Significance Statement
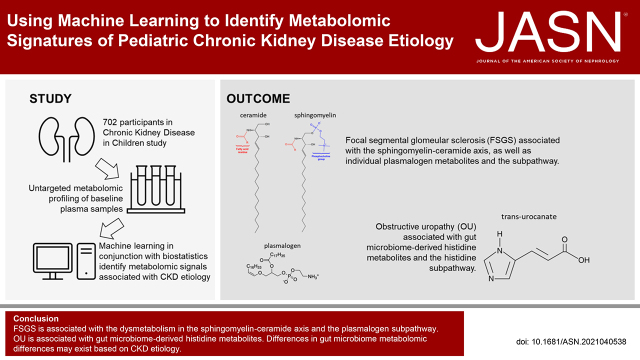
Machine learning used with biostatistics identified metabolomic signatures in the plasma of pediatric patients with CKD, providing clues to cause. Dysmetabolism in the sphingomyelin-ceramide axis is associated with both FSGS and the aplasia/dysplasia/hypoplasia spectrum. Pediatric FSGS is associated with elevated plasmalogen levels, in contrast to reports of associations with plasmalogen deficiencies. Our strategy also revealed associations of obstructive uropathy with gut-derived histidines and of reflux nephropathy with indole-tryptophans.
Keywords: metabolomics, pediatric nephrology, chronic kidney disease, machine learning, machine learning collection
Visual Abstract
Abstract
Background
Untargeted plasma metabolomic profiling combined with machine learning (ML) may lead to discovery of metabolic profiles that inform our understanding of pediatric CKD causes. We sought to identify metabolomic signatures in pediatric CKD based on diagnosis: FSGS, obstructive uropathy (OU), aplasia/dysplasia/hypoplasia (A/D/H), and reflux nephropathy (RN).
Methods
Untargeted metabolomic quantification (GC-MS/LC-MS, Metabolon) was performed on plasma from 702 Chronic Kidney Disease in Children study participants (n: FSGS=63, OU=122, A/D/H=109, and RN=86). Lasso regression was used for feature selection, adjusting for clinical covariates. Four methods were then applied to stratify significance: logistic regression, support vector machine, random forest, and extreme gradient boosting. ML training was performed on 80% total cohort subsets and validated on 20% holdout subsets. Important features were selected based on being significant in at least two of the four modeling approaches. We additionally performed pathway enrichment analysis to identify metabolic subpathways associated with CKD cause.
Results
ML models were evaluated on holdout subsets with receiver-operator and precision-recall area-under-the-curve, F1 score, and Matthews correlation coefficient. ML models outperformed no-skill prediction. Metabolomic profiles were identified based on cause. FSGS was associated with the sphingomyelin-ceramide axis. FSGS was also associated with individual plasmalogen metabolites and the subpathway. OU was associated with gut microbiome–derived histidine metabolites.
Conclusion
ML models identified metabolomic signatures based on CKD cause. Using ML techniques in conjunction with traditional biostatistics, we demonstrated that sphingomyelin-ceramide and plasmalogen dysmetabolism are associated with FSGS and that gut microbiome–derived histidine metabolites are associated with OU.
Metabolomics is a developing field of research within nephrology, with adult studies having already discovered new mechanistic insights into GFR estimation, CKD-associated mortality and morbidity, and CKD cause.1–3 There has been a dearth of metabolomics studies in pediatric CKD.4,5
The Chronic Kidney Disease in Children (CKiD) study is the largest North American multicenter prospective cohort of pediatric CKD.6 Significant novel insights have been gleaned from CKiD that serve to guide clinical care of patients and provide new investigative direction.7 CKD in children is usually the consequence of congenital anomalies of the kidney or urinary tract or primary glomerular disorders, and the underlying disorders have substantial heterogeneity. The untargeted plasma metabolomics data from this cohort represent an exciting opportunity to better understand pathophysiologic biochemical differences on the basis of underlying CKD cause. Identification of such differences may improve our ability to predict disease progression and associated morbidity and may identify potential therapeutic targets. Studying metabolomics and CKD cause in children may be advantageous compared with adults. There may be fewer challenges in interpreting the metabolite associations with primary pathologies as opposed to CKD secondary to pathologies such as hypertension and diabetes.8
High-dimensional metabolomics data pose analytic challenges when implementing established statistical approaches. Metabolites are nonindependent and highly multicollinear, which can be difficult to control for in traditional statistical approaches. Machine learning (ML) has rapidly innovated over the past decade and is increasingly being applied to biomedical sciences.9,10 ML algorithms present opportunities to recognize important metabolites through their handling of high-dimensional, multicollinear, and nonindependent metabolomics data.
The overarching goal of this investigation was to identify both individual metabolites and metabolic pathways associated with pediatric CKD causes in order to gain insights into underlying biochemical pathophysiology while generating hypotheses for continued investigation. ML modeling was used as a complement to traditional biostatistical approaches to identify individual metabolites associated with pediatric CKD cause. The goal in utilizing ML was not to build a predictive or diagnostic tool. Clinician diagnosis of pediatric CKD cause is well established and robust. Rather, we leveraged ML handling of high-dimensional, multicollinear data in identifying important metabolite signals. We examined the four largest diagnosis groups within CKiD: FSGS, obstructive uropathy (OU), aplasia/dysplasia/hypoplasia (A/D/H), and reflux nephropathy (RN).
Methods
CKiD Study
CKiD is an ongoing, multicenter, prospective cohort study of children with CKD. Participants included in the present investigation were enrolled in CKiD between January of 2005 and December of 2014 in two recruitment waves at 54 participating medical centers in the United States and Canada. Children were enrolled if they were between the ages of 6 months and 16 years and had an eGFR of 30–90 ml/min per 1.73 m2 by the Schwartz formula. Exclusion criteria included: history of solid organ or bone marrow transplantation, dialysis within 3 years, malignancy or HIV within 12 months, structural cardiac disease, and genetic syndromes involving the central nervous system. The complete details of study design, methods, and sample characteristics have been published previously.6 Blood samples were collected annually to measure renal function and other biomarkers to observe the natural history of pediatric CKD. Written, informed consent was obtained from all parents or legal guardians, along with assent, when appropriate, from the enrolled children. The CKiD study was approved by the institutional review board of each participating institution.
Covariate data were obtained from the first annual CKiD visit or, when missing, at the 6-month visit after enrollment. Covariate information included demographic characteristics (e.g., age, sex, race), urine protein-creatinine ratio (PCR), body mass index Z-score (BMI-Z), CKD duration, hypertension, eGFR, and angiotensin-converting-enzyme inhibitor or aldosterone receptor blocker (ACEi/ARB) use. Hypertension was defined as systolic or diastolic blood pressure ≥95th percentile for age, sex, and height, or any self-reported antihypertensive medication use. eGFR was calculated from serum creatinine, cystatin C, and blood urea nitrogen concentrations according to the equation developed in the CKiD study.11
Metabolomic Profiling
Metabolite profiling was performed on plasma samples collected at the 6-month visit after enrollment. These samples were stored at −80°C until being processed and assayed with an untargeted ultra-high-performance liquid chromatography–tandem mass spectrometry–based (UPLC-MS/MS) metabolomics quantification protocol by Metabolon, Inc. (Durham, NC).12–14 Samples were excluded if >50% metabolites were missing. Metabolites were excluded if ≥80% of them were missing across all samples (n=45). Missing values were imputed with the minimum measured value of each specific metabolite, and values were normalized to the run-day medians to correct for variations across runs or instruments. Metabolite levels were capped at 5 SD for each metabolite. After logarithmic (base 2) transformation, we excluded metabolites with variance <0.01 or missing variance.5,15
Analytic Approach
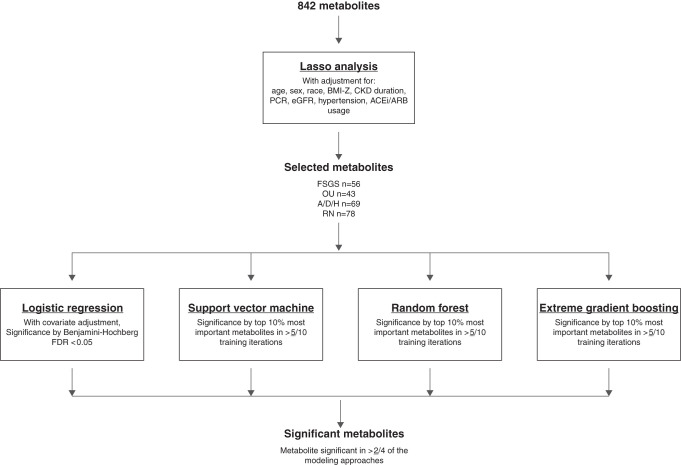
Figure 1 details the analytic schematic we followed to analyze 842 named, nonmissing metabolites. Details for each ML algorithm and performance metrics are expanded upon in the Supplemental Material. All ML models were trained from the R caret package with repeated k-fold crossvalidation for each of the four cause groups examined. In repeated k-fold crossvalidation, the data are randomly divided into k subsets, trained on k−1 subsets, and validated on a holdout subset. Each subset is used as the holdout once, the process is repeated n times, and results are averaged.
Figure 1.
The analytic flow plan for identifying metabolites associated with CKD etiology. The flow of the analytic approach used to identify individual metabolites associated with each CKD cause. Lasso generated panels of metabolites for subsequent analysis while accounting for metabolite and clinical covariate multicollinearity. Second-pass ML classifiers additionally accounted for dimensionality and multicollinearity among the Lasso-selected metabolites.
We first performed a feature-selection analysis with Lasso-penalized logistic regression (LR) with adjustment for: age, sex, race, BMI-Z, CKD duration, log-transformed eGFR, log-transformed urine PCR, hypertension, and ACEi/ARB use. We elected to use Lasso for its ability to account for metabolites’ multicollinearity with clinical covariates and filter down the number of metabolites for further analysis.16
We then applied four methods to further evaluate significance of the Lasso-selected metabolites, including LR and three ML algorithms: support vector machine classification (SVM), random forest (RF), and extreme gradient boosting (XGB). The ML models were selected for their algorithmic differences in accounting for multicollinearity in high-dimensional data. Common signals detected across different mathematic approaches more likely represent the strongest and true patterns in the data. SVM is a widely implemented margin-based classifier.17,18 RF may be the most widely implemented tree-based model in biomedical research, but may have lower generalizability.19,20 XGB is a relatively new tree-based model incorporating boosting that has been adapted for classification tasks and shown to have improved performance compared with older models such as RF.21,22 ML models were trained on 80% subsets with tenfold crossvalidation repeated three times. The 20% holdout subsets were used for validation of the final model. Each training iteration was repeated ten times per model per cause. Hyperparameters were manually set to favor a more generalizable model and limit overfitting. Models were assessed with four evaluation metrics to account for how the skewed distribution of CKD causes may overestimate model performance: receiver-operator area-under-the-curve (ROC-AUC), precision-recall area-under-the-curve (PR-AUC), F1 score, and Matthews correlation coefficient (MCC).23,24 Significant features were designated as metabolites included in the top 10% most-weighted features in ≥5/10 training iterations.
We performed LR to characterize the magnitude and direction of individual Lasso-selected metabolites’ associations with CKD cause, adjusting for the same covariates as Lasso. We reported statistical significance determined by Benjamini–Hochberg false discovery rate (BH-FDR) <0.05 on the basis of the number of Lasso-selected metabolites for each cause. Overall important features were selected on the basis of being significant in at least two of the four modeling approaches (LR, SVM, RF, or XGB).
We also performed pathway enrichment analysis with hypergeometric distribution tests to identify metabolite subpathways associated with CKD causes.25 We generated panels of metabolites associated with each CKD cause through an adjusted LR at P<0.05. We did not utilize Lasso feature-selection for this analysis given a penalized approach may eliminate subpathway enrichment signals. Significant subpathway associations with CKD cause were reported on the basis of BH-FDR<0.05, derived from the number of subpathways tested.
Results
Participant Demographic and Clinical Characteristics
Table 1 shows the clinical characteristics of the 702 CKiD participants according to CKD cause. The FSGS cohort had more ACEi/ARB use, older age, higher BMI-Z, shorter CKD duration, and greater proteinuria. The OU cohort was predominantly male and younger and had less ACEi/ARB usage, lower baseline eGFR, and longer CKD duration. The A/D/H cohort had a greater proportion of female participants, had less ACEi/ARB use, was younger, and had lower BMI-Z, lower GFR, and longer CKD duration. The RN cohort had longer CKD duration and less proteinuria.
Table 1.
Participant characteristics on the basis of CKD cause
| Characteristic | Total Cohort | FSGS | OU | A/D/H | RN |
|---|---|---|---|---|---|
| Number | 702 | 63 | 122 | 109 | 86 |
| Categoric | |||||
| Sex, male | 432 | 35 | 103a | 57a | 49 |
| Hypertension | 95 | 12 | 12 | 17 | 12 |
| ACEi/ARB usage | 396 | 54a | 46a | 36a | 47 |
| Numeric | |||||
| Age, yr | 11.3 (4.3) | 13.6 (3.0)b | 10.1 (4.2)b | 9.8 (4.7)b | 11.6 (4.1) |
| BMI-Z | 0.5 (1.1) | 1.0 (1.3)b | 0.4 (1.2) | 0.3 (1.0)b | 0.4 (1.0) |
| GFR, ml/min per 1.73 m2 | 51.1 (1.4) | 55.1 (1.5) | 47.3 (1.4)b | 47.4 (1.5)b | 50.2 (1.4) |
| CKD duration, yr | 8.5 (4.9) | 5.4 (3.5)b | 10.1 (4.2)b | 9.8 (4.7)b | 11.6 (4.1)b |
| Urine PCR | 0.4 (3.9) | 0.8 (4.6)b | 0.4 (2.9) | 0.3 (3.6) | 0.2 (3.2)b |
Participant characteristics are reported for the total CKiD cohort and on the basis of CKD cause. Significance was on the basis of each major cause group versus all other causes of CKD.
There was a significant difference in categoric variable on the basis of chi-squared test, P<0.05.
There was a significant difference in numeric variable on the basis of Wilcoxon rank-sum test, P<0.05.
Lasso Feature Selection
Lasso generated candidate metabolite subsets of n=56, n=43, n=69, and n=78 and crossvalidated λ 0.01, 0.02, 0.01, and 0.01 for the FSGS, OU, A/D/H, and RN subgroups, respectively (metabolite details are reported in Supplemental Table 1). Lasso performance metrics were: FSGS (ROC-AUC=0.87, PR-AUC=0.47, F1=0.45, MCC=0.42), OU (ROC-AUC=0.82, PR-AUC=0.46, F1=0.51, MCC=0.41), A/D/H (ROC-AUC=0.75, PR-AUC=0.33, F1=0.38, MCC=0.29), and RN (ROC-AUC=0.48, PR-AUC=0.11, F1=0.22, MCC=0.04) (Supplemental Table 2).
ML Training and Performance
ML models were successfully trained to outperform no-skill modeling on the basis of four metrics evaluated in the 20% holdout subsets (Table 2). For all three ML algorithms in each of the four cause groups (total of 12 models), every model performance metric exceeded no-skill modeling. We performed supplemental hyperparameter tuning analyses to determine if tuned hyperparameters significantly affected model performance and feature importance compared with our selected parameters. Results are reported in Supplemental Tables 3 and 4. Model performance and feature importance did not differ significantly with tuned hyperparameters.
Table 2.
ML model performance exceeds no-skill prediction in 20% holdout subsets on the basis of four evaluation metrics
| Cause | Model | ML Model Performance Metric | |||
|---|---|---|---|---|---|
| ROC-AUC | PR-AUC | F1 Score | MCC | ||
| None | No-skill | 0.5 | Prevalence | 0 | 0 |
| FSGS | SVM | 0.94 (0.93, 0.95) | 0.60 (0.57, 0.63) | 0.59 (0.57, 0.61) | 0.59 (0.57, 0.61) |
| RF | 0.89 (0.88, 0.90) | 0.50 (0.48, 0.51) | 0.47 (0.46, 0.48) | 0.45 (0.44, 0.46) | |
| XGB | 0.91 (0.90, 0.91) | 0.54 (0.53, 0.56) | 0.48 (0.47, 0.49) | 0.47 (0.46, 0.48) | |
| OU | SVM | 0.84 (0.84, 0.85) | 0.52 (0.51, 0.53) | 0.54 (0.53, 0.54) | 0.44 (0.44, 0.45) |
| RF | 0.73 (0.73, 0.74) | 0.39 (0.38, 0.40) | 0.42 (0.41, 0.42) | 0.28 (0.27, 0.29) | |
| XGB | 0.79 (0.79, 0.80) | 0.45 (0.43, 0.46) | 0.48 (0.47, 0.48) | 0.37 (0.37, 0.38) | |
| A/D/H | SVM | 0.84 (0.83, 0.85) | 0.51 (0.50, 0.52) | 0.53 (0.51, 0.54) | 0.44 (0.42, 0.45) |
| RF | 0.68 (0.68, 0.69) | 0.30 (0.29, 0.31) | 0.38 (0.38, 0.39) | 0.24 (0.23, 0.25) | |
| XGB | 0.75 (0.75, 0.76) | 0.38 (0.37, 0.39) | 0.43 (0.42, 0.44) | 0.32 (0.31, 0.33) | |
| RN | SVM | 0.80 (0.79, 0.81) | 0.37 (0.36, 0.38) | 0.41 (0.40, 0.42) | 0.34 (0.33, 0.35) |
| RF | 0.66 (0.65, 0.66) | 0.19 (0.19, 0.20) | 0.31 (0.30, 0.31) | 0.20 (0.19, 0.21) | |
| XGB | 0.73 (0.72, 0.73) | 0.25 (0.25, 0.26) | 0.33 (0.33, 0.34) | 0.25 (0.25, 0.26) | |
All 12 iterations of our ML models (three algorithms for four cause subgroups) exceeded no-skill prediction on the basis of four different evaluation metrics in 20% holdout subsets. ROC-AUC is the most traditional and familiar of the four metrics, but may overestimate model performance in samples with low case prevalence rate, as in CKiD. PR-AUC accounts for the skewed case distribution. However, PR-AUC magnitude does not give additional insight into model performance beyond if it surpassed no-skill prediction or not. The F1 score is a harmonic mean of the precision and recall, performing similarly to the PR-AUC. The F1 score magnitude does reflect model performance, with 0 being equivalent to no-skill and 1 being perfect prediction. MCC performs similarly to the F1, but additionally includes true negatives in its calculation, which gives directionality to this metric; perfect negative prediction=−1, perfect positive prediction=0.
Metabolites and Metabolite Subpathways Associated with CKD Cause
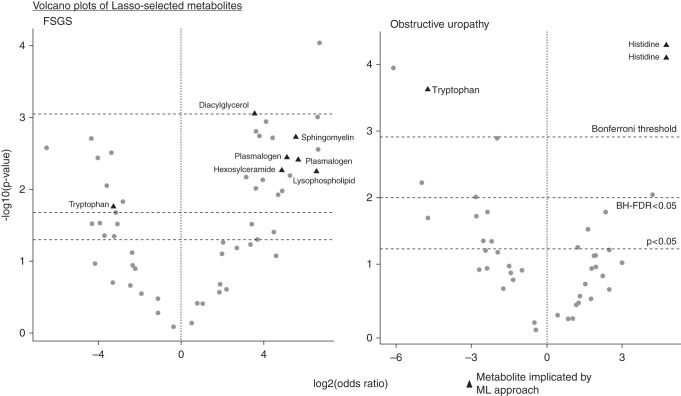
Individual metabolites associated with each CKD cause are reported in Table 3. All metabolites utilized for pathway enrichment analyses are reported in Supplemental Table 5. Metabolite subpathways associated with each CKD cause are reported in Table 4. FSGS was associated with individual lipid metabolites (plasmalogens, hexosylceramides, and sphingomyelins), and FSGS was also associated with the plasmalogen, hexosylceramide, and ceramide subpathways. OU was associated with individual histidine metabolites and the histidine subpathway. A/D/H was associated with individual ceramide and urea cycle metabolites and their subpathways. RN was associated with individual tryptophan, benzoate, and medium chain fatty acid metabolites and their subpathways. Figure 2 visualizes how ML models additionally stratify important metabolites after accounting for potential multicollinearity.
Table 3.
Implicated metabolites on the basis of multiple modeling approaches
| Metabolite | Pathway | Subpathway | Approaches Implicated | Versus CKD Cause | |
|---|---|---|---|---|---|
| Odds Ratio | P | ||||
| FSGS (n=63, 56 metabolites) | |||||
| Palmitoyl-arachidonoyl- glycerol (16:0/20:4) | Lipid | Diacylglycerol | LR, XGB | 11.74 | 0.00089 |
| Sphingomyelin (d18:1/18:1, d18:2/18:0) | Lipid | Sphingomyelins | LR, SVM, RF, XGB | 46.87 | 0.0019 |
| 1-(1-Enyl-palmitoyl)-2- arachidonoyl-GPC (P-16:0/ 20:4) | Lipid | Plasmalogen | LR, SVM, RF, XGB | 34.94 | 0.0036 |
| 1-(1-Enyl-palmitoyl)-2- palmitoyl-GPC (P-16:0/16:0) | Lipid | Plasmalogen | LR, SVM, RF | 50.76 | 0.0039 |
| Glycosyl ceramide (d18:2/ 24:1, d18:1/24:2) | Lipid | Hexosylceramides | LR, SVM | 29.24 | 0.0054 |
| 1-Arachidonoyl-GPI (20:4) | Lipid | Lysophospholipid | LR, RF, XGB | 93.58 | 0.0057 |
| 6-Bromotryptophan | Amino acid | Tryptophan | LR, RF | 0.10 | 0.017 |
| OU (n=122, 43 metabolites) | |||||
| Transurocanate | Amino acid | Histidine | LR, SVM, RF, XGB | 27.41 | 5.30×10−05 |
| Imidazole propionate | Amino acid | Histidine | LR, SVM, RF, XGB | 26.52 | 8.00×10−05 |
| N-acetylkynurenine | Amino acid | Tryptophan | LR, SVM, XGB | 0.037 | 0.00024 |
| A/D/H (n=109, 69 metabolites) | |||||
| Ceramide (d18:1/20:0, d16:1/ 22:0, d20:1/18:0) | Lipid | Ceramides | LR, SVM, RF, XGB | 95.17 | 3.46×10−05 |
| N-δ-acetylornithine | Amino acid | Urea cycle; arginine and proline | LR, SVM, RF, XGB | 8.40 | 0.0082 |
| Sphingomyelin (d18:2/24:2) | Lipid | Sphingomyelins | LR, SVM, RF, XGB | 0.11 | 0.0090 |
| Citramalate | Amino acid | Glutamate metabolism | 4.84 | 0.0091 | |
| Cysteine sulfinic acid | Amino acid | Methionine, cysteine, SAM, and taurine | RF, XGB | 0.16 | 0.061 |
| RN (n=86, 78 metabolites) | |||||
| Dimethylglycine | Amino acid | Glycine, serine, and threonine | LR, SVM | 62.66 | 5.29×10−05 |
| Indolepropionate | Amino acid | Tryptophan | LR, SVM, RF, XGB | 179.75 | 0.00017 |
| Heptanoate (7:0) | Lipid | Medium chain fatty acid | LR, SVM | 24.08 | 0.00091 |
| 4-Vinylphenol sulfate | Xenobiotics | Benzoate | LR, SVM, RF, XGB | 12.05 | 0.0024 |
Significant metabolites are reported for each CKD cause. Fully adjusted odds ratios are reported to indicated association direction. Notably, FSGS is associated with multiple lipid metabolites, involving the sphingomyelin and plasmalogen subpathways. OU is uniquely associated with histidine metabolites. A/D/H is associated with both amino acid and lipid metabolites, notably sharing a sphingomyelin metabolite signal with FSGS.
Table 4.
Metabolite pathway enrichment analysis
| Subpathway | N Significant | N Total | P value |
|---|---|---|---|
| FSGS, 122 metabolites, 24 subpathways | |||
| Endocannabinoid | 7 | 10 | 5.52×10−06 |
| Hexosylceramides | 6 | 8 | 8.12×10−06 |
| Ceramides | 6 | 11 | 0.000228 |
| Plasmalogen | 7 | 15 | 4.14×10−04 |
| Vitamin A | 3 | 7 | 0.01 |
| Tryptophan | 7 | 23 | 0.011 |
| OU, 104 metabolites, 23 subpathways | |||
| Phosphatidylserine | 2 | 2 | 0 |
| Vitamin B6 | 2 | 2 | 0 |
| γ-Glutamyl amino acid | 12 | 18 | 3.91×10−09 |
| Leucine, isoleucine, valine | 11 | 31 | 1.27×10−04 |
| Pentose | 4 | 7 | 4.50×10−04 |
| Glutamate | 5 | 14 | 0.004 |
| Fructose, mannose, and galactose | 2 | 4 | 0.0067 |
| Histidine | 5 | 16 | 0.0087 |
| Pyrimidine, cytidines | 2 | 5 | 0.015 |
| TCA cycle | 3 | 9 | 0.017 |
| Tocopherol | 2 | 12 | 0.018 |
| A/D/H, 69 metabolites, 15 subpathways | |||
| Fructose, mannose, and galactose | 3 | 4 | 4.16×10−05 |
| Glycolysis, gluconeogenesis, and pyruvate | 2 | 5 | 4.68×10−05 |
| Diacylglycerol | 8 | 25 | 7.04×10−05 |
| Ceramides | 5 | 11 | 8.16×10−05 |
| Androgenic steroids | 7 | 24 | 3.45×10−04 |
| Fatty acid, acyl glycine | 2 | 5 | 4.68×10−04 |
| Lactosylceramides | 2 | 3 | 5.29×10−04 |
| TCA cycle | 3 | 9 | 0.0038 |
| Urea cycle, arginine, and proline | 4 | 22 | 0.028 |
| RN, 57 metabolites, 17 subpathways | |||
| Vitamin A | 3 | 7 | 5.69×10−04 |
| γ-Glutamyl amino acid | 4 | 18 | 5.20×10−03 |
| Benzoate | 2 | 7 | 8.50×10−03 |
| Pentose | 2 | 7 | 0.0085 |
| Urea cycle, arginine, and proline | 4 | 22 | 0.012 |
| Tryptophan | 4 | 23 | 0.016 |
| Medium chain fatty acid | 2 | 9 | 0.018 |
Candidate metabolite panels associated with each CKD cause were generated by selecting among all 842 metabolites in fully adjusted LR analyses (P<0.05). Pathway enrichment analysis was performed using the hypergeometric distribution test. We report number of metabolites in each panel associated with CKD cause and the number of subpathways in which >1 metabolite was included in the candidate panel. We report significantly enriched subpathways on the basis of BH-FDR<0.05 determined by the number of subpathways tested.
Figure 2.
Volcano plots demonstrate ML stratification of important metabolite signals. These plots visualize the stratification of Lasso-selected metabolites for FSGS and OU. Implication by Benjamini–Hochberg is the least restrictive. Implication by ML modeling is the most restrictive. The signals detected by ML would meet both Benjamini–Hochberg and Bonferroni thresholds.
Sensitivity Analyses
To determine if the metabolite associations observed were related to a specific cause rather than overarching differences of glomerular versus nonglomerular disease, fully adjusted LR was applied examining FSGS within the glomerular subcohort (n=214) and OU, A/D/H, and RN within the nonglomerular subcohort (n=488). In total, 18 of 19 metabolites remained associated with CKD cause (P<0.05). Only cysteine sulfinic acid was no longer significantly associated with A/D/H (P=0.06).
Feature Selection Comparison
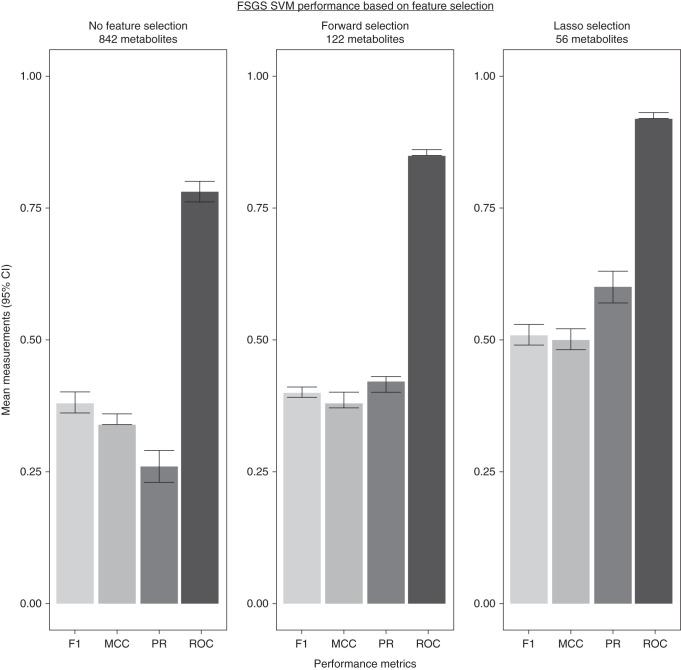
We assessed if Lasso feature selection outperformed no feature selection or forward feature selection. We assessed if there were differences in the signals detected. We used SVM for FSGS for this analysis. Figure 3 shows Lasso feature selection outperformed both no feature selection and forward feature selection on the basis of all four evaluation metrics in 20% holdout subsets. There were no significant differences in metabolite subpathway signals detected. Sphingomyelin (d18:1/18:1, d18:2/18:0), 1-(1-enyl-palmitoyl)-2-palmitoyl-GPC (P-16:0/16:0), and 1-(1-enyl-palmitoyl)-2-arachidonoyl-GPC (P-16:0/20:4) were still associated with FSGS in both no-selection and forward-selection analyses.
Figure 3.
Lasso feature selection improved ML model performance. We performed SVM for FSGS with no feature selection (842 metabolites) and with a forward feature selection (122 metabolites on the basis of LR P<0.05). All three iterations demonstrated better performance than no-skill selection in 20% holdout validation subsets. The SVM with Lasso feature selection outperformed both no-selection and forward-selection models. There were not significant differences in metabolite subpathway signals detected. 95% CI, 95% confidence interval.
Discussion
We identified individual metabolites and metabolite subpathways associated with pediatric CKD cause in the CKiD cohort. Because larger and more complex data are generated by advancing transomic approaches, applying novel tools is necessary to gain more interpretable clinical insights. ML does not replace clinical diagnosis, but rather augments existing analytic methods by aiding in pattern discernment through its processing of high-dimensional, multicollinear data. By identifying novel metabolomic signatures, we provide new directions for future studies of CKD biochemical pathophysiology and potential therapeutic targets.
FSGS and Lipid Associations
FSGS was associated with individual sphingomyelin and plasmalogen metabolites, and the plasmalogen, ceramide, and hexosylceramide subpathways. In a sensitivity analysis within the glomerular CKD subcohort, these lipid metabolites remained associated with FSGS. Our findings corroborate lipid dysmetabolism in FSGS pathophysiology.26–29 Sphingomyelin abnormalities have been previously associated with FSGS and nephrotic syndrome. In one prior study, patients with FSGS who underwent kidney transplant and developed recurrent FSGS had decreases in serum sphingomyelin phosphodiesterase acid–like 3b (SMPDL-3b) protein and acid sphingomyelinase activity.30 In another study examining patients with intractable nephrotic syndrome undergoing rituximab therapy, glomerular SMPDL-3b immunoreactivity and urinary SMPDL-3b excretion increased after rituximab therapy in patients who achieved clinical remission.31 Sphingomyelin metabolites have also been shown to induce neurotoxicity through free radical damage in neurodegenerative disorders such as Alzheimer’s and HIV dementia.32,33 A similar nephrotoxic relationship may exist in the kidney. Ceramides are intermediate products of sphingomyelin metabolism involved in biologic signaling processes, including cellular proliferation, apoptosis, and atherogenesis.34–37 In vitro studies have shown that alterations in sphingomyelin metabolism are associated with ceramide differences and podocyte injury.38,39 We contribute to this existing literature of sphingomyelin dysmetabolism in FSGS by showing evidence of dysregulation in the sphingomyelin-ceramide axis through comprehensive untargeted metabolomic profiling in a large pediatric cohort.
FSGS was also associated with individual plasmalogen metabolites and the plasmalogen subpathway. Plasmalogens represent 20% of total cell membrane phospholipid composition. Imbalances in plasmalogen signaling have been associated with inflammatory disease progression.40 Lower plasmalogen levels are associated with cholesterol dysfunction and cardiovascular mortality in adults with CKD.41,42 Existing literature has focused primarily on plasmalogen deficiencies, and studies have examined plasmalogen replacement therapy in animal models of Alzheimer’s disease and childhood peroxisomal disorders.43,44 Children with FSGS in the study cohort had higher levels of plasmalogen metabolites which warrants further investigation.
OU and Histidine Signals
Another notable signal was the association of histidine metabolites and their subpathway with OU. Imidazole proprionate and transurocanate are both gut microbiome–derived histidine metabolites. These metabolites have been studied in type 2 diabetes mellitus and shown to impair response to metformin at a cellular level.45–48 There is increasing evidence that gut microbiome alterations associated with CKD contribute to comorbidity and disease progression.49–51 Differences in microbiome-derived metabolite profiles on the basis of CKD cause, and how they relate to CKD progression and comorbidity, merit future investigation. Our data suggest that OU is associated with altered gut histidine metabolism. One prior study showed adult patients undergoing surgical correction for OU had increased postoperative urinary histidine excretion, but it was not correlated with renal function recovery.52
A/D/H
The A/D/H and RN metabolomic signals are more difficult to interpret, with both implicating metabolites from multiple subpathways. For A/D/H, this may reflect greater heterogeneity in pathophysiology compared with FSGS and OU. Notably, A/D/H associated with both individual sphingomyelin and ceramide metabolites. A/D/H also associated with the diacylglycerol, lactosylceramide, and ceramide subpathways, all being downstream metabolites of sphingomyelin metabolism. This suggests that dysregulation in the sphingomyelin-ceramide axis may not be unique to FSGS. Another interesting A/D/H association was with N-δ-acetylornithine. N-acetylated peptides have been associated with genomic differences within CKD subpopulations.53,54 N-δ-acetylornthine has also been associated with gut microbiome differences.55,56
RN
RN was associated with an individual tryptophan metabolite and the subpathway, which have been associated with adult CKD progression.57 Indoleproprionate is another gut microbiome–derived metabolite.58 It is interesting to note that each of the major nonglomerular pediatric CKD causes was associated with metabolites linked to different species of gut microbiota: Bacteroides (imidazole proprionate and transurocanate), Lactobacilli (N-δ-acetylornithine), and Clostridiums (indole-tryptophans).48,56,58 This raises an important question for future studies of whether CKD-associated microbiome-metabolomic alterations differ on the basis of primary kidney disease cause.
ML and Study Limitations
An important limitation of our study was the lack of access to a comparable cohort of pediatric patients with CKD with untargeted metabolomic profiling to validate our findings. Recognizing this limitation, we sought to balance reporting the strongest metabolite associations while not missing too many potentially relevant signals. Correction for multiple comparisons is flawed in metabolomics in that metabolites are nonindependent and highly multicollinear.59 However, not accounting for multiple testing can result in reporting false positives. We utilized Lasso feature selection to identify metabolites associated with CKD cause while accounting for collinearity with clinical features and other metabolites. This also serves to make the significance threshold less restrictive in subsequent LR analyses. ML algorithms aided us in stratifying important metabolite signals when examining all Lasso-selected metabolites as a composite panel. Creating 20% holdout validation subsets, repeated k-fold crossvalidation training, repeating training iterations ten times, and selecting metabolites repeatedly identified through multiple algorithms are additional means by which we assessed the strength and stability of metabolite signals.
We recognize our implementation of Lasso feature selection and ML is geared toward identifying the strongest associations of individual metabolites with CKD cause and is likely overly restrictive. These restrictive analyses are complemented by pathway enrichment analyses, leveraging metabolites’ multicollinearity to recognize associated subpathways rather than individual metabolites.
Our study data are cross-sectional and observational, limiting causal inference. The CKiD diagnosis subgroups likely still comprise some pathomechanistic heterogeneity, which may limit metabolite signal detection. However, confounding by comorbid conditions is substantially mitigated compared with analyses of underlying adult CKD cause.
Conclusions
We confirmed that sphingomyelin differences exist in pediatric patients with FSGS. We further showed FSGS is associated with the ceramide subpathway, corroborating the hypothesis that there is sphingomyelin-ceramide dysmetabolism associated with this pathophysiology. Notably, we showed that pediatric FSGS was associated with the plasmalogen subpathway and metabolites. Both the sphingomyelin-ceramide axis and plasmalogens merit further study, having been associated with other pathologies and potential therapeutic targets. We also found associations of gut microbiome–derived metabolites with the nonglomerular CKD causes, OU with histidine metabolites, A/D/H with N-δ-acetylornithine, and RN with indoleproprionate. These associations have not been well characterized previously.
In summary, we identified metabolomic signatures associated with pediatric CKD cause through untargeted metabolomics quantification. ML served as a novel tool to account for nonindependence of metabolites and multicollinearity in recognizing important associations. The CKiD sample size, comprehensive clinical phenotyping, and untargeted plasma metabolomic profiling are strengths of this study. Metabolomics and the application of novel analytic approaches provide interesting directions for gaining new insights into biochemical pathophysiology and hypothesis generation for future studies.
Disclosures
A.G. Abraham reports Consultancy Agreements: Implementation Group, Inc.; Honoraria: National Institutes of Health (NIH) for service on a data monitoring board; Scientific Advisor or Membership: Rare Kidney Stone Disease Study data monitoring board, Associate Editor of the American Journal of Epidemiology, and Population Health Metrics Associate Editor. J. Coresh reports Consultancy Agreements: Healthy.io, Kaleido, Ultragenyx; Ownership Interest: Healthy.io; Research Funding: NIH, National Kidney Foundation (NKF, which receives industry support); and Scientific Advisor or Membership: Healthy.io and the NKF. M. Denburg reports Consultancy Agreements: Trisalus Life Sciences (spouse); Ownership Interest: In-Bore LLC (spouse), Precision Guided Interventions LLC (spouse); Research Funding: Mallinckrodt; Scientific Advisor or Membership: NKF Delaware Valley Medical Advisory Board, Trisalus Life Sciences Scientific Advisory Board (spouse); and Other Interests/Relationships: American Society of Pediatric Nephrology Research and Program Committees, NKF Pediatric Education Planning Committee. H. Feldman reports Consultancy Agreements: DLA Piper, LLP, InMed, Inc., Kyowa Hakko Kirin Co., Ltd (ongoing), NKF (ongoing); Honoraria: Rogosin Institute (invited speaker); Scientific Advisor or Membership: Steering Committee, Chronic Renal Insufficiency Cohort Study, Editor-in-Chief; NKF (Member of Advisory Board); American Journal of Kidney Disease, Editor-in-Chief. P. Kimmel reports Other Interests/Relationships: Co-Editor, Chronic Renal Disease, Academic Press; Co-Editor, Psychosocial Aspects of Chronic Kidney Disease, Academic Press, and Royalties. V. Ramachandran reports Consultancy Agreements: National Institute of Diabetes and Digestive and Kidney Diseases. C. Rebholz reports Scientific Advisor or Membership: Editorial Board member for Diabetes Care. B. Warady reports Consultancy Agreements: Akebia, Amgen, Bayer, Glaxosmithkline, Reata, Relypsa, UpToDate; Research Funding: Baxter Healthcare; Honoraria: Akebia, GlaxoSmithKline, Reata, Relypsa, UpToDate; and Scientific Advisor or Membership: North American Pediatric Renal Trials and Collaborative Studies, NKF, and NTDS Board of Directors.
Funding
This work was supported by Foundation for the National Institutes of Health grant U01DK106982 (CKD Biomarkers Consortium) and National Institute of Diabetes and Digestive and Kidney Diseases grant P50DK114786 (Children's Hospital of Philadelphia Pediatric Center of Excellence in Nephrology).
Supplementary Material
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2021040538/-/DCSupplemental.
Supplemental Methods. Expanded methodologic explanation of machine learning algorithms and metrics.
Supplemental Table 1. All LASSO-selected metabolites on the basis of CKD cause.
Supplemental Table 2. FSGS hyperparameter tuning analysis.
Supplemental Table 3. Obstructive uropathy hyperparameter tuning analysis.
Supplemental Table 4. All metabolites included for pathway enrichment analysis.
Supplemental Table 5. Feature importance rankings in different machine learning approaches.
Supplemental Material. Annotated machine learning sample code.
References
- 1.Hu JR, Coresh J, Inker LA, Levey AS, Zheng Z, Rebholz CM, et al. : Serum metabolites are associated with all-cause mortality in chronic kidney disease. Kidney Int 94: 381–389, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coresh J, Inker LA, Sang Y, Chen J, Shafi T, Post WS, et al. : Metabolomic profiling to improve glomerular filtration rate estimation: a proof-of-concept study. Nephrol Dial Transplant 34: 825–833, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grams ME, Tin A, Rebholz CM, Shafi T, Köttgen A, Perrone RD, et al. : Metabolomic alterations associated with cause of CKD. Clin J Am Soc Nephrol 12: 1787–1794, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanna MH, Brophy PD: Metabolomics in pediatric nephrology: emerging concepts. Pediatr Nephrol 30: 881–887, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denburg MR, Xu Y, Abraham AG, Coresh J, Chen J, Grams ME, et al. ; CKD Biomarkers Consortium : Metabolite biomarkers of CKD progression in children. Clin J Am Soc Nephrol 16: 1178–1189, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furth SL, Cole SR, Moxey-Mims M, Kaskel F, Mak R, Schwartz G, et al. : Design and methods of the Chronic Kidney Disease in Children (CKiD) prospective cohort study. Clin J Am Soc Nephrol 1: 1006–1015, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong CJ, Moxey-Mims M, Jerry-Fluker J, Warady BA, Furth SL: CKiD (CKD in Children) prospective cohort study: a review of current findings. Am J Kidney Dis 60: 1002–1011, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harambat J, van Stralen KJ, Kim JJ, Tizard EJ: Epidemiology of chronic kidney disease in children. Pediatr Nephrol 27: 363–373, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodacre R, Broadhurst D, Silde AK, Kristal BS, Baker JD, Beger R, et al. : Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 3: 231–241, 2007 [Google Scholar]
- 10.Holzinger A, Jurisica I: Knowledge Discovery and Data Mining in Biomedical Informatics: The Future Is in Integrative, Interactive Machine Learning Solutions, Berlin, Heidelberg, Springer, 2014 [Google Scholar]
- 11.Schwartz GJ, Muñoz A, Schneider MF, Mak RH, Kaskel F, Warady BA, et al. : New equations to estimate GFR in children with CKD. J Am Soc Nephrol 20: 629–637, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans AM, Bridgewater, B., Liu, Q., et al. : High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 4(2):1 2014
- 13.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E: Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81: 6656–6667, 2009 [DOI] [PubMed] [Google Scholar]
- 14.Dehaven CD, Evans AM, Dai H, Lawton KA: Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2: 9, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee EP, Waikar SS, Rebholz CM, Zheng Z, Perichon R, Clish CB, et al. ; CKD Biomarkers Consortium : Variability of two metabolomic platforms in CKD. Clin J Am Soc Nephrol 14: 40–48, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tibshirani R: Regression shrinkage and selection via the lasso. J R Stat Soc B Stat Methodol 58: 267–288, 1996 [Google Scholar]
- 17.Corinna Cortes VNV: Support-vector networks. Mach Learn 20: 273–297, 1995 [Google Scholar]
- 18.Noble WS: What is a support vector machine? Nat Biotechnol 24: 1565–1567, 2006 [DOI] [PubMed] [Google Scholar]
- 19.Biau G, Scornet E: A random forest guided tour. TEST 25: 197–227, 2016 [Google Scholar]
- 20.Qi Y: Random forest for bioinformatics. In: Ensemble Machine Learning, edited by Zhang C, Ma Y, Boston, Springer, 2012, pp 307–323 [Google Scholar]
- 21.Chen T, Guestin C: XGBoost: A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, August 13-17, 2016 [Google Scholar]
- 22.Chen X, Huang L, Xie D, Zhao Q: EGBMMDA: Extreme Gradient Boosting Machine for MiRNA-Disease Association prediction. Cell Death Dis 9: 3, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chicco D, Jurman G: The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21: 6, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis J, Goadrich M: The relationship between precision-recall and ROC curves. Presented at the 23rd International Conference of Machine Learning, Pittsburgh, PA, June 25-29, 2006
- 25.Marco-Ramell A, Palau-Rodriguez M, Alay A, Tulipani S, Urpi-Sarda M, Sanchez-Pla A, et al. : Evaluation and comparison of bioinformatic tools for the enrichment analysis of metabolomics data. BMC Bioinformatics 19: 1, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokota R, Bhunu, B, Toba, H, Intapad, S: Sphingolipids and kidney disease: possible role of preeclampsia and intrauterine growth restriction (IUGR). Kidney360 2(3):534-541, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merscher S, Fornoni A: Podocyte pathology and nephropathy—Sphingolipids in glomerular diseases. Front Endocrinol (Lausanne) 5: 127, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abou Daher A, El Jalkh T, Eid AA, Fornoni A, Marples B, Zeidan YH: Translational aspects of sphingolipid metabolism in renal disorders. Int J Mol Sci 18: 2528, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutledge JC, Ng KF, Aung HH, Wilson DW: Role of triglyceride-rich lipoproteins in diabetic nephropathy. Nat Rev Nephrol 6: 361–370, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Fornoni A, Sageshima J, Wei C, Merscher-Gomez S, Aguillon-Prada R, Jauregui AN, et al. : Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci Transl Med 3: 85ra46, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe S, Tsugawa K, Tsuruga K, Imaizumi T, Tanaka H: Urinary excretion of sphingomyelinase phosphodiesterase acid-like 3b in children with intractable nephrotic syndrome. Pediatr Int (Roma) 59: 1112–1115, 2017 [DOI] [PubMed] [Google Scholar]
- 32.Chen SD, Yin JH, Hwang CS, Tang CM, Yang DI: Anti-apoptotic and anti-oxidative mechanisms of minocycline against sphingomyelinase/ceramide neurotoxicity: implication in Alzheimer’s disease and cerebral ischemia. Free Radic Res 46: 940–950, 2012 [DOI] [PubMed] [Google Scholar]
- 33.Haughey NJ, Cutler RG, Tamara A, McArthur JC, Vargas DL, Pardo CA, et al. : Perturbation of sphingolipid metabolism and ceramide production in HIV-dementia. Ann Neurol 55: 257–267, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Kolesnick R: The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J Clin Invest 110: 3–8, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathias S, Peña LA, Kolesnick RN: Signal transduction of stress via ceramide. Biochem J 335: 465–480, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hannun YA, Luberto C, Argraves KM: Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry 40: 4893–4903, 2001 [DOI] [PubMed] [Google Scholar]
- 37.Augé N, Maupas-Schwalm F, Elbaz M, Thiers JC, Waysbort A, Itohara S, et al. : Role for matrix metalloproteinase-2 in oxidized low-density lipoprotein-induced activation of the sphingomyelin/ceramide pathway and smooth muscle cell proliferation. Circulation 110: 571–578, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Mallela SK, Mitrofanova A, Merscher S, Fornoni A: Regulation of the amount of ceramide-1-phosphate synthesized in differentiated human podocytes. Biochim Biophys Acta Mol Cell Biol Lipids 1864: 158517, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitrofanova A, Drexler Y, Merscher S, Fornoni A: Role of sphingolipid signaling in glomerular diseases: focus on DKD and FSGS. J Cell Signal 1: 56–69, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braverman NE, Moser AB: Functions of plasmalogen lipids in health and disease. Biochim Biophys Acta 1822: 1442–1452, 2012 [DOI] [PubMed] [Google Scholar]
- 41.Maeba R, Kojima KI, Nagura M, Komori A, Nishimukai M, Okazaki T, et al. : Association of cholesterol efflux capacity with plasmalogen levels of high-density lipoprotein: A cross-sectional study in chronic kidney disease patients. Atherosclerosis 270: 102–109, 2018 [DOI] [PubMed] [Google Scholar]
- 42.Stenvinkel P, Diczfalusy U, Lindholm B, Heimbürger O: Phospholipid plasmalogen, a surrogate marker of oxidative stress, is associated with increased cardiovascular mortality in patients on renal replacement therapy. Nephrol Dial Transplant 19: 972–976, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Wood PL, Khan MA, Smith T, Ehrmantraut G, Jin W, Cui W, et al. : In vitro and in vivo plasmalogen replacement evaluations in rhizomelic chrondrodysplasia punctata and Pelizaeus-Merzbacher disease using PPI-1011, an ether lipid plasmalogen precursor. Lipids Health Dis 10: 182, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood PL, Smith T, Lane N, Khan MA, Ehrmantraut G, Goodenowe DB: Oral bioavailability of the ether lipid plasmalogen precursor, PPI-1011, in the rabbit: A new therapeutic strategy for Alzheimer’s disease. Lipids Health Dis 10: 227, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarosiek K, Pappan KL, Gandhi AV, Saxena S, Kang CY, McMahon H, et al. : Conserved metabolic changes in nondiabetic and type 2 diabetic bariatric surgery patients: Global metabolomic pilot study. J Diabetes Res 2016: 3467403, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brosnan ME, Brosnan JT: Histidine metabolism and function. J Nutr 150[Suppl 1]: 2570S–2575S, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koh A, Mannerås-Holm L, Yunn NO, Nilsson PM, Ryu SH, Molinaro A, et al. : Microbial imidazole propionate affects responses to metformin through p38γ-dependent inhibitory AMPK phosphorylation. Cell Metab 32: 643–653.e4, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molinaro A, Bel Lassen P, Henricsson M, Wu H, Adriouch S, Belda E, et al. ; MetaCardis Consortium : Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat Commun 11: 5881, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jovanovich A, Isakova T, Stubbs J: Microbiome and cardiovascular disease in CKD. Clin J Am Soc Nephrol 13: 1598–1604, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wing MR, Patel SS, Ramezani A, Raj DS: Gut microbiome in chronic kidney disease. Exp Physiol 101: 471–477, 2016 [DOI] [PubMed] [Google Scholar]
- 51.Shah NB, Allegretti AS, Nigwekar SU, Kalim S, Zhao S, Lelouvier B, et al. : Blood microbiome profile in CKD: A pilot study. Clin J Am Soc Nephrol 14: 692–701, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong B, Jia J, Hu W, et al. : Application of 1H NMR metabonomics in predicting renal function recoverability after the relief of obstructive uropathy in adult patients. Clin Biochem 46: 346–353, 2013 [DOI] [PubMed] [Google Scholar]
- 53.Luo S, Feofanova EV, Tin A, Tung S, Rhee EP, Coresh J, et al. : Genome-wide association study of serum metabolites in the African American Study of Kidney Disease and Hypertension. Kidney Int 100: 430–439, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo S, Surapaneni A, Zheng Z, Rhee EP, Coresh J, Hung AM, et al. : NAT8 variants, N-acetylated amino acids, and progression of CKD. Clin J Am Soc Nephrol 16: 37–47, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagheri M, Shah RD, Mosley JD, Ferguson JF: A metabolome and microbiome wide association study of healthy eating index points to the mechanisms linking dietary pattern and metabolic status. Eur J Nutr 60: 4413–4427, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Tian X, He B, Hoang TK, Taylor CM, Blanchard E, et al. : Lactobacillus reuteri DSM 17938 feeding of healthy newborn mice regulates immune responses while modulating gut microbiota and boosting beneficial metabolites. Am J Physiol Gastrointest Liver Physiol 317: G824–G838, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee EP, Clish CB, Wenger J, Roy J, Elmariah S, Pierce KA, et al. : Metabolomics of chronic kidney disease progression: A case-control analysis in the Chronic Renal Insufficiency Cohort Study. Am J Nephrol 43: 366–374, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menni C, Hernandez MM, Vital M, Mohney RP, Spector TD, Valdes AM: Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut microbiome diversity. Gut Microbes 10: 688–695, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perneger TV: What’s wrong with Bonferroni adjustments. BMJ 316: 1236–1238, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.