Extended Data Figure 8. Distribution of mutations generated during the type III-A CRISPR-Cas immune response against ϕNM1γ6 phage.
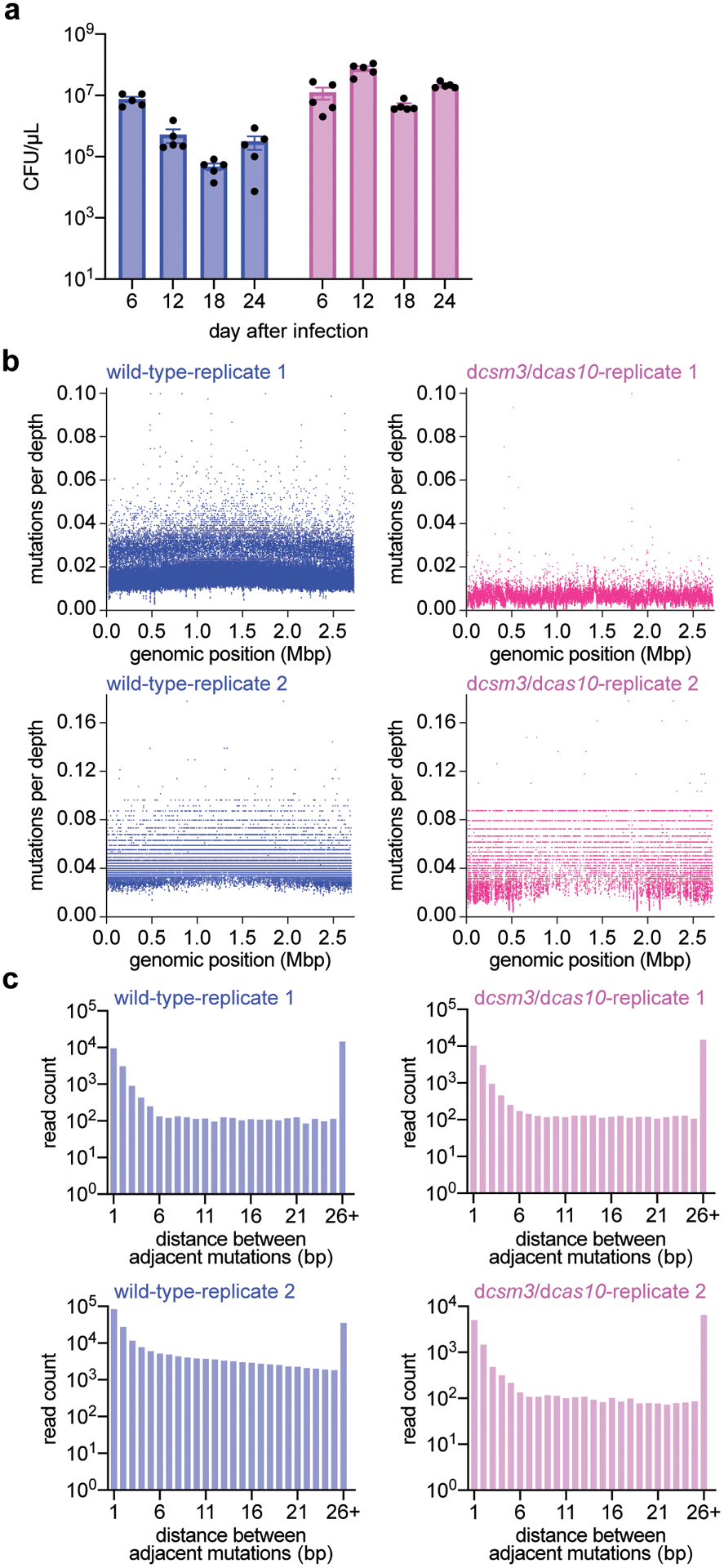
(a) Colony forming units per microliter (CFU/μL) of wild-type and dcas10/dcsm3 cultures after 6, 12, 18 and 24 daily infections with ϕNM1γ6 phage. Bar graphs represent the mean CFU/μL; individual black dots represent the individual values from the 5 independent cultures. (b) Distribution of mutations in the S. aureus TB4 genome of wild-type and dcas10/dcsm3 cells infected daily with ϕNM1γ6 phage after 18 days. Genomic DNA from the cultures was labeled with unique molecular barcodes in order to reduce sequencing errors, subjected to next-generation sequencing and analyzed with the appropriate software for this procedure (https://github.com/Kennedy-Lab-UW/Duplex-Sequencing). Mutations were counted if a particular genomic position possess a minimal depth of 10 reads. The x-axis represents the position within the S. aureus TB4 genome, while the y-axis represents the number of mutations divided by the sequencing depth at that particular position. Replicate 1 shows the data collected from a single wild-type or dcas10/dcsm3 culture, while replicate 2 shows data from the pooled five cultures of each strain. Based on this analysis, the samples do not show an obvious mutational hot spot in the genome. (c) Distance, in base pairs (bp), between adjacent mutations in the S. aureus TB4 genome, calculated from the data obtained in panel (b).
