Abstract
The addition of reagents for assays in digital microfluidic (DMF) systems is traditionally done by merging of droplets containing different analytes or reagents in solution. However, this process significantly increases droplet volume after each step, resulting in dilution of the analyte and reagents. Here, we report a new technique for performing reagent additions to aqueous droplets without significantly increasing the droplet’s volume: volume-less reagent delivery (VRD). VRD is enabled by a physical phenomenon we call “exclusive liquid repellency (ELR), which describes an aqueous/oil/solid 3-phase system where the aqueous phase can be fully repelled from a solid phase (contact angle ~180°). When performing VRD, a reagent of interest in solution is deposited onto the ELR solid surface and allowed to dry. The ELR surface containing the dried reagent is then immersed under oil, followed by introduction of an aqueous droplet. By dragging the aqueous droplet over the spot of dried reagent using paramagnetic particles or via a physical sliding wall, the droplet can then recover and reconstitute the reagent with negligible increase in its total volume, returning the ELR surface to its initial liquid repellent state in the process. We demonstrate that VRD can be performed across a wide range of reagent types including sugars, proteins (antibodies), nucleic acids (DNA), antibiotics, and even complex enzyme/substrate/buffer “kit” mixtures. We believe VRD is a flexible and powerful technique which can further the development of self-contained, multi-step assays in DMF and other microfluidic systems.
Introduction
Digital microfluidics (DMF) is a technique that allows the manipulation of discrete droplets on a planar surface without requiring a sophisticated network of microfluidic channels, valves, or pumps. The manipulation of droplets can be accomplished via a variety of techniques, including, most commonly, electrowetting on dielectric (EWOD),1, 2 dielectrophoresis (DEP),3, 4 magnetic actuation,5, 6 surface acoustic waves,7, 8 among others. Each droplet can serve as an isolated condition for discrete processes or reactions to occur in each droplet. DMF has been utilized in various biological assays or processes such as immunoassays,9, 10 DNA library preparation,11–13 enzymatic assays,14, 15 and a variety of other bioassays. Despite its numerous advantages and potential, the translation of the technology for real-world use has encountered challenges such as reliability,16 biofouling of the DMF surface when transporting protein-rich solutions,17–19 and the significant increase in droplet volumes when performing reagent addition via droplet merging. There have been some efforts in developing DMF systems with dried reagents/analytes deposited on the device surface,20, 21 which mitigates the issue of droplet size increases after reagent delivery and potentially enables the development of fully self-contained DMF systems, which don’t require pipetting of individual reagents by the end user. However, these prior studies have neither demonstrated full recovery of the dried reagents from the DMF device surface, nor shown sequential, multi-step recovery of dried reagents via DMF droplets (only single-step), suggesting that surface fouling-mediated loss of reagents or analytes is still a challenge. Here we report a method which addresses some of these challenges: volumeless reagent delivery (VRD), which enables the sequential delivery and near complete recovery of dried reagents (even high concentration proteins) into aqueous droplets with negligible increase in volume, made possible via a unique 3-phase system we call exclusive liquid repellency (ELR).
ELR describes a physical phenomenon in which an aqueous phase can be completely repelled from a solid phase (contact angle ~180°) in the presence of an oil phase given specific oil and solid surface chemical properties.22 This physical phenomenon is governed by the balance of interfacial energies between the solid, oil, and aqueous phases. Namely ELR will occur if the sum of the solid/oil and aqueous/oil interfacial energies is equal to or less than the solid/aqueous interfacial energy; otherwise adhesion of the aqueous solution to the surface (contact angle < 180°) will occur. We found that this condition can be realized when using a 1,3’dichlorotetramethylsiloxane modified surface as the solid phase paired with silicone oil as the oil phase. The aqueous liquid repellent properties of the ELR 3-phase system allows aqueous solutions to be fully repelled from the solid phase, preventing adsorption or fouling of biomolecules onto the surface and mitigates issues with droplet-to-surface adhesion during operation.
VRD harnesses the liquid repellent properties of ELR to enable delivery and recovery of dried reagents into an aqueous droplet with negligible change in the droplet’s volume (Fig. 1). In VRD, a reagent of interest in solution is deposited onto an ELR surface and allowed to dry under a vacuum. The ELR surface containing the dried reagent is then immersed under oil, followed by introduction of an aqueous droplet (which can contain paramagnetic particles (PMPs)) into the oil. By dragging the aqueous droplet over the spot of dried reagent using a magnet or via a physical “sliding wall” which entraps the droplet, the aqueous droplet can then pick-up and reconstitute the dried reagent with negligible increase in its total volume (Fig. 1), simultaneously returning the device surface back to its original liquid repellent state in the process. Importantly, VRD contrasts with the more traditional reagent addition method in DMF systems accomplished via merging of two discrete droplets, which results in a large increase in droplet volume.23
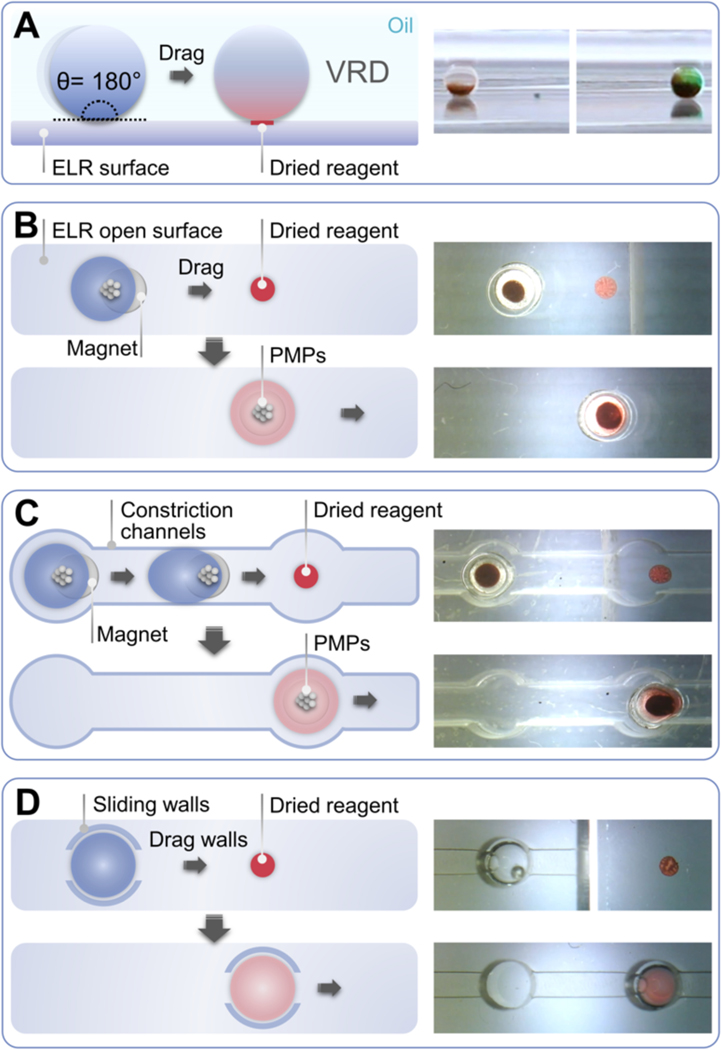
Fig. 1.
Operation principle and different configurations of VRD. (A) Side view of VRD process. An aqueous droplet is dragged across an ELR surface and into contact with a dried reagent spot on the surface (green food coloring for right panel image), which picks up and reconstitutes the dried reagent. (B) The “open surface” configuration of VRD. An aqueous droplet containing PMPs is added to the device above a magnet, which holds the droplet in place. The droplet is then dragged across the ELR surface and into contact with a spot of dried reagent using the magnet. The droplet can pick up and reconstitute the dried reagent, returning the surface back to its original liquid repellent state. (C) The “constriction channel” configuration of VRD. To prevent the movement of droplets after removal of the magnetic field, we designed a device insert consisting of an array of wells with interconnecting constriction channels which are narrower than the diameter of the droplet. The constriction channel insert is also treated with ELR chemistry, which prevents aqueous droplets from sticking to the device walls and impeding droplet movement. (D) The “sliding wall” configuration of VRD. This configuration employs a sliding ELR-treated physical “wall” insert which entraps the aqueous droplet and allows the droplet to be pushed along the surface of the device by physically sliding the insert. The wall has a 200 μm tall channel cutout to provide clearance for the dried reagent to pass under during droplet manipulation.
The VRD technique is made possible primarily due to the highly aqueous-repellent characteristic of ELR, which prevents fouling-mediated sample/reagent loss caused by nonspecific adsorption of molecules to the device surface. Importantly, the near complete reagent recovery capability of VRD allows the operation to be performed multiple times sequentially, enabling the multi-step addition of different reagents in a row via a simple droplet dragging operation, all while keeping the total volume of the droplet mostly unchanged. We’ve also demonstrated that VRD is compatible with a wide range of different biomolecule types, including sugars (dextran), proteins (IgG antibodies), nucleic acids (DNA), small molecule antibiotics (gentamicin), and complex enzyme/substrate/buffer “kit” mixtures (CellTiter-Glo). The device for performing VRD (modified from an OmniTray well plate) benefits from having a thin, transparent glass bottom construction, enabling optical assay readouts including microscopy and luminescence, as well as efficient magnetic droplet manipulation.
We propose that VRD is a highly flexible and widely applicable technique which helps solve some of the issues plaguing traditional DMF systems, potentially enabling the development of self-contained microfluidic systems which can perform complex, multi-step assays with simplicity and ease of operation.
Experimental
Fabrication of VRD devices
A 43 mm x 60 mm rectangular hole was cut out of the bottom of an OmniTray single-well plate (Thermo Scientific Nunc) using a computer numerical control (CNC) 3-axis mill (PCNC 770, Tormach) and the plastic cutout discarded. Medical grade double-sided tape (ARCare 90106, Adhesives Research, thickness: 0.14 mm) was cut using a laser engraver (350–60W CO2 Laser, Automation Technology). After removal of the protective backing on the double-sided tape, a 48 mm x 65 mm x 0.17 mm coverslip (Gold Seal Cover Glass) was aligned and adhered to the double-sided tape. The coverslip with the attached double-sided tape was then aligned and adhered to the bottom of the OmniTray cutout. To further prevent oil leakage, the outer edges along the coverslip were sealed using Duro Super Glue. The coverslip bottom OmniTrays were then treated with oxygen plasma for 2 min at 100 W (Diener Electronic Femto, Plasma Surface Technology). After plasma treatment, the OmniTray was placed in a vacuum desiccator with 2 trays (40 μL each) of an alkyl silane (1,3’dichlorotetramethylsiloxane, Gelest, SID3372.0). The desiccator was then pumped down to vaporize and condense the silane onto the device surface at RT for at least 1 hour to functionalize the device surface. The OmniTray was then thoroughly rinsed with 100% isopropyl alcohol then dried using an air gun to remove residual unattached silane. An image of the finished device containing dried spots of food dye on the device surface is shown in Fig. S1. To form the ELR surface, a drop of silicone oil (viscosity: 5 cSt, Sigma Aldrich) was deposited on the device surface, then blown off using an air gun to form a thin coating of silicone oil on the surface. The constriction channel inserts and the sliding wall inserts were milled out of 2.4 mm thick polycarbonate sheets (LEXAN 9034, United States Plastic Corporation) using a 3-axis CNC mill (PCNC 770, Tormach). Following milling, the polycarbonate device inserts were thoroughly washed with 100% isopropyl alcohol, dried using an air gun, and plasma and silane treated in the same way as above. The constriction channel or sliding wall inserts were then placed on top of the coverslip in the OmniTray VRD device. For creating patterned hydrophilic spots on the ELR surface of the device, which can be used as “traps” for capturing and immobilizing aqueous droplets, we placed a PDMS mask with a column of 2 mm through-holes on top of the silane-treated coverslip and oxygen plasma-treated the device (2 min at 100 W) to selectively etch away the silane on the exposed areas of the PDMS mask, followed by removal of the PDMS mask.
Fabrication of magnetic manipulator
A 4 mm polystyrene (PS) sheet (Goodfellow) was CNC milled (PCNC 770, Tormach) to fabricate the magnetic manipulator for performing PMP manipulation. The sliding magnetic manipulator was fitted with individual rare-earth circular magnets (diameter 1.6 mm, thickness 0.8 mm, Magcraft) with a pitch of 4.5 mm between each magnet. An image of the magnetic manipulator is shown in Fig. S1.
VRD of Texas Red dextran
1 μL droplets of Texas Red dextran (10,000 MW, Invitrogen) at various concentrations (5, 1, 0.2, and 0 μM) were deposited onto the VRD device surface using a pipet, then dried in a vacuum desiccator till all the spots appear fully dry. The device was then placed on the magnetic manipulator, filled with silicone oil (viscosity: 5 cSt, Sigma Aldrich), followed by introduction of 5 μL droplets of water containing 1:10 diluted silica PMPs (MagneSil, Promega) via a pipet. The droplets were pipetted directly on top of each magnet to allow the droplets to be immobilized in place via the PMPs. The droplets were then dragged into contact with the Texas Red dextran spots by sliding the magnetic manipulator and allowed to incubate for ~1 min (incubation time may vary for different types and concentrations of reagent) to reconstitute the Texas Red dextran. 2 μL of the reconstituted droplet was then transferred to a fluorospectrometer (NanoDrop 3300, Thermo Scientific) to perform fluorescence intensity measurements at 610 nm (emission). The input Texas Red dextran solution was diluted 5-fold (to account for the 5-fold dilution when performing VRD with 5 μL droplets on a 1 μL reagent spot) and also measured on the fluorospectrometer to determine the reagent recovery efficiency of VRD.
VRD of IgG antibody
2 μL droplets of Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) at a concentration of 2, 0.2, and 0 mg/mL in 60 mM Trehalose in ddH2O were deposited onto the VRD device surface and dried in a vacuum desiccator as described above. The dried IgG spots were imaged on an inverted epifluorescence microscope (Nikon Ti Eclipse) before and after VRD reconstitution to assess if any residual IgG remained. VRD was performed similarly as described above using 2 μL droplets containing 1:10 diluted silica PMPs. The fluorescence of both the reconstituted droplet and input solution containing Alexa Fluor 488 IgG was measured using a fluorospectrometer (NanoDrop 3300, Thermo Scientific) at 520 nm (emission) to assess recovery efficiency.
VRD of DNA
2 μL droplets of LNCaP cell DNA in buffer AE (Qiagen) at a concentration of 200 and 0 ng/μL were deposited onto the VRD device surface and dried in a vacuum desiccator as described. VRD was performed similarly as described above with 2 μL droplets containing 1:10 diluted silica PMPs. The absorbance of the reconstituted droplet and input solution containing LNCaP DNA was measured using a UV/Visible spectrophotometer (NanoDrop 1000, Thermo Scientific) at 260 nm to assess DNA recovery efficiency.
Antibiotic dosing of bacteria using VRD
A 1 μL droplet of gentamicin at various concentrations in 2 mM Trehalose (as a carrier) was pipetted onto the ELR device surface as described above. After drying the gentamicin droplets in vacuum, the device was filled with silicone oil, and placed on the magnetic manipulator. 2 μL droplets containing Pseudomonas aeruginosa CFP (strain PA01) at an OD 600 nm of 0.001, 1:10 diluted silica PMPs, and Mueller Hinton broth were then pipetted into the ELR device on top of the individual magnets. The 2 μL droplets were then magnetically transported onto the dried gentamicin spots to perform VRD by sliding the magnetic manipulator. The droplets containing bacteria were then allowed to incubate at room temperature for 24 h (while sitting on the magnetic manipulator), followed by magnetically dragging the droplets onto a column of patterned hydrophilic spots on the device surface for droplet immobilization and fluorescence microscope imaging on an inverted epifluorescence microscope (Nikon Ti Eclipse). For sequential antibiotic dosing experiments, gentamicin was diluted to 1X, 1/3X, and 1/5X final concentrations and spotted onto the device surface (a single 1 μL gentamicin spot + 4 blank vehicle spots for the 1X concentration condition, 3 gentamicin spots + 2 blank vehicle spots for the 1/3X concentration condition, and 5 gentamicin spots for the 1/5X concentration condition). After drying the gentamicin spots and filling with oil, droplets containing bacteria were introduced and sequentially magnetically dragged from 1 antibiotic spot to another to perform VRD (with 1 h interval between each VRD addition). Following the 5 additions, the droplets were allowed to incubate at room temperature for 24~48 h, followed by dragging them onto the hydrophilic patterned spots for immobilization and imaging as described above. Fluorescence intensity of the droplets were quantified using Fiji (https://fiji.sc/) with a circular region of interest (ROI).
Antibiotic sequential dosing in well plate
100 μL of P. aeruginosa CFP at an OD 600 nm of 0.001 in Mueller Hinton broth was added to a 96-well plate, followed by introduction of 1 μL gentamicin or blank vehicle control at 1 h intervals for a total of 5 additions. The plate was incubated at room temperature for 24~48 h, and bacterial growth was measured via absorbance using an ELISA reader (SpectraMax Plus 384, Molecular Devices) at 600 nm.
VRD of CellTiter-Glo
2 μL droplets of CellTiter-Glo were pipetted onto the ELR device surface. After drying the CellTiter-Glo droplets in vacuum, a constriction channel insert was placed onto the glass coverslip bottom of the VRD device then filled with silicone oil. 2 μL silica PMPs diluted 1:10 in PBS were pipetted into column 1 wells of the device insert. 2 μL droplets containing 10000, 1000, 100, 10, and 0 LNCaP cells in RPMI cell media were pipetted into column 2 wells of the device insert. The dried CellTiter-Glo reagents reside within column 3 wells of the device insert. The “extraction channels” between columns 1 and 2 have a cross section of 0.8 mm (width) and 0.3 mm (height), which only allows PMPs to traverse and not 2 μL aqueous droplets. The “constriction channels” between columns 2 and 3 have a cross section of 1.2 mm (width) and 2.4 mm (height), which allows 2 μL aqueous droplets to traverse if a dragging force was applied to the PMPs within the droplet. The VRD device was then placed on the magnetic manipulator, and the PMPs were magnetically extracted from column 1 wells into column 2 wells containing the LNCaP cell droplets. The 2 μL cell droplets were then magnetically transported from column 2 wells into column 3 wells onto the dried CellTiter-Glo spots to perform VRD. The cell droplets were then allowed to incubate at room temperature for 10 min, followed by magnetically removing the PMPs from the cell droplets. Luminescence intensity of the droplets were then immediately measured from the bottom of the plate using a plate reader (PHERAstar FS, BMG Labtech).
Results and discussion
Operation principle of VRD
The VRD system consists of reagents that are deposited and dried onto an ELR surface. The ELR surface containing dried spots of reagents are then immersed under silicone oil, followed by introduction of aqueous droplets containing analytes of interest which are dragged over the dried reagent spot to perform VRD (Fig. 1A, Fig. S2). Although straightforward in principle, we discovered that this operation is challenging to achieve without the ELR chemistry. For instance, a droplet/PMP ratio that works well for droplet actuation in ELR conditions failed to actuate droplets in a hydrophobic, but non-ELR condition (fluoro silane modified glass with mineral oil) as shown in Fig. S2. This is due to the lower contact angle in the fluoro silane/mineral oil 3-phase system resulting in increased surface adhesion of the droplet. We’ve developed 3 different configurations of VRD, each with its advantages and disadvantages to suit the desired use case scenario. In the “open surface” configuration of VRD, aqueous droplets containing PMPs are pipetted onto the VRD device surface to enable contact-less droplet manipulation via an external magnet (Fig. 1B, Movie S1). This configuration allows for maximal degrees of freedom in droplet movement across all directions on the device surface. However, the open surface configuration also requires that the PMPs be introduced together with the aqueous droplet and pipetted directly on top of a magnet to prevent premature droplet movement on the hydrophobic ELR surface. If droplet immobilization without a magnet is desired (i.e., for microscope imaging), the device surface can be additionally patterned with hydrophilic spots to allow droplets to be “trapped” in a given region of the device. However, once a droplet is trapped in a hydrophilic spot it can no longer be magnetically transported freely on the surface, and hence, is more suited for the last step of an assay. The antibiotic dosing experiments in Fig. 4 and Fig. 5A employs this configuration. To enable droplet immobilization without the need for a constant external magnetic force or an irreversible hydrophilic droplet trap, we designed a device insert consisting of an array of wells with interconnecting “constriction channels” to physically confine the droplets in the wells (Fig. 1C, Movie S1). Each well has a diameter of 2 mm and height of 2.4 mm, which can accommodate a 2 μL droplet and allow for easy pipette access. The pitch between each well is 4.5 mm corresponding to that of a standard 384-well plate to allow for compatibility with conventional plate readers. The wells are interconnected with 1.2 mm wide, 2.4 mm tall constriction channels, which is narrower than the diameter of a 2 μL spherical droplet (~1.56 mm) to provide physical confinement, but still sufficiently wide to allow passage of droplets from one well to another following the application of dragging force via an external magnet (the droplets deform and elongate when traveling through the constriction channels (Movie S1)). We tested various widths of the constriction channels ranging from 1 mm to 1.6 mm and found that a width of 1.2 mm is a good balance between good droplet confinement and ease of dragging droplets from one well to another. We’ve also further designed “extraction channels” with a width of 0.8 mm and height of 0.3 mm which permits the passage of PMPs but blocks the movement of the droplet, which can be used for removal of PMPs from droplets. This configuration is employed in the CellTiter-Glo VRD experiments in Fig. 6. Importantly, the constriction channel insert is also ELR-treated, which is essential for preventing the droplets from adhering-to and getting stuck to the channel walls during droplet movement. As such, droplet immobilization in this configuration relies solely on physical confinement, with no hydrophilic surface interactions/wetting involved. The disadvantage of the constriction channel configuration is that it has fewer degrees of freedom in terms of droplet movement compared to the open surface configuration. In both the open surface and constriction channel configurations of VRD, the movement of droplets rely on the use of PMPs in the droplet plus an external magnetic manipulator on the bottom of the device. However, the presence of PMPs can interfere with assays requiring optical readouts (such as microscopy, absorbance, fluorescence, or luminescence measurements) and potentially even the reactions or analytes in the droplet itself. Thus, it would be advantageous to have an alternative droplet manipulation method for performing VRD that is not dependent on magnetic actuation. Thus, we further developed the “sliding wall” configuration, which adds a device insert consisting of a sliding ELR-treated physical “wall” which entraps the aqueous droplet and allows the droplet to be pushed along the surface of the device by physically sliding the insert (Fig. 1D, Movie S1). The walls are 2.4 mm tall with a 200 μm tall, 800 μm wide channel cutout to provide clearance for the dried reagent to pass under during droplet movement, but the cutout is shallow enough to prevent aqueous droplets from escaping into the channels. Importantly, the silicone oil also serves as a liquid sealant and lubricant between the sliding wall insert and the bottom ELR device surface, thus preventing any aqueous liquid from seeping into the gap between the insert and the device bottom. Worth noting is that the ELR treatment for the sliding wall insert is also essential: it prevents droplets from adhering to the walls or wicking into the channel cutouts, and facilitates the formation of a robust oil seal between the sliding wall insert and the device bottom (owing to the high affinity of silicone oil to ELR treated surfaces).
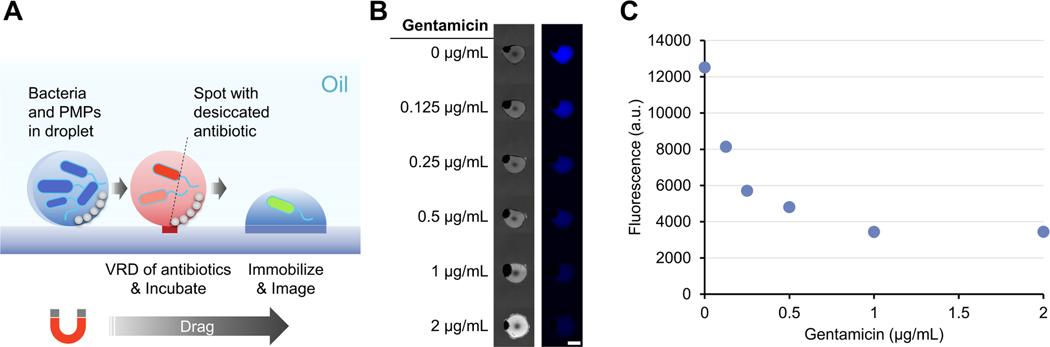
Fig. 4.
VRD of antibiotics (gentamicin) into droplets containing bacteria (P. aeruginosa CFP). (A) Schematic of VRD addition of gentamicin into droplets containing P. aeruginosa CFP, followed by a 24 h incubation, then immobilization onto patterned hydrophilic spots for microscopic imaging. (B) Bright field (left) and fluorescence (right) image of immobilized droplet containing P. aeruginosa CFP after gentamicin treatment with VRD. Scale bar: 1 mm. (C) Quantified fluorescence of immobilized droplets containing P. aeruginosa CFP after gentamicin treatment.
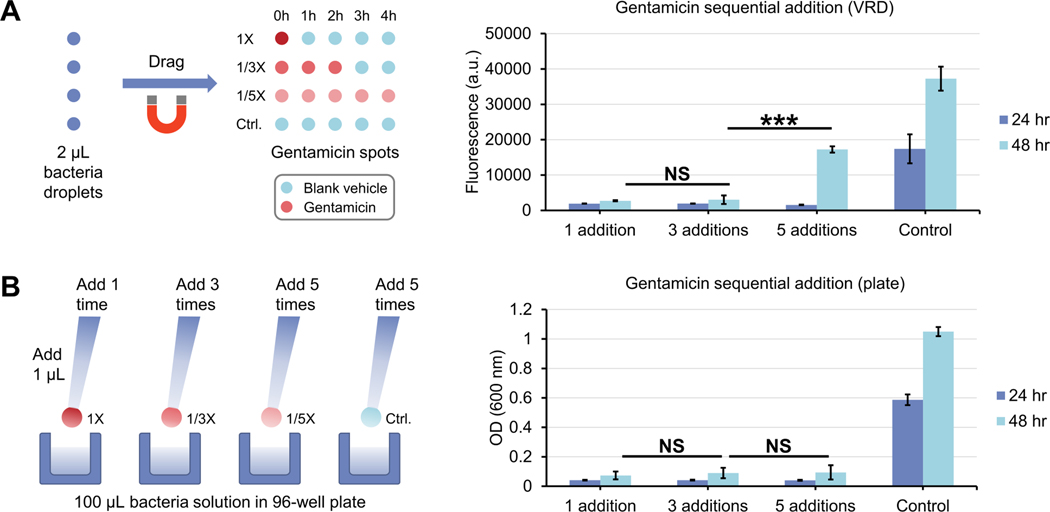
Fig. 5.
Sequential antibiotic dosing of bacteria using VRD compared to a conventional macroscale well plate format. (A) Workflow schematic (left) and results (right) for sequential antibiotic dosing of bacteria (P. aeruginosa CFP) using VRD. (B) Workflow schematic (left) and results (right) for sequential antibiotic dosing of P. aeruginosa CFP using a traditional 96-well plate workflow. Error bars denote the standard deviation from 3 technical replicates. Statistical significance as determined by Student’s t-test is represented by *** p ≤ 0.001, NS = not significant.
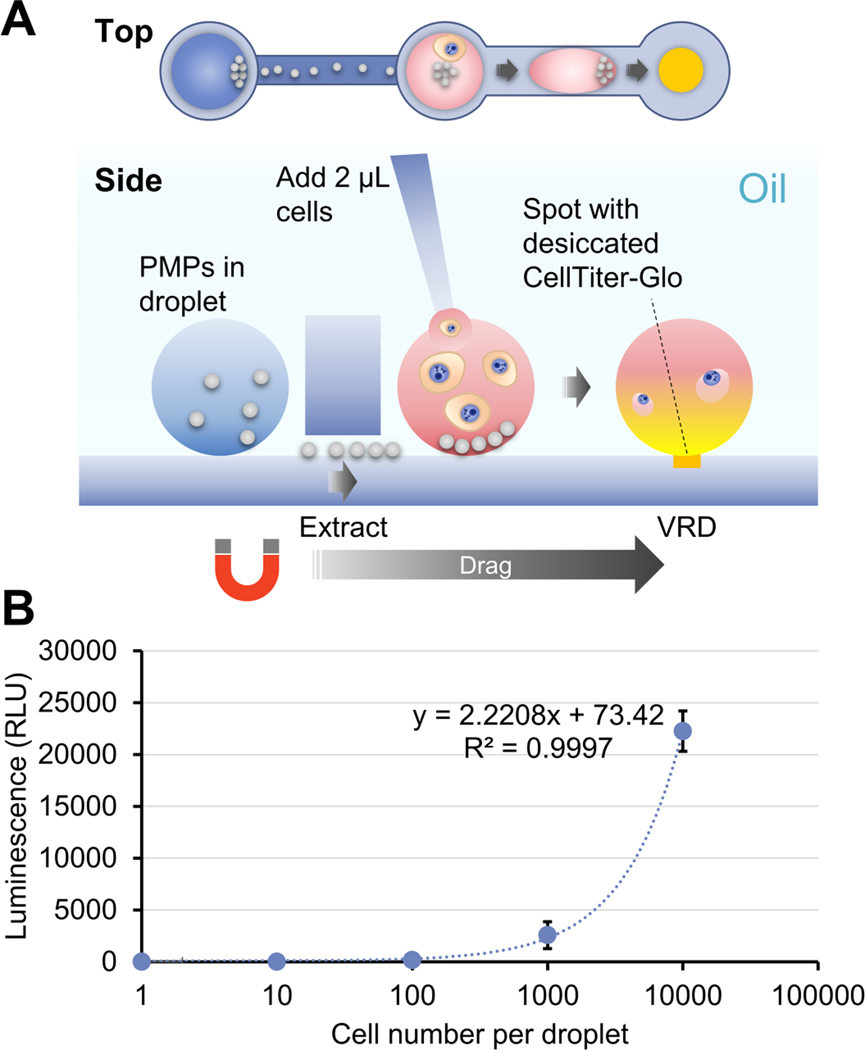
Fig. 6.
VRD of CellTiter-Glo into droplets containing LNCaP cells. (A) Schematic of device and operation process for performing VRD of CellTiter-Glo, an enzymatic assay reagent for quantifying viable cells via luminescence, into droplets containing LNCaP cell suspension. (B) Measured luminescence of droplets after VRD of CellTiter-Glo with the indicated number of input LNCaP cells. Error bars denote the standard deviation from 2 technical replicates.
Characterization of VRD performance
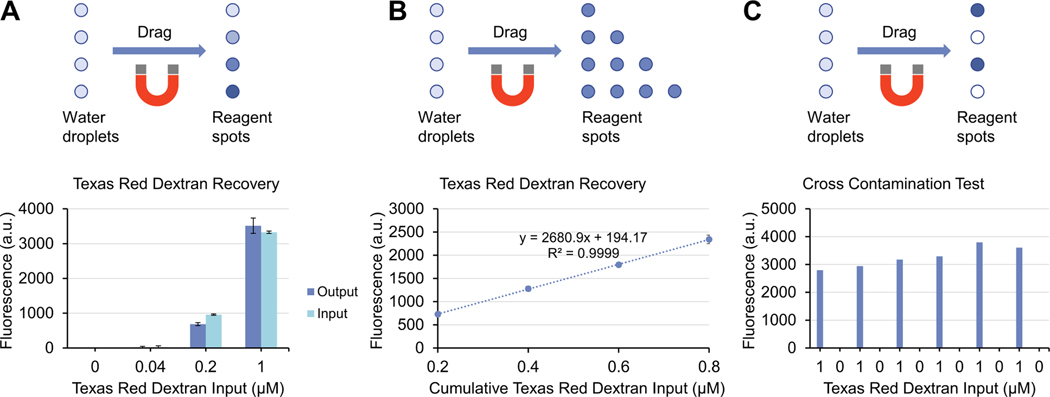
To evaluate the reagent recovery performance of VRD, we chose a fluorescently labeled sugar (Texas Red dextran, 10,000 MW) as a model reagent. Aqueous droplets containing Texas Red dextran at various concentrations were spotted onto the device surface (using the “open surface” configuration device), then dried in a vacuum desiccator. The device was then placed on the magnetic manipulator, filled with silicone oil, then introduced with droplets of water containing 1:10 diluted PMPs via a pipet. The droplets were then dragged into contact with the dried Texas Red dextran spots by sliding the magnetic manipulator and allowed to incubate for ~1 min to reconstitute the Texas Red dextran. The droplet containing the reconstituted Texas Red dextran was then transferred to a fluorospectrometer to measure its fluorescence intensity compared to the input Texas Red dextran solution (prior to drying) to evaluate the recovery efficiency of VRD. Results show that the fluorescence intensities of the input and output droplets is very similar for both the 1 and 0.2 μM input concentrations (the fluorescence of the 0.04 μM condition is too low and close to the background to be accurately measured), suggesting very high reagent recovery efficiency (Fig. 2A). Encouraged by this result, we further performed a sequential VRD experiment to evaluate the ability of the VRD method to perform sequential, multi-step reagent additions. Rows containing 1, 2, 3, and 4 spots of Texas Red dextran were sequentially recovered using VRD to measure the reagent recovery performance ranging from a single addition to 4 sequential additions (Fig. 2B). Results show that the VRD process can not only successfully perform sequential reagent additions, but also has high reliability as indicated by R2 value of the regression line (Fig. 2B), further suggesting near 100% recovery efficiency for each VRD step. To ensure that no cross contamination occurs between adjacent reagent spots, we spotted Texas Red dextran and blank vehicle control in alternating rows and measured the fluorescence of the recovered droplets. As shown in Fig. 2C, no detectable cross contamination was observed. The variation in fluorescence signal for the 1 μM output droplets in Fig. 2C may be attributed to pipetting inconsistencies when dispensing 1 μL droplets onto the device surface, or variability in measuring the fluorescence of microscale droplets using a fluorescent NanoDrop.
Fig. 2.
Reagent recovery performance of VRD. (A) Recovery of Texas Red dextran at various input concentrations using VRD. (B) Sequential, multi-step recovery of Texas Red dextran using VRD. (C) Blank vehicle and 1 μM Texas Red dextran were spotted in alternating sequence and recovered using VRD to demonstrate that cross contamination between adjacent spots does not occur. Error bars denote the standard deviation from 3 technical replicates.
Protein and DNA recovery using VRD
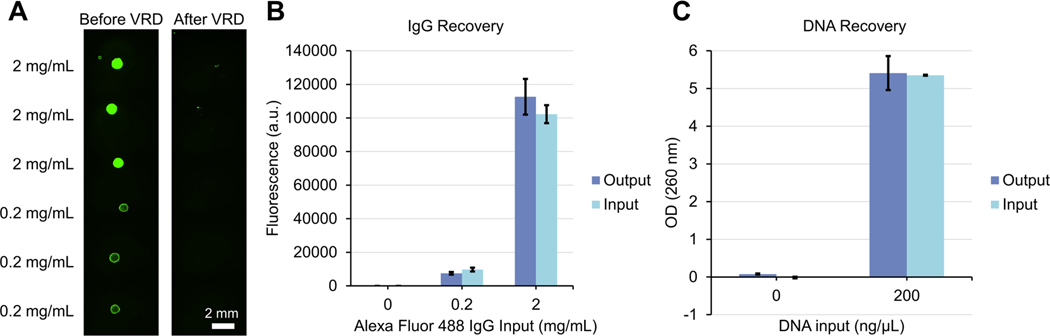
Proteins, especially antibodies, are one of the most widely used molecules in biological assays such as immunoassays and chromatin immunoprecipitation. However, they are also one of the most challenging molecules to work with in DMF systems, especially at high concentrations, often leading to biofouling of the DMF surface which results in process failures stemming from droplets getting stuck at the fouled area,24 or cross-contamination when a droplet is moved across an already used region of the DMF surface.25, 26 Protein surface absorption is already a significant issue when actuating an aqueous droplet containing higher concentrations of proteins across a DMF surface, let alone trying to recover a high concentration dried protein spot deposited on the DMF surface. To test the possibility of recovering proteins using VRD, we spotted a fluorescent antibody solution (Alexa Fluor 488 goat anti-rabbit IgG) at a concentration of 2, 0.2, and 0 mg/mL onto the ELR surface of the device and dried the protein spots in a vacuum desiccator as described above. Trehalose, a commonly used protein stabilizer27 was included in the antibody solution at a concentration of 60 mM as previously reported28 for use as a stabilizer and also as a carrier to provide extra “dry mass” for the dried reagent spot, especially for lower amounts of reagent. We imaged the dried IgG spots on an inverted epifluorescence microscope before and after VRD reconstitution to assess if any residual IgG remained on the surface, and also measured the fluorescence intensity of both the input solution and reconstituted droplet using a fluorospectrometer to assess the protein recovery efficiency. Quantified fluorescence images before and after VRD recovery show that there was no significant difference in the recovered Alexa Fluor 488 IgG spots (both 0.2 and 2 mg/mL) compared to the recovered blank input spots (0 mg/mL) or image background, suggesting full recovery of the dried IgG was achieved (Fig. 3A, Fig. S3), even with a high concentration of antibody (2 mg/mL). The measured fluorescence of the input solution and the reconstituted droplet was also very similar (Fig. 3B). This, to the best of our knowledge, was not previously achieved in DMF systems. Similarly, we also tested the ability of VRD to reconstitute DNA, a molecule ubiquitously used in molecular bioassays and also explored as a data storage medium in DMF systems.21 We spotted droplets of LNCaP cell DNA dissolved in buffer AE (Qiagen) at a concentration of 200 and 0 ng/μL onto the device surface and dried the spots under a vacuum as described. After VRD recovery of the DNA spots, the absorbance of the input and reconstituted droplet was measured using a UV/Visible spectrophotometer at 260 nm to assess DNA recovery efficiency, which showed no significant difference in absorbance values for the input solution and reconstituted output droplet (Fig. 3C), suggesting near complete recovery of the dried DNA with VRD.
Fig. 3.
Protein (IgG) and DNA recovery efficiency of VRD. (A) Fluorescence microscope images of dried Alexa Fluor 488 IgG spots before (left) and after (right) VRD reconstitution. Scale bar: 2 mm. (B) Fluorescence of the input Alexa Fluor 488 IgG solution (without drying) compared to the output droplet (after VRD reconstitution of the dried reagent) as measured on a fluorospectrometer. (C) Absorbance of the input DNA solution (without drying) compared to output droplet (after VRD reconstitution of the dried reagent) as measured on a UV/Visible spectrophotometer at 260 nm. Error bars denote the standard deviation from 3 technical replicates.
Antibiotic recovery and susceptibility testing of bacteria using VRD
To explore the utility of the VRD method for performing bioassays with live biological specimens, we performed an antibiotic dosing experiment with droplets containing fluorescent bacteria (P. aeruginosa CFP). Gentamicin, an antibiotic known to be effective against P. aeruginosa, was spotted and dried onto an “open surface” VRD device which contains a patterned column of hydrophilic spots for droplet immobilization during fluorescence microscope imaging. The device was filled with silicone oil (enough to fully cover the 2 μL droplets), followed by introduction of 2 μL droplets containing P. aeruginosa CFP and silica PMPs in Mueller Hinton broth. The bacterial droplets were then magnetically dragged onto the dried gentamicin spots to perform VRD antibiotic dosing, then allowed to incubate at room temperature for 24 h (while sitting on the magnetic manipulator). After the 24 h incubation, the droplets were magnetically dragged onto the hydrophilic patterned spots for droplet immobilization and fluorescence microscope imaging (Fig. 4A). The incubation period (which allows the bacteria to grow) was not performed with the droplet immobilized on the hydrophilic spots as we wanted to test the ability of the ELR surface to resist fouling during bacterial growth, especially since P. aeruginosa is well known for its ability to form biofilms on mucosal surfaces and medical implants.29 Results show that the ELR surface was highly effective in resisting biofouling by P. aeruginosa, whereas a hydrophobic but non-ELR condition (fluoro silane modified glass with mineral oil) exhibited severe fouling after the same 24 h culture (Fig. S4). Additionally, we were also successful in achieving an antibiotic dose response using this VRD setup (Fig. 4B and 4C), suggesting both successful VRD recovery of the gentamicin antibiotics and culture of P. aeruginosa under silicone oil without surface biofouling. Also worth mentioning is that the silicone oil, in addition to providing ELR properties which enables the VRD process, also greatly reduces liquid evaporation when working with small volumes of liquid over extended periods of time (we found that a 1 μL droplet can be kept under silicone oil for over a week at 37 °C).
Antibiotic sequential dosing of bacteria
After successfully showing both sequential reagent delivery and antibiotic dosing experiments without bacteria-induced biofouling in the VRD system, we then used the VRD system to test a simple biological question to illustrate the assay capabilities of the method: will a higher concentration of antibiotics delivered within a short period of time have the same effect as a lower concentration of antibiotics delivered over a longer period of time, provided the total cumulative dose of the antibiotic remains the same? To test this, we spotted a VRD device with gentamicin at 1X concentration (1 gentamicin spot + 4 blank vehicle spots), 1/3X concentration (3 gentamicin spots + 2 blank vehicle spots), and 1/5X concentration (5 gentamicin spots). After drying the antibiotic spots and filling the device with oil, 2 μL droplets containing P. aeruginosa CFP were introduced and sequentially dragged from 1 antibiotic spot to another to perform VRD (with 1 h interval between each VRD addition). After the 5 VRD additions, the bacterial droplets were allowed to incubate at room temperature for 24~48 h, then dragged onto a column of hydrophilic patterned spots for droplet immobilization and imaging as described above (Fig. 5A). For comparison, we also performed a similar experiment in a macroscale 96-well plate. For the macroscale experiment, 100 μL of P. aeruginosa CFP bacterial suspension was added to a 96-well plate, followed by introduction of 1 μL gentamicin (to a final concentration of 1X, 1/3X, and 1/5X) or blank vehicle control at 1 h intervals for a total of 5 additions (Fig. 5B). The plate was similarly incubated at room temperature for 24~48 h, and bacterial growth was measured via absorbance on a plate reader. Interestingly, although no significant difference in growth inhibition was observed for all 3 antibiotic addition conditions (1X dose at 0 h, 1/3X over 2 hours, and 1/5X over 4 hours) in the macroscale well-plate experiment (Fig. 5B), we saw a significant reduction in antibiotic growth inhibition for the 1/5X dose over 4 hours condition with the VRD method, which only became apparent after a 48-h culture (Fig. 5A). These results suggest that there are differences in bacterial growth dynamics in microscale droplets in the VRD method compared to a conventional macroscale well plate, which become more apparent under antibiotic stress, but may not show up under normal growth conditions (the control group in the VRD and well plate methods appear very similar as shown in Fig. 5). Such differences in growth dynamics have also been previously documented for microscale vs. macroscale culture conditions, attributed to differences such as diffusion dynamics.30
Recovery of complex enzyme assay mixtures using VRD
To test the ability of the VRD method in recovering more complex assay reagent mixtures, we attempted to recover an enzyme assay “kit” mixture: CellTiter-Glo (Promega), a luminescence-based enzyme assay reagent used for quantifying viable cells via measurement of cellular ATP, into droplets containing live mammalian cells. Although the exact composition of the reagent is a commercial secret, at a minimum it contains a mixture of luciferase enzyme, the enzyme substrate (luciferin), enzyme reaction buffer, salts, and potentially other additives. Success in performing this assay with the VRD system will require all the following to be accomplished: drying and recovery of all the essential ingredients in the CellTiter-Glo reagent via VRD, maintenance of luciferase enzyme activity following drying and reconstitution, successful lysis of cells via the recovered CellTiter-Glo reagent, successful reaction of the released cellular ATP with the enzyme assay reagents, and lastly, direct measurement of luminescence signal from the droplets in the device. We spotted CellTiter-Glo onto the VRD device and vacuum dried the reagent as described above, followed by placing a constriction channel insert (this version also contains a shallow “extraction channel” for removal of PMPs) onto the glass bottom of the VRD device (Fig. 6A). The device was then filled with silicone oil, then added with 2 μL droplets of silica PMPs into column 1 wells of the device insert. Separately, 2 μL droplets containing 10000, 1000, 100, 10, and 0 LNCaP cells in RPMI cell media were added into column 2 wells of the device insert, and the dried CellTiter-Glo reagents reside within column 3 wells of the device insert. This device design allows the PMPs to be kept separate from the primary reaction droplet except during droplet actuation.
To start the assay, the PMPs were magnetically extracted from column 1 wells into column 2 wells containing the LNCaP cell droplets. The LNCaP cell droplets were then magnetically transported via the PMPs from column 2 wells, through the constriction channels and into column 3 wells which contain the dried CellTiter-Glo reagent spots to perform VRD (Fig. 6A). The cell droplets were then allowed to incubate for 10 min for cell lysis and the enzyme reaction to occur, followed by magnetically removing the PMPs from the reaction droplets to enable better luminescence measurements to be performed via a plate reader. Results show that we were able to achieve a highly linear luminescent signal as a function of input cell number (Fig. 6B), suggesting that both the VRD recovery and the enzyme assay of CellTiter-Glo were successful.
Conclusions
Here we report a versatile and powerful reagent delivery method: VRD, for adding reagents to microscale aqueous droplets without significantly increasing volume. Compared to conventional droplet merging in DMF systems, VRD affords on-device long-term storage of dried reagents and simpler operation because of the fewer droplets to pipet and manipulate (Table 1). VRD can robustly recover of a wide range of biomolecule reagents, including sugars, proteins, DNA, and complex enzyme assay mixtures. The liquid repellency afforded by the ELR 3-phase system prevents device surface biofouling caused by protein molecules and biofilm forming bacteria, and the oil overlay enables assays involving long-term culture to be performed in microscale droplets without evaporation concerns. These combined advantages highlight the potential of VRD to be employed in a wide range of microscale, digital, and droplet-based bioassays. However, VRD does have trade-offs compared to droplet merging methods including slower reagent delivery (it takes time for dried reagents to be reconstituted), more upfront device preparation work, and incompatibility with reagents that cannot survive a drying and reconstitution cycle (Table 1). Also, due to the extremely high contact angle (~180°) achieved in the ELR system, we hypothesize that the VRD technique is likely not compatible with DMF droplet actuation methods which rely on electrowetting (EWOD), owing to the lack of aqueous liquid wetting that occurs on the ELR surface, although we have yet to prove this. Nevertheless, it will be of interest to explore the compatibility of the VRD method with other DMF droplet actuation methods such as dielectrophoresis (DEP) in the future to allow for broader flexibility in integrating the technology with existing DMF systems.
Table 1:
Comparison of droplet merging with VRD for delivery of reagents into microscale droplets
| Droplet merging | VRD | |
|---|---|---|
| Increases droplet volume | Yes | Minimal |
| On-device reagent storage | Difficult (liquid) | Easy (dry solid) |
| Reagent shelf life | Short (liquid evaporation, limited stability of many reagents in liquid form) | Long (no evaporation, higher stability of dried reagents |
| Reagent delivery speed | Faster (droplet merging is near instantaneous) | Slower (needs time to allow reagents to dissolve) |
| Reagent compatibility | High (biological reagents are in native liquid form) | Moderate (reagent has to survive drying and reconstitution) |
| Setup complexity (manufacturer) | Simple (no pre-depositing and drying of reagents) | More complex (requires pre-depositing and drying of reagents) |
| Operation complexity (user) | Complex (multiple reagents to pipet, multi-droplet operation) | Simple (very few reagents to pipet, single droplet operation) |
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Scott Berry, Chao Li, Jamie Sperger, and Brian Johnson for their helpful suggestions on the project. This study was funded by the National Institutes of Health (NIH 1R01CA247479, NIH P30CA014520) and a UW Institute for Clinical and Translational Research pilot grant (NIH UL1TR002373).
Footnotes
Conflicts of interest
The authors declare the following conflicts of interest. David J. Beebe holds equity in Bellbrook Labs LLC, Tasso Inc., Salus Discovery LLC, Lynx Biosciences Inc., Stacks to the Future LLC, Turba LLC, Onexio Biosystems LLC, and Flambeau Diagnostics LLC. David J. Beebe is also a consultant for Abbott Laboratories. Joshua M Lang holds equity in Salus Discovery LLC. Duane S. Juang holds equity in Turba LLC. A patent application (US-2020–0406260) which covers some of the concepts described in this work was filed through the Wisconsin Alumni Research Foundation.
References
- 1.Pollack MG, Fair RB and Shenderov AD, Appl Phys Lett, 2000, 77, 1725–1726. [Google Scholar]
- 2.Pollack MG, Shenderov AD and Fair RB, Lab on a Chip, 2002, 2, 96–101. [DOI] [PubMed] [Google Scholar]
- 3.Velev OD, Prevo BG and Bhatt KH, Nature, 2003, 426, 515–516. [DOI] [PubMed] [Google Scholar]
- 4.Hunt TP, Issadore D. and Westervelt RM, Lab on a Chip, 2008, 8, 81–87. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y. and Nguyen NT, Lab on a Chip, 2017, 17, 994–1008. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann U, Hadjidj S, Parashar VK, Vandevyver C, Rida A. and Gijs MAM, Sensor Actuat B-Chem, 2006, 117, 457–463. [Google Scholar]
- 7.Rezk AR, Manor O, Friend JR and Yeo LY, Nat Commun, 2012, 3, 1167. [DOI] [PubMed] [Google Scholar]
- 8.Renaudin A, Tabourier P, Camart JC and Druon C, J Appl Phys, 2006, 100, 116101. [Google Scholar]
- 9.Miller EM, Ng AHC, Uddayasankar U. and Wheeler AR, Anal Bioanal Chem, 2011, 399, 337–345. [DOI] [PubMed] [Google Scholar]
- 10.Huang CY, Tsai PY, Lee IC, Hsu HY, Huang HY, Fan SK, Yao DJ, Liu CH and Hsu W, Biomicrofluidics, 2016, 10, 011901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neoprep library prep system, Illumina Inc., 2018, https://support.illumina.com/content/dam/illumina-marketing/documents/products/brochures/brochure-neoprep.pdf. [Google Scholar]
- 12.VolTRAX. Automated sample prep for nanopore analyses, Oxford Nanopore Technologies Inc., 2016, https://nanoporetech.com/products/voltrax. [Google Scholar]
- 13.Li J. and Kim CJ, Lab on a Chip, 2020, 20, 1705–1712. [DOI] [PubMed] [Google Scholar]
- 14.Miller EM and Wheeler AR, Anal Chem, 2008, 80, 1614–1619. [DOI] [PubMed] [Google Scholar]
- 15.Sista RS, Eckhardt AE, Wang T, Graham C, Rouse JL, Norton SM, Srinivasan V, Pollack MG, Tolun AA, Bali D, Millington DS and Pamula VK, Clin Chem, 2011, 57, 1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robison K, Illumina Drops NeoPrep, Omics! Omics!, 2017, http://omicsomics.blogspot.com/2017/02/illumina-drops-neoprep.html.
- 17.Jebrail MJ, Bartsch MS and Patel KD, Lab on a Chip, 2012, 12, 2452–2463. [DOI] [PubMed] [Google Scholar]
- 18.Samiei E, Tabrizian M. and Hoorfar M, Lab on a Chip, 2016, 16, 2376–2396. [DOI] [PubMed] [Google Scholar]
- 19.Yoon JY and Garrell RL, Anal Chem, 2003, 75, 5097–5102. [DOI] [PubMed] [Google Scholar]
- 20.Yang H, Luk VN, Abeigawad M, Barbulovic-Nad I. and Wheeler AR, Anal Chem, 2009, 81, 1061–1067. [DOI] [PubMed] [Google Scholar]
- 21.Newman S, Stephenson AP, Willsey M, Nguyen BH, Takahashi CN, Strauss K. and Ceze L, Nat Commun, 2019, 10, 1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Yu JQ, Schehr J, Berry SM, Leal TA, Lang JM and Beebe DJ, Acs Appl Mater Inter, 2018, 10, 17065–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho SK, Moon HJ and Kim CJ, J Microelectromech S, 2003, 12, 70–80. [Google Scholar]
- 24.Ng AHC, Choi K, Luoma RP, Robinson JM and Wheeler AR, Anal Chem, 2012, 84, 8805–8812. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y. and Chakrabarty K, 2009 Design, Automation & Test in Europe Conference & Exhibition, 2009, 1290–1295. [Google Scholar]
- 26.Lin CCY and Chang YW, Design Automation Conference, 2010, 641–646. [Google Scholar]
- 27.Mensink MA, Frijlink HW, Maarschalk KV and Hinrichs WLJ, Eur J Pharm Biopharm, 2017, 114, 288–295. [DOI] [PubMed] [Google Scholar]
- 28.Cleland JL, Lam X, Kendrick B, Yang J, Yang TH, Overcashier D, Brooks D, Hsu C. and Carpenter JF, J Pharm Sci, 2001, 90, 310–321. [DOI] [PubMed] [Google Scholar]
- 29.Maurice NM, Bedi B. and Sadikot RT, Am J Resp Cell Mol, 2018, 58, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu HM, Meyvantsson I, Shkel IA and Beebe DJ, Lab on a Chip, 2005, 5, 1089–1095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.