Significance Statement
Volumetric measurements are needed to characterize kidney structural findings on CT images to evaluate and test their potential utility in clinical decision making. Deep learning can enable this task in a scalable and reliable manner. Although automated kidney segmentation has been previously explored, methods for distinguishing cortex from medulla have never been done before. In addition, automated methods are typically evaluated at a single institution, without testing generalizability and robustness across different institutions. The tool developed in this study performs at the level of human readers and could enable large diverse population studies to evaluate how kidney, cortex, and medulla volumes can be used in various clinical settings, and establish normative values at large scale.
Keywords: kidney cortex, kidney medulla, kidney volume, deep learning, segmentation, computed tomography, machine learning collection
Abstract
Background
In kidney transplantation, a contrast CT scan is obtained in the donor candidate to detect subclinical pathology in the kidney. Recent work from the Aging Kidney Anatomy study has characterized kidney, cortex, and medulla volumes using a manual image-processing tool. However, this technique is time consuming and impractical for clinical care, and thus, these measurements are not obtained during donor evaluations. This study proposes a fully automated segmentation approach for measuring kidney, cortex, and medulla volumes.
Methods
A total of 1930 contrast-enhanced CT exams with reference standard manual segmentations from one institution were used to develop the algorithm. A convolutional neural network model was trained (n=1238) and validated (n=306), and then evaluated in a hold-out test set of reference standard segmentations (n=386). After the initial evaluation, the algorithm was further tested on datasets originating from two external sites (n=1226).
Results
The automated model was found to perform on par with manual segmentation, with errors similar to interobserver variability with manual segmentation. Compared with the reference standard, the automated approach achieved a Dice similarity metric of 0.94 (right cortex), 0.90 (right medulla), 0.94 (left cortex), and 0.90 (left medulla) in the test set. Similar performance was observed when the algorithm was applied on the two external datasets.
Conclusions
A fully automated approach for measuring cortex and medullary volumes in CT images of the kidneys has been established. This method may prove useful for a wide range of clinical applications.
In renal imaging, phenotypic features such as kidney volume have been shown to be useful in many clinical situations. These include following progression and effectiveness of treatment interventions for polycystic kidney disease,1–4 assessing candidate kidney donor suitability for renal transplantation,5,6 and detecting structural manifestations of CKD.7,8 Although most studies have focused on total kidney volume, separate assessment of the cortex and medulla volumes may be more useful. A disproportionate loss of cortical volume relative to medulla is a characteristic finding of both aging7 and CKD (i.e., cortical thinning on ultrasound).9,10 When considering a candidate living kidney donor for renal transplantation, a contrast-enhanced computed tomography (CT) scan is obtained to detect any subclinical pathology in the kidney and to identify the number of renal arteries in both kidneys (a kidney with fewer arteries is preferred for donation).
Recent work from the Aging Kidney Anatomy study has evaluated kidney volumes, cortical volumes, medullary volumes, and kidney surface roughness using the image processing tool (ITK-snap).11 Although this technique has a small interobserver variability, it requires significant manual involvement that is time consuming and impractical for clinical care, and thus, these measures are not available during donor evaluations. However, this work has shown that age, kidney function, and CKD risk factors associate differently with cortical volume compared with medullary volume in the kidney.7 The imaging characteristics of low cortex volume and low cortex to medulla volume ratio have been shown to reflect severity of underlying nephrosclerosis (glomerulosclerosis, arteriosclerosis, and interstitial fibrosis/tubular atrophy) on biopsy of donor kidneys.12 Further, a larger cortex is a marker of larger nephron size (larger glomerular volumes, more cortex per glomerulus, and larger tubular cross-sectional areas) on biopsy of donor kidneys.12 There may be prognostic utility in knowing the cortical and medullary volumes of donor kidneys. At a decade after donation, donors with nephron number (calculated from cortical volumes) below their age-specific fifth percentile were 3.4 times more likely to have developed an eGFR <45 ml/min per 1.72 m2, independent of baseline kidney function and CKD risk factors.13 Recipients of living donor kidneys with smaller medullary volume were also at higher risk for graft failure, independent of donor and recipient kidney function and CKD risk factors.14 Ideally, automated analysis tools are needed to characterize these kidney structural findings on CT images to evaluate and test their potential utility in clinical decision making.
Previous renal segmentation efforts in CT imaging have focused on whole kidney15,16 or, in some patients, renal cortex segmentation.17,18 Many of these studies propose deep-learning methods such as convolutional neural networks to identify and segment kidney boundaries from surrounding tissue in structurally complex kidney disease (e.g., polycystic kidney disease),16 in addition to patients with little to no structural abnormalities.15 Along with whole kidney segmentation, there is a strong interest in further segmentation of the major kidney regions. Some studies have shown the performance of different algorithms applied to magnetic resonance (MR) imaging. Cai et al. proposed a semiautomatic method for the segmentation of renal cortex using dynamic MR images from pigs. The method required manual placing of a bounding box on each kidney followed by the subtraction of the noncontrast phase with the parenchymal phase images.19 Will et al. presented a thresholding method using T1- and T2-weighted MR images from healthy volunteers to segment the renal cortex, medulla, and pelvis.20 More recently, Huang et al. described a method to segment the renal cortex, medulla, and pelvis regions on dynamic contrast-enhanced MR images from healthy subjects by automatically detecting seed regions obtained from adaptive thresholding to later build a supervised classifier using the temporal information from the seed regions to finally classify the unseeded voxels.21 In parallel, similar algorithms have been applied to CT imaging. Some studies used a manual thresholding approach to segment the cortex and medulla from transverse CT images obtained during the angiogram phase.7,17,18 Wang et al. applied a model-based approach that achieved an average renal cortex segmentation accuracy (Dice) of 97.5% when comparing the manual method with the automatic model.22 In the machine-learning domain, Jin et al. used a combination of random forests and random ferns to segment contrast-enhanced CT images, and Chen et al. used a three-dimensional (3D) shape-constrained graph-cut method to segment the renal cortex in contrast-enhanced CT images.23
Although there have been prior efforts to automate the segmentation of kidneys, a fully automated method to separately segment each kidney’s cortex and medulla in CT images does not yet exist. Deep-learning approaches within the field of artificial intelligence (AI) are providing computational solutions to a wide range of automated classification and organ delineation tasks, particularly in radiology.24,25 These approaches are even being integrated into clinical systems to aid in diagnosis and enhance patient care.26 This study took advantage of an established large database of reference standard segmentations of each kidney’s cortex and medulla from CT imaging data obtained across multiple scanners and sites. Our primary objective was to develop and evaluate a fully automated approach for segmenting the kidney volumes (cortex and medulla) from abdominal CT scans. Our secondary objective was to determine whether clinical characteristics associated with kidney volumes by the automated method were similar or different from those by the manual (reference) method.
Methods
Study Cohort
This study has been reviewed and approved by our institution’s Institutional Review Board with a waiver of informed consent for the repurposing of existing clinical CT scans for this study. The Aging Kidney Anatomy study has collected extensive data on living kidney donors who underwent a CT scan with intravenous contrast and obtained angiogram phase images (cortex but not medulla become radiopaque). In this study, we used data from predonation scans from 2001 to 2015 to train, validate, and test our fully automated method. All CT exams were acquired on either a four-channel MDCT scanner (Qxi, GE Medical Systems, Milwaukee, WI) or a 64-channel MDCT scanner (Sensation 64, Siemens Medical Solutions) at the Mayo Clinic in Rochester and Arizona, and Cleveland Clinic (Sensation 64, Siemens Medical Solutions). The data from Mayo Clinic in Minnesota were used to train, validate, and test the algorithm. Data from Mayo Clinic in Arizona and Cleveland Clinic were used as separate external test sets to evaluate model generalizability. Clinical characteristics including age, sex, body mass index (BMI), measured GFR by iothalamate clearance, estimated GFR with the creatinine-based Chronic Kidney Disease Epidemiology Collaboration equation, family history of ESKD, hypertension, serum uric acid, and 24-hour urine albumin were obtained from the medical record. Microstructural features of % globally sclerotic glomeruli, glomerular volume, and mean tubular area were obtained from a kidney biopsy at the time of donation as previously described.12
Reference Standard (Manual Segmentation)
Supplemental Figure 1 depicts the manual segmentation process utilized to create the ground truth for training and evaluating the deep-learning algorithm. All of the exams were segmented by an expert technologist using ITK-SNAP (version 2.2). A subregion from the axial plane is manually identified to include both kidneys. Active contour evolution is applied to the subregion for kidney segmentation. Cortex and medulla were on the basis of intensity value thresholds. Manual correction was utilized for over- and undersegmentation errors.
Deep-learning Methods (Automated Segmentation)
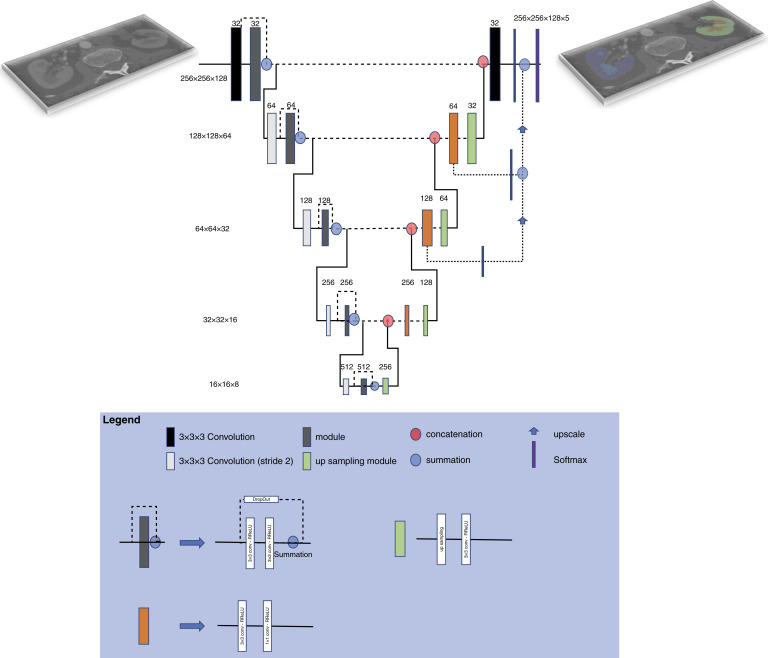
A modified 3D U-Net architecture (Figure 1)27 was used to segment the four classes (right cortex, right medulla, left cortex, and left medulla) considered in this study. The Tversky loss function was utilized, whereas Adam was the optimizer. The test set made up 20% of the entire Mayo Clinic Minnesota dataset, and the remaining training/validation set was then split 80%/20%. The selection of patients for the training, validation, and test sets was random. The validation set was used to optimize the network hyperparameters whereas the test set was used to evaluate the final model selected. The network generated four labels corresponding to the left and right medulla and cortex volumes. The code for the model is available in github (https://github.com/potis/MedCorSeg).
Figure 1.
Schematic illustration of the 3D U-Net architecture utilized in this study. The U-Net architecture consists of two pathways. The first combined downsampling with convolutional layers to encode the input. The second recombined these representations with shallower features transferred via skip connections to generate the final segmentation mask.
Statistical Analysis
To evaluate the segmentation algorithm, we calculated the Dice coefficient against the reference standard. The Dice coefficient measures the relative volumetric overlap between the automated algorithm and the reference manual segmentations (0, No overlap; 1, Perfect Overlap). Bland–Altman analysis was further performed to compare the automated and manual segmentations with bias and precision indicating the observed mean difference and variance (i.e., SD) of segmentation volumes and 95% limits of agreements. Subgroup analysis compared by BMI > or ≤25 kg/m2 and by sex. Differences in automated and manual volumes were compared with a Wilcoxon signed rank test. To assess interobserver variability with manual segmentation, all four regions were segmented by nine different readers for 10 subjects. The association of BMI and sex with the automated segmentations versus manual segmentations was compared. Donors’ clinical characteristics were correlated with kidney cortical and medullary volumes obtained by either manual or automated methods (using Spearman correlations coefficients). Two-sided Steiger’s test for dependent correlations was used to determine whether the correlation between clinical characteristics and automated kidney volumes differed from the correlation between clinical characteristics and manual kidney volumes in the test set.
Results
Donor Characteristics
A total of 1930 donors with CT images were used from Mayo Clinic Minnesota; 1544 donors were selected for training and validation (1238 and 306 donors, respectively) set, whereas 386 (out of 1930) donors were retained for testing purposes. Additional external test sets included CT images from 830 donors from the Mayo Clinic Arizona and 396 donors from the Cleveland Clinic. Table 1 summarizes the clinical characteristics of the donors studied.
Table 1.
Clinical characteristics of study subjects for the training validation and test sets considered in this study
| Clinical Characteristics | Training Set | Validation Set | MCM Test Set | MCA Test Set | CC Test Set |
|---|---|---|---|---|---|
| (1238) | (n=306) | (n=386) | (n=830) | (n=396) | |
| Agea (yr) | 44.5±11.9 | 43.6±12.1 | 44.6±12.3 | 43.1±12.3 | 41.3±10.6 |
| Sex, n (%) | |||||
| Men | 514 (41) | 127 (41) | 169 (44) | 295 (36) | 153 (39) |
| Women | 724 (59) | 179 (59) | 217 (56) | 535 (64) | 243 (61) |
| BMIa | 28.2±5 | 28.0±5.0 | 28.2±5.0 | 26.7±5.1 | 26.7±4.0 |
| Volumea (reference) | |||||
| Right cortex | 102.1±22.2 | 103.7±22.6 | 104.5±23.7 | 101.5±22.0 | 105.5±20.4 |
| Right medulla | 42.2±11.1 | 43.3±11.7 | 39.5±10.6 | 38.2±8.7 | 39.9±9.7 |
| Left cortex | 104.1±21.7 | 104.2±22.2 | 103.5±22.5 | 101.7±22.4 | 104.9±21.6 |
| Left medulla | 39.2±10.3 | 39.8±10.5 | 42.7±11.1 | 41.1±9.6 | 41.7±10.1 |
MCM, Mayo Clinic Minnesota (Rochester); MCA, Mayo Clinic Arizona; CC, Cleveland Clinic.
All data expressed as mean±SD or n (%).
Performance of Deep-learning Network in Test Sets
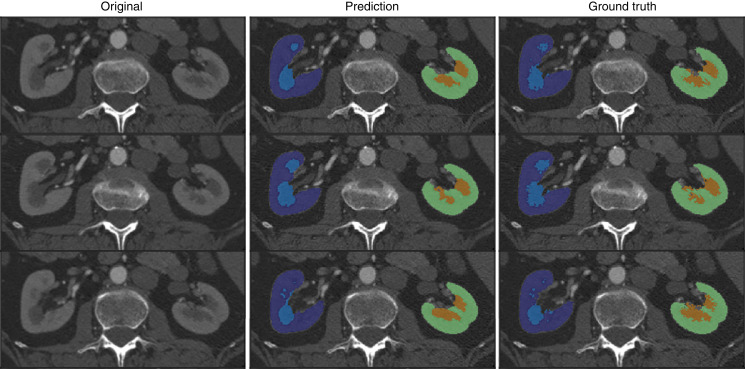
Table 2 captures the performance of the proposed network compared with the reference standard with respect to Dice and volume similarity in the Mayo Clinic Minnesota test set. The Dice coefficient was 0.94 for right or left cortex, and 0.90 for right or left medulla. Figure 2 is an example of automated segmentation in a Mayo Clinic Minnesota test set donor. The automated algorithm tended to segment areas with smoothed borders, which differ from the rough border generated by the ground truth manual method.
Table 2.
Algorithm performance analysis (Mayo Clinic Minnesota test set) on the basis of Dice coefficient. Metrics are reported for the entire cohort and with respect to the BMI and sex for the four structures considered in this study. The statistics reported are mean±SD and range (min/max)
| Category | Right Cortex | Left Cortex | Right Medulla | Left Medulla |
|---|---|---|---|---|
| Overall Testing Set n=386 |
0.94±0.02 (0.68/0.97) |
0.94±0.01 (0.81/0.97) |
0.90±0.03 (0.57/0.94) |
0.90±0.03 (0.56/0.94) |
| BMI ≤25 n=113 (29.1%) |
0.94±0.01 (0.86/0.97) |
0.95±0.01 (0.90/0.96) |
0.91±0.02 (0.76/0.94) |
0.91±0.02 (0.79/0.94) |
| BMI>25 n=275 (70.9%) |
0.94±0.02 (0.68/0.97) |
0.94±0.01 (0.81/0.97) |
0.90±0.03 (0.57/0.94) |
0.90±0.03 (0.56/0.94) |
| Female n=219 (56.4%) |
0.94±0.01 (0.85/0.97) |
0.94±0.01 (0.81/0.97) |
0.91±0.02 (0.71/0.94) |
0.90±0.03 (0.56/0.94) |
| Male n=169 (43.6%) |
0.94±0.02 (0.68/0.97) |
0.94±0.01 (0.87/0.97) |
0.89±0.04 (0.57/0.94) |
0.89±0.03 (0.64/0.94) |
Figure 2.
Examples of automated segmentations. Red arrows indicate differences between the prediction and the ground truth. The dark blue and light blue masks correspond to the right cortex and medulla, respectively. The green and orange masks correspond to the left cortex and medulla, respectively. In the case depicted, the Dice coefficient calculated was approximately 0.90 for the cortex area and approximately 0.81 for the medulla region.
The Dice coefficients did not meaningfully differ by BMI or by sex (P>0.05 for all). The Dice coefficient was slightly lower for cortex at the Mayo Clinic Arizona (0.92 right and 0.93 left) and at the Cleveland Clinic (0.93 right and 0.93 left). The Dice coefficient was also lower for the medulla at the Mayo Clinic Arizona (0.88 right and 0.88 left) and at the Cleveland Clinic (0.85 right and 0.84 left) (Supplemental Table 1).
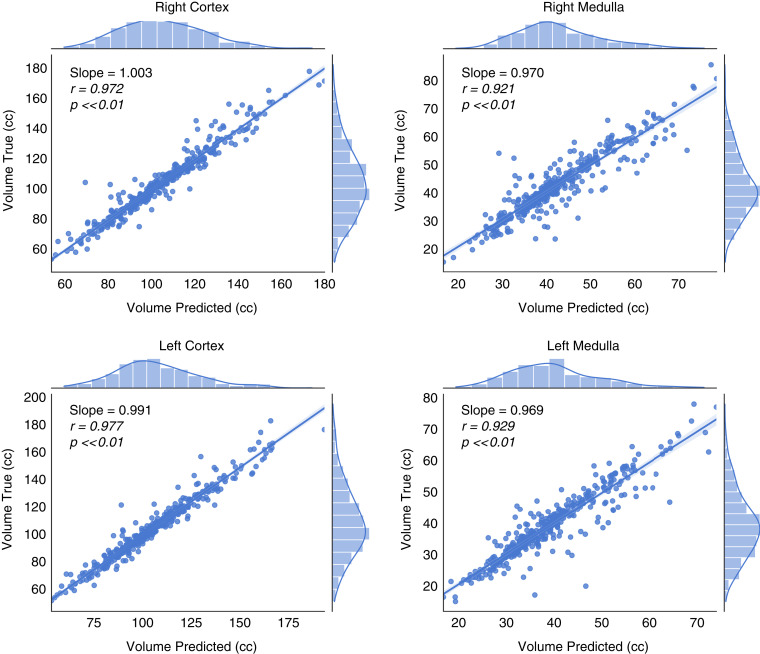
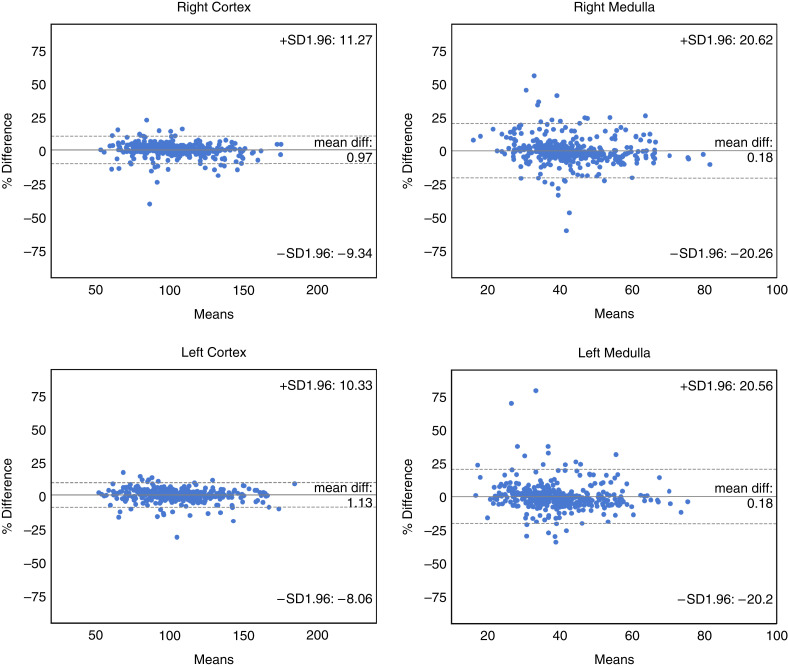
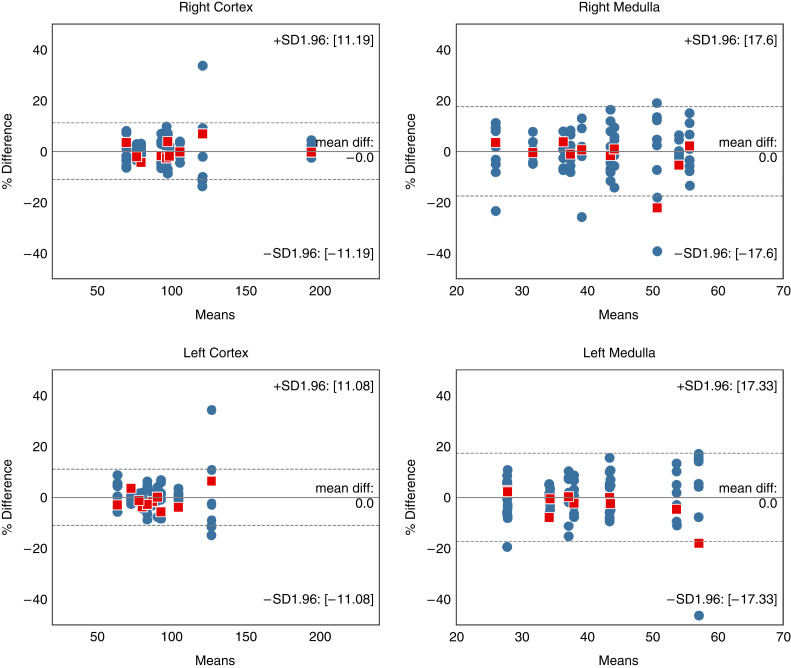
Comparison of the volumes of the manual versus automated segmentations for the four structures for the Mayo Clinic Minnesota test set are shown in Table 3. Differences between manual/reference and automated segmentation were not evident (left cortex P=0.44; right cortex P=0.85; left medulla P=0.11; right medulla P>0.99). Correlation plots and Bland–Altman analyses comparing the manual method with the automated method for the cortex and medulla are shown in Figures 3 and 4. The Pearson correlation coefficient between manual and automated were 0.97–0.98 for cortex and 0.92–0.93 for medulla (P>0.05 for all). The percent difference bias±precision for the right cortex was 0.97±5.2%, for the left cortex was 1.1±4.6%, for the right medulla was 0.2±10.2%, and for the left medulla was 0.2±10.2%.
Table 3.
Reference standard and predicted volumes for each structure considered in this study for the patients in the Mayo Clinic Minnesota test and validation set. The statistics reported are mean±SD and range (min/max) and the P values for difference
| Label | Manual Segmentation | Automated Segmentation | P Value | |
|---|---|---|---|---|
| Right cortex, cc | Test n=386 |
103.5±22.4 (53.3/177.7) |
104.4±21.7 (53.8/180.2) |
>0.05 |
| Validation n=306 |
103.7±22.6 (58.0/174.9) |
104.7±21.9 (61.4/181.0) |
>0.05 | |
| Left cortex, cc | Test n=386 |
104.5±23.6 (51.5/182.4) |
105.6±23.3 (53.0/193.6) |
>0.05 |
| Validation n=306 |
104.2±22.2 (49.8/184.2) |
105.2±21.5 (51.1/187.6) |
>0.05 | |
| Right medulla, cc | Test n=386 |
42.6±11.1 (15.3/85.5) |
42.5±10.5 (16.7/78.6) |
>0.05 |
| Validation n=306 |
43.3±11.7 (11.7/90.7) |
43.05±11.3 (19.5/81.4) |
>0.05 | |
| Left medulla, cc | Test n=386 |
39.5±10.6 (15.2/77.8) |
39.4±10.1 (16.8/74.0) |
>0.05 |
| Validation n=306 |
39.8±10.5 (16.8/80.5) |
39.5±10.4 (15.2/80.4) |
>0.05 |
cc, cubic centimeters.
Figure 3.
High correlation and agreement for volume measurements of all four regions (right cortex, right medulla, left cortex, left medulla) obtained by the automated approach and manual segmentation in the Mayo Clinic Minnesota test set. The slope, Pearson coefficient, and P values are provided. Pearson coefficients close to zero imply no correlation.
Figure 4.
The agreement assessed using Bland–Altman, between the automated approach and manual segmentation of all four regions (right cortex, right medulla, left cortex, left medulla) in the Mayo Clinic Minnesota test set. Mean volumes along the x-axis are represented in cubic centimeters. The solid line represents the actual mean difference (bias), and the dotted lines show 95% limits of agreements (LoAs).
Multi-reader Interobserver Variability Analysis
The percent difference between each reader and the mean of all nine manual readers and the difference between the automated method and the mean of all nine manual readers was plotted for 10 random donors (Figure 5). As shown graphically, the automated variability was substantially less than the variability between the nine manual segmentations.
Figure 5.
Bland–Altman analysis of nine readers (blue circles) and the automated method (red squares) compared with the reference standard (mean of nine readers) for all four regions. Mean volumes along the x-axis are represented in cubic centimeters. The solid line represents the actual mean difference (bias), and the dotted lines show 95% LoAs. Both are calculated from the nine readers.
Correlation of Donors’ Clinical Characteristics with Cortex and Medullary Volumes
We assessed correlation between clinical characteristics and the total cortex or total medulla volumes using both automated and manual approaches to segmentation in the test sets from all three sites (n=1612). These correlations were generally similar between manual and automated methods, however, some differences were evident (Table 4). An age-related decline in cortical volume was stronger by the automated method and a modest increase in medullary volume with age was only detected by the automated method. The correlation of taller height with larger cortex did not differ between manual and automated methods, but the correlation with larger medulla was weaker by the automated method. The correlation of obesity (higher BMI) with larger cortical volume and with larger medulla was weaker by the automated method. Although measured GFR correlated with cortex and medulla volume to a similar extent by manual and automated methods, estimated GFR differed in its correlation with cortex and medulla volumes between manual and automated methods. A decrease in cortical volume with hypertension was detected by the automated method but not the manual method. Biopsy measures of average cortical nephron size (glomerular volume and mean profile tubular area) were more strongly correlated with manual than automated cortical volume. These associations in each of the three sites seperately is shown for the Mayo Clinic Minnesota (Supplemental Table 2), the Mayo Clinic Arizona (Supplemental Table 3), and the Cleveland Clinic (Supplemental Table 4).
Table 4.
Spearman’s correlation coefficients between clinical characteristic and cortical and medullary volumes obtained by manual or AI-based approaches from the test sets combining all three sites.
| N=1612 kidney donors | Cortical volume | Medullary volume | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Manual | Automated | Manual | Automated | ||||||||
| Characteristic | rs | P value | rs | P value | P value | rs | P value | rs | P value | P value | |
| Age, years | -0.220 | <0.001 | -0.246 | <0.001 | 0.002 | 0.032 | 0.20 | 0.087 | <0.001 | < 0.001 | |
| Male | 0.529 | <0.001 | 0.517 | <0.001 | 0.11 | 0.196 | <0.001 | 0.188 | <0.001 | 0.62 | |
| Donor’s height, cm | 0.519 | <0.001 | 0.511 | <0.001 | 0.28 | 0.310 | <0.001 | 0.230 | <0.001 | < 0.001 | |
| BMI, kg/m2 | 0.429 | <0.001 | 0.378 | <0.001 | < 0.001 | 0.151 | <0.001 | 0.221 | <0.001 | < 0.001 | |
| Measured GFR (ml/min)* | 0.568 | <0.001 | 0.560 | <0.001 | 0.31 | 0.325 | <0.001 | 0.336 | <0.001 | 0.51 | |
| Measured GFR (ml/min/1.73m2)* | 0.256 | <0.001 | 0.273 | <0.001 | 0.06 | 0.170 | <0.001 | 0.155 | <0.001 | 0.39 | |
| Estimated GFR (ml/min/1.73m2)¥ | 0.276 | <0.001 | 0.309 | <0.001 | < 0.001 | 0.062 | 0.01 | -0.020 | 0.43 | < 0.001 | |
| Family history of ESRD† | 0.124 | <0.001 | 0.119 | 0.02 | 0.60 | 0.022 | 0.42 | 0.033 | 0.23 | 0.54 | |
| Hypertension | -0.037 | 0.14 | -0.055 | 0.03 | 0.04 | 0.032 | 0.20 | 0.049 | 0.05 | 0.30 | |
| Uric acid, mg/dlⱡ | 0.356 | <0.001 | 0.340 | <0.001 | 0.05 | 0.034 | 0.18 | 0.054 | 0.03 | 0.22 | |
| 24hr Albumin‡ | 0.130 | <0.001 | 0.108 | 0.003 | 0.09 | 0.087 | 0.02 | 0.094 | 0.01 | 0.77 | |
| Glomerular volume** | 0.253 | <0.001 | 0.207 | <0.001 | < 0.001 | -0.058 | 0.06 | 0.000 | 0.99 | 0.004 | |
| Mean tubular area** | 0.124 | 0.002 | 0.100 | 0.001 | 0.03 | 0.024 | 0.44 | 0.023 | 0.46 | 0.96 | |
| % Globally sclerotic glomeruli** | -0.071 | 0.02 | -0.079 | 0.01 | 0.46 | 0.010 | 0.74 | -0.009 | 0.78 | 0.34 | |
Measured GFR available in 1361 donors.
Estimated GFR available in 1604 donors.
Family history of ESRD available in 1339 donors.
Uric acid available in 1581 donors.
24hr Albumin available in 747 donors.
Biopsy measures available in 1066 kidney donors.
Discussion
An image-processing method on the basis of a deep-learning network allows for automated, comprehensive, and noninvasive structural kidney volume analysis from high-resolution contrast-enhanced CT images. Many of the clinical cases in which this type of analysis could be easily implemented already include contrast-enhanced CT imaging in the protocol. The proposed algorithm can yield results in <5 minutes and has the potential to reduce differences in kidney donor evaluations arising from inter- and intraobserver variability. It is also reassuring that clinical characteristics generally had similar associations with kidney volumes whether the volumes were determined using a manual or automated method. Therefore, integrating this automatic analysis into clinical systems poses low risk/disruption to the practice and potentially high reward.
Because this method does not require unique CT imaging acquisitions, there also exists an opportunity to apply it to large existing datasets across numerous clinical settings.
Jin et al.28 reported accuracies of 0.89 and 0.86 in terms of Dice for the cortex and medulla compartments on a validation dataset consisting of 37 contrast enhanced CT scans utilizing random forest segmentation, whereas Chen et al.23 reported accuracy of 0.90 in terms of true positive fraction in a similar-sized dataset using a 3D shape–constrained graph cut method. The proposed algorithm yielded higher accuracy in terms of Dice coefficient: 0.93 for cortex and 0.90 for medulla, on a significantly larger test set (n=386). The model’s performance was also evaluated on two external test sites yielding similar Dice coefficients for cortex (0.93) and only a slightly lower Dice coefficient for medulla (0.88).
In general, the agreement between the cortex regions was better than the agreement between the medulla regions, as shown through Dice and regression analysis. On average, the volume of the cortex is more than double the volume of the medulla, and this may explain a smaller margin of error. In addition, the medulla is inherently more difficult to manually segment because, unlike the cortex, it is not radiopaque with angiogram phase images, it is adjacent to vessels and sinus fat that may be difficult to distinguish from medulla, and it is composed of medullary pyramids that are both contiguous and noncontiguous with each other. This thresholding can be sensitive to noise and the automated method may be more precise as it agreed well with the mean value across nine observers in the multireader analysis, particular for the medulla.
Finally, it was reassuring to see that cortical and medullary volumes obtained by deep learning showed similar associations with clinical characteristics, as did manually obtained volumes from multiple institutions. There were some differences, however. Of note, the cortical nephron size measures (glomerular volume and mean profile tubular area) represent a measure of size at the microstructural level and were slightly more correlated with the manual method of determining cortical volume. Better distinction between cortex and medulla with the automated method could have led to stronger BMI correlation with medulla than cortex with the automated method. It is also possible the deep-learning network is detecting other age-related features on the CT scans (possibly age-related sinus fat) that contribute to a stronger correlation between age and kidney volumes by the automated method. Because eGFR is calculated with age, this may also explain the discrepancy between differing correlations between manual and automated with estimated GFR but not with measured GFR.
The proposed algorithm can yield results in <5 minutes depending on the CT image size and the compute resources available. When a human observer executes the same task, the time required is 30–90 minutes. One of the main advantages of the AI solution is that, due to its automatic nature, clinical implementation of this system can be achieved. Involving human observers to perform the segmentation in a clinical workflow can lead to delays. Figure 5 captures the variability of volume measures occurring when nine readers segment the structure of interest in ten patients. AI has the potential to reduce differences in kidney donor evaluations arising from inter- and intraobserver variability. It is also reassuring that clinical characteristics generally had similar associations with kidney volumes, whether the volumes were determined using a manual or automated method. Therefore, integrating this automatic analysis into clinical systems poses low risk/disruption to the practice and potentially high reward.
There are potential limitations to this method. First, is that deep-learning approaches (e.g., with U-net architectures) require large amounts of training data to generalize well in unknown test cases (e.g., unique anatomy characteristics or poor image quality due to noise of metal artifact). In addition, the proposed algorithm was developed assuming that a cropped region around the kidneys is used as an input. Cropping close to the area of interest reduces the anatomic noise and enabled us to utilize 3D models and larger batch sizes alleviating the need for high GPU memory during training. To ensure clinical applicability, the algorithm needs to incorporate a kidney region identifier. This is the subject for future work. The method was developed in a healthy population of potential living kidney donors. Performance in less healthy populations or populations with overt kidney disease requires further assessment. Even then, there are concerns for contrast nephropathy with the use of intravenous contrast in patients with CKD29 that may limit utilization in populations at highest risk for kidney failure. An additional limitation of the study is the lack of training examples from multiple institutions, which can affect the model generalizability. To account for this, augmentation techniques including noise simulations was utilized. In the testing set, however, we have shown the performance of the algorithm on images from two external sites is on par with the performance on images from the Mayo Clinic in Minnesota, highlighting the generalizability of the proposed algorithm. Figure 5 is indicative of the interobserver variability of human readers performing the segmentations for this task. Although the algorithm’s performance was within the interobserver variability, the study would have benefited from multiple readers generating the segmentations utilized to train the algorithm. Future work could also focus on multisite training of the algorithm; however, in this study we were interested in highlighting the issues surrounding generalization to external datasets where the imaging and population characteristics differ from the training set.
Fully automated image-processing techniques can be significantly leveraged to allow fast, accurate, and reproducible segmentation of kidney structures within routine CT images. Automated segmentation techniques aid in the development of texture-based metrics that may be superior to or complement volume measurements. The next step will be to evaluate texture features within these various regions of the kidney.
A fully automated segmentation method has been developed that segments the kidneys, and the cortex and medullary regions in contrast-enhanced CT images. The method is not only significantly faster than the manual approach, but it also performs better in terms of associations with clinical characteristics. The method may be useful for both research and the clinical practice to rapidly evaluate renal donor suitability, and study the associated implications these image-derived biomarkers have on donor and recipient outcomes.
Disclosures
A. Rule reports being a scientific advisor or member of the National Institute of Diabetes and Digestive and Kidney Diseases CKD Biomarker Consortium External Expert Panel, JASN Associate Editor, and Mayo Clinic Proceedings Section Editor; and reports having other interests/relationships with UpToDate. All remaining authors have nothing to disclose.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under awards K01DK110136, R03DK125632, and R01DK090358.
Supplementary Material
Acknowledgments
Because Dr. A. Rule is an editor of JASN, he was not involved in the peer review process for this manuscript. A guest editor oversaw the peer review and decision-making process for this manuscript. The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2021030404/-/DCSupplemental.
Supplemental Figure 1. Workflow overview for manual segmentation of kidneys in abdominal CT scans (used in this study as the reference standard).
Supplemental Table 1. Algorithm performance analysis on the basis of Dice coefficient for the two additional datasets originating from two sites (MCA, Mayo Clinic Arizona; CC, Cleveland Clinic).
Supplemental Table 2. Spearman’s correlation coefficients between clinical characteristic and cortical and medullary volumes obtained by manual or AI-based approaches from the test set at Mayo Clinic Minnesota.
Supplemental Table 3. Spearman’s correlation coefficients between clinical characteristic and cortical and medullary volumes obtained by manual or AI-based approaches from the test set at Mayo Clinic Arizona.
Supplemental Table 4. Spearman’s correlation coefficients between clinical characteristic and cortical and medullary volumes obtained by manual or AI-based approaches from the test set at Cleveland Clinic.
References
- 1.Chapman AB, Wei W: Imaging approaches to patients with polycystic kidney disease. Semin Nephrol 31: 237–244, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liebau MC, Serra AL: Looking at the (w)hole: Magnet resonance imaging in polycystic kidney disease. Pediatr Nephrol 28: 1771–1783, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Fick-Brosnahan GM: Endothelial dysfunction and angiogenesis in autosomal dominant polycystic kidney disease. Curr Hypertens Rev 9: 32–36, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Grantham JJ, Torres VE, Chapman AB, Guay-Woodford LM, Bae KT, King BF Jr, et al. ; CRISP Investigators : Volume progression in polycystic kidney disease. N Engl J Med 354: 2122–2130, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Dias J, Malheiro J, Almeida M, Dias L, Silva-Ramos M, Martins LS, et al. : CT-based renal volume and graft function after living-donor kidney transplantation: Is there a volume threshold to avoid? Int Urol Nephrol 47: 851–859, 2015 [DOI] [PubMed] [Google Scholar]
- 6.Fananapazir G, Benzl R, Corwin MT, Chen LX, Sageshima J, Stewart SL, et al. : Predonation volume of future remnant cortical kidney helps predict postdonation renal function in live kidney donors. Radiology 288: 153–157, 2018 [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Vrtiska TJ, Avula RT, Walters LR, Chakkera HA, Kremers WK, et al. : Age, kidney function, and risk factors associate differently with cortical and medullary volumes of the kidney. Kidney Int 85: 677–685, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu ASL, Shen C, Landsittel DP, Harris PC, Torres VE, Mrug M, et al. ; Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) : Baseline total kidney volume and the rate of kidney growth are associated with chronic kidney disease progression in autosomal dominant polycystic kidney disease. Kidney Int 93: 691–699, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita SR, von Atzingen AC, Iared W, Bezerra AS, Ammirati AL, Canziani ME, et al. : Value of renal cortical thickness as a predictor of renal function impairment in chronic renal disease patients. Radiol Bras 48: 12–16, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snoek R, de Heus R, de Mooij KJ, Pistorius LR, Lilien MR, Lely AT, et al. : Assessing nephron hyperplasia in fetal congenital solitary functioning kidneys by measuring renal papilla number. Am J Kidney Dis 72: 465–467, 2018 [DOI] [PubMed] [Google Scholar]
- 11.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. : User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31: 1116–1128, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Denic A, Alexander MP, Kaushik V, Lerman LO, Lieske JC, Stegall MD, et al. : Detection and clinical patterns of nephron hypertrophy and nephrosclerosis among apparently healthy adults. Am J Kidney Dis 68: 58–67, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merzkani MA, Denic A, Narasimhan R, Lopez CL, Larson JJ, Kremers WK, et al. : Kidney microstructural features at the time of donation predict long-term risk of chronic kidney disease in living kidney donors. Mayo Clin Proc 96: 40–51, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Issa N, Lopez CL, Denic A, Taler SJ, Larson JJ, Kremers WK, et al. : Kidney structural features from living donors predict graft failure in the recipient. J Am Soc Nephrol 31: 415–423, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thong W, Kadoury S, Piché N, Pal CJ: Convolutional networks for kidney segmentation in contrast-enhanced CT scans. Comput Methods Biomech Biomed Eng Imaging Vis 6: 277–282, 2018 [Google Scholar]
- 16.Sharma K, Rupprecht C, Caroli A, Aparicio MC, Remuzzi A, Baust M, et al. : Automatic segmentation of kidneys using deep learning for total kidney volume quantification in autosomal dominant polycystic kidney disease. Sci Rep 7: 2049, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan X, Rule AD, Elsherbiny H, Vrtiska TJ, Avula RT, Alexander MP, et al. : Automated assessment of renal cortical surface roughness from computerized tomography images and its association with age. Acad Radiol 21: 1441–1445, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rule AD, Sasiwimonphan K, Lieske JC, Keddis MT, Torres VE, Vrtiska TJ: Characteristics of renal cystic and solid lesions based on contrast-enhanced computed tomography of potential kidney donors. Am J Kidney Dis 59: 611–618, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Q, Geng P, Li H, Sun H, Kang Y: Semi-automatic segmentation of renal cortex and medulla based on dynamic magnetic resonance images. 2010. 3rd International Conference on Biomedical Engineering and Informatics. IEEE, 2010 pp 555–559
- 20.Will S, Martirosian P, Würslin C, Schick F: Automated segmentation and volumetric analysis of renal cortex, medulla, and pelvis based on non-contrast-enhanced T1- and T2-weighted MR images. MAGMA 27: 445–454, 2014 [DOI] [PubMed] [Google Scholar]
- 21.Huang W, Li H, Wang R, Zhang X, Wang X, Zhang J: A self-supervised strategy for fully automatic segmentation of renal dynamic contrast-enhanced magnetic resonance images. Med Phys 46: 4417–4430, 2019 [DOI] [PubMed] [Google Scholar]
- 22.Xiang D, Bagci U, Jin C, Shi F, Zhu W, Yao J, et al. : CorteXpert: A model-based method for automatic renal cortex segmentation. Med Image Anal 42: 257–273, 2017 [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Summers RM, Cho M, Bagci U, Yao J: An automatic method for renal cortex segmentation on CT images: Evaluation on kidney donors. Acad Radiol 19: 562–570, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenspan H, Ginneken BV, Summers RM: Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35: 1153–1159, 2016 [Google Scholar]
- 25.van Gastel MDA, Edwards ME, Torres VE, Erickson BJ, Gansevoort RT, Kline TL: Automatic measurement of kidney and liver volumes from MR images of patients affected by autosomal dominant polycystic kidney disease. J Am Soc Nephrol 30: 1514–1522, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasaka K, Abe O: Deep learning and artificial intelligence in radiology: Current applications and future directions. PLoS Med 15: e1002707, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weston AD, Korfiatis P, Philbrick KA, Conte GM, Kostandy P, Sakinis T, et al. : Complete abdomen and pelvis segmentation using U-net variant architecture. Med Phys 47: 5609–5618, 2020 [DOI] [PubMed] [Google Scholar]
- 28.Jin C, Shi F, Xiang D, Zhang L, Chen X: Fast segmentation of kidney components using random forests and ferns. Med Phys 44: 6353–6363, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Davenport MS, Khalatbari S, Cohan RH, Dillman JR, Myles JD, Ellis JH: Contrast material-induced nephrotoxicity and intravenous low-osmolality iodinated contrast material: Risk stratification by using estimated glomerular filtration rate. Radiology 268: 719–728, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.