in active clinical case diagnosis and can be applied in pathogenesis and pathogenicity studies of P. multocida.
The probe pmhyb449 is recommended, as our results suggest that the probe might be applied with advantage to studies of P. multocida pathogenesis and its infections in poultry, pigs, and maybe other animals. Furthermore, results can be obtained quickly, since pure cultures of the bacteria are not needed. It can also be used to confirm and differentiate P. multocida from other Pasteurellaceae in culture. The test is simple and can be applied in most research and diagnostic laboratories.
ACKNOWLEDGMENTS
We thank A. R. Pedersen, U. L. Andreasen, Tony Bønnelycke, Gitte Frederiksen, and Pia Mortensen for technical assistance. Pig lung tissues were kindly provided by T. K. Jensen, Danish Veterinary Laboratory, Copenhagen, Denmark. The University of Nairobi administration is acknowledged for granting Paul Gichohi Mbuthia study leave in Denmark.
Financial support of this project was further provided through the ENRECA program (Improving the Health and Productivity of the Rural Poultry in Africa) and the Danish Agricultural and Veterinary Research Council (grant no. 9702797).
REFERENCES
- 1.Alm E W, Oerther D W, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for the determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R, Springer N, Ludwig W, Gortz H-D, Schleifer K-H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991;351:161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- 5.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angen Ø, Mutters R, Caugant D A, Olsen J E, Bisgaard M. Taxonomic relationships of the [Pasteurella] haemolytica complex as evaluated by DNA-DNA hybridizations and 16S rRNA sequencing with proposal of Mannheimia haemolytica gen. nov., comb. nov., Mannheimia granulomatis comb. nov., Mannheimia glucosida sp. nov., Mannheimia ruminalis sp. nov. and Mannheimia varigena sp. nov. Int J Syst Bacteriol. 1999;49:67–86. doi: 10.1099/00207713-49-1-67. [DOI] [PubMed] [Google Scholar]
- 7.Bisgaard M. Ecology and significance of Pasteurellaceae in animals. Zentbl Bakteriol. 1993;279:7–26. doi: 10.1016/s0934-8840(11)80487-1. [DOI] [PubMed] [Google Scholar]
- 8.Bisgaard M, Abdullahi M Z, Gilmour N J L. Further studies on the identification of Pasteurellaceae from cattle lungs. Vet Rec. 1991;128:428–429. doi: 10.1136/vr.128.18.428. [DOI] [PubMed] [Google Scholar]
- 9.Bisgaard M, Houghton S B, Mutters R, Stenzel A. Reclassification of German, British and Dutch isolates of so-called Pasteurella multocida obtained from pneumonic calf lungs. Vet Microbiol. 1991;26:115–124. doi: 10.1016/0378-1135(91)90048-k. [DOI] [PubMed] [Google Scholar]
- 10.Boye M, Jensen T K, Møller K, Leser T D, Jorsal S E. Specific detection of Lawsonia intracellularis in porcine proliferative enteropathy inferred from fluorescent rRNA in situ hybridisation. Vet Pathol. 1998;35:153–156. doi: 10.1177/030098589803500212. [DOI] [PubMed] [Google Scholar]
- 11.Boye M, Jensen T K, Moller K, Leser T D, Jorsal S E. Specific detection of the genus Serpulina, S. hyodysenteriae, and S. pilosicoli in porcine intestines by fluorescent rRNA in situ hybridisation. Mol Cell Probes. 1998;12:323–330. doi: 10.1006/mcpr.1998.0193. [DOI] [PubMed] [Google Scholar]
- 12.Boye M, Feenstra A A, Tegtmeier C, Andresen L O, Rasmussen S R, Bille-Hansen V. Detection of Streptococcus suis by in situ hybridisation, indirect immunofluorescence, and peroxidase-antiperoxidase assays in formalin-fixed, paraffin-embedded tissue sections from pigs. J Vet Diagn Investig. 2000;12:224–232. doi: 10.1177/104063870001200305. [DOI] [PubMed] [Google Scholar]
- 13.Brown C. In situ hybridization with riboprobes: an overview for veterinary pathologists. Vet Pathol. 1998;35:159–167. doi: 10.1177/030098589803500301. [DOI] [PubMed] [Google Scholar]
- 14.Cameron R D A, O‘Boyle D, Frost A J, Fregan N. An outbreak of haemorrhagic septicaemia associated with Pasteurella multocida subsp. gallicida in a large pig herd. Aust Vet J. 1996;73:27–29. doi: 10.1111/j.1751-0813.1996.tb09949.x. [DOI] [PubMed] [Google Scholar]
- 15.Christensen H, Hansen M, Sørensen J. Counting and size classification of active soil bacteria by fluorescence in situ hybridization with rRNA oligonucleotide probe. Appl Environ Microbiol. 1999;65:1753–1761. doi: 10.1128/aem.65.4.1753-1761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen J P, Bisgaard M. Fowl cholera. Rev Sci Tech Off Int Epiz. 2000;19:626–637. doi: 10.20506/rst.19.2.1236. [DOI] [PubMed] [Google Scholar]
- 17.Christensen J P, Dietz H H, Bisgaard M. Phenotypic and genotypic characters of isolates of Pasteurella multocida obtained from back-yard poultry and two outbreaks of avian cholera in avifauna in Denmark. Avian Pathol. 1998;27:373–381. doi: 10.1080/03079459808419354. [DOI] [PubMed] [Google Scholar]
- 18.Delong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single microbial cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 19.Dewhirst F E, Paster B J, Olsen I, Fraser G J. Phylogeny of Pasteurellaceae as determined by comparison of 16S ribosomal ribonucleic acid sequences. Zentbl Bakteriol. 1993;279:35–44. doi: 10.1016/s0934-8840(11)80489-5. [DOI] [PubMed] [Google Scholar]
- 20.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 21.Frederiksen W. Ecology and significance of Pasteurellaceae in man—an update. Zentbl Bakteriol. 1993;279:27–34. doi: 10.1016/s0934-8840(11)80488-3. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R I. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fussing V, Paster B J, Dewhirst F E, Poulsen L K. Differentiation of Actinobacillus pleuropneumoniae strains by sequence analysis of 16S rDNA and ribosomal intergenic regions, and development of a species specific oligonucleotide for in situ detection. Syst Appl Microbiol. 1998;21:408–418. doi: 10.1016/S0723-2020(98)80050-7. [DOI] [PubMed] [Google Scholar]
- 24.Giovannoni S J, Delong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligonucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen G J, Mooibroek M, Idema J, Harmsen H J M, Welling G W, Degener J E. Rapid identification of bacteria in blood cultures by using fluorescently labeled oligonucleotide probes. J Clin Microbiol. 2000;38:814–817. doi: 10.1128/jcm.38.2.814-817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen T K, Boye M, Hagedorn-Olsen T, Riising H J, Angen Ø. Actinobacillus pleuropneumoniae osteomyelitis in pigs demonstrated by fluorescent in situ hybridisation. Vet Pathol. 1999;36:258–261. doi: 10.1354/vp.36-3-258. [DOI] [PubMed] [Google Scholar]
- 27.Jensen T K, Møller K, Boye M, Leser T D, Jorsal S E. Scanning electron microscopy and fluorescent in situ hybridisation of experimental Brachyspira (Serpulina) pilosicoli infection in growing pigs. Vet Pathol. 2000;37:22–32. doi: 10.1354/vp.37-1-22. [DOI] [PubMed] [Google Scholar]
- 28.Kemp P F, Lee S, Laroche J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 1993;59:2594–2601. doi: 10.1128/aem.59.8.2594-2601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempf V A J, Trebesius K, Autenrieth I B. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J Clin Microbiol. 2000;38:830–838. doi: 10.1128/jcm.38.2.830-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause T, Bertschinger H U, Corboz L, Mutters R. V-factor dependent strains of Pasteurella multocida subsp. multocida. Zentbl Bakteriol Hyg A. 1993;266:255–260. doi: 10.1016/s0176-6724(87)80039-1. [DOI] [PubMed] [Google Scholar]
- 31.Krimmer V, Merkert H, von Eiff C, Frosch M, Eulert J F J, Löhr J F, Hacker J, Ziebuhr W. Detection of Staphylococcus aureus and Staphlococcus epidermidis in clinical samples by 16S rRNA-directed in situ hybridization. J Clin Microbiol. 1999;37:2667–2673. doi: 10.1128/jcm.37.8.2667-2673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Licht T R, Krogfelt K A, Cohen P S, Poulsen L K, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestines by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackie J T, Barton M, Kettlewell J. Pasteurella multocida septicaemia in pigs. Aust Vet J. 1992;69:227–228. doi: 10.1111/j.1751-0813.1992.tb09931.x. [DOI] [PubMed] [Google Scholar]
- 34.Madsen E B, Bisgaard M, Mutters R, Pedersen K B. Characterization of Pasteurella species isolated from lungs of calves with pneumonia. Can J Comp Med. 1985;49:63–67. [PMC free article] [PubMed] [Google Scholar]
- 35.Muhairwa A P, Christensen J P, Bisgaard M. Investigations on the carrier rate of Pasteurella multocida in healthy commercial poultry flocks and flocks affected by fowl cholera. Avian Pathol. 2000;29:133–145. doi: 10.1080/03079450094162. [DOI] [PubMed] [Google Scholar]
- 36.Mutters R, Piechulla K, Hinz K-H, Mannheim W. Pasteurella avium (Hinz and Kunjara 1977) comb. nov. and Pasteurella volantium sp. nov. Int J Syst Bacteriol. 1985;35:5–9. [Google Scholar]
- 37.Mutters R, Ihm P, Pohl S, Frederiksen W, Mannheim W. Reclassification of the genus Pasteurella Trevisan 1887 on the basis of deoxyribonucleic acid homology, with proposals for the new species Pasteurella dagmatis, Pasteurella canis, Pasteurella stomatis, Pasteurella anatis, and Pasteurella langaa. Int J Syst Bacteriol. 1985;35:309–322. [Google Scholar]
- 38.Mutters R, Mannheim W, Bisgaard M. Taxonomy of the group. In: Adlam C, Rutter J M, editors. Pasteurella and pasteurellosis. London, United Kingdom: Academic Press; 1989. pp. 3–34. [Google Scholar]
- 39.Nordentoft S, Christensen H, Wegener H C. Evaluation of a fluorescence-labeled oligonucleotide probe targeting 23S rRNA for in situ detection of Salmonella serovars in paraffin-embedded tissue sections and their rapid identification in bacterial smears. J Clin Microbiol. 1997;35:2642–2648. doi: 10.1128/jcm.35.10.2642-2648.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuovo G J. In situ hybridisation. In: Nuovo G J, editor. PCR in situ hybridisation. Protocols and applications. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 123–192. [Google Scholar]
- 41.Pijoan, C. 1999. Pneumonic pasteurellosis, p. 511–520. In B. E. Straw, S. D‘Alliare, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames, Iowa.
- 42.Poulsen L K, Lan F, Kristensen C S, Hobolth P, Molin S, Krogfelt K A. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun. 1994;62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rimler R B, Glisson J R. Fowl cholera. In: Calnek B W, Barnes H J, Beard C W, McDougald L R, Saif Y M, editors. Diseases of poultry. 10th ed. Ames, Iowa: Iowa state University press; 1997. pp. 143–161. [Google Scholar]
- 44.Tolker-Nielsen T, Larsen M H, Kyed H, Molin S. Effect of stress treatments on the detection of Salmonella typhimurium by in situ hybridization. Int J Food Microbiol. 1997;35:251–258. doi: 10.1016/s0168-1605(97)01242-7. [DOI] [PubMed] [Google Scholar]
- 45.Townsend K M, O‘Boyle D, Phan T T, Hanh T X, Wijewardana T G, Wilke I, Trung N T, Frost A J. Acute septicaemic pasteurellosis in Vietnamese pigs. Vet Microbiol. 1998;63:205–215. doi: 10.1016/s0378-1135(98)00233-8. [DOI] [PubMed] [Google Scholar]