Abstract
Introduction
Secondary analysis of hospital-hosted clinical data can save time and cost compared with prospective clinical trials for neuroimaging biomarker development. We present such a study for Sturge-Weber syndrome (SWS), a rare neurovascular disorder that affects 1 in 20 000–50 000 newborns. Children with SWS are at risk for developing neurocognitive deficit by school age. A critical period for early intervention is before 2 years of age, but early diagnostic and prognostic biomarkers are lacking. We aim to retrospectively mine clinical data for SWS at two national centres to develop presymptomatic biomarkers.
Methods and analysis
We will retrospectively collect clinical, MRI and neurocognitive outcome data for patients with SWS who underwent brain MRI before 2 years of age at two national SWS care centres. Expert review of clinical records and MRI quality control will be used to refine the cohort. The merged multisite data will be used to develop algorithms for abnormality detection, lesion-symptom mapping to identify neural substrate and machine learning to predict individual outcomes (presence or absence of seizures) by 2 years of age. Presymptomatic treatment in 0–2 years and before seizure onset may delay or prevent the onset of seizures by 2 years of age, and thereby improve neurocognitive outcomes. The proposed work, if successful, will be one of the largest and most comprehensive multisite databases for the presymptomatic phase of this rare disease.
Ethics and dissemination
This study involves human participants and was approved by Boston Children’s Hospital Institutional Review Board: IRB-P00014482 and IRB-P00025916 Johns Hopkins School of Medicine Institutional Review Board: NA_00043846. Participants gave informed consent to participate in the study before taking part. The Institutional Review Boards at Kennedy Krieger Institute and Boston Children’s Hospital approval have been obtained at each site to retrospectively study this data. Results will be disseminated by presentations, publication and sharing of algorithms generated.
Keywords: sturge-weber syndrome, anti-epilepsy drugs, neuroimaging biomarker, pre-symptomatic treatment, machine learning
Strengths and limitations of this study.
We will retrospectively gather multisite neuroimaging and comprehensive data from >100 subjects with Sturge-Weber syndrome, a rare disease, compared with prior single-site studies with typically only a dozen or so patients.
This will be the first rigorous multisite neuroimaging biomarker study in the presymptomatic phase of Sturge-Weber syndrome, compared with most studies focusing on the postsymptomatic phase.
We will develop and use statistically rigorous lesion-symptom mapping to predict neurocognitive outcomes in Sturge-Weber syndrome and rigorously test whether combining clinical and MRI information outperforms brain MRI alone in outcome prediction.
Heterogenous brain MRI protocols (scanner, vendor, scanning site, age at MRI, etc) will be used; however, this heterogeneity increases the generalisability of the results.
Introduction
About 1/1000 infants are born with port-wine birthmarks (PWB) on the face or neck. Of those infants, 5% develop a vascular neurocutaneous disorder called Sturge-Weber syndrome (SWS).1 This makes SWS patients 1 in 20 000 to 1 in 50 000 newborns, and, therefore, a rare disease.2 3 SWS is usually caused by an activating somatic mutation in the gene GNAQ, encoding the Gαq subunit in the affected tissue.1 Diagnosis is based on the classic PWB at birth and vascular findings in brain MRI that are routinely acquired during infancy. The disease is not commonly fatal but is incurable.4 5 Neurocognitive impairment by school age (6–10 years of age) is a primary issue in SWS.
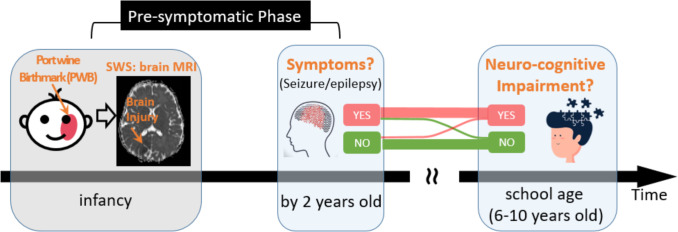
A landmark event in the progression of SWS is the presence or absence of seizures by 2 years of age (figure 1). About 15% of patients with SWS do not develop seizures by 2 years and they often enjoy good school-age neurocognitive outcomes6 7 Because of this, a critical time window to treat or intervene in patients with SWS is during infancy up to 2 years of age, the so-called presymptomatic phase. In this phase, very early treatment with antiepilepsy drugs (AEDs, such as levetiracetam, with or without low-dose aspirin)8–10 may delay or even avoid seizure symptoms by 2 years of age and thus improve school-age outcomes.6 7 The evidence of the early onset of seizures resulting in worse neurologic outcome6 has led to a demand for early diagnosis and aggressive intervention for those at increased risk of developing seizures.
Figure 1.
Need for the early prediction of risk to develop epilepsy/seizure symptoms by 2 years of age among patients with SWS in the ‘pre-symptomatic phase’. SWS, Sturge-Weber syndrome.
However, two questions remain open. For one, how to accurately identify who is at risk to develop the seizure/epilepsy symptoms by 2 years of age? These at-risk patients should be the ideal target patients to be included in future trials for testing AEDs in the critical ‘presymptomatic’ time window. For the other, what are the underlying neuroanatomic mechanisms that drive infants with SWS to develop epilepsy/seizure symptoms before 2 years of age? The neuromechanisms should be the targets to design the next-generation AEDs or other presymptomatic treatment strategies.4 11
Infant brain MRI is suggested by recent expert consensus (20184; 2019)5 in the field to address these two open questions. This is because infant brain MRI is routinely acquired during the clinical care of patients with SWS and is non-invasive. Besides, MRI can probe 3D neuroanatomy, with both diagnostic value (commonly seen presymptomatic brain abnormalities including leptomeningeal, parenchymal and choroidal capillary venous malformations with congenitally abnormal and ectatic vessels)12 and prognostic value (abnormal neuronal excitation,13 transient diffusion abnormalities14 15 and bilateral hemispheric injuries1 as seen in MRI may suggest an adverse outcome). However, these findings were based on single-site data, small sample sizes (single-digit or only a dozen or so). Moreover, expert interpretations of MRIs are largely qualitative or descriptive and may not be replicated across sites. This issue is further complicated by the limited clinical radiology expertise on this rare disease, especially in less developed regions or countries.4 16
To address these issues, this study aims to collect a relatively large multisite cohort for this rare disease and develop machine learning biomarkers. Existing MRI studies for SWS usually had ≤155 17–19 or dozens11 16 20 21 of patients with SWS in single sites. We aim to merge existing clinical data from Boston Children’s Hospital (BCH) and Kennedy Krieger Institute (KKI) to achieve an sample size at least five times bigger. Existing MRI studies for SWS mostly used expert reviews. We will leverage MRI analytics and machine learning algorithms, which have had success in characterising subtle yet complex injury patterns22 23 and in predicting outcomes for stroke,24 tumour,25 autism26 and other neurologic diseases,27 but have not yet been used in SWS. Our hypothesis is that machine learning-driven MRI and clinical biomarkers can predict the onset or absence of seizure symptoms by 2 years at a sensitivity and specificity higher than random guess (ie, 50%). Setting the random guess as the bar is because the lack of such a presymptomatic biomarker for SWS.
Methods and analysis
Overview
This retrospective study was approved by institutional review boards at BCH and KKI. Written consent was waived for the retrospective analysis of existing data that are anonymised. Figure 2 outlines the major components of our study. We plan to start the study in April or December of 2022 (pending funding from National Institute of Health (NIH)). The study period will be for ~2 years.
Figure 2.
Outline of our study protocol.
Part 1. Retrospective data acquisition
Find candidate patients
We plan to collect data for all patients who (a) were born between 2009 and 2020, (b) were under 2 years of age at the first MRI and (c) had SWS brain involvement confirmed by contrast-enhanced MRI before 2 years of age.
Patients will be primarily identified by clinical registries, in the neurology departments from patients cared by Dr. Pinto at BCH and Dr. Comi at KKI.
A secondary source will be the hospital-wide database. We will search clinical database and picture achieving and communication system (PACS) imaging systems by the ICD codes for SWS (table 1). Candidates missed in the clinical registry will be further vetted by their clinical records to confirm eligibility to be included in our database.
Table 1.
ICD-9/10 codes for SWS as a secondary source to query candidate patients beyond the clinical registry as the primary source
| ICD-9 | Meaning | ICD-10 | Meaning |
| 759.6 | Other hamartoses, not elsewhere classified | Q85.8 | Other phakomatoses, not elsewhere classified |
| 757.32 | Angiomatosis | Q85.02 | Other congenital malformations |
| 759.6 | Sturge (-Weber) (-Dimitri) (encephalocutaneous angiomatosis) |
SWS, Sturge-Weber syndrome.
Query and fetch clinical data
We will review patients’ electronic health records (EHRs) and gathered the following clinical variables.
Demographics: sex, the extent of brain involvement, the extent of PWB (bilateral, unilateral in the left or right side), age at first MRI, year of MRI, age at electroencephalogram (EEG) and clinical result, year of birth and family history of epilepsy
Treatment: whether the patient was treated presymptomatically (ie, before the first onset of seizures).
Outcome by 2 years: the primary neurological outcome will be the presence or absence of seizure onset by 2 years, and the age at seizure onset if present. The secondary outcome is the SWS neuroscore (including seizure score). The neuroscore is a scale that measures seizure frequency, hemiparesis, visual field cut and cognitive function (points range from 0 to 15, with lower scores indicating better neurologic status.28–31 Patients treated presymptomatically were assigned a seizure score of 0 at the time of treatment initiation. Subjects with a seizure score of 0 at 2 years of age are patients who have achieved 2 years of age without seizure onset.
Gather brain MRI data and quality control
We will use patients’ medical record numbers to search and download their brain MRIs in the clinical PACS. The Computational Health Research Integration System (ChRIS) platform offers this function,32 similar to other retrospective paediatric brain MRI projects at BCH.27 33 KKI also hosts the i2b234 platform for this function.
MRI sequences to be downloaded include: T1-weighted (T1w; spin echo vs 3D gradient echo), T2-weighted (T2w) spin echo, susceptibility-weighted imaging (SWI) including the phase and magnitude images, or T2*-weighted gradient echo, fluid-attenuated inversion recovery (FLAIR), diffusion-weighted imaging or diffusion tensor imaging (DTI) and postgadolinium-contrast T1w (spin echo or 3D gradient-echo T1 contrast (T1c) images).
For quality control, MRIs will be visually reviewed by imaging experts (Dr. Ou for BCH data and Dr. Lin for KKI data). MRIs with severe motion, artefacts or part of the brain missing will be discarded.
For the kept MRIs, we will record scanner vendors, scanner models, field of strength, in addition to the available sequences among those listed above.
Manage data
We will use REDCap35 to manually enter and maintain clinical variables from EHR. REDCap is Health Insurance Portability and Accountability Act (HIPAA)-complicant, secure and user-friendly and has been used in similar multisite database by this team.27
Part 2. Identify presymptomatic injury patterns for seizure outcome (population analysis)
MRI preprocessing
Each patient’s T1w brain MRI will undergo our extensively validated brain MRI analysis pipeline. Steps include N4 bias correction,36 field of view normalisation,37 multiatlas skull stripping,38 atlas-based regional segmentation39 as adapted and validated in infants 0–2 years of age.38 40 41 The computed brain mask in T1w MRI will automatically skull strip other MRI sequences when rigidly aligned with the T1w MRI.2 To compare across patients, their T1w MRIs will be non-rigidly aligned into our atlas space33 by our validated42 and public deformable image registration algorithm.43 DTIs will be used to derive the fractional anisotropy (FA) and apparent diffusion coefficient (ADC) maps, after motion correction and tensor reconstruction using Functional Magnetic Resonance Imaging of the Brain Software Library (FSL),44 by factoring out the effects of scanners and sites, while controlling for other covariates such as sex, age, race, etc. We will use the validated45 ComBat tool to harmonise MRI metrics extracted from multisite data.46
Abnormality detection
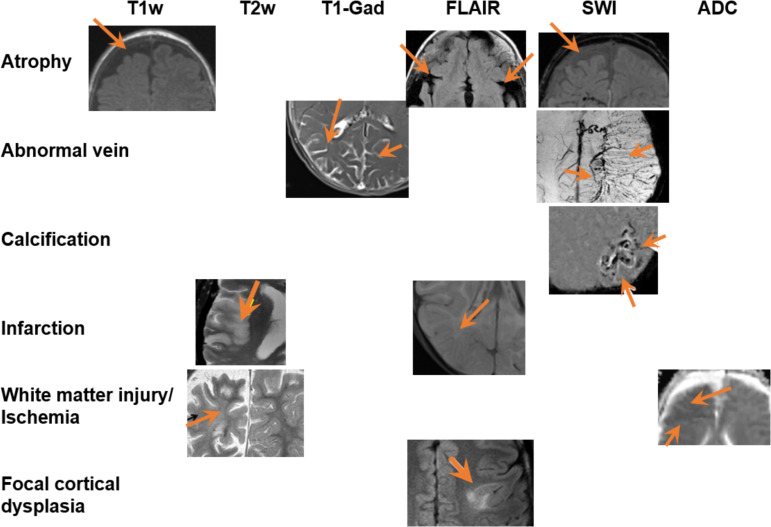
Figure 3 shows different types of abnormalities associated with SWS that manifest in multiparametric brain MRI:
Figure 3.
Typical abnormalities (different rows) found in the brain MRI of patient with SWS, in multiple MRI sequences (different columns). Different figure panels are from different patients. Orange arrows point out the abnormal regions. ADC, apparent diffusion coefficient; FLAIR, fluid-attenuated inversion recovery; SWI, susceptibility-weighted imaging; SWS, Sturge-Weber syndrome.
atrophy, infarction and focal cortical dysplasia or polymicrogyria are often observed in T1w 11 14 21 30 47–49 and T2w 50–52, FLAIR 50–52 and SWI53;
abnormal veins (leptomeningeal venous malformation, enlarged deep medullary and ependymal veins) are often observed in T1w with Gadolinium-contrast enhancement (T1-Gad 12 53–55), and SWI 54 56;
calcification is best visualised on SWI53);
abnormal diffusion, accelerated myelination or ischaemic injury are often observed in ADC map,57–59 a parameter map from diffusion MRI sequences; and
damages in the white matter microstructures or disruptions in fibre tracts are often observed inFA map60–62 of the DTI.
To automate the abnormality detection, we will extend our recently-developed algorithm that quantifies normal and detect abnormalities as outliers to normal.
The first major component in this algorithm is to quantify normal through constructing age-specific normal brain atlases. The atlases will quantify the normal range of signals (mean and SD) at each voxel in a normal control cohort. We recently developed age-specific normal ADC atlases, based on ADC maps from 201 normal participants who were scanned during 0–6 years of age, divided into 10 age groups (first 2 weeks, every quarter in the first year, then every year till 6 years).33 The atlases in each age group quantified voxel-wise normal range of ADC values, and characterised regional and hemispheric differences in early-childhood brain development patterns.40 The atlases are unbiased to any subject’s anatomy,63 64 based on our extensively-validated42 group-wise deformable image registration algorithm.43 We plan to build such age-specific atlases for each MRI sequence (T1w, T2w, FLAIR, SWI, ADC), based on our recently-gathered >200 multi-parametric brain MRIs on normal participants. These normal brain MRIs are from published data acquired from Massachusetts General Hospital and BCH (see references for more details on demographics and imaging protocols)33 40 and they are distinctive from the SWS patient data we plan to merge from KKI and BCH.
The second step is to detect abnormalities. This will be done by converting the patient’s MRI signal in each sequence into a Z score map. The Z map will quantify voxel-wise deviation (eg, how many SD away) from the normal mean intensity at the corresponding voxel in the age-matched atlas of the same sequence. We recently used this strategy and successfully identified pre-symptomatic ischaemia lesions in ADC maps of 7 SWS patients.59 Another study used a similar strategy on T1w and T2w images (after histogram normalisation) and identified white matter abnormalities in preterm infant’s brain MRIs.65 We will do this for each MRI sequence, generating ZT1, ZT2, ZFLAIR, ZSWI and ZADC maps, and we will call abnormalities when Z values are lower than −2.
Accuracy for abnormality detection will be quantified by calculating the Dice overlap, sensitivity and specificity compared with expert-annotated abnormality regions. However, expert annotations of exact abnormality boundary in patients with SWS will be subjected to intrareader/inter-reader variability. Therefore, an alternative approach is to indirectly evaluate the accuracy of abnormality detection in the context of pattern analysis and individual outcome prediction (later sections).
Mapping brain injury patterns to outcome
We will focus on the group differences between those who had MRI and no symptom till 2 years (group A) and those whose seizure occurred after MRI and before 2 years (group B). The postsymptomatic patients (first MRI occurred after seizures but before 2 years of age) will form a group C that serves as a validation cohort.
The key question is the ‘neural substrate’ underlying outcome—namely, patterns of brain injuries in the presymptomatic MRI that underpin the subsequent development of seizure by 2 years? This maps to group analysis between group A and group B.
We will use the lesion-symptom mapping (LSM) framework66 67 to identify the neural substrate of the outcomes (seizure onset or not) by 2 years. To reduce false positives, we will control for patient sex, age at MRI and whole-brain abnormality volume as covariates.68 69 Three versions of LSM models will be used and compared. Voxel-wise LSM (V-LSM) compares whether the presence or absence of abnormality at each voxel is statistically associated with the binary outcomes across patients.68 A limitation of V-LSM is that it overlooks the potential interaction among voxels. To address this, multivariate LSM (M-LSM) jointly considers voxels and voxel clusters and uses multivariate machine learning (support vector machine (SVM)) to find a subset of voxels that jointly associate with outcomes.70 71 A potential limitation of M-LSM is that the autoselected voxel clusters may not have anatomic explanations. To address this, the latest generation of LSM, known as connectome-based LSM or C-LSM,72–74 builds on findings in NIH’s human connectome project75–77 and NIH’s Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) initiative77 and quantifies which fibre bundles or functional brain circuits, if injured, are associated with outcomes.72 The assumption is that patients may differ in exact anatomic locations that are injured, but their injuries may intersect with the same fibre bundles or brain circuit, which will explain why they experience similar outcomes.
LSM successfully identified neural substrates of survival and language outcomes after adult brain tumor78; LSM identified neural substrates of language,79–81 motor,82 word comprehension,83 voice recognition,84 attention,85 computational ability82 and somatosensory outcomes86 after adult stroke lesions, etc. These studies used one LSM model, not three, and mostly V-LSM, the basic LSM model. We plan to use all three LSM models, with different assumptions, for the first time in SWS, based on the identified abnormality regions from the previous task (abnormality detection).
We will use two methods to validate the identified neural substrate for the outcome. One indirect validation will be by comparison with group C (postsymptomatic cohort). We hypothesise that the injury patterns that differentiate between group A and group B are the early and subtle stage of abnormalities that will also manifest, to a more severe extent, in group C. The other validation is a direct quantification of the accuracy in individual predictions, as described next.
Part 3. Outcome prediction (individual precision)
The previous part 2 focuses on the group contrast. However, patients in the same outcome group are different in their clinical and MRI characteristics. The key question in this part 3 is: how to use the presymptomatic brain MRI to predict which patient is at risk to develop seizures after MRI and by 2 years? This is individual prediction.
Traditional machine learning models
SVM87 88 or random forest (RF),89 90 represent each patient by a feature vector. One set of features will come from the presence or absence (1/0) of abnormality in the key neuroanatomic regions as identified by V-LSM, M-LSM and C-LSM neural substrate pattern analysis. This set of features assumes that the spatial patterns of abnormalities can infer outcome. Our second set of features will be imaging appearance features, as listed in table 2. We will concatenate features in neural substrates (spatial features) and imaging appearance features into a long feature vector to represent a patient. We will use our iterative forward inclusion and backward elimination (iFIBE) feature selection algorithm to automatically identify the subset of features that optimally predict outcomes.43 91–94 Here, optimality is defined as the sensitivity, specificity and area under the receiver operating characteristic curve (AUC) in k-fold (we will choose k=5) cross validations.
Table 2.
MRI appearance features characterising abnormalities for outcome prediction.
| Categories | Details of features |
| I. Anatomy of abnormalities | I.1. Mass centre in standard atlas space; I.2. Percentage of the whole-brain volume and the volume of each of the 61 auto-segmented brain structures being injured39 41; I.3. Ratios of volumetric injury in the same brain structures between the left and right hemisphere; I.4. Percentage and distribution of abnormalities in 28 major fibre tracts as defined in the JHU atlas.145 |
| II. Geometry of abnormalities | II.1. Volume of abnormal regions; II.2. Maximum diameter along different orthogonal directions, maximum surface of abnormal regions, geometric compactness, spherecity, surface-to-volume ratio in the abnormal regions. |
| III. Heterogeneity of abnormalities | III.1. Histogram analysis (0, 25, 50, 75 and 100-percentile) of T1w, T2w, T1-Gad, FLAIR, ADC, SWI, ZT1w, ZT2w, ZFLAIR, ZADC, ZSWI signal values within the abnormal regions; III.2. Skewness (asymmetry), kurtosis (flatness), and randomness (entropy, SD) of T1w, T2w, T1-Gad, FLAIR, ADC, SWI, ZT1w, ZT2w, ZFLAIR, ZADC, ZSWI signal values within abnormal regions; |
| IV. Texture of abnormalities | IV.1. Gray-level co-occurrence matrix features and gray-level run-length matrix of T1w, T2w, T1-Gad, FLAIR, ADC, SWI, ZT1w, ZT2w, ZT1-Gad, ZFLAIR, ZADC, ZSWI signal values within abnormal regions; IV.2. fractal analysis, Minkowski functionals, wavelet transform and Laplacian transforms of Gaussian-filtered images for the abnormal regions. |
ADC, apparent diffusion coefficient; FLAIR, fluid-attenuated inversion recovery; SWI, susceptibility-weighted imaging.
Not all features are equally useful. Some features may be redundant. To select the best feature combinations, we developed and implemented into software an iFIBE feature selection tool.43 91 Usually, iFIBE automatically selects 5–70 features out of hundreds to thousands of features. It automatically decides how many features to select, based on tasks. It starts from the single most predictive feature (by AUC in cross-validation) and iteratively adds features that increase the AUC than adding any other feature. When no more features can be added that further improves AUC, the algorithm removes features from the selected subset, one at a time, such that the removed feature will increase the AUC of the remaining feature subset more than removing any other feature. The iFIBE algorithm iterates between adding and removing features, until no more features can be added or removed to increase the AUC. Using our iFIBE feature selector43 91 with Gaussian-kernel SVM,87 88 we selected 12 texture features out of ~200 features for deformable brain MRI registration42 43 95; we selected 7 from >3 00 000 features for histology-based tumour staging (AUC=0.96) and grading (AUC=0.83),91 from 83 patients; we selected 53 features among >2000 brain MRI features for classifying bipolar disorder, major depression and normal controls (AUC=0.61),92 from 113 subjects; and, we selected 69 from >2000 brain MRI features for diagnosing schizophrenia (AUC=0.75),93 from 124 subjects.
These MRI features and the SVM/RF prediction models are commonly used radiomics features and models for predicting brain disease progression and outcomes. The use of SVM and RF is because of their wide use in radiomics to predict brain disease progression and outcomes, and also because they are the baseline model on which we built the iFIBE feature selection algorithm (ie, our iFIBE feature selection is at the best performance when coupled with SVM and RF). Using some of these MRI features in prediction models has led to 0.6–1.0 sensitivity/0.76–0.99 specificity classifying original or metastatic brain tumor,96 97 0.77–0.93 sensitivity/0.76–0.92 specificity predicting good/poor outcomes after acute brain haemorrhage,98 99 0.91–0.94 sensitivity/0.91–1.0 specificity grading brain tumor100 0.86 sensitivity/0.92 specificity predicting psychomotor outcome in preterm neonates,101 and 0.71–0.82 sensitivity/0.68–0.92 specificity predicting multiple sclerosis worsening or not102 or predicting relapsing-remitting multiple sclerosis (MS) patients.103
Other clinical features include patient sex, the extent of PWB (left/right/bilateral), age at first MRI, the extent of brain involvement and EEG report (normal/not). Our hypothesis is that combining these clinical variables with MRI features will improve the sensitivity and specificity of outcome prediction compared with using MRI features alone. Our feature selection algorithm will be used to select the subset of clinical and MRI features that together best predicts the outcome.
Deep learning
Deep learning convolutional neural network (CNN) takes the whole image or image patches as input for outcome prediction. We will use the transfer learning strategy,104 using the pretrained 2D CNN models that were trained in the ImageNet database (>14 million natural images).105–107 The big data-powered pretrained CNN model will be refined in our SWS cohort, on 2D slices that contain the abnormal regions. This transfer learning strategy is widely used to enable and stabilise deep learning in small sample studies. Successful applications included image-based classification and outcome prediction in 2D chest X-ray images,104 108 2D retina images,109–112 2D histological images113–118 and recently in 3D brain MRI for tumour classification.11 Prediction results on various slices of the same patient can be combined through majority voting into a final prediction.116 This plan avoids training a 3D CNN directly on our data, where the sample size may lead to overfitting. The accuracy of deep learning outcome prediction will also be quantified by sensitivity, specificity and AUC in fivefold cross-validations and be compared with those metrics by the traditional machine learning.
The transfer learning strategy has been commonly used to deal with small sample problems. For 2D images, we can initialise with the benchmark artificial intelligence (AI) model from imageNet, and refine the model by samples in a specific task, for each specific AI study.104 The benchmark AI model can initialise studies (1) across sample size: trained in >14 million images in ImageNet, it can initialise other AI studies with only hundreds to thousands of samples104 114 119; (2) across tasks: trained to recognise natural objects (eg, cats, dogs, balloons, etc), it can initialise clinical AI tasks, such as disease diagnosis,104 110 111 113 115 120 severity evaluation,109 117 118 treatment evaluation3 and prediction of prognosis108 112 114 121 and (3) across imaging modalities: trained in camera-acquired natural images, it can initialise studies involving chest X-ray images,104 108 retina images110 111 and histological images.113 114 119 In these Nature, Cell, Lancet, JAMA, PNAS studies, initialisation with the benchmark model and refinement with samples in each specific AI task has led superior accuracies than training from scratch in specific tasks.104 122 In 3D medical images, we just trained a 3D benchmark AI model from 16 705 brain MRIs across the lifespan for predicting the continuously valued age.123 124 This offers one of the first benchmark AI models that can be transferred to small sample size studies such as in this SWS case. We will test the prediction accuracy in SWS with and without transferring the benchmark model.
Integrate MRI and non-MRI information
We hypothesise that such integration can more accurately predict outcomes than using MRI alone. In traditional machine learning models, clinical features can be concatenated into the feature vector for each patient. The remaining steps (classification, feature selection and cross-validation) stay the same. In deep learning models, clinical features can be added into each feature layer in the CNN architecture,125 and the rest is the same. In both cases, the learning process will automatically parse the interaction between MRI and clinical information and find the best combination. We will compare the sensitivity, specificity and AUC between the traditional and deep learning models after combining MRI and clinical information.
Expected sample size and power analysis
From clinical registries, we have identified 98 patients (62 from KKI and 36 from BCH) with brain MRIs before 2 years of age, of which 63 (42 from KKI and 21 from BCH) had the first MRI before the onset of seizure. We expect a more thorough search of clinical registries and ICD-based hospital-wide search can bring us to the expected 80 sample size for infants who had the first MRI before the first onset of seizure. This sample size will be >5 times bigger than most existing presymptomatic studies for SWS.
In the single-variate analysis, with power=0.8 and alpha=0.05, correlating a predictive variable and the neuroscore outcome variable will need 194, 85, 47, 29 subjects to call significance if the correlation is 0.2, 0.3, 0.4, 0.5. Between patient groups presenting and not presenting seizure symptoms by 2 years of age, we need 393, 99 or 63 patients to call significant group differences if a feature has a shared SD between two groups and the difference in this feature between two groups is 20%, 40%, or 50% of the mean of one group. With our expected sample size (BCH+KKI), we will be able to call a predictive variable significantly associated with outcome if the correlation is greater than 0.41, and the group difference is more than 50% of the mean in one group.
In multivariate machine learning, our expected sample size of 80 will allow our algorithm to select up to eight MRI and clinical variables for outcome prediction, according to the ‘rule of 10’ in multivariate ML prediction.126–128 Our algorithm typically selects 5–10 variables from hundreds of variables extracted from MRI and clinical variabels.11 14 25 48 49 Thus, overfitting is not a big concern in this multisite MRI data for SWS.
Patient and public involvement
Patient advocacy groups will be involved in the distribution of the summary of the findings after publication of results. The development of the research question was informed by the unmet needs determined during the Sturge-Weber Foundation Clinical Care Network conference in 2018. Patients or the public were not involved in the design, or conduct, or reporting or dissemination plans of our research.
Discussion and dessemination
Secondary use of hospital-hosted data can be time-efficient and cost-efficient. This efficiency is further amplified in rare diseases. A typical clinical trial often requires significant funding and years to collect data from single digits or dozens of patients with SWS. Our study protocol, on the other than, plans to retrospectively collect presymptomatic data for ~80 patients with SWS who had neuroimaging under 2 years of age and were cared in BCH and KKI, two large nationwide centres for SWS care, including presymptomatic SWS care.
The importance of identifying reliable biomarkers for early diagnosis of SWS relies on the potential for presymptomatic intervention impacting neurological outcomes of SWS. The first goal of this study is to further elucidate the pathogenesis of early epileptogenesis and brain injury in SWS by quantifying and comparing neuroimaging patterns, which reflect the integrity and reorganisation of neural networks. Comparing brain MRIs in patients with SWS with or without seizures after the first MRI will identify injury patterns that may aid early diagnosis of SWS brain involvement, and guide treatment to improve the prognosis of those at high risk for seizures before 2 years of age and subsequent cognitive and neurologic impairment.
SWS is a developmental disorder and the epileptogenesis mechanism during early ages will likely differ from the mechanisms in the mature brain. Prior studies have repeatedly associated early age of seizure onset in SWS with worse neurologic outcome, more severe epilepsy and worse cognitive outcome.6 48 129 Thus, delaying or preventing seizure onset in SWS has become a main treatment goal driving recent efforts to enhance early diagnosis of brain involvement.9 Standard imaging techniques and assessments result in only about a 25% sensitivity in early imaging130 resulting in the loss of opportunity to intervene early in many children and slowing efforts to determine the most effective presymptomatic treatment. The proposed quantitative and objective study may improve early diagnosis. More infants with SWS brain involvement may benefit from presymptomatic studies, anticipatory guidance and close monitoring for neurologic and developmental impairments. Zallman et al showed that infants with PWBs and evidence of SWS brain involvement on imaging were more likely to receive appropriate early care, compared with those who were not diagnosed before the onset of seizures.130
SWS is a spectrum disorder with a well-identified pathological process allowing precise diagnosis, and it is an excellent model for LSM to quantify neuroanatomic basis of outcomes. The brain involvement ranging in extent from single lobe (usually occipital) to extensive bilateral brain involvement of both hemispheres.131 As a result, the most important factor determining the neurologic and cognitive outcome, other than the age of seizure onset, is the extent of brain involvement. The greater extent of SWS brain involvement is correlated with earlier age of seizure onset.131–133 Early age of seizure onset, particularly less than a year of age, has been associated with a significant impact on intelligence quotient (IQ),48 worse hemiparesis129 and lower cognitive function quality of life.28 Therefore, the full extent of SWS brain involvement, as determined by later MRI imaging with contrast and appropriate imaging, after a year of age, is needed; early imaging, even when abnormal, can underestimate the extent of brain involvement. The extent of brain involvement (on imaging after a year of age) will be an important clinical factor in the AI analysis.
Adding clinical variables to neuroimaging is important. Males are associated with worse cognitive outcomes, and with a greater risk of suicidality in SWS.134 135 Whether gender impacts the early evolution of SWS as manifested in neuroimaging remains to be studied. A family history of seizures or epilepsy is associated with an early age of seizure onset in SWS.136 This suggests that genetic factors, beyond the underlying somatic mutation in GNAQ, have a role in determining the timing of seizure onset in SWS.1 The extent of skin involvement with PBW may also be a prognostic factor. A greater extent of PWB (bilateral vs unilateral/none) is associated with worse neurologic status.137 Bilateral skin involvement is associated with bilateral brain involvement. Therefore, the extent of skin involvement is to some extent a proxy for the extent of brain involvement that impacts the neurologic outcome. However, it has also been shown that patients with no PWB present later and have a better outcome than patients with a similar extent of brain involvement.138 139 This observation suggests that more extensive SWS (in terms of more structures (brain, skin and/or eye) involved is in some way different with regards to the evolution of brain injury, from less extensive SWS (brain only) involvement. Gender, family history of epilepsy and the extent of skin involvement are, therefore, also important predictive clinical factors to include in the AI neuroimaging modelling and analysis.
During early stage SWS, the extent of neuroimaging alteration is similar to that of other small diffuse lesions; our sample size is similar to other published datasets (including chronic stroke lesions140 141 and multiple sclerosis).142–144 Identificatication of biomarkers is a complex process but has proved to result in rapid bench-to-bedside translation in many other pathological conditions. The identification of reliable presymptomatic biomarkers will ultimately be used to predict response to therapies and influence ‘go’ or ‘no go’ decisions before advancing clinical trials for SWS.
This protocol describes a multicentre effort to validate emerging techniques in a rare potentially devastating disease. The centres involved have the expertise and the combined number of cases, indicating the strong feasibility of this study. The results will benefit future prospective trials.
Ethics and dissemination
The Institutional Review Boards at Kennedy Krieger Institute and Boston Children’s Hospital approval have been obtained at each site to retrospectively study this data. Results will be disseminated by presentations, publication and sharing of algorithms generated.
Supplementary Material
Footnotes
Collaborators: BCH-KKI SWS Pre-symptomatic Biomarker Working Group members include: Mustafa Sahin, MD, Boston Children’s Hospital. P. Ellen Grant, MD, Boston Children’s Hospital. Katrina Boyer, PhD, Boston Children’s Hospital. Masanori Takeoka, MD, Boston Children’s Hospital. Bernard Cohen MD, Johns Hopkins Dermatology. Joshua Ewen MD, Kennedy Krieger Institute Epilepsy. Eric Kossoff MD, Johns Hopkins Child Neurology and Epilepsy. Teressa Garcia Reidy, MS, OTR/L, Kennedy Krieger Institute Occupational Therapy. Stacy Suskauer MD, Kennedy Krieger Institute Medical Rehabilitation. Andrew Zabel PhD, Kennedy Krieger Institute Neuropsychology.
Contributors: Study design: ARLP, AMC, YO. Data acquisition: PV, ARLP, DDML, AMC, YO. Data analysis plan: ARLP, DDML, AMC, YO. Paper drafting and editing: PV, ARLP, DDML, AMC, YO.
Funding: Work leading to this proposal was partly funded by the Celebrate Hope Foundation (Comi), Harvard Medical School/ Boston Children’s Hospital Faculty Career Development Award (Ou), St. Baldrick Foundation (Ou), and NIH R03 HD091464 (Ou). Application to further NIH funding is under review.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Contributor Information
BCH-KKI SWS Pre-symptomatic Biomarker Working Group:
Mustafa Sahin, P. Ellen Grant, Katrina Boyer, Masanori Takeoka, Bernard Cohen, Joshua Ewen, Eric Kossoff, Teressa Garcia Reidy, Stacy Suskauer, and Andrew Zabel
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Shirley MD, Tang H, Gallione CJ, et al. Sturge–Weber Syndrome and Port-Wine Stains Caused by Somatic Mutation in GNAQ. N Engl J Med Overseas Ed 2013;368:1971–9. 10.1056/NEJMoa1213507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jacobs AH, Walton RG. The incidence of birthmarks in the neonate. Pediatrics 1976;58:218–22. 10.1542/peds.58.2.218 [DOI] [PubMed] [Google Scholar]
- 3. Comi AM. Update on Sturge-Weber syndrome: diagnosis, treatment, quantitative measures, and controversies. Lymphat Res Biol 2007;5:257–64. 10.1089/lrb.2007.1016 [DOI] [PubMed] [Google Scholar]
- 4. De la Torre AJ, Luat AF, Juhász C, et al. A multidisciplinary consensus for clinical care and research needs for Sturge-Weber syndrome. Pediatr Neurol 2018;84:11–20. 10.1016/j.pediatrneurol.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luat AF, Juhász C, Loeb JA, et al. Neurological complications of Sturge-Weber syndrome: current status and unmet needs. Pediatr Neurol 2019;98:31–8. 10.1016/j.pediatrneurol.2019.05.013 [DOI] [PubMed] [Google Scholar]
- 6. Sujansky E, Conradi S. Sturge-Weber syndrome: age of onset of seizures and glaucoma and the prognosis for affected children. J Child Neurol 1995;10:49–58. 10.1177/088307389501000113 [DOI] [PubMed] [Google Scholar]
- 7. Sujansky E, Conradi S. Outcome of Sturge-Weber syndrome in 52 adults. Am J Med Genet 1995;57:35–45. 10.1002/ajmg.1320570110 [DOI] [PubMed] [Google Scholar]
- 8. Glauser TA, Mitchell WG, Weinstock A, et al. Pharmacokinetics of levetiracetam in infants and young children with epilepsy. Epilepsia 2007;48:1117–22. 10.1111/j.1528-1167.2007.01090.x [DOI] [PubMed] [Google Scholar]
- 9. Day AM, Hammill AM, Juhász C, et al. Hypothesis: presymptomatic treatment of Sturge-Weber syndrome with aspirin and antiepileptic drugs may delay seizure onset. Pediatr Neurol 2019;90:8–12. 10.1016/j.pediatrneurol.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Juhász C. Predicting and preventing epilepsy in Sturge-Weber syndrome? Pediatr Neurol Briefs 2016;30:43. 10.15844/pedneurbriefs-30-11-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luat AF, Behen ME, Chugani HT, et al. Cognitive and motor outcomes in children with unilateral Sturge-Weber syndrome: effect of age at seizure onset and side of brain involvement. Epilepsy Behav 2018;80:202–7. 10.1016/j.yebeh.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bar C, Pedespan J-M, Boccara O, et al. Early magnetic resonance imaging to detect presymptomatic leptomeningeal angioma in children with suspected Sturge-Weber syndrome. Dev Med Child Neurol 2020;62:227–33. 10.1111/dmcn.14253 [DOI] [PubMed] [Google Scholar]
- 13. Juhász C, Hu J, Xuan Y, et al. Imaging increased glutamate in children with Sturge-Weber syndrome: association with epilepsy severity. Epilepsy Res 2016;122:66–72. 10.1016/j.eplepsyres.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bosnyák E, Behen ME, Guy WC, et al. Predictors of cognitive functions in children with Sturge-Weber syndrome: a longitudinal study. Pediatr Neurol 2016;61:38–45. 10.1016/j.pediatrneurol.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kobylecki C, Jones M, Williams T, et al. Reversible increases in cortical diffusion-weighted Mr signal in a patient with Sturge-Weber syndrome and subacute hemiplegia. J Neurol 2011;258:2095–6. 10.1007/s00415-011-6073-2 [DOI] [PubMed] [Google Scholar]
- 16. Jülich K, Neuberger I, Sahin M, et al. Yield of emergent neuroimaging in patients with Sturge-Weber syndrome presenting with acute neurologic symptoms. J Child Neurol 2019;34:17–21. 10.1177/0883073818801635 [DOI] [PubMed] [Google Scholar]
- 17. Wu J, Tarabishy B, Hu J, et al. Cortical calcification in Sturge-Weber syndrome on MRI-SWI: relation to brain perfusion status and seizure severity. J Magn Reson Imaging 2011;34:791–8. 10.1002/jmri.22687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dutkiewicz A-S, Ezzedine K, Mazereeuw-Hautier J, et al. A prospective study of risk for Sturge-Weber syndrome in children with upper facial port-wine stain. J Am Acad Dermatol 2015;72:473–80. 10.1016/j.jaad.2014.11.009 [DOI] [PubMed] [Google Scholar]
- 19. Arulrajah S, Ertan G, M Comi A, et al. Mri with diffusion-weighted imaging in children and young adults with simultaneous supra- and infratentorial manifestations of Sturge-Weber syndrome. J Neuroradiol 2010;37:51–9. 10.1016/j.neurad.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 20. Kim J-A, Jeong J-W, Behen ME, et al. Metabolic correlates of cognitive function in children with unilateral Sturge-Weber syndrome: evidence for regional functional reorganization and crowding. Hum Brain Mapp 2018;39:1596–606. 10.1002/hbm.23937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pinto AL, Chen L, Friedman R, et al. Sturge-Weber Syndrome: Brain Magnetic Resonance Imaging and Neuropathology Findings. Pediatr Neurol 2016;58:25–30. 10.1016/j.pediatrneurol.2015.11.005 [DOI] [PubMed] [Google Scholar]
- 22. Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441–6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winzeck S, Hakim A, McKinley R, et al. Isles 2016 and 2017-Benchmarking ischemic stroke lesion outcome prediction based on multispectral MRI. Front Neurol 2018;9:679. 10.3389/fneur.2018.00679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rodriguez Gutierrez D, Manita M, Jaspan T, et al. Serial Mr diffusion to predict treatment response in high-grade pediatric brain tumors: a comparison of regional and voxel-based diffusion change metrics. Neuro Oncol 2013;15:981–9. 10.1093/neuonc/not034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uddin LQ, Dajani DR, Voorhies W, et al. Progress and roadblocks in the search for brain-based biomarkers of autism and attention-deficit/hyperactivity disorder. Transl Psychiatry 2017;7:e1218. 10.1038/tp.2017.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Weiss RJ, Bates SV, Song Ya'nan, et al. Mining multi-site clinical data to develop machine learning MRI biomarkers: application to neonatal hypoxic ischemic encephalopathy. J Transl Med 2019;17:385. 10.1186/s12967-019-2119-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hatfield LA, Crone NE, Kossoff EH, et al. Quantitative EEG asymmetry correlates with clinical severity in unilateral Sturge-Weber syndrome. Epilepsia 2007;48:191–5. 10.1111/j.1528-1167.2006.00630.x [DOI] [PubMed] [Google Scholar]
- 29. Lin DDM, Barker PB, Hatfield LA, et al. Dynamic Mr perfusion and proton MR spectroscopic imaging in Sturge-Weber syndrome: correlation with neurological symptoms. J Magn Reson Imaging 2006;24:274–81. 10.1002/jmri.20627 [DOI] [PubMed] [Google Scholar]
- 30. Kelley TM, Hatfield LA, Lin DDM, et al. Quantitative analysis of cerebral cortical atrophy and correlation with clinical severity in unilateral Sturge-Weber syndrome. J Child Neurol 2005;20:867–70. 10.1177/08830738050200110201 [DOI] [PubMed] [Google Scholar]
- 31. Kavanaugh B, Sreenivasan A, Bachur C, et al. [Formula: see text]Intellectual and adaptive functioning in Sturge-Weber Syndrome. Child Neuropsychol 2016;22:635–48. 10.1080/09297049.2015.1028349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pienaar R, Rannou N, Bernal J, et al. ChRIS--A web-based neuroimaging and informatics system for collecting, organizing, processing, visualizing and sharing of medical data. Annu Int Conf IEEE Eng Med Biol Soc 2015;2015:206–9. 10.1109/EMBC.2015.7318336 [DOI] [PubMed] [Google Scholar]
- 33. Ou Y, Zöllei L, Retzepi K, et al. Using clinically acquired MRI to construct age-specific ADC atlases: quantifying spatiotemporal ADC changes from birth to 6-year old. Hum Brain Mapp 2017;38:3052–68. 10.1002/hbm.23573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30. 10.1136/jamia.2009.000893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patridge EF, Bardyn TP. Research electronic data capture (REDCap). J Med Libr Assoc 2018;106:142–4. 10.5195/jmla.2018.319 [DOI] [Google Scholar]
- 36. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010;29:1310–20. 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ou Y, Zöllei L, Da X, et al. Field of view normalization in multi-site brain MRI. Neuroinformatics 2018;16:431–44. 10.1007/s12021-018-9359-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Srinivasan D, Erus G, Doshi J, et al. A comparison of Freesurfer and multi-atlas MUSE for brain anatomy segmentation: findings about size and age bias, and inter-scanner stability in multi-site aging studies. Neuroimage 2020;223:117248. 10.1016/j.neuroimage.2020.117248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doshi J, Erus G, Ou Y, et al. Muse: MUlti-atlas region segmentation utilizing ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage 2016;127:186–95. 10.1016/j.neuroimage.2015.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sotardi S, Gollub RL, Bates SV, et al. Voxelwise and regional brain apparent diffusion coefficient changes on MRI from birth to 6 years of age. Radiology 2021;298:415–24. 10.1148/radiol.2020202279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morton SU, Vyas R, Gagoski B, et al. Maternal dietary intake of omega-3 fatty acids correlates positively with regional brain volumes in 1-month-old term infants. Cereb Cortex 2020;30:2057–69. 10.1093/cercor/bhz222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ou Y, Akbari H, Bilello M, et al. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE Trans Med Imaging 2014;33:2039–65. 10.1109/TMI.2014.2330355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ou Y, Sotiras A, Paragios N, et al. DRAMMS: deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal 2011;15:622–39. 10.1016/j.media.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jenkinson M, Beckmann CF, Behrens TEJ, et al. Fsl. Neuroimage 2012;62:782–90. 10.1016/j.neuroimage.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 45. Fortin J-P, Parker D, Tunç B, et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage 2017;161:149–70. 10.1016/j.neuroimage.2017.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- 47. Martí-Bonmatí L, Menor F, Mulas F. The Sturge-Weber syndrome: correlation between the clinical status and radiological CT and MRI findings. Childs Nerv Syst 1993;9:107–9. 10.1007/BF00305319 [DOI] [PubMed] [Google Scholar]
- 48. Juhasz C, Lai C, Behen ME, et al. White matter volume as a major predictor of cognitive function in Sturge-Weber syndrome. Arch Neurol 2007;64:1169–74. 10.1001/archneur.64.8.1169 [DOI] [PubMed] [Google Scholar]
- 49. Arigliani M, Bravar G, Crichiutti G, et al. A cerebral infarction in a girl with Sturge-Weber syndrome. Pediatr Neurol 2016;64:99–100. 10.1016/j.pediatrneurol.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 50. Adamsbaum C, Pinton F, Rolland Y, et al. Accelerated myelination in early Sturge-Weber syndrome: MRI-SPECT correlations. Pediatr Radiol 1996;26:759–62. 10.1007/BF01396195 [DOI] [PubMed] [Google Scholar]
- 51. George U, Rathore S, Nittala P. Mr demonstration of accelerated myelination in early sturge Weber syndrome. Neurol India 2010;58:336. 10.4103/0028-3886.63769 [DOI] [PubMed] [Google Scholar]
- 52. Moritani T, Kim J, Sato Y, et al. Abnormal hypermyelination in a neonate with Sturge-Weber syndrome demonstrated on diffusion-tensor imaging. J Magn Reson Imaging 2008;27:617–20. 10.1002/jmri.21248 [DOI] [PubMed] [Google Scholar]
- 53. Pilli VK, Behen ME, Hu J, et al. Clinical and metabolic correlates of cerebral calcifications in Sturge-Weber syndrome. Dev Med Child Neurol 2017;59:952–8. 10.1111/dmcn.13433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pilli VK, Chugani HT, Juhász C. Enlargement of deep medullary veins during the early clinical course of Sturge-Weber syndrome. Neurology 2017;88:103–5. 10.1212/WNL.0000000000003455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaseka ML, Bitton JY, Décarie J-C, et al. Predictive factors for epilepsy in pediatric patients with Sturge-Weber syndrome. Pediatr Neurol 2016;64:52–8. 10.1016/j.pediatrneurol.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 56. John F, Maqbool M, Jeong J-W, et al. Deep cerebral vein expansion with metabolic and neurocognitive recovery in Sturge-Weber syndrome. Ann Clin Transl Neurol 2018;5:502–6. 10.1002/acn3.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Juhász C, Haacke EM, Hu J, et al. Multimodality imaging of cortical and white matter abnormalities in Sturge-Weber syndrome. AJNR Am J Neuroradiol 2007;28:900–6. [PMC free article] [PubMed] [Google Scholar]
- 58. Alkonyi B, Chugani HT, Behen M, et al. The role of the thalamus in neuro-cognitive dysfunction in early unilateral hemispheric injury: a multimodality imaging study of children with Sturge-Weber syndrome. Eur J Paediatr Neurol 2010;14:425–33. 10.1016/j.ejpn.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pinto ALR, Ou Y, Sahin M, et al. Quantitative apparent diffusion coefficient mapping may predict seizure onset in children with Sturge-Weber syndrome. Pediatr Neurol 2018;84:32–8. 10.1016/j.pediatrneurol.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kamson DO, Juhász C, Shin J, et al. Patterns of structural reorganization of the corticospinal tract in children with Sturge-Weber syndrome. Pediatr Neurol 2014;50:337–42. 10.1016/j.pediatrneurol.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jeong J-won, Tiwari VN, Shin J, et al. Assessment of brain damage and plasticity in the visual system due to early occipital lesion: comparison of FDG-PET with diffusion MRI tractography. J Magn Reson Imaging 2015;41:431–8. 10.1002/jmri.24556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alkonyi B, Govindan RM, Chugani HT, et al. Focal white matter abnormalities related to neurocognitive dysfunction: an objective diffusion tensor imaging study of children with Sturge-Weber syndrome. Pediatr Res 2011;69:74–9. 10.1203/PDR.0b013e3181fcb285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guimond A, Meunier J, Thirion J-P. Average brain models: a convergence study. Computer Vision and Image Understanding 2000;77:192–210. 10.1006/cviu.1999.0815 [DOI] [Google Scholar]
- 64. Fonov V, Evans AC, Botteron K, et al. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 2011;54:313–27. 10.1016/j.neuroimage.2010.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O'Muircheartaigh J, Robinson EC, Pietsch M, et al. Modelling brain development to detect white matter injury in term and preterm born neonates. Brain 2020;143:467–79. 10.1093/brain/awz412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Buschman TJ, Siegel M, Roy JE, et al. Neural substrates of cognitive capacity limitations. Proc Natl Acad Sci U S A 2011;108:11252–5. 10.1073/pnas.1104666108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pagonabarraga J, Kulisevsky J, Strafella AP, et al. Apathy in Parkinson's disease: clinical features, neural substrates, diagnosis, and treatment. Lancet Neurol 2015;14:518–31. 10.1016/S1474-4422(15)00019-8 [DOI] [PubMed] [Google Scholar]
- 68. Bates E, Wilson SM, Saygin AP, et al. Voxel-Based lesion-symptom mapping. Nat Neurosci 2003;6:448–50. 10.1038/nn1050 [DOI] [PubMed] [Google Scholar]
- 69. Rorden C, Karnath H-O, Bonilha L. Improving lesion-symptom mapping. J Cogn Neurosci 2007;19:1081–8. 10.1162/jocn.2007.19.7.1081 [DOI] [PubMed] [Google Scholar]
- 70. Zhang Y, Kimberg DY, Coslett HB, et al. Multivariate lesion-symptom mapping using support vector regression. Hum Brain Mapp 2014;35:5861–76. 10.1002/hbm.22590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. DeMarco AT, Turkeltaub PE. A multivariate lesion symptom mapping toolbox and examination of lesion-volume biases and correction methods in lesion-symptom mapping. Hum Brain Mapp 2018;39:4169–82. 10.1002/hbm.24289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fox MD. Mapping symptoms to brain networks with the human connectome. N Engl J Med 2018;379:2237–45. 10.1056/NEJMra1706158 [DOI] [PubMed] [Google Scholar]
- 73. Gleichgerrcht E, Fridriksson J, Rorden C, et al. Connectome-based lesion-symptom mapping (CLSM): a novel approach to map neurological function. Neuroimage Clin 2017;16:461–7. 10.1016/j.nicl.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yourganov G, Fridriksson J, Rorden C, et al. Multivariate Connectome-Based symptom mapping in post-stroke patients: networks supporting language and speech. J Neurosci 2016;36:6668–79. 10.1523/JNEUROSCI.4396-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sporns O. The human connectome: a complex network. Ann N Y Acad Sci 2011;1224:109–25. 10.1111/j.1749-6632.2010.05888.x [DOI] [PubMed] [Google Scholar]
- 76. Van Essen DC, Smith SM, Barch DM, et al. The WU-Minn human connectome project: an overview. Neuroimage 2013;80:62–79. 10.1016/j.neuroimage.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci 2011;31:15775–86. 10.1523/JNEUROSCI.3539-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu TT, Achrol AS, Mitchell LA, et al. Computational identification of tumor anatomic location associated with survival in 2 large cohorts of human primary glioblastomas. AJNR Am J Neuroradiol 2016;37:621–8. 10.3174/ajnr.A4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hope TMH, Leff AP, Price CJ. Predicting language outcomes after stroke: is structural disconnection a useful predictor? Neuroimage Clin 2018;19:22–9. 10.1016/j.nicl.2018.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xing S, Lacey EH, Skipper-Kallal LM, et al. Right hemisphere grey matter structure and language outcomes in chronic left hemisphere stroke. Brain 2016;139:227–41. 10.1093/brain/awv323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mirman D, Chen Q, Zhang Y, et al. Neural organization of spoken language revealed by lesion-symptom mapping. Nat Commun 2015;6:6762. 10.1038/ncomms7762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Karnath H-O, Rennig J. Investigating structure and function in the healthy human brain: validity of acute versus chronic lesion-symptom mapping. Brain Struct Funct 2017;222:2059–70. 10.1007/s00429-016-1325-7 [DOI] [PubMed] [Google Scholar]
- 83. Bonilha L, Hillis AE, Hickok G, et al. Temporal lobe networks supporting the comprehension of spoken words. Brain 2017;140:2370–80. 10.1093/brain/awx169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Roswandowitz C, Kappes C, Obrig H, et al. Obligatory and facultative brain regions for voice-identity recognition. Brain 2018;141:234–47. 10.1093/brain/awx313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liu J, Wang C, Diao Q, et al. Connection disruption underlying attention deficit in subcortical stroke. Radiology 2018;288:186–94. 10.1148/radiol.2018171730 [DOI] [PubMed] [Google Scholar]
- 86. Meyer S, Kessner SS, Cheng B, et al. Voxel-Based lesion-symptom mapping of stroke lesions underlying somatosensory deficits. Neuroimage Clin 2016;10:257–66. 10.1016/j.nicl.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;20:273–97. 10.1007/BF00994018 [DOI] [Google Scholar]
- 88. Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2011;2:Article 27. [Google Scholar]
- 89. Breiman L. Random forests. Mach Learn 2001;45:5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 90. Hemant I, Udaya BK, Eugene HB. Random survival forests. The Annals of Applied Statistics 2008;2:841–60. [Google Scholar]
- 91. Ou Y, Shen D, Zeng J, et al. Sampling the spatial patterns of cancer: optimized biopsy procedures for estimating prostate cancer volume and Gleason score. Med Image Anal 2009;13:609–20. 10.1016/j.media.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Serpa MH, Ou Y, Schaufelberger MS, et al. Neuroanatomical classification in a population-based sample of psychotic major depression and bipolar I disorder with 1 year of diagnostic stability. Biomed Res Int 2014;2014:1–9. 10.1155/2014/706157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zanetti MV, Schaufelberger MS, Doshi J, et al. Neuroanatomical pattern classification in a population-based sample of first-episode schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry 2013;43:116–25. 10.1016/j.pnpbp.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 94. Da X, Toledo JB, Zee J, et al. Integration and relative value of biomarkers for prediction of MCI to AD progression: spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. Neuroimage Clin 2014;4:164–73. 10.1016/j.nicl.2013.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ou Y, Weinstein SP, Conant EF, et al. Deformable registration for quantifying longitudinal tumor changes during neoadjuvant chemotherapy. Magn Reson Med 2015;73:2343–56. 10.1002/mrm.25368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen C, Ou X, Wang J, et al. Radiomics-Based machine learning in differentiation between glioblastoma and metastatic brain tumors. Front Oncol 2019;9:806. 10.3389/fonc.2019.00806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Artzi M, Bressler I, Ben Bashat D. Brain metastasis and subtypes using radiomics analysis. J Magn Reson Imaging 2019;50:519–28. [DOI] [PubMed] [Google Scholar]
- 98. Nawabi J, Kniep H, Elsayed S, et al. Imaging-Based outcome prediction of acute intracerebral hemorrhage. Transl Stroke Res 2021;12:958–67. 10.1007/s12975-021-00891-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Xu X, Zhang J, Yang K, et al. Prognostic prediction of hypertensive intracerebral hemorrhage using CT radiomics and machine learning. Brain Behav 2021;11:e02085. 10.1002/brb3.2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chu H, Lin X, He J, et al. Value of MRI Radiomics based on enhanced T1WI images in prediction of meningiomas grade. Acad Radiol 2021;28:687–93. 10.1016/j.acra.2020.03.034 [DOI] [PubMed] [Google Scholar]
- 101. Shin Y, Nam Y, Shin T, et al. Brain MRI radiomics analysis may predict poor psychomotor outcome in preterm neonates. Eur Radiol 2021;31:6147–55. 10.1007/s00330-021-07836-7 [DOI] [PubMed] [Google Scholar]
- 102. Zhao Y, Healy BC, Rotstein D, et al. Exploration of machine learning techniques in predicting multiple sclerosis disease course. PLoS One 2017;12:e0174866. 10.1371/journal.pone.0174866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Doyle A, Precup D, Arnold D. Predicting future disease activity and treatment responders for multiple sclerosis patients using a Bag-of-Lesions brain representation. Lecture Notes in Computer Science 2017:186–94. 10.1007/978-3-319-66179-7_22 [DOI] [Google Scholar]
- 104. Kermany DS, Goldbaum M, Cai W, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018;172:1122–31. 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 105. Deng J, Dong W, Socher R. ImageNet: a large-scale hierarchical image database. 2009 IEEE conference on computer vision and pattern recognition 2009:248–55.
- 106. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM 2017;60:84–90. 10.1145/3065386 [DOI] [Google Scholar]
- 107. Russakovsky O, Deng J, Su H, et al. ImageNet large scale visual recognition challenge. Int J Comput Vis 2015;115:211–52. 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 108. Toba S, Mitani Y, Yodoya N, et al. Prediction of pulmonary to systemic flow ratio in patients with congenital heart disease using deep Learning-Based analysis of chest radiographs. JAMA Cardiol 2020;5:449–57. 10.1001/jamacardio.2019.5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Burlina PM, Joshi N, Pekala M, et al. Automated grading of age-related macular degeneration from color fundus images using deep Convolutional neural networks. JAMA Ophthalmol 2017;135:1170–6. 10.1001/jamaophthalmol.2017.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus Photographs. JAMA 2016;316:2402–10. 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 111. Ting DSW, Cheung CY-L, Lim G, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 2017;318:2211–23. 10.1001/jama.2017.18152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus Photographs via deep learning. Nat Biomed Eng 2018;2:158–64. 10.1038/s41551-018-0195-0 [DOI] [PubMed] [Google Scholar]
- 113. Campanella G, Hanna MG, Geneslaw L, et al. Clinical-Grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019;25:1301–9. 10.1038/s41591-019-0508-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018;24:1559–67. 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hollon TC, Pandian B, Adapa AR, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med 2020;26:52–8. 10.1038/s41591-019-0715-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Buda M, AlBadawy EA, Saha A, et al. Deep Radiogenomics of Lower-Grade gliomas: Convolutional neural networks predict tumor genomic subtypes using Mr images. Radiol Artif Intell 2020;2:e180050. 10.1148/ryai.2019180050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bulten W, Pinckaers H, van Boven H, et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol 2020;21:233–41. 10.1016/S1470-2045(19)30739-9 [DOI] [PubMed] [Google Scholar]
- 118. Ström P, Kartasalo K, Olsson H, et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol 2020;21:222–32. 10.1016/S1470-2045(19)30738-7 [DOI] [PubMed] [Google Scholar]
- 119. Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054–6. 10.1038/s41591-019-0462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–8. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A 2018;115:E2970–9. 10.1073/pnas.1717139115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Huh M, Agrawal P, Efros A. What makes ImageNet good for transfer learning? 2016.
- 123. He S, Pereira D, David Perez J, et al. Multi-Channel attention-fusion neural network for brain age estimation: accuracy, generality, and interpretation with 16,705 healthy MRIs across lifespan. Med Image Anal 2021;72:102091. 10.1016/j.media.2021.102091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. He S, Grant PE, Ou Y. Global-Local transformer for brain age estimation. IEEE Trans Med Imaging 2022;41:213–24. 10.1109/TMI.2021.3108910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Huang S-C, Pareek A, Seyyedi S, et al. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med 2020;3:136. 10.1038/s41746-020-00341-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Harrell FE, Lee KL, Califf RM, et al. Regression modelling strategies for improved prognostic prediction. Stat Med 1984;3:143–52. 10.1002/sim.4780030207 [DOI] [PubMed] [Google Scholar]
- 127. Harrell FE, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361–87. [DOI] [PubMed] [Google Scholar]
- 128. Riley RD. Correction to: minimum sample size for developing a multivariable prediction model: part II-binary and time-to-event outcomes by Riley RD, Snell Ki, Ensor J, et al. Stat Med 2019;38:5672. 10.1002/sim.8409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kossoff EH, Ferenc L, Comi AM. An infantile-onset, severe, yet sporadic seizure pattern is common in Sturge-Weber syndrome. Epilepsia 2009;50:2154–7. 10.1111/j.1528-1167.2009.02072.x [DOI] [PubMed] [Google Scholar]
- 130. Zallmann M, Mackay MT, Leventer RJ, et al. Retrospective review of screening for Sturge-Weber syndrome with brain magnetic resonance imaging and electroencephalography in infants with high-risk port-wine stains. Pediatr Dermatol 2018;35:575–81. 10.1111/pde.13598 [DOI] [PubMed] [Google Scholar]
- 131. Bebin EM, Gomez MR. Prognosis in Sturge-Weber disease: comparison of unihemispheric and bihemispheric involvement. J Child Neurol 1988;3:181–4. 10.1177/088307388800300306 [DOI] [PubMed] [Google Scholar]
- 132. Alkonyi B, Chugani HT, Karia S, et al. Clinical outcomes in bilateral Sturge-Weber syndrome. Pediatr Neurol 2011;44:443–9. 10.1016/j.pediatrneurol.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zabel TA, Reesman J, Wodka EL, et al. Neuropsychological features and risk factors in children with Sturge-Weber syndrome: four case reports. Clin Neuropsychol 2010;24:841–59. 10.1080/13854046.2010.485133 [DOI] [PubMed] [Google Scholar]
- 134. Harmon KA, Day AM, Hammill AM, et al. Quality of life in children with Sturge-Weber syndrome. Pediatr Neurol 2019;101:26–32. 10.1016/j.pediatrneurol.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sebold AJ, Ahmed AS, Ryan TC, et al. Suicide screening in Sturge-Weber syndrome: an important issue in need of further study. Pediatr Neurol 2020;110:80–6. 10.1016/j.pediatrneurol.2020.03.013 [DOI] [PubMed] [Google Scholar]
- 136. Day AM, McCulloch CE, Hammill AM, et al. Physical and family history variables associated with neurological and cognitive development in Sturge-Weber syndrome. Pediatr Neurol 2019;96:30–6. 10.1016/j.pediatrneurol.2018.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Dymerska M, Kirkorian AY, Offermann EA, et al. Size of facial port-wine birthmark may predict neurologic outcome in Sturge-Weber syndrome. J Pediatr 2017;188:205–9. 10.1016/j.jpeds.2017.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Comi AM, Fischer R, Kossoff EH. Encephalofacial angiomatosis sparing the occipital lobe and without facial nevus: on the spectrum of Sturge-Weber syndrome variants? J Child Neurol 2003;18:35–8. 10.1177/08830738030180010601 [DOI] [PubMed] [Google Scholar]
- 139. Aylett SE, Neville BG, Cross JH, et al. Sturge-Weber syndrome: cerebral haemodynamics during seizure activity. Dev Med Child Neurol 1999;41:480–5. [PubMed] [Google Scholar]
- 140. Liew S-L, Anglin JM, Banks NW, et al. A large, open source dataset of stroke anatomical brain images and manual lesion segmentations. Sci Data 2018;5:180011. 10.1038/sdata.2018.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Chen X, You S, Tezcan KC, et al. Unsupervised lesion detection via image restoration with a normative prior. Med Image Anal 2020;64:101713. 10.1016/j.media.2020.101713 [DOI] [PubMed] [Google Scholar]
- 142. Carass A, Roy S, Jog A, et al. Longitudinal multiple sclerosis lesion segmentation: resource and challenge. Neuroimage 2017;148:77–102. 10.1016/j.neuroimage.2016.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Commowick O, Istace A, Kain M, et al. Objective evaluation of multiple sclerosis lesion segmentation using a data management and processing infrastructure. Sci Rep 2018;8:13650. 10.1038/s41598-018-31911-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Karimi D, Dou H, Warfield SK, et al. Deep learning with noisy labels: exploring techniques and remedies in medical image analysis. Med Image Anal 2020;65:101759. 10.1016/j.media.2020.101759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Oishi K, Mori S, Donohue PK, et al. Multi-contrast human neonatal brain atlas: application to normal neonate development analysis. Neuroimage 2011;56:8–20. 10.1016/j.neuroimage.2011.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.