Abstract
Programmed cell death is an active extinction process, including autophagy, ferroptosis, pyroptosis, apoptosis, and necroptosis. m6A is a reversible RNA modification which undergoes methylation under the action of methylases (writers), and is demethylated under the action of demethylases (erasers). The RNA base site at which m6A is modified is recognized by specialized enzymes (readers) which regulate downstream RNA translation, decay, and stability. m6A affects many aspects of mRNA metabolism, and also plays an important role in promoting the maturation of miRNA, the translation and degradation of circRNA, and the stability of lncRNA. The regulatory factors including writers, erasers and readers promote or inhibit programmed cell death via up-regulating or down-regulating downstream targets in a m6A-dependent manner to participate in the process of disease. In this review, we summarize the functions of m6A with particular reference to its role in programmed cell death.
Keywords: m6A, autophagy, ferroptosis, pyroptosis, apoptosis, necroptosis
Cell Death
Programmed cell death (PCD) is the active process of cell death, including autophagy, ferroptosis, pyroptosis, apoptosis, and necroptosis (Cabon et al., 2013). These forms of cell death have different and independent regulatory pathways (Supplementary Table S1) (Kaiser et al., 2011; Newton et al., 2014; Broz and Dixit, 2016; New and Thomas, 2019; Uzdensky, 2019; Capelletti et al., 2020; Qiu et al., 2020; Beroske et al., 2021; Bhardwaj et al., 2021; Tsuchiya, 2021; Yuan et al., 2021a). However, there are interconnections and crosstalk between these pathways (Bedoui et al., 2020; Wang and Kanneganti, 2021). For example, Bcl-2 is not only an inhibitor of apoptosis but also a fundamental regulator of autophagy (Mukhopadhyay et al., 2014). Bcl-2 inhibits autophagy induced by Beclin-1 by binding to the autophagy-related protein Beclin-1 to form a complex (Mukhopadhyay et al., 2014). Caspase-8 acts as a molecular switch and is activated in response to cell death signals to play a key role in apoptosis and necroptosis (Fritsch et al., 2019; Schwarzer et al., 2020). A study of lysosomal storage disease has shown that lysosomal dysfunction affects the accumulation of autophagosomes to induce autophagy-dependent ferroptosis (Pierzynowska et al., 2021). A previous study of the pathological mechanisms underlying renal tubular necrosis found that necroptosis initiated the cell death process via ferroptosis (Belavgeni et al., 2020). Interestingly, it was proved that these forms of cell death can be mediated by post-transcriptional regulation, including N6-methyladenosine (m6A) (Song et al., 2019).
Overview of m6A
More than 100 different modifications have been identified in coding RNA and non-coding RNA (ncRNA) (Boccaletto et al., 2018). Methylation modification is one of the most common form of RNA modification, usually including N1-methyladenosine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (5hmC), m6A and 7-methylguanine (m7G) (Boccaletto and Bagiński, 2021). m6A was first discovered by Desrosiers et al. in the mRNA of liver cancer cells in 1974 and acted as the most prominent and abundant internal RNA modification in mammalian RNA (Desrosiers et al., 1974; Coker et al., 2019; Muthusamy, 2020). Adenosine can be chemically modified by adding methyl groups to the adenine bases in RNA. m6A occurs when the N6 position of adenosine is methylated (Oerum et al., 2021). This process is dynamically regulated by methylase and demethylase, and plays its functional role under the action of corresponding recognition enzymes (Fu et al., 2014).
Enzymes Involved in m6A
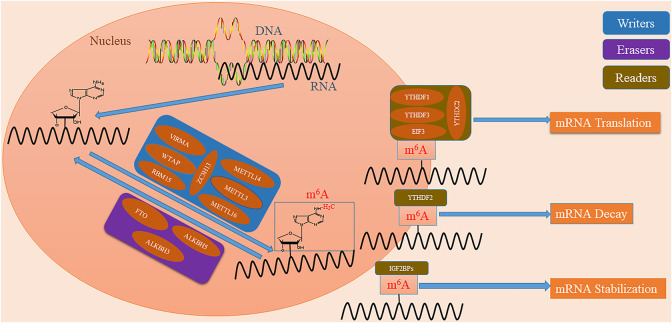
Installation of m6A is a reversible process regulated by the methylases (writers) and demethylases (erasers) (Supplementary Table S2) (Figure 1) (Karthiya and Khandelia, 2020). For one thing, the writers form the methyltransferase complex (MTC) and catalyze the process for m6A (Bokar et al., 1994; Śledź and Jinek, 2016). Methyltransferase-like protein 3 (METTL3) is an S-adenosylmethionine (SAM) binding protein which catalyzes the transfer of methyl groups in SAM (Wang et al., 2016). Methyltransferase-like protein 14 (METTL14) is another active component of MTC and forms a stable complex with METTL3 (in a ratio of 1:1) to stabilize the structure of MTC (Wang et al., 2016). WT1 associated protein (WTAP) recruits METTL3 and METL14 to nuclear areas, and RNA binding motif protein 15 (RBM15) guides METTL3 and WTAP to RNA sites (Patil et al., 2016; Schöller et al., 2018). VIRMA/KIAA1429 recruits MTC and mediates the methylation of adenine bases near the 3′- untranslated region (UTR) (Yue et al., 2018). Zinc finger CCCH domain-containing protein 13 (ZC3H13) enhances its catalytic function by interacting with WTAP (Knuckles et al., 2018). Methyltransferase-like protein 16 (METTL16) is a newly discovered independent RNA methylase that catalyzes the installation of m6A (Warda et al., 2017). For another, m6A undergoes demethylation under the action of erasers (Jia et al., 2011). FTO that proves the reversibility of the methylation process is the first protein identified to catalyze the demethylation of m6A (Jia et al., 2011). ALKBH5 is the second demethylase identified to reverse m6A, catalyzing the demethylation of m6A in a Fe(II) and α-ketoglutarate-dependent manner (Aik et al., 2014). In addition, a recent study identified ALKBH3 as a new demethylase involved in this process with a similar mechanism (Ueda et al., 2017).
FIGURE 1.
Mechanisms underlying the m6A modification. m6A modification is a reversible process. Writers represent methylases which include METTL3, METTL14, WTAP, VIRMA/KIAA1429, RBM15, ZC3H13, and METTL16, to make the N6 position of adenosine methylated. Erasers represent demethylases including FTO, ALKBH5 and ALKBH3, to make m6A demethylated. Readers represent specialized enzymes, including YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, IGF2BPs, and EIF3, which recognize the RNA base site at which m6A is modified, to regulate downstream mRNA translation, degradation, and stabilization.
Interestingly, the RNA base site at which m6A is modified is recognized by specialized enzymes (readers) which regulate downstream mRNA translation, decay, and stability (Supplementary Table S2) (Figure 1) (Meyer et al., 2015; Huang et al., 2018; Shi et al., 2021). YTHDF1 recruits initiation factors to promote mRNA translation and protein synthesis (Wang et al., 2015). YTHDF2 recruits mRNA modified by the selective binding of m6A to the mRNA decay site to induce the degradation of transcripts (Wang et al., 2015). YTHDF3 has been shown to combine with YTHDF1 and YTHDF2 to enhance their function (Lasman et al., 2020). YTHDC1 promotes RNA splicing and export by recruiting mRNA splicing factors (Woodcock et al., 2020). YTHDC2 interacts with RNA helicase to improve the translation efficiency of target RNA (Kim and Siddiqui, 2021). IGF2BPs and EIF3 play important roles in improving the stability of mRNA and the efficiency of the translation initiation complex (Lee et al., 2016; Müller et al., 2019). Heterogenous nuclear ribonucleoproteins (HNRNPs) are a complex and functionally diverse family of RNA binding proteins (Low et al., 2021). HNRNPA2/B1 as a member of the HNRNPs family, is identified as a nuclear reader of m6A and regulates the alternative splicing of exons in a set of transcripts (Alarcón et al., 2015).
The Function of m6A
m6A is one of the most prominent RNA modification for mRNA and ncRNA and plays a variety of biological functions by regulating these forms of RNA (Supplementary Table S3(Roignant and Soller, 2017; Dai et al., 2018; He et al., 2020; Zhang et al., 2020; Han et al., 2021).
m6A in mRNA Metabolism
Transcription of mRNA is the first step which leads to the production of protein, and the post-transcriptional control of mRNA is regulated by multiple mechanisms (Ivanov and Anderson, 2013). m6A modification affects many aspects of mRNA metabolism, including processing, export, translation, and decay (Roignant and Soller, 2017; Dai et al., 2018). mRNA processing promotes mRNA maturation through 5′capping, 3′polyadenylation and splicing (Dai et al., 2018). FTO depletion or METTL3 overexpression is known to increases m6A levels and promote the splicing of arginine-rich splicing factor (SRSF) (Zhao et al., 2014). YTHDC1 promotes RNA splicing and export by recruiting SRSF (Woodcock et al., 2020). HNRNP may affect the abundance and alternative splicing of target mRNA in a manner regulated by the “m6A switch” (Alarcón et al., 2015).
The nuclear export and translation of mRNA are key steps in the regulation of gene expression; m6A modification is known to facilitate this process (Dai et al., 2018). The knockdown of METTL3 was shown to reduce mRNA export to the cytoplasm, while inhibition of ALKBH5 had the opposite effect (Song et al., 2019). EIF3 plays an important role in improving the efficiency of mRNA translation initiation complex; YTHDF1 and METTL3 can recruit EIF3 to promote the translation of m6A-modified mRNA (Wang et al., 2015; Lee et al., 2016). Interestingly, YTHDF3 promotes mRNA translation in an m6A-dependent manner by enhancing the function of YTHDF1 (Lasman et al., 2020).
RNA decay is characterized by deadenylation which regulated by m6A modification (Du et al., 2016). YTHDF2 directly interacts with the SH domain of CCR4-NOT transcription complex subunit 1 (CNOT1) to mediate mRNA deadenylation (Du et al., 2016). YTHDF2 has also been shown to recruit mRNA modified by the m6A to mRNA decay sites to induce decay (Wang et al., 2015).
m6A in ncRNA
ncRNA includes microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA). m6A modification regulates cell proliferation, apoptosis, and the cell cycle by promoting the maturation of miRNA, the translation and degradation of circRNA, and the stability of lncRNA (He et al., 2020; Zhang et al., 2020; Han et al., 2021). Cigarette smoke condensate (CSC) mediates METTL3 to promote the maturation of miRNA-25-3 to enhance pancreatic ductal adenocarcinoma (Zhang et al., 2019). In addition, METTL3 can enhance the binding ability of pri-miRNA-221/222 and DGCR8 to promote bladder cancer (Han et al., 2019). METTL14 promotes the maturation of pri-miRNA-126 to inhibit the invasion of hepatocellular carcinoma (Ma et al., 2017). Some circRNAs have potential protein coding capabilities, and the process can be driven by m6A (Zhang et al., 2020). This process is initiated by YTHDF3, enhanced by METTL3/14, and inhibited by FTO (Zhang et al., 2020). Surprisingly, m6A modification can also promote circRNA degradation (Zhang et al., 2020). HRSP12 is an adaptor protein that can connect YTHDF2 and RNase P/MRP (endoribonuclease). circRNA is cleaved by endoribonuclease through the YTHDF2-HRSP12-RNase P/MRP axis; m6A promotes this process (Dong et al., 2019). m6A modification is known to affect the carcinogenic role of MALAT1 by promoting the binding ability with HNRNP C (Yang et al., 2013). Moreover, m6A modification can increase the stability of FAM225A and up-regulate its level to promote the proliferation and invasion of nasopharyngeal carcinoma (Zheng et al., 2019). Silencing METTL3 will reduce the m6A level and stability of total RNA of FAM225A (Zheng et al., 2019).
The Role of m6A in PCD
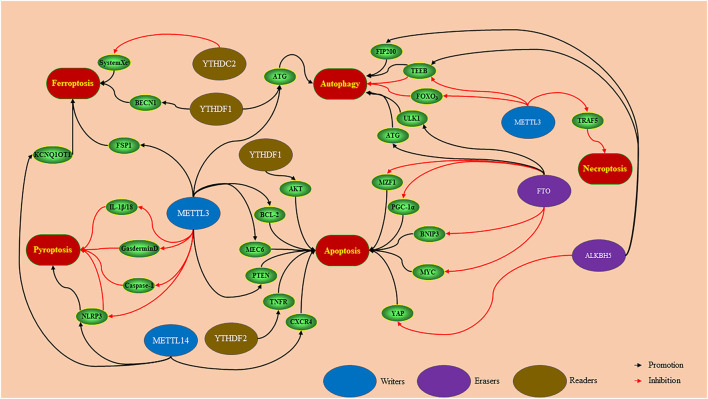
PCD works a key role in the pathogenesis of cancer, neurodegenerative diseases, and inflammation (Florean et al., 2019; Heckmann et al., 2019). m6A is controlled by regulatory factors (writers and erasers) and recognition factors (readers) to mediate downstream targets to regulate PCD (Figure 2) (Song et al., 2019; Liu et al., 2020; Wang et al., 2020a; Tsuruta et al., 2020).
FIGURE 2.
Writers, erasers and readers mediates downstream targets to regulate PCD in a m6A-dependent manner. METTL3 promotes ferroptosis and apoptosis, and inhibits necroptosis, via regulating downstream targets. According to the different functions of downstream targets, METTL3 can inhibit or promote pyroptosis and autophagy. METTL14 regulates downstream targets to promote ferroptosis, pyroptosis, and apoptosis. FTO and ALKBH5 mediate downstream targets to promote autophagy and apoptosis. The YTH family plays a role in promoting ferroptosis, autophagy, and apoptosis according to mediate different target proteins.
m6A and Autophagy
Autophagy is a basic pathway of cellular degradation with a lysosomal-assisted degradation mechanism, which is characterized by the formation of autophagosomes (Frankel et al., 2017). Autophagy-related genes (ATG), uncoordinated 51-like kinase 1 (ULK1) and transcription factor EB (TFEB) are important regulators of autophagy (Bhardwaj et al., 2021). m6A modification is known to exert effects by strictly regulating these regulators in different disease backgrounds, which include direct inhibition, the formation of autophagosomes, initiation, and enhancement on autophagy (Song et al., 2019; Liu et al., 2020; Wang et al., 2020a). TFEB is a key transcription factor that regulates the function of lysosome and expression of ATG (Corà et al., 2021). The expression of METTL3 is known to increase in mouse models of ischemic heart disease and inhibits autophagy by down-regulating TFEB; but ALKBH5 can make the opposite effect by up-regulating TFEB (Song et al., 2019). FTO up-regulates ULK1 that is a positive regulator of autophagy to initiate autophagy in a m6A-dependent manner (Jin et al., 2018). FTO is identified to increase the expression of ATG5 and ATG7, and promote the autophagosome formation to promote both autophagy and adipogenesis (Jin et al., 2018; Wang et al., 2020a). In addition, the overexpression of METTL3 is proved to increase the level of m6A modification of ATG5 while the knockdown of ATG5 reduces the autophagy induced by METTL3, indicating that ATG5 is a key target to induce autophagy (Chen et al., 2021). YTHDF1 promotes the translation of the ATG gene by combining with m6A-modified ATG2 and ATG14 mRNA, thereby promoting autophagy in human hepatocellular carcinoma (Li et al., 2021).
Forkhead box O3 (FOXO3) is a key transcription factor in a variety of carcinogenic pathways (Lin et al., 2020). METTL3 affects FOXO3 RNA stability in a YTHDF1-dependent manner to inhibit the expression of FOXO3 to inhibit autophagy; The reduced expression of METTL3 increases the number of autophagosomes to activate autophagy in liver cancer cells (Lin et al., 2020). FIP200 is an essential autophagy gene, and ALKBH5 enhances autophagy by mediating the demethylation of FIP200 mRNA (Li et al., 2020a). Surprisingly, one study found that reduced levels of FTO can weaken the activation of the mTORC1 pathway to enhance autophagy (Gulati et al., 2013). FTO regulates mTOR signaling by mediating m6A demethylation and activates autophagy to promote cell proliferation in melanoma (Yang et al., 2019). In short, m6A modification has different roles in the regulation of autophagy in different disease backgrounds.
m6A and Ferroptosis
Ferroptosis is a newly discovered type of programmed cell death that involves iron-dependent lipid peroxidation along with the loss of glutathione peroxidase 4 and dense mitochondrial membrane (Dixon and Stockwell, 2014). m6A modification, as a new form of post-transcriptional regulatory mechanism, is known to play an important role in ferroptosis (Shen et al., 2021; Zhuang et al., 2021). During the ferroptosis of liver fibrosis, the expression of METTL4 was shown to up-regulated; in addition, the level of m6A was shown to increase and was enhanced by ferroptosis inducers (Shen et al., 2021). Previous researchers investigated the mechanisms underlying the effect of doxorubicin on cardiotoxicity and found that doxorubicin induced the up-regulation of METTL14 expression and catalyzed the m6A modification of RNA KCNQ1OT1 to participate in the ferroptosis of cardio myocytes (Zhuang et al., 2021). Moreover, METTL3 acts as the target of miR-4443, and regulates the expression of FSP1 to mediate the ferroptosis of non-small cell lung cancer (Song et al., 2021). Autophagy may also represent a targeted pathway by which to regulate the sensitivity of cells to ferroptosis (Gryzik et al., 2021). BECN1 is a key protein that regulates autophagy and promotes ferroptosis by regulating the activity of the cysteine/glutamate antiporter (also referred to as system Xc-) (Kang et al., 2018). RNA-seq analysis has shown that m6A modification triggers the activation of autophagy by stabilizing BECN1 mRNA and by inducing ferroptosis; YTHDF1 can promote this process by recognizing the m6A binding site in the coding region of BECN1 (Shen et al., 2021). Studies have also found that YTHDC2 is a powerful endogenous inducer of ferroptosis and plays a predominant role in the treatment of lung adenocarcinoma by targeting the SLC3A2 subunit of system Xc− (Ma et al., 2021).
m6A and Pyroptosis
Pyroptosis is a cell death pathway that is characterized by the activation of various caspases including caspase-1 which is mediated by inflammasomes and causes cell perforation (Mulay, 2019). Studies have suggested that m6A plays a key role in the regulatory pathway responsible for pyroptosis in cells (Zha et al., 2020; Liu et al., 2021). NOD-like receptor protein 3 (NLRP3) is a key component of the inflammasome that causes pyroptosis and is known to promote pyroptosis and increase the levels of pro-inflammatory cytokines (Mulay, 2019). High glucose levels can induce the increased expression of pyroptosis-related proteins (Caspase-1, Gasdermin D, NLRP3, IL-1β, and IL-18) to lead to cell death (Zha et al., 2020). The overexpression of METTL3 reduces the expression of these proteins and reduces the extent of damage while the knockdown of METTL3 will aggravate cellular damage (Zha et al., 2020). Previous study investigated the manner by which interferon regulatory factor-1 (IRF-1) can promote the pyroptosis of macrophages in patients with acute coronary syndrome and detected increased levels of m6A and METTL3 in the macrophages (Guo et al., 2020). The overexpression of IRF-1 can induce an increase in the levels of m6A and METTL3 to promote acute coronary syndrome (Guo et al., 2020). A research on intervertebral disc degeneration found that METTL14 specifically induced an increase in the m6A modification of NLRP3 mRNA and increased the expression of NLRP3 protein (Yuan et al., 2021b).
m6A and Apoptosis
Apoptosis is the spontaneous and orderly death of cells triggered by the endogenous mitochondrial pathway (BCL-2 pathway) and the exogenous death receptor pathway (Clavier et al., 2016). Recent studies have shown that apoptosis is regulated by m6A (Dominissini et al., 2012). More and more studies have shown that m6A plays a role in the occurrence of cancer by regulating apoptosis-related proteins to promote or inhibit apoptosis (Supplementary Table S4(Liu et al., 2018; Su et al., 2018; Niu et al., 2019; Tang et al., 2019; Zhuang et al., 2019). METTL3 is an important methylase for m6A modification, and it shows important performance in lung cancer, breast cancer, ovarian cancer, gastrointestinal cancer, and leukemia by regulating downstream targets BCL-2, MEC6, and PTEN to inhibit apoptosis (Vu et al., 2017; Wei et al., 2019; Wang et al., 2020b; Wang et al., 2020c; Bi et al., 2021). METTL3 is up-regulated in breast cancer cells (Wang et al., 2020b). The knockdown of METTL3 can reduce methylation levels, target Bcl-2, reduce proliferation, accelerate cell apoptosis, and inhibit tumor growth (Wang et al., 2020b). Similarly, METTL3 has been shown to mediate apoptosis by regulating Bcl-2 in lung cancer (Wei et al., 2019). In a mouse model of leukemia, the depletion of METTL3 was shown to induce cell differentiation and apoptosis via the same mechanism to prevent the progression of leukemia in mice (Vu et al., 2017). Moreover, METTL14 is elevated in breast cancer and regulates chemokine receptor 4 (CXCR4) to inhibit apoptosis (Sun et al., 2020). Interestingly, as an important demethylase, FTO promotes apoptosis and participates in the occurrence of cancer by regulating downstream targets BNIP3, MZF1, MYC, and PGC-1α (Liu et al., 2018; Su et al., 2018; Niu et al., 2019; Zhuang et al., 2019). BNIP3 is considered to be a pro-apoptotic member of the Bcl-2 apoptotic protein family (Gorbunova et al., 2020). In breast cancer, the expression of FTO is up-regulated while that of BNIP3 is down-regulated (Niu et al., 2019). Silencing FTO promotes an increase in the levels of BNIP3 mRNA and protein (Niu et al., 2019). FTO demethylates m6A in the 3′-UTR of BNIP3 and reduces its expression; this promotes the proliferation, colony formation, and metastasis of breast cancer cells (Niu et al., 2019). Another demethylase ALKBH5 plays a role in osteosarcoma by regulating YAP (Yuan et al., 2021c). In addition, the methylation recognition enzyme YTHDF1/2 has also received important attention in cancer, and studies have found that they affect the occurrence of liver cancer and leukemia by regulating AKT and TNFR (Bian et al., 2020; Li et al., 2020b).
m6A and Necroptosis
Necroptosis is a lysed form of PCD that can cause inflammation (Negroni et al., 2020). Tumor necrosis factor (TNF), Toll-like receptor (TOLLR) family members, interferon, and other mediators, are the main factors that can lead to necroptosis (Shirjang et al., 2019). TRAF5 is a member of the TNF receptor-related factor family and can exert influence on cell survival, proliferation, differentiation, and death, by mediating a variety of downstream signaling pathways (Potter et al., 2007). Previous researchers investigated the mechanisms underlying resistance to oxaliplatin in colorectal cancer and found that the mRNA and protein levels of TRAF5 increased when METTL3 was knocked out; the overexpression of METTL3 had the opposite result (Lan et al., 2021). METTL3 is known to inhibit necroptosis by inhibiting the levels of TRAF5 m6A (Lan et al., 2021). There have been few studies of m6A modification in necroptosis; consequently, the specific regulatory mechanisms involved remain unclear.
Conclusion and Future Perspectives
Collectively, these findings suggest that m6A is essential for regulating PCD. The promotion or inhibition of PCD by m6A mainly depends on the level of m6A (dynamically regulated by writers and erasers), the function of downstream targets, and the changes in target RNA after methylation (depending on readers). Although some progress has been made in identifying the regulatory mechanisms involved, further research is still needed to explore the exact link between m6A modification and PCD in various pathological conditions.
First of all, the current studies mainly focus on the regulatory factors of m6A, and the direct influence mechanisms of m6A on downstream targets are still unclear. For example, in the study of cell death caused by high glucose, METTL3 mediates pyroptosis by regulating NLRP3 (Zha et al., 2020). Whether METTL3 can directly affect the expression of NLRP3 without an m6A-dependent manner and how the changes of m6A regulates NLRP3, it still needs further research. Secondly, the regulation of m6A is affected by many factors. As mentioned in this paper, METTL3 promotes the installation of m6A and FTO inhibits the installation. However, studies have shown that METTL3 up-regulates downstream targets to promote apoptosis, and FTO down-regulates downstream targets to promote apoptosis (Niu et al., 2019; Wang et al., 2020b). This seems to indicate the dual role of m6A in apoptosis, which provides a direction for future cancer treatment. Finally, the most challenging issue is that there are often multiple forms of cell death in the same disease. m6A may act a “double-edged sword”. For example, m6A can not only inhibit the occurrence of cancer by promoting apoptosis and ferroptosis, but also promote the occurrence of cancer by inhibiting apoptosis, pyroptosis, and autophagy. The regulation of m6A in the various stages of cell death and the intricate connections among them still need to be further explored.
m6A is extremely important for mRNA metabolism at different stages. Except for m6A, other chemical modifications also are irreplaceable, such as m1A and m5C. What is the regulation mechanism of these chemical modifications on PCD? Is it synergistic with m6A? These issues still need to be resolved. In general, understanding the link between m6A and PCD will not only promote the understanding of the underlying mechanisms of certain diseases, but may also discover new therapeutic strategies.
Acknowledgments
Thank EditSprings (https://www.editsprings.com) for editing English language of our manuscript.
Author Contributions
Conceptualization, FT and DX; software, FT, DX, LC, and XL; resources, FT, DX, HG, and XL; writing-original draft preparation, FT and LC; writing-review and editing, FT, DX, LC, HG, and XL; visualization, LC and XL; supervision, DX and XL; All authors read and approved the final manuscript.
Funding
The present study was supported by the National Science Foundation of China (Grant Nos 82071353, 82001593, 81330016, 81630038, and 81771634), the National Key R&D Program of China (Grant Nos 2017YFA 0104201 and 2017YFA 0104200), and the Science and Technology Bureau of Chengdu City (Grant No. 2015-HM01-00424-SF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.817112/full#supplementary-material
The regulatory pathways of different forms of PCD.
The classification and function of enzymes involved in m6A.
The function of m6A in mRNA and ncRNA.
The roles of m6A by regulating apoptosis in the occurrence of cancer.
Abbreviations
ALKBH, AlkB homolog; ATG, autophagy-related gene; circRNA, circular RNA; EIF3, eukaryotic initiation factor 3; FOXO3, Forkhead box O3; FTO, fat mass and obesity-associated protein; IGF2BPs, insulin-like growth factor 2; IRF-1, interferon regulatory factor-1; lncRNA, long non-coding RNA; m6A, N6–methyladenosine; METTL, methyltransferase-like protein; miRNA, microRNA; MTC, methyltransferase complex; mTOR, mammalian target of rapamycin; ncRNA, non-coding RNA; NLRP3, NOD-like receptor protein 3; PCD, programmed cell death; RBM15, RNA binding motif protein 15; SAM, S-adenosylmethionine; SRSF, splicing of arginine-rich splicing factor; TFEB, transcription factor EB; TNF, Tumor necrosis factor; TOLLR, Toll-like receptor; ULK1, uncoordinated 51-like kinase 1; VIRMA, Vir-like m6A methyltransferase; WTAP, WT1 associated protein; ZC3H13, zinc finger CCCH-type contains 13.
References
- Aik W., Scotti J. S., Choi H., Gong L., Demetriades M., Schofield C. J., et al. (2014). Structure of Human RNA N6-Methyladenine Demethylase ALKBH5 Provides Insights into its Mechanisms of Nucleic Acid Recognition and Demethylation. Nucleic Acids Res. 42 (7), 4741–4754. 10.1093/nar/gku085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015). HNRNPA2B1 Is a Mediator of m6A-dependent Nuclear RNA Processing Events. Cell 162 (6), 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoui S., Herold M. J., Strasser A. (2020). Emerging Connectivity of Programmed Cell Death Pathways and its Physiological Implications. Nat. Rev. Mol. Cel Biol 21 (11), 678–695. 10.1038/s41580-020-0270-8 [DOI] [PubMed] [Google Scholar]
- Belavgeni A., Meyer C., Stumpf J., Hugo C., Linkermann A. (2020). Ferroptosis and Necroptosis in the Kidney. Cel Chem. Biol. 27 (4), 448–462. 10.1016/j.chembiol.2020.03.016 [DOI] [PubMed] [Google Scholar]
- Beroske L., Van den Wyngaert T., Stroobants S., Van der Veken P., Elvas F. (2021). Molecular Imaging of Apoptosis: The Case of Caspase-3 Radiotracers. Int. J. Mol. Sci. 22 (8), 3948. 10.3390/ijms22083948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N., Tripathi N., Goel B., Jain S. (2021). Anticancer Activity of Diosgenin and its Semi-synthetic Derivatives: Role in Autophagy Mediated Cell Death and Induction of Apoptosis. Mini Rev. Med. Chem. 21 (13), 1646–1665. 10.2174/1389557521666210105111224 [DOI] [PubMed] [Google Scholar]
- Bi X., Lv X., Liu D., Guo H., Yao G., Wang L., et al. (2021). METTL3-mediated Maturation of miR-126-5p Promotes Ovarian Cancer Progression via PTEN-Mediated PI3K/Akt/mTOR Pathway. Cancer Gene Ther. 28, 335–349. 10.1038/s41417-020-00222-3 [DOI] [PubMed] [Google Scholar]
- Bian S., Ni W., Zhu M., Song Q., Zhang J., Ni R., et al. (2020). Identification and Validation of the N6-Methyladenosine RNA Methylation Regulator YTHDF1 as a Novel Prognostic Marker and Potential Target for Hepatocellular Carcinoma. Front. Mol. Biosci. 7, 604766. 10.3389/fmolb.2020.604766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P., Bagiński B. (2021). MODOMICS: An Operational Guide to the Use of the RNA Modification Pathways Database. Methods Mol. Biol. (Clifton, NJ) 2284, 481–505. 10.1007/978-1-0716-1307-8_26 [DOI] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M. A., Purta E., Piątkowski P., Bagiński B., Wirecki T. K., et al. (2018). MODOMICS: a Database of RNA Modification Pathways. 2017 Update. Nucleic Acids Res. 46, D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J. A., Rath-Shambaugh M. E., Ludwiczak R., Narayan P., Rottman F. (1994). Characterization and Partial Purification of mRNA N6-Adenosine Methyltransferase from HeLa Cell Nuclei. Internal mRNA Methylation Requires a Multisubunit Complex. J. Biol. Chem. 269 (26), 17697–17704. 10.1016/s0021-9258(17)32497-3 [DOI] [PubMed] [Google Scholar]
- Broz P., Dixit V. M. (2016). Inflammasomes: Mechanism of Assembly, Regulation and Signalling. Nat. Rev. Immunol. 16 (7), 407–420. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Cabon L., Martinez-Torres A.-C., Susin S. A. (2013). La mort cellulaire programmée ne manque pas de vocabulaire. Med. Sci. (Paris) 29 (12), 1117–1124. 10.1051/medsci/20132912015 [DOI] [PubMed] [Google Scholar]
- Capelletti M. M., Manceau H., Puy H., Peoc'h K. (2020). Ferroptosis in Liver Diseases: An Overview. Int. J. Mol. Sci. 21 (14), 4908. 10.3390/ijms21144908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xiang Y., Yin Y., Peng J., Peng D., Li D., et al. (2021). The m6A Methyltransferase METTL3 Regulates Autophagy and Sensitivity to Cisplatin by Targeting ATG5 in Seminoma. Transl Androl. Urol. 10 (4), 1711–1722. 10.21037/tau-20-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavier A., Rincheval-Arnold A., Colin J., Mignotte B., Guénal I. (2016). Apoptosis in Drosophila: Which Role for Mitochondria. Apoptosis 21 (3), 239–251. 10.1007/s10495-015-1209-y [DOI] [PubMed] [Google Scholar]
- Coker H., Wei G., Brockdorff N. (2019). m6A Modification of Non-coding RNA and the Control of Mammalian Gene Expression. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1862 (3), 310–318. 10.1016/j.bbagrm.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Corà D., Bussolino F., Doronzo G. (2021). TFEB Signalling-Related MicroRNAs and Autophagy. Biomolecules 11 (7), 985. 10.3390/biom11070985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D., Wang H., Zhu L., Jin H., Wang X. (2018). N6-methyladenosine Links RNA Metabolism to Cancer Progression. Cell Death Dis 9 (2), 124. 10.1038/s41419-017-0129-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., Rottman F. (1974). Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. 71 (10), 3971–3975. 10.1073/pnas.71.10.3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J., Stockwell B. R. (2014). The Role of Iron and Reactive Oxygen Species in Cell Death. Nat. Chem. Biol. 10 (1), 9–17. 10.1038/nchembio.1416 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485 (7397), 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Dong W., Bi J., Liu H., Yan D., He Q., Zhou Q., et al. (2019). Circular RNA ACVR2A Suppresses Bladder Cancer Cells Proliferation and Metastasis through miR-626/EYA4 axis. Mol. Cancer 18 (1), 95. 10.1186/s12943-019-1025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., et al. (2016). YTHDF2 Destabilizes m6A-Containing RNA through Direct Recruitment of the CCR4-Not Deadenylase Complex. Nat. Commun. 7, 12626. 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florean C., Song S., Dicato M., Diederich M. (2019). Redox Biology of Regulated Cell Death in Cancer: A Focus on Necroptosis and Ferroptosis. Free Radic. Biol. Med. 134, 177–189. 10.1016/j.freeradbiomed.2019.01.008 [DOI] [PubMed] [Google Scholar]
- Frankel L. B., Lubas M., Lund A. H. (2017). Emerging Connections between RNA and Autophagy. Autophagy 13 (1), 3–23. 10.1080/15548627.2016.1222992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch M., Günther S. D., Schwarzer R., Albert M.-C., Schorn F., Werthenbach J. P., et al. (2019). Caspase-8 Is the Molecular Switch for Apoptosis, Necroptosis and Pyroptosis. Nature 575 (7784), 683–687. 10.1038/s41586-019-1770-6 [DOI] [PubMed] [Google Scholar]
- Fu Y., Dominissini D., Rechavi G., He C. (2014). Gene Expression Regulation Mediated through Reversible m6A RNA Methylation. Nat. Rev. Genet. 15 (5), 293–306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- Gorbunova A. S., Denisenko T. V., Yapryntseva M. A., Pivnyuk A. D., Prikazchikova T. A., Gogvadze V. G., et al. (2020). BNIP3 as a Regulator of Cisplatin-Induced Apoptosis. Biochem. Mosc. 85 (10), 1245–1253. 10.1134/s0006297920100120 [DOI] [PubMed] [Google Scholar]
- Gryzik M., Asperti M., Denardo A., Arosio P., Poli M. (2021). NCOA4-mediated Ferritinophagy Promotes Ferroptosis Induced by Erastin, but Not by RSL3 in HeLa Cells. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1868 (2), 118913. 10.1016/j.bbamcr.2020.118913 [DOI] [PubMed] [Google Scholar]
- Gulati P., Cheung M. K., Antrobus R., Church C. D., Harding H. P., Tung Y.-C. L., et al. (2013). Role for the Obesity-Related FTO Gene in the Cellular Sensing of Amino Acids. Proc. Natl. Acad. Sci. 110 (7), 2557–2562. 10.1073/pnas.1222796110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Yan R., Ji Q., Yao H., Sun M., Duan L., et al. (2020). IFN Regulatory Factor-1 Induced Macrophage Pyroptosis by Modulating m6A Modification of Circ_0029589 in Patients with Acute Coronary Syndrome. Int. immunopharmacology 86, 106800. 10.1016/j.intimp.2020.106800 [DOI] [PubMed] [Google Scholar]
- Han J., Wang J.-z., Yang X., Yu H., Zhou R., Lu H.-C., et al. (2019). METTL3 Promote Tumor Proliferation of Bladder Cancer by Accelerating Pri-miR221/222 Maturation in m6A-dependent Manner. Mol. Cancer 18 (1), 110. 10.1186/s12943-019-1036-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Guo J., Fan Z. (2021). Interactions between m6A Modification and miRNAs in Malignant Tumors. Cel Death Dis 12 (6), 598. 10.1038/s41419-021-03868-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R.-Z., Jiang J., Luo D.-X. (2020). The Functions of N6-Methyladenosine Modification in lncRNAs. Genes Dis. 7 (4), 598–605. 10.1016/j.gendis.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann B. L., Tummers B., Green D. R. (2019). Crashing the Computer: Apoptosis vs. Necroptosis in Neuroinflammation. Cell Death Differ 26 (1), 41–52. 10.1038/s41418-018-0195-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cel Biol 20 (3), 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., Anderson P. (2013). Post-transcriptional Regulatory Networks in Immunity. Immunol. Rev. 253 (1), 253–272. 10.1111/imr.12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7 (12), 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Zhang X., Miao Y., Liang P., Zhu K., She Y., et al. (2018). m6A RNA Modification Controls Autophagy through Upregulating ULK1 Protein Abundance. Cell Res 28 (9), 955–957. 10.1038/s41422-018-0069-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W. J., Upton J. W., Long A. B., Livingston-Rosanoff D., Daley-Bauer L. P., Hakem R., et al. (2011). RIP3 Mediates the Embryonic Lethality of Caspase-8-Deficient Mice. Nature 471 (7338), 368–372. 10.1038/nature09857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Zhu S., Zeh H. J., Klionsky D. J., Tang D. (2018). BECN1 Is a New Driver of Ferroptosis. Autophagy 14 (12), 2173–2175. 10.1080/15548627.2018.1513758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthiya R., Khandelia P. (2020). m6A RNA Methylation: Ramifications for Gene Expression and Human Health. Mol. Biotechnol. 62 (10), 467–484. 10.1007/s12033-020-00269-5 [DOI] [PubMed] [Google Scholar]
- Kim G.-W., Siddiqui A. (2021). N6-methyladenosine Modification of HCV RNA Genome Regulates Cap-independent IRES-Mediated Translation via YTHDC2 Recognition. Proc. Natl. Acad. Sci. USA 118 (10), e2022024118. 10.1073/pnas.2022024118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P., Lence T., Haussmann I. U., Jacob D., Kreim N., Carl S. H., et al. (2018). Zc3h13/Flacc Is Required for Adenosine Methylation by Bridging the mRNA-Binding Factor Rbm15/Spenito to the m6A Machinery Component Wtap/Fl(2)d. Genes Dev. 32, 415–429. 10.1101/gad.309146.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H., Liu Y., Liu J., Wang X., Guan Z., Du J., et al. (2021). Tumor-Associated Macrophages Promote Oxaliplatin Resistance via METTL3-Mediated m6A of TRAF5 and Necroptosis in Colorectal Cancer. Mol. Pharmaceutics 18 (3), 1026–1037. 10.1021/acs.molpharmaceut.0c00961 [DOI] [PubMed] [Google Scholar]
- Lasman L., Krupalnik V., Viukov S., Mor N., Aguilera-Castrejon A., Schneir D., et al. (2020). Context-dependent Functional Compensation between Ythdf m6A Reader Proteins. Genes Dev. 34, 1373–1391. 10.1101/gad.340695.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. S. Y., Kranzusch P. J., Doudna J. A., Cate J. H. D. (2016). eIF3d Is an mRNA Cap-Binding Protein that Is Required for Specialized Translation Initiation. Nature 536 (7614), 96–99. 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Song Y., Liao Z., Wang K., Luo R., Lu S., et al. (2020). Bone-derived Mesenchymal Stem Cells Alleviate Compression-Induced Apoptosis of Nucleus Pulposus Cells by N6 Methyladenosine of Autophagy. Cel Death Dis 11 (2), 103. 10.1038/s41419-020-2284-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wu L., Pei M., Zhang Y. (2020). YTHDF2, a Protein Repressed by miR-145, Regulates Proliferation, Apoptosis, and Migration in Ovarian Cancer Cells. J. Ovarian Res. 13 (1), 111. 10.1186/s13048-020-00717-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ni Y., Zhang L., Jiang R., Xu J., Yang H., et al. (2021). HIF-1α-induced Expression of m6A Reader YTHDF1 Drives Hypoxia-Induced Autophagy and Malignancy of Hepatocellular Carcinoma by Promoting ATG2A and ATG14 Translation. Sig Transduct Target. Ther. 6 (1), 76. 10.1038/s41392-020-00453-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Niu Y., Wan A., Chen D., Liang H., Chen X., et al. (2020). RNA M 6 A Methylation Regulates Sorafenib Resistance in Liver Cancer through FOXO 3‐mediated Autophagy. Embo J. 39 (12), e103181. 10.15252/embj.2019103181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.-H., Tu Y., Ni G.-X., Yan J., Yue L., Li Z.-L., et al. (2021). Total Flavones of Abelmoschus Manihot Ameliorates Podocyte Pyroptosis and Injury in High Glucose Conditions by Targeting METTL3-dependent m6A Modification-Mediated NLRP3-Inflammasome Activation and PTEN/PI3K/Akt Signaling. Front. Pharmacol. 12, 667644. 10.3389/fphar.2021.667644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ren D., Du Z., Wang H., Zhang H., Jin Y. (2018). m 6 A Demethylase FTO Facilitates Tumor Progression in Lung Squamous Cell Carcinoma by Regulating MZF1 Expression. Biochem. biophysical Res. Commun. 502 (4), 456–464. 10.1016/j.bbrc.2018.05.175 [DOI] [PubMed] [Google Scholar]
- Liu S., Li Q., Li G., Zhang Q., Zhuo L., Han X., et al. (2020). The Mechanism of m6A Methyltransferase METTL3-Mediated Autophagy in Reversing Gefitinib Resistance in NSCLC Cells by β-elemene. Cel Death Dis 11 (11), 969. 10.1038/s41419-020-03148-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low Y.-H., Asi Y., Foti S. C., Lashley T. (2021). Heterogeneous Nuclear Ribonucleoproteins: Implications in Neurological Diseases. Mol. Neurobiol. 58 (2), 631–646. 10.1007/s12035-020-02137-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. z., Yang F., Zhou C. c., Liu F., Yuan J. h., Wang F., et al. (2017). METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N 6 ‐methyladenosine‐dependent Primary MicroRNA Processing. Hepatology 65 (2), 529–543. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- Ma L., Zhang X., Yu K., Xu X., Chen T., Shi Y., et al. (2021). Targeting SLC3A2 Subunit of System XC− Is Essential for m6A Reader YTHDC2 to Be an Endogenous Ferroptosis Inducer in Lung Adenocarcinoma. Free Radic. Biol. Med. 168, 25–43. 10.1016/j.freeradbiomed.2021.03.023 [DOI] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163 (4), 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Panda P. K., Sinha N., Das D. N., Bhutia S. K. (2014). Autophagy and Apoptosis: where Do They Meet. Apoptosis 19 (4), 555–566. 10.1007/s10495-014-0967-2 [DOI] [PubMed] [Google Scholar]
- Mulay S. R. (2019). Multifactorial Functions of the Inflammasome Component NLRP3 in Pathogenesis of Chronic Kidney Diseases. Kidney Int. 96 (1), 58–66. 10.1016/j.kint.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Müller S., Glaß M., Singh A. K., Haase J., Bley N., Fuchs T., et al. (2019). IGF2BP1 Promotes SRF-dependent Transcription in Cancer in a m6A- and miRNA-dependent Manner. Nucleic Acids Res. 47 (1), 375–390. 10.1093/nar/gky1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy S. (2020). m6A mRNA Methylation: A Pleiotropic Regulator of Cancer. Gene 736, 144415. 10.1016/j.gene.2020.144415 [DOI] [PubMed] [Google Scholar]
- Negroni A., Colantoni E., Cucchiara S., Stronati L. (2020). Necroptosis in Intestinal Inflammation and Cancer: New Concepts and Therapeutic Perspectives. Biomolecules 10 (10), 1431. 10.3390/biom10101431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J., Thomas S. M. (2019). Autophagy-dependent Secretion: Mechanism, Factors Secreted, and Disease Implications. Autophagy 15 (10), 1682–1693. 10.1080/15548627.2019.1596479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K., Dugger D. L., Wickliffe K. E., Kapoor N., de Almagro M. C., Vucic D., et al. (2014). Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343 (6177), 1357–1360. 10.1126/science.1249361 [DOI] [PubMed] [Google Scholar]
- Niu Y., Lin Z., Wan A., Chen H., Liang H., Sun L., et al. (2019). RNA N6-Methyladenosine Demethylase FTO Promotes Breast Tumor Progression through Inhibiting BNIP3. Mol. Cancer 18 (1), 46. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerum S., Meynier V., Catala M., Tisné C. (2021). A Comprehensive Review of m6A/m6Am RNA Methyltransferase Structures. Nucleic Acids Res. 49 (13), 7239–7255. 10.1093/nar/gkab378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D. P., Chen C.-K., Pickering B. F., Chow A., Jackson C., Guttman M., et al. (2016). m6A RNA Methylation Promotes XIST-Mediated Transcriptional repressionA RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 537 (7620), 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierzynowska K., Rintz E., Gaffke L., Węgrzyn G. (2021). Ferroptosis and its Modulation by Autophagy in Light of the Pathogenesis of Lysosomal Storage Diseases. Cells 10 (2), 365. 10.3390/cells10020365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C., Eyre S., Cope A., Worthington J., Barton A. (2007). Investigation of Association between the TRAF Family Genes and RA Susceptibility. Ann. Rheum. Dis. 66 (10), 1322–1326. 10.1136/ard.2006.065706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Zheng Y., Grace C. R. R., Liu X., Klionsky D. J., Schulman B. A. (2020). Allosteric Regulation through a Switch Element in the Autophagy E2, Atg3. Autophagy 16 (1), 183–184. 10.1080/15548627.2019.1688550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant J.-Y., Soller M. (2017). m 6 A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. 33 (6), 380–390. 10.1016/j.tig.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Schöller E., Weichmann F., Treiber T., Ringle S., Treiber N., Flatley A., et al. (2018). Interactions, Localization, and Phosphorylation of the m6A Generating METTL3-METTL14-WTAP Complex. Rna 24 (4), 499–512. 10.1261/rna.064063.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer R., Laurien L., Pasparakis M. (2020). New Insights into the Regulation of Apoptosis, Necroptosis, and Pyroptosis by Receptor Interacting Protein Kinase 1 and Caspase-8. Curr. Opin. Cel. Biol. 63, 186–193. 10.1016/j.ceb.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Shen M., Li Y., Wang Y., Shao J., Zhang F., Yin G., et al. (2021). N6-methyladenosine Modification Regulates Ferroptosis through Autophagy Signaling Pathway in Hepatic Stellate Cells. Redox Biol. 47, 102151. 10.1016/j.redox.2021.102151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Ying S., Li Y., Zhu L., Wang X., Jin H. (2021). Linking the YTH Domain to Cancer: the Importance of YTH Family Proteins in Epigenetics. Cel Death Dis 12 (4), 346. 10.1038/s41419-021-03625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirjang S., Mansoori B., Asghari S., Duijf P. H. G., Mohammadi A., Gjerstorff M., et al. (2019). MicroRNAs in Cancer Cell Death Pathways: Apoptosis and Necroptosis. Free Radic. Biol. Med. 139, 1–15. 10.1016/j.freeradbiomed.2019.05.017 [DOI] [PubMed] [Google Scholar]
- Śledź P., Jinek M. (2016). Structural Insights into the Molecular Mechanism of the m6A Writer Complex. eLife 5, e18434. 10.7554/eLife.18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Feng X., Zhang H., Luo Y., Huang J., Lin M., et al. (2019). METTL3 and ALKBH5 Oppositely Regulate m6A Modification of TFEB mRNA, Which Dictates the Fate of Hypoxia/reoxygenation-Treated Cardiomyocytes. Autophagy 15 (8), 1419–1437. 10.1080/15548627.2019.1586246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Jia G., Ma P., Cang S. (2021). Exosomal miR-4443 Promotes Cisplatin Resistance in Non-small Cell Lung Carcinoma by Regulating FSP1 m6A Modification-Mediated Ferroptosis. Life Sci. 276, 119399. 10.1016/j.lfs.2021.119399 [DOI] [PubMed] [Google Scholar]
- Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 172, 90–105. 10.1016/j.cell.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Wu Z., Wang X., Wang Y., Hu X., Qin W., et al. (2020). LNC942 Promoting METTL14-Mediated m6A Methylation in Breast Cancer Cell Proliferation and Progression. Oncogene 39 (31), 5358–5372. 10.1038/s41388-020-1338-9 [DOI] [PubMed] [Google Scholar]
- Tang X., Liu S., Chen D., Zhao Z., Zhou J. (2019). The Role of the Fat Mass and Obesity-Associated Protein in the Proliferation of Pancreatic Cancer Cells. Oncol. Lett. 17 (2), 2473–2478. 10.3892/ol.2018.9873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya K. (2021). Switching from Apoptosis to Pyroptosis: Gasdermin-Elicited Inflammation and Antitumor Immunity. Int. J. Mol. Sci. 22 (1), 426. 10.3390/ijms22010426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruta N., Tsuchihashi K., Ohmura H., Yamaguchi K., Ito M., Ariyama H., et al. (2020). RNA N6-Methyladenosine Demethylase FTO Regulates PD-L1 Expression in colon Cancer Cells. Biochem. biophysical Res. Commun. 530 (1), 235–239. 10.1016/j.bbrc.2020.06.153 [DOI] [PubMed] [Google Scholar]
- Ueda Y., Ooshio I., Fusamae Y., Kitae K., Kawaguchi M., Jingushi K., et al. (2017). AlkB Homolog 3-mediated tRNA Demethylation Promotes Protein Synthesis in Cancer Cells. Sci. Rep. 7, 42271. 10.1038/srep42271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzdensky A. B. (2019). Apoptosis Regulation in the Penumbra after Ischemic Stroke: Expression of Pro- and Antiapoptotic Proteins. Apoptosis 24, 687–702. 10.1007/s10495-019-01556-6 [DOI] [PubMed] [Google Scholar]
- Vu L. P., Pickering B. F., Cheng Y., Zaccara S., Nguyen D., Minuesa G., et al. (2017). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of normal Hematopoietic and Leukemia Cells. Nat. Med. 23 (11), 1369–1376. 10.1038/nm.4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xu B., Shi J. (2020). N6-methyladenosine METTL3 Promotes the Breast Cancer Progression via Targeting Bcl-2. Gene 722, 144076. 10.1016/j.gene.2019.144076 [DOI] [PubMed] [Google Scholar]
- Wang P., Doxtader K. A., Nam Y. (2016). Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cel. 63 (2), 306–317. 10.1016/j.molcel.2016.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Geng W., Guo H., Wang Z., Xu K., Chen C., et al. (2020). Emerging Role of RNA Methyltransferase METTL3 in Gastrointestinal Cancer. J. Hematol. Oncol. 13 (1), 57. 10.1186/s13045-020-00895-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wu R., Liu Y., Zhao Y., Bi Z., Yao Y., et al. (2020). m6A mRNA Methylation Controls Autophagy and Adipogenesis by Targeting Atg5 and Atg7. Autophagy 16 (7), 1221–1235. 10.1080/15548627.2019.1659617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161 (6), 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kanneganti T.-D. (2021). From Pyroptosis, Apoptosis and Necroptosis to PANoptosis: A Mechanistic Compendium of Programmed Cell Death Pathways. Comput. Struct. Biotechnol. J. 19, 4641–4657. 10.1016/j.csbj.2021.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warda A. S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., et al. (2017). Human METTL16 Is a N 6 ‐methyladenosine (M 6 A) Methyltransferase that Targets pre‐mRNAs and Various Non‐coding RNAs. EMBO Rep. 18 (11), 2004–2014. 10.15252/embr.201744940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Huo B., Shi X. (2019). miR-600 Inhibits Lung Cancer via Downregulating the Expression of METTL3. Cancer Manag. Res. 11, 1177–1187. 10.2147/cmar.s181058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C. B., Horton J. R., Zhou J., Bedford M. T., Blumenthal R. M., Zhang X., et al. (2020). Biochemical and Structural Basis for YTH Domain of Human YTHDC1 Binding to Methylated Adenine in DNA. Nucleic Acids Res. 48 (18), 10329–10341. 10.1093/nar/gkaa604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Yi F., Han X., Du Q., Liang Z. (2013). MALAT-1 Interacts with hnRNP C in Cell Cycle Regulation. FEBS Lett. 587 (19), 3175–3181. 10.1016/j.febslet.2013.07.048 [DOI] [PubMed] [Google Scholar]
- Yang S., Wei J., Cui Y.-H., Park G., Shah P., Deng Y., et al. (2019). m6A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 10 (1), 2782. 10.1038/s41467-019-10669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Pratte J., Giardina C. (2021). Ferroptosis and its Potential as a Therapeutic Target. Biochem. Pharmacol. 186, 114486. 10.1016/j.bcp.2021.114486 [DOI] [PubMed] [Google Scholar]
- Yuan X., Li T., Shi L., Miao J., Guo Y., Chen Y. (2021). Human Umbilical Cord Mesenchymal Stem Cells Deliver Exogenous miR-26a-5p via Exosomes to Inhibit Nucleus Pulposus Cell Pyroptosis through METTL14/NLRP3. Mol. Med. 27 (1), 91. 10.1186/s10020-021-00355-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Yan G., He M., Lei H., Li L., Wang Y., et al. (2021). ALKBH5 Suppresses Tumor Progression via an m6A-dependent Epigenetic Silencing of Pre-miR-181b-1/YAP Signaling axis in Osteosarcoma. Cel Death Dis 12 (1), 60. 10.1038/s41419-020-03315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., et al. (2018). VIRMA Mediates Preferential m6A mRNA Methylation in 3′UTR and Near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov 4, 10. 10.1038/s41421-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha X., Xi X., Fan X., Ma M., Zhang Y., Yang Y. (2020). Overexpression of METTL3 Attenuates High-Glucose Induced RPE Cell Pyroptosis by Regulating miR-25-3p/PTEN/Akt Signaling cascade through DGCR8. Aging 12 (9), 8137–8150. 10.18632/aging.103130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Bai R., Li M., Ye H., Wu C., Wang C., et al. (2019). Excessive miR-25-3p Maturation via N6-Methyladenosine Stimulated by Cigarette Smoke Promotes Pancreatic Cancer Progression. Nat. Commun. 10 (1), 1858. 10.1038/s41467-019-09712-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hou C., Chen C., Guo Y., Yuan W., Yin D., et al. (2020). The Role of N6-Methyladenosine (m6A) Modification in the Regulation of circRNAs. Mol. Cancer 19 (1), 105. 10.1186/s12943-020-01224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Yang Y., Sun B.-F., Shi Y., Yang X., Xiao W., et al. (2014). FTO-dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cel Res 24 (12), 1403–1419. 10.1038/cr.2014.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z.-Q., Li Z.-X., Zhou G.-Q., Lin L., Zhang L.-L., Lv J.-W., et al. (2019). Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 79 (18), 4612–4626. 10.1158/0008-5472.can-19-0799 [DOI] [PubMed] [Google Scholar]
- Zhuang C., Zhuang C., Luo X., Huang X., Yao L., Li J., et al. (2019). N6-methyladenosine Demethylase FTO Suppresses clear Cell Renal Cell Carcinoma through a Novel FTO-PGC-1α Signalling axis. J. Cel Mol Med 23 (3), 2163–2173. 10.1111/jcmm.14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang S., Ma Y., Zeng Y., Lu C., Yang F., Jiang N., et al. (2021). METTL14 Promotes Doxorubicin-Induced Cardiomyocyte Ferroptosis by Regulating the KCNQ1OT1-miR-7-5p-TFRC axis. Cell Biol Toxicol. 10.1007/s10565-021-09660-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The regulatory pathways of different forms of PCD.
The classification and function of enzymes involved in m6A.
The function of m6A in mRNA and ncRNA.
The roles of m6A by regulating apoptosis in the occurrence of cancer.