SUMMARY
We report an inborn error of metabolism caused by an expansion of a GCA-repeat tract in the 5′ untranslated region of the gene encoding glutaminase (GLS) that was identified through detailed clinical and biochemical phenotyping, combined with whole-genome sequencing. The expansion was observed in three unrelated patients who presented with an early-onset delay in overall development, progressive ataxia, and elevated levels of glutamine. In addition to ataxia, one patient also showed cerebellar atrophy. The expansion was associated with a relative deficiency of GLS messenger RNA transcribed from the expanded allele, which probably resulted from repeat-mediated chromatin changes upstream of the GLS repeat. Our discovery underscores the importance of careful examination of regions of the genome that are typically excluded from or poorly captured by exome sequencing.
Over the past decade, the accelerated discovery of disease-causing genes and an increased frequency of diagnostic success in patients with rare mendelian diseases (from approximately 10% with the use of traditional genetic testing to approximately 40% with the use of genomewide testing) have been facilitated by innovation in high-throughput sequencing, as well as integration of multidisciplinary approaches into accurate data interpretation.1,2
Despite these advances, establishing a diagnosis for such conditions remains a challenge. In most phenotypic categories, more than 50% of the patients with a rare disease lack a molecular diagnosis.1 One reason for this shortcoming may be that exome sequencing, the preferred method to date, captures only a small portion (<2%) of the genome and is very limited in its capacity to detect copy-number variants, insertions, translocations, inversions, tandem repeat expansions, non-coding and deep-intronic variants, and variants within complex regions of the genome. Indeed, emerging studies have shown that unlike exome sequencing, genome sequencing has the potential to detect all classes of genetic variation3–8 and thus to increase the diagnostic yield above that of exome sequencing.5,7
However, the immense amounts of data generated by genome sequencing and limitations of reference genomes impede the detection of such variations. The interpretation of genome-sequencing data can be strengthened through the incorporation of a targeted approach, which harnesses information obtained from phenotypic and functional characterization of patients to focus on specific families of genes. Here, we describe detailed clinical and biochemical phenotyping and genome sequencing to identify a novel repeat expansion disorder that causes a deficiency of glutaminase, an enzyme important for neurogenesis and neurotransmission.
CASE REPORT
Three unrelated patients, all of whom were born to nonconsanguineous white parents of European ancestry after uncomplicated pregnancy and delivery, presented at the age of 3 years (Patients 1 and 2, from the United Kingdom) and 2 years (Patient 3, from Canada) with early-onset delay in both gross and fine motor skills, as well as delayed speech (Fig. 1A). In all three patients, ataxia developed, along with dependency on the use of a wheelchair or walker. Early brain imaging did not reveal abnormalities; however, at 11 years of age, Patient 3 was found to have cerebellar atrophy on repeated magnetic resonance imaging. Additional clinical details are provided in Figure S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org.
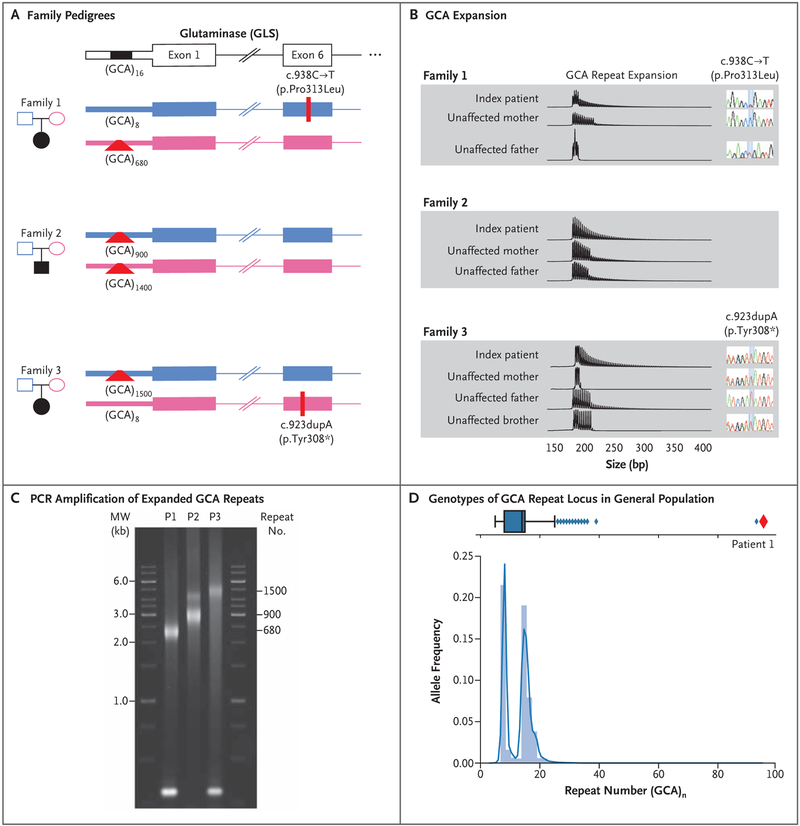
Figure 1 (facing page). Genotyping of GLS Probands.
Panel A shows the pedigrees of the three affected families. The presence of c.938C→T (p.Pro313Leu) in Family 1 and c.923dupA (p.Tyr308⋆) in Family 3 was confirmed on Sanger sequencing. Polymerase-chain-reaction (PCR) amplification of the 5′ region containing the GCA repeat and subsequent Sanger sequencing allowed for the determination of the number of GCA repeats in nonexpanded alleles. In family members in whom a large expansion was observed, the number of GCA repeats was estimated on PCR amplification, followed by agarose gel electrophoresis. Panel B shows the results of triplet repeat–primed PCR assay and subsequent capillary electrophoresis to confirm the presence of the GCA expansion. Each vertical line represents a single GCA repeat. Thus, larger expansions are represented by a greater number of vertical lines. The results of Sanger sequencing that correspond to the missense and frameshift variants are shown next to each member of Families 1 and 3. Panel C shows PCR amplification of the expanded GCA repeats in blood samples obtained from the three patients (P), followed by agarose electrophoresis. The approximately 300-bp PCR product visible in the samples from Patients 1 and 3 reflects the nonexpanded alleles in these patients, who are heterozygous for an expansion and a point mutation. Panel D shows the genotypes of the GCA repeat locus in an untargeted population, indicating the number of GCA repeats for 8295 persons (16,590 total genotypes) as determined with the use of Expansion Hunter run on PCR-free genomesequencing data sets. The blue box plot shows the distribution of alleles with different numbers of GCA repeats in the general population. The vertical line in the box indicates the median, the left side of the box indicates quartile 1 (Q1), and the right side of the box indicates quartile 3 (Q3), with the interquartile range (IQR) defined as Q1 to Q3. The left and right whiskers represent the closest genotype to Q1 minus 1.5 × IQR and Q3 plus 1.5 × IQR, respectively. Any genotypes falling outside this range are considered to be outliers and are represented by diamonds. The repeat number ranged from 5 to 26 in the majority of persons who were assayed, with a bimodal distribution centered at 8 and 16 and 12 outliers with 26 to 38 repeats. Patient 1 is represented by a red diamond, and only one unaffected person was detected with a large expansion similar to that of Patient 1 (far-right outlier at more than 90 repeat units).
METHODS
CLINICAL DATA AND SPECIMEN COLLECTION
Families 1 and 2 were enrolled through the TIDEX study, which was approved by the institutional review board of the Faculty of Medicine at the University of British Columbia. Family 3 was enrolled in the Care4Rare study, approved by the institutional review board of the Children’s Hospital of Eastern Ontario. The patients’ parents provided written informed consent for their participation in the study, specimen collection, and publication of the results. During the consenting process, risks and benefits of research-based exome sequencing or genome sequencing were explained, and an option for disclosure of medically actionable incidental findings was provided.
SEQUENCING AND GENOMIC ANALYSIS
Genomic DNA was isolated from peripheral blood, and trio-exome sequencing (mother, father, and proband) was performed in samples obtained from Families 1 and 3, whereas samples obtained from Family 2 underwent duo-exome sequencing (mother and proband). In addition, singleton-genome sequencing of a sample obtained from the proband in Family 1 was performed. We used an in-house, open-source bioinformatics pipeline — similar to the unbiased semiautomated approach described previously2 — to analyze the sequencing data. Sanger sequencing was used to confirm the missense and frameshift variants, and triplet repeat–primed polymerasechain-reaction (PCR) and GLS repeat PCR assays were used to confirm the presence of the repeat expansions. Details regarding these analyses are provided in the Supplementary Appendix.
BIOCHEMICAL ANALYSIS
The GLS activity in fibroblasts and peripheral-blood mononuclear cells was determined in a reaction mixture containing glutamine. The amount of glutamate, produced by GLS from glutamine, was subsequently determined by means of spectrophotometry at 340 nm with the use of glutamate dehydrogenase and NAD. Quantification of the amount of glutamate was performed by comparison with external glutamate standards.
FUNCTIONAL ANALYSIS
We performed chromatin analysis and pyrosequencing and cloning of the GLS promoter into a luciferase reporter vector to investigate the effect of the expanded repeat on transcription and translation. Enzyme assays, immunoblotting, and flux and complementary DNA (cDNA) analyses were performed on samples obtained from the patients. Functional analysis of the recombinantly expressed GLS mutants was performed in a GLS-deficient HEK293-Flp-In cell line and by crystal structure analysis. The technical details of all methods that were used are described in the Supplementary Appendix.
RESULTS
METABOLIC FINDINGS
We performed metabolic evaluations in all three patients according to the published diagnostic algorithm for the identification of treatable conditions in intellectual developmental disorder, which includes an analysis of plasma amino acids among others.9 Metabolic testing revealed a persistent elevation in plasma glutamine of 1800 to 2209 μmol per liter, which approximately equals or exceeds the upper limit of the normal range (405 to 781 μmol per liter) by a factor of 2.5, despite normal plasma ammonia levels. Urea-cycle defects were ruled out, and treatment with sodium phenylbutyrate at a dose of 250 mg per kilogram of body weight per day was ineffective in all three patients. The parents and siblings of the three patients were unaffected and had plasma glutamine levels within the normal range.
EXOME SEQUENCING
We excluded urea-cycle disorders using clinical gene-panel sequencing (see the Supplementary Appendix). Of the 550 patients in our genomic discovery studies who had met the criteria for intellectual developmental disorder plus unexplained metabolic phenotype2 according to the diagnostic algorithm for treatable conditions,9 the 3 patients described here were the only ones with persistently elevated glutamine levels of unknown cause.
On the basis of the hypothesis that these patients had a novel inborn error of metabolism, we performed exome sequencing with a combined analysis of Family 1 (proband and both parents) and Family 2 (proband and one parent). We analyzed the data from Family 3 (proband and both parents) independently.
The standard bioinformatics approach that assumes mendelian modes of inheritance with full penetrance did not identify any leading gene candidates in any of the three families. We then evaluated the exome sequences of only the probands in Family 1 and Family 2; rare, deleterious variants in the heterozygous state were identified in 345 and 320 genes, respectively, of which 24 and 29 were associated with intellectual disability or encephalopathy in the Human Phenotype Ontology or Medical Subject Heading Overrepresentation Profiles (Table S1 in the Supplementary Appendix).10,11 The heterozygous, paternally inherited variant c.938C→T (p.Pro313Leu; GenBank accession number, NM_014905) in GLS (encoding glutaminase) was of interest, given the excellent match between the encoded phosphate-activated glutaminase and the biochemical phenotype (elevated levels of glutamine). In Family 2, we identified no candidate variants that were consistent with the clinical or biochemical phenotype.
In a separate study, exome analysis revealed a maternally inherited heterozygous GLS variant, c.923dupA (p.Tyr308⋆; NM_014905), in Patient 3 (Fig. 1A). Both the missense and frameshift GLS variants that were identified in Patients 1 and 3, respectively, were predicted by all tested in silico metrics to be damaging. The variant p.Tyr308⋆ has an allele frequency of 3.984×10−6 in the Genome Aggregation Database (gnomAD, rs1212883982); the p.Pro313Leu variant was absent from this database.12
BIOCHEMICAL PHENOTYPE
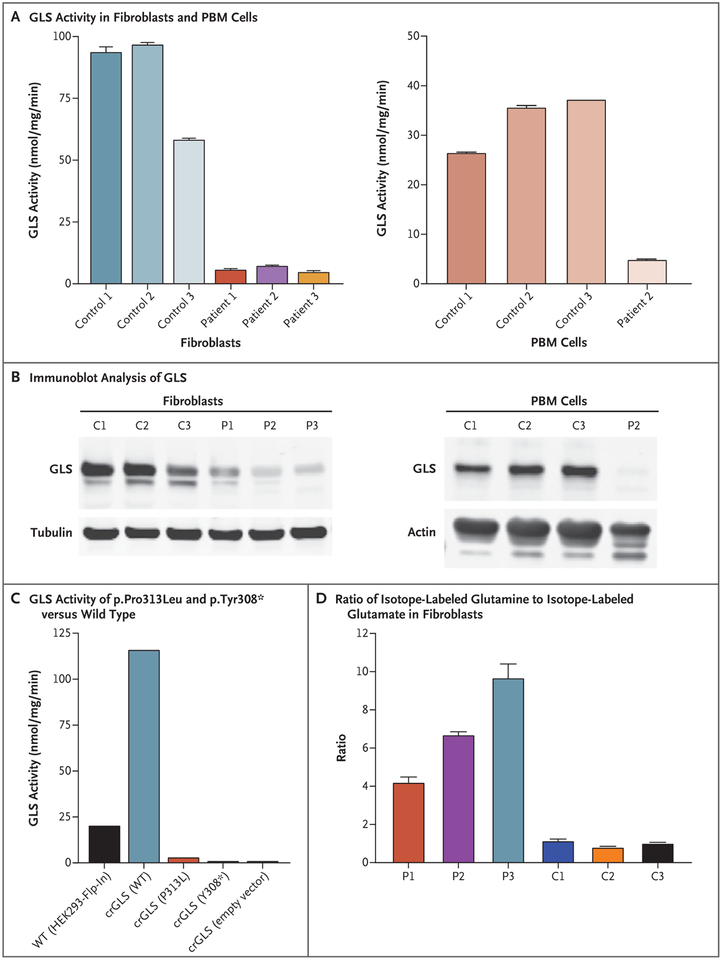
The results of exome analysis were suggestive of a GLS deficiency but were not conclusive, so we performed further biochemical phenotyping. Functional analyses in lymphocytes and fibroblasts showed markedly reduced GLS activity in all three patients (Fig. 2A). Immunoblot analysis showed reduced levels of glutaminase in fibroblasts obtained from all three patients and virtually no glutaminase in the lymphocytes of Patient 2 (Fig. 2B).
Figure 2 (facing page). Biochemical Characterization of GLS Deficiency.
Panel A shows GLS activity in fibroblasts obtained from all three study patients and from three controls and in peripheral-blood mononuclear (PBM) cells obtained from Patient 2 and from three controls. The GLS activity is measured in nanomoles of glutamate per milligram of protein per minute. Panel B shows the results of immunoblot analysis of GLS in fibroblasts and PBM cells obtained from the same patients (P) and controls (C) shown in Panel A. Panel C shows GLS activity of recombinantly expressed p.Pro313Leu and p.Tyr308⋆ GLS variants, as compared with wild-type (WT) GLS enzyme, expressed in GLS-deficient HEK293-Flp-In cells. Panel D shows the ratio of stable isotope-labeled glutamine to stable isotope-labeled glutamate in fibroblasts obtained from the three patients and three controls. In Panels A and D, the T bars indicate the standard deviation.
For functional analyses, stable-isotope-labeled glutamine, glucose, and oleate were used for the in situ analysis of the GLS deficiency and its metabolic consequences. In samples obtained from all three patients, we observed markedly decreased levels of isotope-labeled glutamate in fibroblasts that had been incubated with isotope-labeled glutamine, a finding that indicates impaired GLS activity (Fig. 2D). In addition, a decreased flux of glutamine into the citric acid cycle was observed in the fibroblasts, which was compensated, to some extent, by an increased flux from glucose and fatty-acid oxidation (Figs. S2, S3, and S4 in the Supplementary Appendix). Recombinantly expressed mutant GLS (carrying the p.Pro313Leu variant) showed residual enzymatic activity of 2.8%, whereas no activity could be detected for the recombinantly expressed p.Tyr308⋆ variant (Fig. 2C). An analysis of available crystal structures of GLS isoforms showed that the p.Pro313Leu mutation probably compromises dimerization and thus the formation of catalytically active tetramers (Fig. S5 in the Supplementary Appendix). For the p.Tyr308⋆ variant, molecular modeling suggests that the truncation would prevent the proper formation of the active site and the subunit interfaces. This would probably render the protein catalytically inactive, even if it was expressed and stable (Fig. S6 in the Supplementary Appendix).
A glutamate deficit in the neurons of the patients could not be examined, since the induction of pluripotent stem cells from GLS-deficient fibroblasts for differentiation into neurons was not successful, perhaps because GLS has been shown to be essential for the differentiation, proliferation, and survival of human neural progenitor cells.13 Knockdown of the GLS orthologues glsa, glsl, or both in zebrafish was associated with a smaller body size, curved body, and cardiac edema (Fig. S7 in the Supplementary Appendix).
GENOME SEQUENCING
We hypothesized that the missing heritability in each of the three patients was due to variation at the GLS locus refractory to detection on exome sequencing. We based this hypothesis on the results of the biochemical GLS and flux assays indicating the presence of a GLS deficiency, which was consistent with elevated levels of plasma glutamine. Sanger sequencing of samples of cDNA generated from messenger RNA (mRNA) obtained from members of Family 1 supported this hypothesis, since the proportion of the c.938T allele relative to the second allele was much higher in the patient than in the father, a finding that was consistent with an imbalance in allelic expression (Figs. S8 and S9 in the Supplementary Appendix). Thus, we sequenced the genome of Patient 1. Manual analyses of cisregulatory regions around GLS revealed the presence of a GCA repeat in the 5′ untranslated region of GLS. Applying Expansion Hunter,14 which detects expansions in genome-sequence data in a locus-specific manner, we identified a trinucleotide expansion consisting of more than 90 GCAs on the chromosome that the proband had inherited from the mother. The paternally inherited allele had eight GCA copies (Fig. 1A).
TRINUCLEOTIDE REPEAT ANALYSIS
We then performed a triplet repeat–primed PCR assay (Fig. 1B) and a GLS repeat PCR assay (Fig. 1C). Triplet repeat–primed PCR assays with a primer complementary to a stretch of GCA repeats indicated that all three patients had large GCA repeat expansions, as did one or both of their parents (Fig. 1B). The proband in Family 1 had compound heterozygosity for the paternally inherited c.938C→T variant and a maternally inherited GCA repeat expansion allele. The proband in Family 2 was homozygous for GCA repeat expansion alleles inherited from both parents, and the proband in Family 3 had compound heterozygosity for the maternally inherited c.923dupA variant and a paternally inherited GCA repeat expansion allele (Fig. 1A). Repeat PCR assays with the use of primers flanking the GCA repeat showed major expansion products of approximately 680, 900, and 1500 repeats, respectively, in blood samples obtained from the three probands (Fig. 1C). We detected expansions with smaller numbers of trinucleotide repeats (400 to 1100) in the fibroblasts of the probands in Families 1, 2, and 3 (Fig. S10 in the Supplementary Appendix), which was consistent with somatic heterogeneity that has been reported for many other repeat expansion disorders.15
Analysis of this GCA repeat expansion locus in an untargeted collection consisting of 8295 genomes showed that this short tandem repeat had a median size of 14 repeats (i.e., 42 bp) and a bimodal prevalence at 8 and 16 repeats. Of the 8295 analyzed genomes, 1 was heterozygous for an allele with more than 90 repeats, making the allele frequency of this repeat 6.03×10−5 (Fig. 1D).
REPEAT-MEDIATED EFFECTS
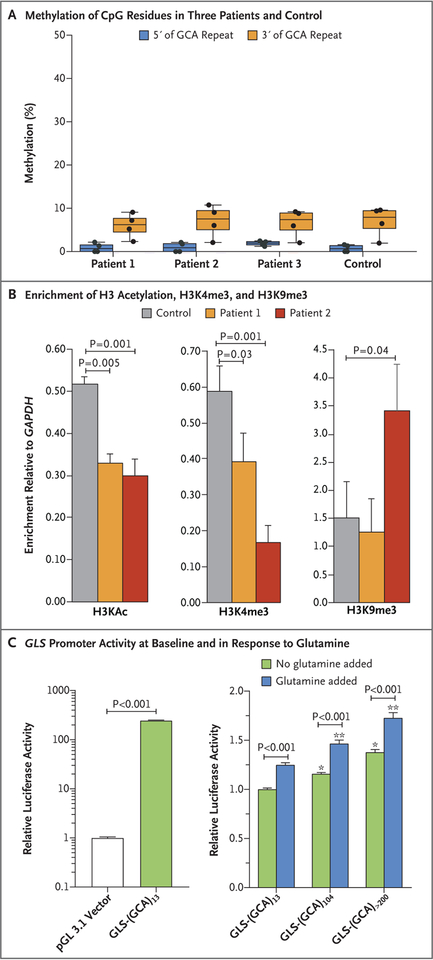
To investigate the mechanism of the allele-biased mRNA expression (Fig. S9 in the Supplementary Appendix), which is consistent with the trinucleotide-repeat expansion suppressing GLS expression, we carried out pyrosequencing of DNA (upstream and downstream of the trinucleotide-repeat expansion) purified from blood cells; no evidence of increased DNA methylation was seen in samples obtained from any of the patients (Fig. 3A). The absence of methylation in the patients’ fibroblasts was confirmed by the loss of almost all the GLS PCR product when the genomic DNA was predigested with a methylation-sensitive restriction enzyme (Fig. S10 in the Supplementary Appendix).
Figure 3. Repeat-Associated Effects.
Panel A shows the percentage methylation of CpG residues in the upstream genomic region (four sites) and the downstream genomic region (four sites) of the repeat in the three study patients and in a control. CpG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases from 5′ to 3′. Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosines. Panel B shows the results of chromatin immunoprecipitation experiments using fibroblasts obtained from Patients 1 and 2 and from a control, indicating the levels of enrichment of H3 acetylation (H3KAc), H3K4me3, and H3K9me3 on the GLS promoter relative to the gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Differences between the control sample and those obtained from the two patients were significant for H3KAc, H3K4me3, and H3K9me3 in Patient 2. Differences between enrichment for different histone modifications were evaluated with a t-test to determine significance. The enrichment was calculated as the percentage of input. The y axis shows the enrichment for GLS as the factor change over enrichment for GAPDH. The results are from two independent chromatin immunoprecipitation experiments. Panel C shows the results of cloning of DNA fragments containing the GLS promoter with 13, 104, and approximately 240 GCA repeats — wild-type GLS-(GCA)13, mutant GLS-(GCA)104, and GLS-(GCA)>200 — into luciferase promoter reporter constructs with or without glutamine supplementation at 48 hours. For vector normalization, activity of two luciferases, firefly and Renilla, were measured in the same cells or lysate in three independent experiments. Two-way analyses of variance with Bonferroni post-tests were used for analysis. One asterisk indicates P<0.001 for the comparison with GLS-(GCA)13 with no glutamine supplementation, and two asterisks indicate P<0.001 for the comparison with GLS-(GCA)13 with glutamine supplementation. In Panel A, the data are shown in box plots with the same measures as described in Figure 1D; the T bars indicate the standard deviation of the mean in Panel B and standard error in Panel C.
Chromatin immunoprecipitation assays were performed to determine whether the expansion affected histone modifications of the adjacent GLS promoter. The patients’ alleles showed reduced levels of histone modifications that are characteristic of transcriptionally active regions (H3 acetylation and H3K4 trimethylation) and were enriched for a histone modification characteristic of some transcriptionally silenced regions (H3K9me3) (Fig. 3B).16 This effect was more marked in Patient 2, who carried two expanded repeat alleles. These data suggest that the repeat expansion causes a change in the chromatin configuration, which results in decreased transcription.
To test for the effect of the expanded repeat on transcription initiation and elongation (i.e., the elongation of the nascent RNA strand with the addition of nucleotides during transcription) or translation, we cloned DNA fragments containing the GLS promoter with 13, 104, and approximately 240 GCA repeats into a luciferase reporter construct (pGL3.1) (Fig. S11 in the Supplementary Appendix). A comparison of the empty vector (pGL3.1) and vector containing the 13-repeat construct confirmed robust GLS promoter activity at baseline and in response to glutamine supplementation. We observed no negative effects of the repeat expansions on luciferase activity (Fig. 3C). These assays are based on the transient transfection of nonreplicating plasmids containing GLS fragments outside their normal chromosomal and chromatin context. They therefore address some of the direct effects of the cloned repeats on transcription and the efficiency of translation of the resultant transcript, rather than any epigenetic effects that the repeat may have in situ. Thus, taken together, our data suggest that the predominant effect of the repeats is at the level of histone modifications, causing the glutaminase deficiency by promoting the formation of repressive heterochromatin and thereby reducing GLS transcription.
DISCUSSION
In this study, we describe the identification of an inborn error of metabolism caused by a novel trinucleotide GCA repeat expansion. The expansion in the 5′ untranslated region of GLS, which encodes glutaminase, results in reduced expression and glutaminase deficiency. The neurodegenerative course of the patients’ development resembles that of another sibling pair of patients with a GLS deficiency caused by a homozygous 8-kb duplication spanning exon 1.17
Alternative splicing of GLS generates two protein isoforms: glutaminase C and kidney-type glutaminase.18 Both isoforms are localized in mitochondria and expressed in multiple tissues, with high expression levels of kidney-type glutaminase in the brain (i.e., cerebral cortex) and kidney.18,19 Glutamate is the major excitatory neurotransmitter in the brain and can be synthesized from α-ketoglutarate20 or glutamine (the most abundant amino acid)21 through the action of GLS. Studies in model organisms have shown that the ablation of GLS results in partially impaired glutamatergic synaptic transmission in mice and early death from respiratory problems.22 The small residual GLS activity that was detected in the fibroblasts and lymphocytes of our patients may account for the comparatively milder phenotypes. Mice that are heterozygous for a GLS deficiency had hippocampal hypoactivity.23 Patients with other causes of glutamate deficiency have ataxia, as did our patients.24 Whether elevated glutamine levels affect the phenotype in the three patients we describe here is unclear.
Our study underscores the importance of examining noncoding regions of the genome. Repetitive elements represent an estimated 50% of the human genome,25,26 and if such elements are unstable, they may have multiple deleterious effects. Short (1 to 6 bp) tandem repeats (of which the GCA repeat is one) make up more than 3% of the human genome,27 yet relatively few repeat expansion disorders have been described to date. Those that have been reported predominantly affect the nervous system.26 Current emphasis on the use of short-read exome-sequencing methods has made it difficult to systematically identify repetitive elements as a source of genetic deficit in rare mendelian diseases, but repetitive elements may represent missing heritability in the cause of disease.
Supplementary Material
Acknowledgments
Supported by the BC Children’s Hospital Foundation (First Collaborative Area of Innovation), Alberta Children’s Hospital Research Institute Foundation, a grant (301221) from the Canadian Institutes of Health Research, the Rare Diseases Foundation, the Canadian Rare Disease Models and Mechanisms Network, a Health Canada Platform Support Grant (PSG14–3505, to Dr. Wen) from the Brain Canada Foundation, a grant (GPIN 05389–14, to Dr. Wen) from the Natural Sciences and Engineering Research Council of Canada, the Michael Smith Foundation for Health Research, the National Ataxia Foundation, a grant (DK057808, to Dr. Usdin) from the National Institutes of Health, a grant (to Dr. Scherer) from Autism Speaks, and a grant (to Dr. van Karnebeek) from Foundation Metakids. The informatics infrastructure was supported by Genome BC and Genome Canada (ABC4DE Project).
We thank the patients and their families for participating in this study and their local physicians and health care teams for providing medical reports; X. Han, F. Miao, and M. Higginson for performing DNA extraction, triple repeat–primed polymerase-chain-reaction assays, and Sanger sequencing; and A. Ghani, L. Muttumacoroe, E. Lomba, and D. Pak for enrollment of patients, study administration, and logistic support.
APPENDIX
The authors’ full names and academic degrees are as follows: André B.P. van Kuilenburg, Ph.D., Maja Tarailo-Graovac, Ph.D., Phillip A. Richmond, B.A., Britt I. Drögemöller, Ph.D., Mahmoud A. Pouladi, Ph.D., René Leen, B.Sc., Koroboshka Brand-Arzamendi, B.Sc., Doreen Dobritzsch, Ph.D., Egor Dolzhenko, Ph.D., Michael A. Eberle, Ph.D., Bruce Hayward, Ph.D., Meaghan J. Jones, Ph.D., Farhad Karbassi, Ph.D., Michael S. Kobor, Ph.D., Janet Koster, B.Sc., Daman Kumari, Ph.D., Meng Li, Ph.D., Julia MacIsaac, Ph.D., Cassandra McDonald, Ph.D., Judith Meijer, B.Sc., Charlotte Nguyen, B.Sc., Indhu-Shree Rajan-Babu, Ph.D., Stephen W. Scherer, Ph.D., Bernice Sim, M.Sc., Brett Trost, Ph.D., Laura A. Tseng, M.D., Marjolein Turkenburg, B.Sc., Joke J.F.A. van Vugt, Ph.D., Jan H. Veldink, M.D., Ph.D., Jagdeep S. Walia, M.D., Youdong Wang, M.D., Michel van Weeghel, Ph.D., Galen E.B. Wright, Ph.D., Xiaohong Xu, M.D., Ph.D., Ryan K.C. Yuen, Ph.D., Jinqiu Zhang, M.D., Ph.D., Colin J. Ross, Ph.D., Wyeth W. Wasserman, Ph.D., Michael T. Geraghty, M.D., Saikat Santra, M.D., Ronald J.A. Wanders, Ph.D., Xiao-Yan Wen, M.D., Ph.D., Hans R. Waterham, Ph.D., Karen Usdin, Ph.D., and Clara D.M. van Karnebeek, M.D., Ph.D.
The authors’ affiliations are as follows: Amsterdam University Medical Centers, University of Amsterdam, Departments of Clinical Chemistry, Pediatrics, and Clinical Genetics, Emma Children’s Hospital, Amsterdam Gastroenterology and Metabolism (A.B.P.K., R.L., J.K., J. Meijer, L.A.T., M.T., M.W., R.J.A.W., H.R.W., C.D.M.K.), and United for Metabolic Diseases (A.B.P.K., R.J.A.W., H.R.W., C.D.M.K.), Amsterdam, and the Department of Neurology, Brain Center Rudolf Magnus, University Medical Center Utrecht (J.J.F.A.V., J.H.V.), and the Project MinE ALS Sequencing Consortium (J.J.F.A.V., J.H.V.), Utrecht — all in the Netherlands; the Departments of Biochemistry and Molecular Biology and Medical Genetics, Cumming School of Medicine, and Alberta Children’s Hospital Research Institute, University of Calgary, Calgary (M.T.-G.), Centre for Molecular Medicine and Therapeutics, BC Children’s Hospital Research Institute (P.A.R., M.J.J., M.S.K., J. MacIsaac, W.W.W., C.D.M.K.), the Faculty of Pharmaceutical Sciences (B.I.D., G.E.B.W., C.J.R.), and the Departments of Medical Genetics (C.M., I.-S.R.-B., W.W.W.) and Pediatrics (C.D.M.K.), University of British Columbia, Vancouver, the Zebrafish Centre for Advanced Drug Discovery, St. Michael’s Hospital and University of Toronto (K.B.-A., F.K., M.L., Y.W., X.-Y.W.), the Centre for Applied Genomics, Genetics and Genome Biology, the Hospital for Sick Children (C.N., S.W.S., B.T., R.K.C.Y.), and the Department of Molecular Genetics (C.N., S.W.S., R.K.C.Y.), the McLaughlin Centre (S.W.S.), and the Departments of Medicine, Physiology, and Laboratory Medicine and Pathobiology, Institute of Medical Science (X.-Y.W.), University of Toronto, Toronto, and the Division of Medical Genetics, Department of Pediatrics, Children’s Hospital Eastern Ontario, University of Ottawa, Ottawa (J.S.W., M.T.G.) — all in Canada; the Departments of Medicine and Physiology, National University of Singapore (M.A.P.), and the Translational Laboratory in Genetic Medicine, Agency for Science, Technology, and Research (M.A.P., B.S., X.X., J.Z.) — both in Singapore; Uppsala University, Department of Chemistry–Biomedical Center, Uppsala, Sweden (D.D.); Illumina, San Diego, CA (E.D., M.A.E.); Gene Structure and Disease Section, Laboratory of Cell and Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD (B.H., D.K., K.U.); and the Department of Clinical Inherited Metabolic Disorders, Birmingham Children’s Hospital, Birmingham, United Kingdom (S.S.).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Wright CF, FitzPatrick DR, Firth HV. Paediatric genomics: diagnosing rare disease in children. Nat Rev Genet 2018; 19: 253–68. [DOI] [PubMed] [Google Scholar]
- 2.Tarailo-Graovac M, Shyr C, Ross CJ, et al. Exome sequencing and the management of neurometabolic disorders. N Engl J Med 2016; 374: 2246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–7. [DOI] [PubMed] [Google Scholar]
- 4.Tarailo-Graovac M, Drögemöller BI, Wasserman WW, et al. Identification of a large intronic transposal insertion in SLC17A5 causing sialic acid storage disease. Orphanet J Rare Dis 2017; 12: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfares A, Aloraini T, Subaie LA, et al. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet Med 2018; 20: 1328–33. [DOI] [PubMed] [Google Scholar]
- 6.Guéant JL, Chéry C, Oussalah A, et al. APRDX1 mutant allele causes a MMACHC secondary epimutation in cblC patients. Nat Commun 2018; 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lionel AC, Costain G, Monfared N, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med 2018; 20: 435–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Kuilenburg ABP, Tarailo-Graovac M, Meijer J, et al. Genome sequencing reveals a novel genetic mechanism underlying dihydropyrimidine dehydrogenase deficiency: a novel missense variant c.1700G>A and a large intragenic inversion in DPYD spanning intron 8 to intron 12. Hum Mutat 2018; 39: 947–53. [DOI] [PubMed] [Google Scholar]
- 9.van Karnebeek CD, Shevell M, Zschocke J, Moeschler JB, Stockler S. The metabolic evaluation of the child with an intellectual developmental disorder: diagnostic algorithm for identification of treatable causes and new digital resource. Mol Genet Metab 2014; 111: 428–38. [DOI] [PubMed] [Google Scholar]
- 10.Köhler S, Vasilevsky NA, Engelstad M, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res 2017; 45: D865–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung WA, Ouellette BF, Wasserman WW. Quantitative biomedical annotation using Medical Subject Heading Over-representation Profiles (MeSHOPs). BMC Bioinformatics 2012; 13: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Huang Y, Zhao L, Li Y, Zheng J. Glutaminase 1 is essential for the differentiation, proliferation, and survival of human neural progenitor cells. Stem Cells Dev 2014; 23: 2782–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolzhenko E, van Vugt JJFA, Shaw RJ, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res 2017; 27: 1895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, et al. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol 2013; 12: 978–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat Rev Mol Cell Biol 2015; 16: 499–513. [DOI] [PubMed] [Google Scholar]
- 17.Lynch DS, Chelban V, Vandrovcova J, Pittman A, Wood NW, Houlden H. GLS loss of function causes autosomal recessive spastic ataxia and optic atrophy. Ann Clin Transl Neurol 2018; 5: 216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgadi KM, Meguid RA, Qian M, Souba WW, Abcouwer SF. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol Genomics 1999; 1: 51–62. [DOI] [PubMed] [Google Scholar]
- 19.Thul PJ, Åkesson L, Wiking M, et al. A subcellular map of the human proteome. Science 2017; 356(6340): eaal3321. [DOI] [PubMed] [Google Scholar]
- 20.Takeda K, Ishida A, Takahashi K, Ueda T. Synaptic vesicles are capable of synthesizing the VGLUT substrate glutamate from α-ketoglutarate for vesicular loading. J Neurochem 2012; 121: 184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer 2016; 16: 749. [DOI] [PubMed] [Google Scholar]
- 22.Masson J, Darmon M, Conjard A, et al. Mice lacking brain/kidney phosphate-activated glutaminase have impaired glutamatergic synaptic transmission, altered breathing, disorganized goal-directed behavior and die shortly after birth. J Neurosci 2006; 26: 4660–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaisler-Salomon I, Miller GM, Chuhma N, et al. Glutaminase-deficient mice display hippocampal hypoactivity, insensitivity to pro-psychotic drugs and potentiated latent inhibition: relevance to schizophrenia. Neuropsychopharmacology 2009; 34: 2305–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guergueltcheva V, Azmanov DN, Angelicheva D, et al. Autosomal-recessive congenital cerebellar ataxia is caused by mutations in metabotropic glutamate receptor 1. Am J Hum Genet 2012; 91: 553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 2009; 25(1): 4.10.1–4.10.4. [DOI] [PubMed] [Google Scholar]
- 26.Hannan AJ. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet 2018; 19: 286–98. [DOI] [PubMed] [Google Scholar]
- 27.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol 2003; 4: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.