Abstract
The clinical microbiology laboratory plays a critical role in the detection of Staphylococcus aureus with decreased susceptibility to vancomycin. Staff education and rapid laboratory response are of utmost importance. We report on our laboratory's experience and provide recommendations for the identification and confirmation of vancomycin-intermediate S. aureus.
Despite the low incidence of vancomycin-intermediate Staphylococcus aureus (VISA) worldwide, VISA remains a significant concern. The VISA strains isolated at the Medical Center of the University of California at Los Angeles (UCLA) mark the fifth of eight sets of strains confirmed by the Centers for Disease Control and Prevention (CDC) (Fred Tenover, personal communication) in the United States (1, 2, 2a, 3, 6). Internationally, cases have been reported in Japan, Korea, and Europe (3, 4, 5). Expedient confirmation of VISA isolates from clinical specimens is critical to patient care and infection control. Clinical microbiologists must be knowledgeable about procedures that can accurately detect VISA. In addition, when a VISA strain is found, results must be effectively communicated to health care providers and public health officials. Here we report on our laboratory's experience with the isolation, detection, and confirmation of two VISA isolates from a single specimen. Our goal is to highlight specific issues that must be considered when VISA is suspected in a clinical specimen.
On 8 June 2000, a biliary drainage specimen was submitted to the Clinical Microbiology Laboratory of the UCLA Medical Center for bacterial culture. The specimen was obtained from a transhepatic biliary drainage catheter from a home health care patient with multiple hepatic abscesses who had received long-term vancomycin therapy for methicillin-resistant S. aureus (MRSA) infection.
Following overnight incubation, examination of the primary plates revealed large colonies of S. aureus (isolate 1) and pinpoint colonies of lactose-negative, gram-negative rods which were subsequently identified as Stenotrophomonas maltophilia. Broth microdilution susceptibility testing of S. aureus isolate 1 demonstrated oxacillin and vancomycin MICs of >16 and 2 μg/ml, respectively.
On the second day of incubation of the primary plates, pinpoint, beta-hemolytic, staphylococcus-like colonies were seen. A positive slide coagulase test confirmed the identification of these colonies as S. aureus (isolate 2), and susceptibility testing was performed. The results of the broth microdilution test read the next morning (18 h) revealed an oxacillin MIC of >16 μg/ml and a vancomycin MIC of 4 μg/ml. These results remained unchanged after 24 h of incubation. In our laboratory it is policy to inoculate a purity plate (blood agar plate) at the time that broth microdilution MIC test plates are inoculated. The purity plate for S. aureus (isolate 2) demonstrated multiple colony types resembling staphylococcus and was considered to possibly contain a mixture of isolates. A Gram stain of the well that contained 2 μg of vancomycin per ml was performed and showed only gram-positive cocci in clusters. The contents of this well and the positive control well were subcultured onto blood agar plates to check for purity. The susceptibilities of the two distinct colonial phenotypes, one comprising small yellow-white isolates (S. aureus isolate 2a) and the other comprising pinpoint grey-white isolates (S. aureus isolate 2b), were determined (Fig. 1). At 18 h, the vancomycin MIC was 4 μg/ml for isolates of both phenotypes. At 24 h, the vancomycin MIC had increased to 8 μg/ml for isolates of both phenotypes. Interestingly, the oxacillin MICs for isolates 2a and 2b were >16 and 0.5 μg/ml, respectively. Vancomycin MICs were determined by the E-test (AB BIODISK, Solna, Sweden) and were 6 μg/ml for both isolates. In addition, both isolates grew on vancomycin at 6 μg/ml on brain heart infusion screen agar (Hardy, Santa Barbara, Calif.).
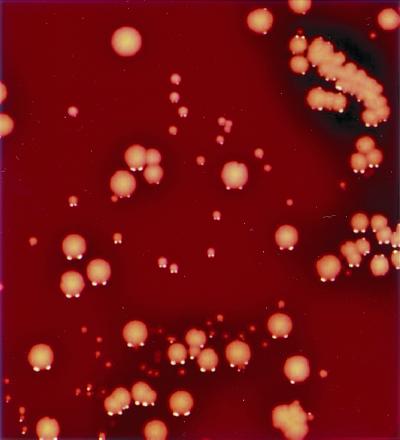
FIG. 1.
Photo of S. aureus morphotypes from biliary culture. Isolates of S. aureus isolate 1 (MRSA) appear as classical large colonies that are yellow-white and domed with a zone of beta-hemolysis. The smaller colonies were identified as VISA. S. aureus isolate 2a (VISA) appears as a small yellow-white colony with a narrow zone of beta-hemolysis. S. aureus isolate 2b (VISA) appears as a pinpoint grey-white colony with a barely detectable zone of beta-hemolysis. Both isolate 2a and isolate 2b were domed and distinct.
Laboratory personnel notified hospital infection control, the Los Angeles County Public Health Department (LACPHD), and CDC. Isolates were subcultured onto Trypticase soy agar slants (BBL Microbiology Systems, Sparks, Md.) for shipment to LACPHD and CDC. The initial isolates sent to CDC were not confirmed to be VISA isolates. After further investigation at our laboratory, it was discovered that in order to expedite the confirmation process, the slants had been incubated for only 4 h prior to shipping. An important feature of VISA is its slow growth (8). It was suspected that this 4-h incubation period might not have been adequate and thus contributed to the conflicting results. The isolates were again subcultured and incubated on chocolate agar slants (BBL Microbiology Systems) for a full 24 h before they were shipped to CDC. Subsequently, CDC confirmed that the isolates were VISA. Pulsed-field gel electrophoresis performed at both UCLA and CDC showed that the two VISA isolates were closely related and similar to other previously confirmed VISA isolates (Hageman et al., unpublished data).
As this report demonstrates, the detection of VISA can be challenging. VISA isolates may demonstrate multiple colony morphologies and appear to be mixed (Fig. 1). Detection of the VISA isolates in our laboratory was dependent upon identification of pinpoint colonies from the primary plate at day 2 and then testing of the various colony types from the purity plate subcultures as MIC test inocula. Like MRSA, VISA may demonstrate heteroresistance or there may be subpopulations that are resistant (7; CDC, http://www.cdc.gov/ncidod/hip/Lab/FactSheet/gisa.htm). To date all CDC-confirmed clinical VISA isolates have been MRSA; however, for one of the VISA strains isolated at UCLA, the oxacillin MIC was 0.5 μg/ml and the strain was mecA negative by PCR (9).
Not all commonly used susceptibility test methods have been able to detect VISA. The NCCLS disk diffusion (Kirby-Bauer) method is not an appropriate method. Broth microdilution, with incubation for a full 24 h, remains the method of choice for detection of decreased susceptibility to vancomycin. Conventional MicroScan panels and the E-test can detect this resistance when the cultures are held for a full 24 h (8; CDC, http://www.cdc.gov/ncidod/hip/Lab/FactSheet/gisa.htm and http://www.cdc.gov/ncidod/HIP/aresist/search.htm). The method with the Vitek system is also acceptable (8; CDC, http://www.cdc.gov/ncidod/hip/Lab/FactSheet/gisa.htm). For a review, see the report by Tenover et al. (8).
On the basis of our experience, we recommend the following. (i) Be aware that VISA may not appear on the primary culture plate until day 2 of incubation (48 h). (ii) VISA grows more slowly than typical vancomycin-sensitive S. aureus. Thus, it is recommended that isolates for testing and shipping be incubated for a full 24 h. (iii) Be aware that VISA isolates may demonstrate variable colony morphologies on the primary culture and subsequent subcultures. (iv) Be cognizant of the fact that patients with VISA infection are likely to respond poorly to vancomycin therapy. Thus, receipt of information that a patient is responding poorly to vancomycin should warrant appropriate tests for VISA. (v) Determine if the susceptibility method in your laboratory is likely to detect VISA. If not, determine if a backup method is warranted. (vi) Define a mechanism that can be used to communicate the results indicating probable isolation of a VISA isolate to appropriate health care workers in a timely manner.
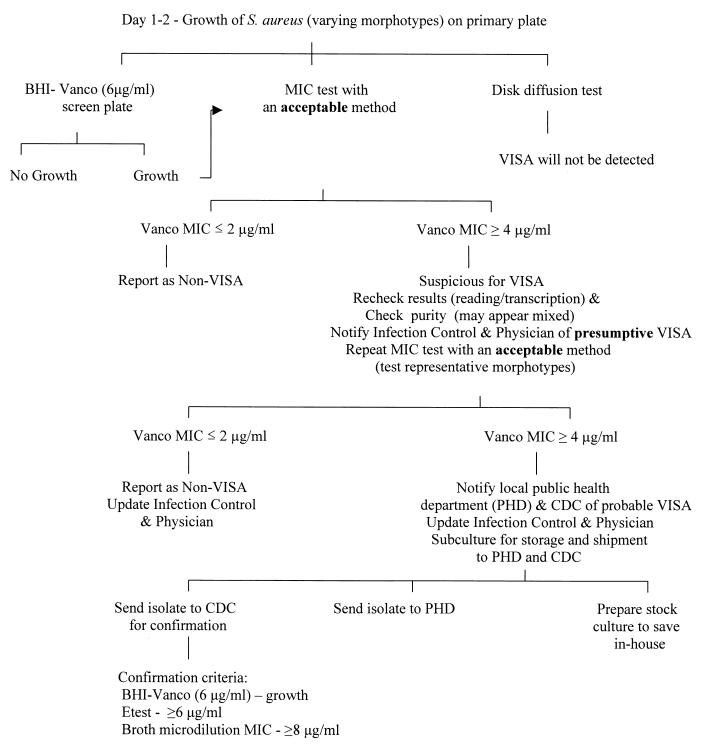
Algorithms for VISA detection can be designed to best fit the institution and the patient population (Fig. 2). Information on how to detect and report VISA is available through the CDC website (www.cdc.gov/ncidod/hip/lab/factsheet/gisa.htm and www.CDC.gov/ncidod/hip/ARESIST/search.htm).
FIG. 2.
Algorithm for detection and confirmation of VISA. This algorithm is a general guideline for workup of VISA isolates in the clinical laboratory. Algorithms should be designed for the specific hospital and patient population. Abbreviations: BHI, brain heart infusion; Vanco, vancomycin.
Acknowledgments
We thank Fred Tenover for comments and assistance. We also thank CDC for aid in the confirmation of the identities of the isolates.
REFERENCES
- 1.Centers for Disease Control and Prevention. Update: Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. Morb Mortal Wkly Rep. 1997;46:813–815. [PubMed] [Google Scholar]
- 2.Fridkin S K. Vancomycin-intermediate and -resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin Infect Dis. 2001;32:108–115. doi: 10.1086/317542. [DOI] [PubMed] [Google Scholar]
- 2a.Hageman, J. C., D. Peques, C. Jepson, R. Bell, M. Guinan, K. Ward, M. Cohen, J. Hindler, F. C. Tenover, S. McAllister, M. Kellum, and S. Fridkin. Case report of vancomycin-intermediate Staphylococcus aureus in a home healthcare patient. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 3.Hiramatsu K, Artiaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 4.Kim M, Pai C H, Woo J H, Ryu J S, Hiramatsu K. Vancomycin-intermediate Staphyloccus aureus in Korea. J Clin Microbiol. 2000;38:3879–3881. doi: 10.1128/jcm.38.10.3879-3881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchese A, Balistreti G, Tonoli E, Debbia E A, Schito G C. Heterogeneous vancomycin resistance in methicillin-resistant Staphylococcus aureus strains isolated in a large Italian hospital. J Clin Microbiol. 2000;38:866–869. doi: 10.1128/jcm.38.2.866-869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith T L, Pearson M L, Wilcox K R, Cruz C, Lancaster M V, Robinson-Dunn B, Tenover F C, Zeros M J, Band J D, White E, Jarvis W R. Emergence of vancomycin resistance in Staphylococcus aureus. N Engl J Med. 1999;340:493–501. doi: 10.1056/NEJM199902183400701. [DOI] [PubMed] [Google Scholar]
- 7.Tenover F C. VRSA, VISA, and GISA: the dilemma behind the name game. Clin Microbiol Newsl. 2000;22:49–53. [Google Scholar]
- 8.Tenover F C, Lancaster M V, Hill B C, Steward C D, Stocker S A, Hancock G A, O'Hara C M, Clark N C, Hiramatsu K. Characterization of staphylococci with reduced susceptibilities to vancomycin and other glycopeptides. J Clin Microbiol. 1998;36:1020–1027. doi: 10.1128/jcm.36.4.1020-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vannuffel P, Gigi J, Ezzedine H, Vanderman B, Delmee M, Wauters G, Gala J L. Specific detection of methicillin-resistant Staphylococcus aureus species by multiplex PCR. J Clin Microbiol. 1995;33:2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]