Abstract
Modern neuroimaging studies frequently combine data collected from multiple scanners and experimental conditions. Such data often contain substantial technical variability associated with image intensity scale (image intensity scales are not the same in different images) and scanner effects (images obtained from different scanners contain substantial technical biases). Here we evaluate and compare results of data analysis methods without any data transformation (RAW), with intensity normalization using RAVEL, with regional harmonization methods using ComBat, and a combination of RAVEL and ComBat. Methods are evaluated on a unique sample of 16 study participants who were scanned on both 1.5T and 3T scanners a few months apart. Neuroradiological evaluation was conducted for 7 different regions of interest (ROI’s) pertinent to Alzheimer’s disease (AD). Cortical measures and results indicate that: (1) RAVEL substantially improved the reproducibility of image intensities; (2) ComBat is preferred over RAVEL and the RAVEL-ComBat combination in terms of regional level harmonization due to more consistent harmonization across subjects and image-derived measures; (3) RAVEL and ComBat substantially reduced bias compared to analysis of RAW images, but RAVEL also resulted in larger variance; and (4) the larger root mean square deviation (RMSD) of RAVEL compared to ComBat is due mainly to its larger variance.
Keywords: MRI, Scanner effects, Normalization, Harmonization, Alzheimer’s disease
1. Introduction
Over the past few decades, neuroimaging studies of structural magnetic resonance imaging (MRI) have provided invaluable insights into the changes underlying neurodegenerative diseases, such as Alzheimer’s disease (AD) (Frisoni et al., 2010), Parkinson disease (Politis, 2014) and many others. Large-scale studies obtained by aggregating already collected multi-site neuroimaging databases hold the promise to: (1) increase the power to detect important biological or clinical associations; and (2) provide resources for confirmatory analyses and for hypothesis generation for more targeted specialized studies. However, these aggregated datasets often contain hidden technical variability and biases due to differences in sites, scanners, and image acquisition protocols. This problem is widely recognized in genetics under the name of batch effects (Lander, 1999; Leek and Storey, 2007), and is now increasingly recognized in neuroimaging as scanner effects (Fortin et al., 2018). Scanner effects refer to both within- and between-scanner variability and harmonization refers to removal of such variability.
In neuroimaging, it has been shown that scanner effects can affect downstream analyses of derived measures of regional healthy tissue or brain lesion volumes (Jovicich et al., 2013; Schnack et al., 2010; Schwartz et al., 2019). These effects can be very large and exceed the biological variations of interest. For example, Shinohara et al. (2014a) provided evidence of striking differences among the raw image intensities at the different study sites of the ADNI (Mueller et al., 2005) and AIBL (Ellis et al., 2009) studies. Heinen et al. (2016) and Shinohara et al. (2017) demonstrated the difference in manual image volume measures when using 1.5T vs. 3T MRI data. With the increase of multi-site neuroimaging studies in neuroimaging, there is a pressing need to develop harmonization pipelines designed to ensure reproducibility and increase trust in downstream analyses (Karayumak et al., 2019; Ning et al., 2020; Yu et al., 2018).
A different source of variability is due to the arbitrary nature of MRI intensity scale (Nyúl and Udupa, 1999; Shinohara et al., 2011; 2014b). This type of variability makes direct quantitative analysis of image intensities difficult (Shah et al., 2011). Indeed, consider the case when a study participant is scanned twice in an interval that is inconsistent with biological changes. Even if the two images are perfectly registered, their differences should not be expected to be zero (or close to zero) due to such variability. We refer to this type of variability as the intensity unit effects. It was defined as the unwanted technical variability both within- and between-scanners due to the arbitrary units of MRI scans. Intensity normalization refers to normalization of intensities both in terms of location and scale (Wrobel et al., 2020).
Intensity normalization is usually addressed during preprocessing steps such as: registration (Hellier, 2003), harmonization (Fortin et al., 2016), and segmentation (Sweeney et al., 2013). A comprehensive review of the initial intensity normalization methods (Jäger et al., 2006; Leung et al., 2010; Madabhushi and Udupa, 2006; Nyúl and Udupa, 1999; Nyúl et al., 2000; Weisenfeld and Warfteld, 2004) can be found in (Shah et al., 2011). Histogram matching is a popular intensity normalization approach proposed by (Nyúl and Udupa, 1999) and refined by (Nyúl et al., 2000; Shah et al., 2011), though the approach is prone to removing wanted variation, such as false erosion of gray matter (GM) or elimination of white matter lesions (Shinohara et al., 2014b). Shinohara et al. (2014b) formalized the principle of intensity normalization in a seven-rule framework: the statistical principles of image normalization (SPIN). In that work, they also proposed White Stripe, an intensity normalization approach designed to use patches of normal-appearing white matter to estimate the latent sub-distributions of the z-score transformation. Variants of z-score normalization, i.e., the finding of a scan-specific scale and shift parameter to normalize intensities, are common. Fortin and colleagues (Fortin et al., 2016) proposed RAVEL to remove the inter-subject technical variability left after White Stripe intensity normalization. This variability is then extracted as latent variables from the cerebrospinal fluid (CSF) voxels, where intensities are known to be unassociated with disease status and clinical covariates, but might be associated with technical variability. Accordingly, RAVEL estimates the latent variables using singular value decomposition (SVD).
A distinction between various methods is that some operate at the subject level, while others operate at the population level. Subject-level methods are methods that use information from a single subject to derive subject specific estimates, such as White Stripe; while population level methods use information from multiple subject to derive estimates, such as ComBat harmonization. ComBat is a popular location and scale adjustment method that was developed for genomics data (Johnson et al., 2007). It was initially adapted to neuroimaging use for harmonizing Diffusion Tensor Imaging (DTI) measurements across two scanners (Fortin et al., 2017) and cortical thickness measurements obtained from structural MRIs across 11 scanners in two large multi-site studies (Fortin et al., 2018). ComBat was subsequently used for the harmonization of multi-site fMRI datasets (Nielson et al., 2018; Yu et al., 2018) and to address the covariance of multivariate image measurements across sites, named CovBat (Chen et al., 2019). Pomponio et al. (2020) proposed ComBat-GAM, which is an extension of ComBat to non-linear associations covariates (e.g., age) and image summaries (e.g., volumes). Beer et al. (2020) proposed longitudinal-ComBat, an extension of ComBat that used mixed effects models to account for the within-study participant variability. Furthermore, Wachinger et al. (2020) addresses the concept of confounding bias in more detail and applied ComBat to explicitly address non-biological variables in the model.
Both intensity unit and scanner effects are sources of technical variability and should be addressed in multi-site/scanner studies. This type of variability can be expected as within-subject variability in a scan-rescan sample. In this study, we investigated the separate and joint effects of RAVEL and ComBat on regional level harmonization in a study of 16 healthy elderly study participants who were scanned on two different MRI scanners, GE 1.5T and Siemens 3T, at most three months apart. The within-subject variability is assumed as technical variability in this scan-rescan sample. The effects of these methods were assessed in terms of: (1) reproducibility of scanner intensities; (2) comparison of automatic and manual segmentations of the hippocampus, a region used extensively in evaluation of AD progression (Minhas et al., 2020; Schwarz et al., 2016); and (3) reproducibility of MRI derived summary measures that are pertinent to Alzheimer’s disease (Schwarz et al., 2016). Results indicate that (1) RAVEL significantly removed intensity unit effects between and within scanners; (2) RAVEL preserved the segmentation accuracy of hippocampus; (3) RAVEL, ComBat, and the combination of both achieved regional level harmonization to different extents; and (4) ComBat is preferred over RAVEL and RAVEL-ComBat combination due to more consistent harmonization across subjects and image-derived measures.
2. Materials and methods
2.1. Study population and image acquisition
The sample used in this study consists of 16 subjects that are part of an ongoing project (Normal aging, RF1 AG025516 to W.E. Klunk). These 16 subjects were scanned on both GE 1.5T and Siemens 3T MRI scanners, separated by at most 3 months to evaluate scanner differences. The median age in the sample was 77.5 years (range=70 – 79 years) and 25% (n = 4) were males. T1-weighted (T1-w) MRIs were acquired coronally on a GE Signa 1.5T MRI scanner with a birdcage volume coil (TE = 5 ms; TR = 25 ms; Flip Angle = 40°; Pulse Sequence = SPGR) and sagittally on a Siemens MAGNETOM Prisma 3T MRI scanner (TE = 2.22 ms; TI = 1000 ms; TR = 2400 ms; Flip Angle = 8°; Pulse Sequence = MPRAGE). No scanner-specific non-uniformity correction was applied to the 1.5T MRI. Siemen’s Prescan Normalize was applied to the 3T MRI. Image matrix and voxel sizes were 256 × 256 × 124mm and 0.94 × 0.94 × 1.5mm, respectively, for the 1.5T T1-w MRI and 240 × 256 × 160mm and 1.0 × 1.0 × 1.2mm, respectively, for the 3T T1-w MRI.
2.2. Image preprocessing
All images were preprocessed in R (R Core Team, 2020) following the exact preprocessing steps1 prescribed before using RAVEL. Accordingly, all images were first registered to a high-resolution T1-w image atlas (Oishi et al., 2009) using the non-linear symmetric diffeomorphic image registration algorithm proposed in (Avants et al., 2008). Then, the N4 bias correction was applied (Tustison et al., 2010) to each of the images to correct for spatial intensity inhomogeneity. Images were then skull-stripped using the brain mask provided in (Fortin et al., 2016). Throughout this manuscript, these preprocessed but not intensity normalized images will be referred to as RAW images.
2.3. Intensity normalization: RAVEL
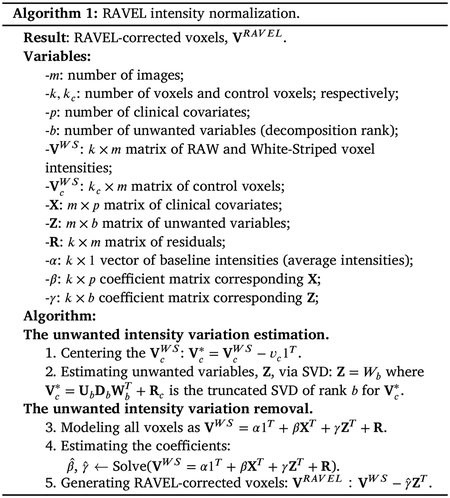
RAVEL is a voxel-wise intensity normalization technique (Fortin et al., 2016). This technique takes the RAW set of MRIs as input and: (1) applies the White Stripe intensity normalization; and (2) estimates and removes the remaining unwanted intensity variation, detailed in Algorithm 1. The second step extracts the singular value decomposition of the observed variability in control voxels (e.g., cerebrospinal fluid voxels) from the population of participants and selects the first b components of the decomposition as an estimation of the unwanted intensity variation. These steps can be found in Algorithm 1 lines 1:2. Once the unwanted variation is estimated from the control voxels, RAVEL then models the intensity values of all the image voxels as a linear combination of the unwanted variables and the clinical covariates (Algorithm 1 line 3). Using this model, the technical variability is estimated for each voxel and then removed from the original voxel intensity values (Algorithm 1 line 4:5). Henceforth, the set of RAVEL intensity normalized images and MRI derived summary measures will be referred to as RAVEL-normalized.
Although adjusting for clinical covariates are optional in RAVEL normalization, we investigated the effects of age and gender on density plots of the tissue types. While controlling for gender did not change the plots, age widened them. Moreover, it was observed that the higher rank resulted in greater overlap of the plots. Since better overlap and narrower density plots are desired, we fit RAVEL to our data by setting the decomposition rank (b) to three and controlling for no clinical variables. More details on the fitting process were provided in the Supplementary File 1 and Supplementary Figures 1 and 2.
2.4. Harmonization: ComBat
ComBat2 (Johnson et al., 2007) is an empirical location (mean) and scale (variance) adjustment based on empirical Bayes (EB) estimation. Data are first modeled as a linear combination of the biological variables of interest and scanner effects (as additive and multiplicative effects). Data adjustments are then made to harmonize across sites/scanners. Our focus is on regional level harmonization, harmonization of the summary measures (i.e, cortical thicknesses and volumes) extracted from RAW images, henceforth called features. Using ComBat, the value for each feature f, i.e. Yijf, for subject j for site/scanner i is first modeled as follows:
| (1) |
Here αf is the average value for feature f, Xij is the design vector of biological variables, βf is the vector of regression coefficients corresponding to Xij, and γif and δif are the additive and multiplicative terms for site/scanner i and feature f, respectively. The error terms, ϵijf, are assumed to be independent with distribution . The estimated parameters of scanner effects are and , respectively. Data are harmonized as follows:
| (2) |
where represents the average over the values of feature f for all subjects, and is estimated using a feature-wise ordinary least-squares approach. See (Johnson et al., 2007) for details on derivation of Eq. (2) and the non-parametric framework. Henceforth, the summary measures harmonized using ComBat will be referred to as ComBat-harmonized.
Here we have used the parametric EB framework and did not adjust ComBat for age and gender. We tested the addition of age, gender or age and gender to our model using F-tests. None of the F-tests were significant, therefore no age, gender or age and gender effects were added to the ComBat model. The summary measures (will be explained in Section 2.6) consist of cortical thickness and volume values, for which separate ComBat models were prepared.
2.5. Normalization and harmonization: RAVEL-ComBat
RAVEL and ComBat were used together in this order to harmonize imaging measures based on intensity normalized images. Figure 1 summarizes the pipeline, which we refer to as RAVEL-ComBat. Henceforth, the summary measures harmonized using RAVEL-ComBat will be referred to as RAVEL-ComBat-harmonized.
Fig. 1.
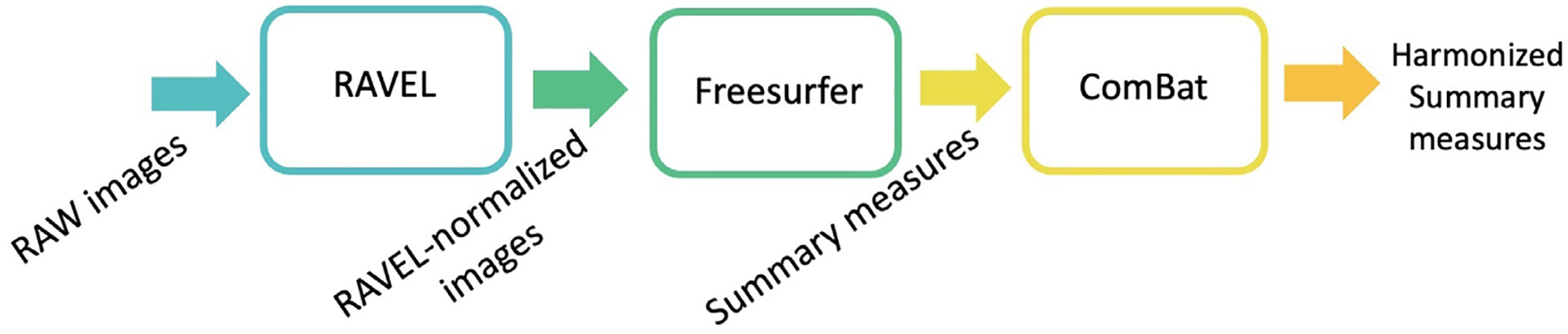
RAVEL-ComBat pipeline.
2.6. Data analysis
Analyses were performed on brain images obtained from two different scanners, a GE 1.5T and Siemens 3T, for 16 healthy study participants described in Section 2.1. Every 1.5T scan was followed by the 3T MR scans within three months, which would not be enough for naturally biologically meaningful changes between the scans. Thus, the observed variability is assumed to be technical variability.
First, to assess the effect of intensity normalization (i.e., RAVEL), we compared voxel intensity density plots before and after normalization for three brain tissue types, using the tissue mask provided in the EveTemplate package Oishi et al. (2009): gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF).
Second, we estimated the scanner effects on summary measures derived using FreeSurfer 7.1.1 (FS) (Fischl, 2012), and evaluated regional-level harmonization effect of RAVEL, ComBat, and the combination of both. FS was consistently run on native-space MRIs. For the RAVEL pipeline, non-linear registration to template space was performed specifically for skull-stripping and RAVEL processing, and inverse transformations were consistently applied to RAVEL images to return them to native space prior to running FS. The full FS pipeline includes its own internal bias correction and skull-stripping steps. Skull-stripping is specifically performed using a watershed algorithm, different from the template-based masking skull-strip in the RAVEL pre-processing pipeline. In examining RAW FS vs RAVEL FS volume and cortical thickness measures, with and without ComBat, we did not want to confound comparisons with inconsistent pre-processing steps, e.g., bias correction and skull-stripping. As such, pre-processing involved: (1) non-linear registration to a common template; (2) N4 bias correction; and (3) skull-stripping for RAW, RAVEL, ComBat, and RAVEL-ComBat pipelines. Subsequently, as stated previously, for both the RAW and RAVEL pipelines, pre-processed MRIs were returned to native space prior to running FS, and subsequently ComBat.
For FS summary measures, we used the cortical thickness for entorhinal, fusiform, inferior parietal, inferior temporal, and middle temporal regions, as well as the volume measure of the entorhinal, inferior temporal, middle temporal, amygdala, and hippocampus. This set is identified as the set of FS-derived measures pertinent to AD in (Schwarz et al., 2016) and were extracted for both hemispheres of images. We then computed the descriptive statistics, including means and standard deviations (SD) of these measures within each of the scanners, before and after applying methods. We also measured scanner effects and evaluated harmonization using two different metrics: 1) bias: the mean of cross-scanner differences (Siemens 3T - GE 1.5T), compared using paired t-tests with p < 0.05 indicating statistical significance, and 2) variance: the root mean square deviation (RMSD) of measures across scanners.
Lastly, to evaluate segmentation accuracy, a neuroradiologist visually rated FS-derived hippocampal segmentations of RAW and RAVEL-normalized MRIs. The RAW brain images were used to evaluate these segmentations of RAW and RAVEL-normalized MRIs. A four-point scale was used for rating the accuracy of the segmentations (1 = poor, 2 = some errors, 3 = good, 4 = excellent). The rater was blinded to subject demographics, segmentation method and image preprocessing and normalization. We did not have the ground truth segmentation, thus we presented the relative accuracy.
3. Results
We first show the unwanted variability in the preprocessed MRIs without intensity normalization (RAW) in Section 3.1. We then present the results of applying other methods, including RAVEL, ComBat, and the RAVEL-ComBat pipeline to data in Sections 3.2, 3.3, and 3.4; respectively.
3.1. Unwanted variability in RAW data
Figure 2 displays the intensity density plots for each of the three brain tissues (CSF in column one, GM in column two, and WM in column three) and different levels of data processing (RAW: panel a, White Stripe: panel b, and RAVEL: panel c). Densities are shown in orange for the 1.5T scanner and in cyan for the 3T scanner. Results indicate that: (1) the distribution of RAW image intensities vary substantially between- and within-scanners; (2) White Stripe substantially improves the distance between densities, especially in white matter; and (3) RAVEL further improves the distance between densities, especially in CSF and GM.
Fig. 2.
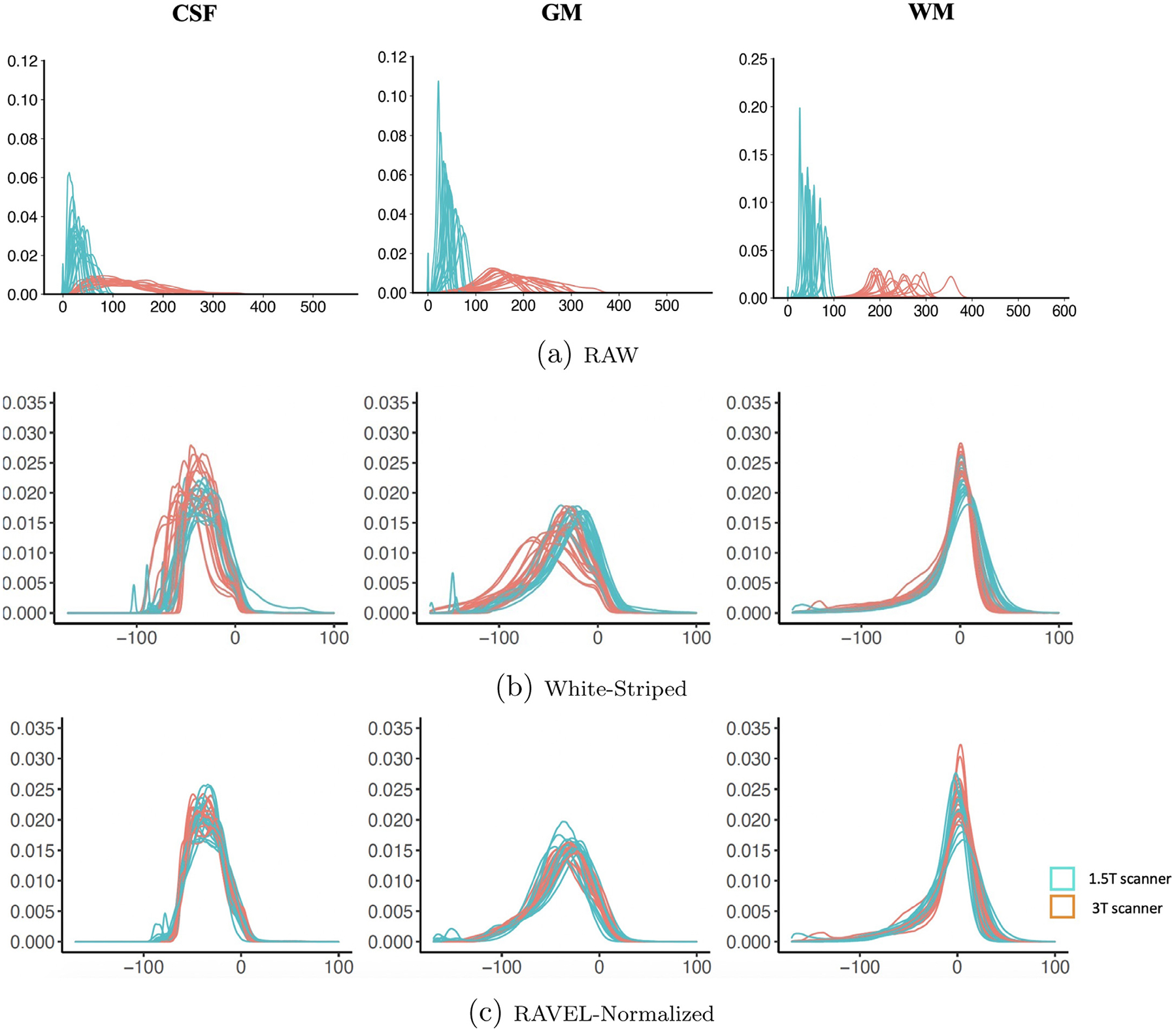
Density plots of MRI voxel intensities by tissue type (cerebrospinal fluid (CSF), grey matter (GM), and white matter (WM)) across scanners (GE 1.5T (orange) and Siemens 3T (cyan)) for (a) RAW, (b) White-Striped, and (c) RAVEL-normalized MRIs. Note that White Stripe increases the overlap of the densities greatly for WM, which was the intent of the method, but there is still some non-overlapping regions for GM and CSF, which RAVEL improves. Initially referenced in Section 3.1. For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
Table 1, provides bias (mean of cross-scanner differences) and variance (RMSD values) of the 20 summary measures extracted for all 4 methods. Also, for each method in Table 1 the statistically significant biases and increased RMSDs (compared to their corresponding values in RAW data) were highlighted and presented in bold, respectively. Focusing on these two metrics for RAW data in Table 1, we observed scanner effects as: (1) statistically significant bias for 11 summary measures, and (2) deviation of values across scanners for all summary measures. In addition, Supplementary Table 1 shows the within-scanner mean and SD of the summary measures (cortical thicknesses and volumes) in both hemispheres for RAW, RAVEL, ComBat, and RAVEL-ComBat data.
Table 1.
Mean (SD) of cross-scanner differences (Siemens 3T - GE 1.5T) as well as cross-scanner RMSDs, for cortical thickness and volume measures relevant to AD. These statistics were prepared for each of the RAW, RAVEL, ComBat, and RAVEL-ComBat methods. For each method, the increased RMSD values (compared to RAW) were reported in bold and the statistical significant differences (p < 0.05) were highlighted. Information on confidence intervals of the t-tests is reported in Supplementary Table 2.
| RAW | RAVEL | ComBat | RAVEL-ComBat | |||||
|---|---|---|---|---|---|---|---|---|
| Mean(SD) | RMSD | Mean(SD) | RMSD | Mean(SD) | RMSD | Mean(SD) | RMSD | |
| ROIs | Cortical Thickness (mm) | |||||||
| Left | ||||||||
| Entorhinal | 0.22 (0.23) | 0.31 | 0.19 (0.42) | 0.45 | 0.08 (0.22) | 0.23 | 0.18 (0.40) | 0.43 |
| Fusiform | 0.24 (0.10) | 0.26 | 0.10 (0.23) | 0.25 | 0.11 (0.09) | 0.14 | 0.10 (0.22) | 0.23 |
| Inferior Parietal | −0.05 (0.10) | 0.11 | −0.15 (0.20) | 0.25 | −0.04 (0.10) | 0.10 | −0.04 (0.19) | 0.19 |
| Inferior Temporal | 0.25 (0.17) | 0.30 | 0.06 (0.27) | 0.27 | 0.11 (0.16) | 0.19 | 0.08 (0.24) | 0.25 |
| Middle Temporal | 0.08 (0.15) | 0.17 | −0.01 (0.19) | 0.18 | 0.02 (0.15) | 0.15 | 0.03 (0.19) | 0.18 |
| Right | ||||||||
| Entorhinal | 0.23 (0.45) | 0.49 | 0.17 (0.34) | 0.37 | 0.08 (0.43) | 0.43 | 0.15 (0.32) | 0.35 |
| Fusiform | 0.22 (0.11) | 0.25 | 0.05 (0.20) | 0.20 | 0.10 (0.10) | 0.14 | 0.06 (0.20) | 0.20 |
| Inferior Parietal | −0.02 (0.07) | 0.07 | −0.07 (0.15) | 0.16 | −0.02 (0.07) | 0.07 | −0.01 (0.14) | 0.14 |
| Inferior Temporal | 0.26 (0.11) | 0.28 | 0.09 (0.16) | 0.18 | 0.11 (0.10) | 0.15 | 0.08 (0.15) | 0.17 |
| Middle Temporal | 0.05 (0.16) | 0.16 | −0.01 (0.13) | 0.13 | 0.01 (0.16) | 0.15 | 0.03 (0.13) | 0.13 |
| ROIs | Volume (cm)3 | |||||||
| Left | ||||||||
| Entorhinal | 0.08 (0.25) | 0.26 | 0.19 (0.41) | 0.44 | 0.01 (0.25) | 0.24 | 0.12 (0.40) | 0.40 |
| Inferior Temporal | 0.79 (1.09) | 1.09 | 0.78 (0.84) | 0.92 | 0.31 (0.65) | 0.70 | 0.26 (0.73) | 0.75 |
| Middle Temporal | 0.16 (0.84) | 0.83 | −0.35 (1.05) | 1.07 | −0.16 (0.74) | 0.73 | −0.23 (0.98) | 0.98 |
| Amygdala | 0.13 (0.20) | 0.27 | 0.09 (0.15) | 0.17 | 0.06 (0.19) | 0.19 | 0.04 (0.14) | 0.14 |
| Hippocampus | −0.08 (0.15) | 1.25 | −0.19 (0.19) | 0.27 | −0.03 (0.14) | 0.14 | −0.05 (0.18) | 0.20 |
| Right | ||||||||
| Entorhinal | 0.09 (0.27) | 0.91 | 0.12 (0.33) | 0.34 | 0.01 (0.26) | 0.25 | 0.07 (0.32) | 0.32 |
| Inferior Temporal | 0.98 (0.80) | 0.23 | 0.67 (0.89) | 1.09 | 0.42 (0.71) | 0.80 | 0.41 (0.85) | 0.92 |
| Middle Temporal | 0.21 (0.92) | 0.16 | 0.06 (0.85) | 0.82 | −0.10 (0.79) | 0.77 | 0.02 (0.78) | 0.76 |
| Amygdala | 0.05 (0.09) | 0.10 | 0.02 (0.08) | 0.08 | 0.03 (0.09) | 0.09 | 0.01 (0.08) | 0.08 |
| Hippocampus | −0.09 (0.15) | 0.17 | −0.10 (0.24) | 0.26 | −0.03 (0.14) | 0.14 | −0.04 (0.24) | 0.24 |
3.2. RAVEL
3.2.1. Segmentation accuracy result
Neuroradiological ratings comparing hippocampal segmentation of RAW to RAVEL-normalized images revealed that RAVEL neither significantly improve nor deteriorate the FS segmentations. In fact, ratings of the left hemisphere segmentation in Fig. 3a showed that RAVEL performed slightly worse than RAW, by having a greater number of erroneous (5 to 1) and fewer good (26 to 29) and excellent (1 to 2) segmentations. However, for the right hemisphere segmentations (Fig. 3b), RAVEL performed similarly to RAW, by having similar erroneous (2 to 2), one more good (29 to 28), and one less excellent segmentations (1 to 2). The Wilcoxon hypothesis testing on collected ratings of the left and right hemispheres resulted in (W-value = 10.0, p-value = 0.096) and (W-value = 12.0, p-value = 0.705), respectively.
Fig. 3.
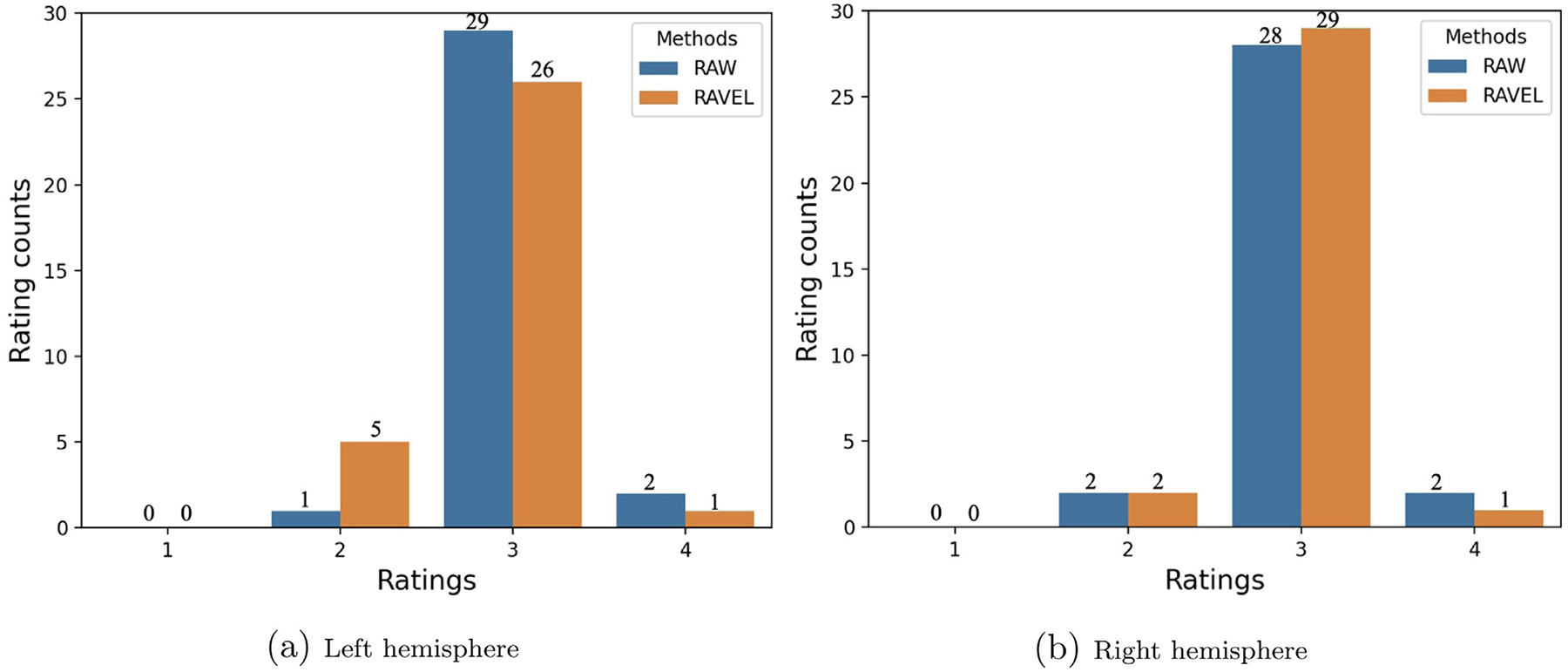
Visual ratings of FS-based hippocampal segmentations for RAW and RAVEL-normalized (RAVEL) MRIs, using a four-point rating scale (1 = poor, 2 = some errors, 3 = good, and 4 = excellent). Initially referenced in Section 3.2.1.
Two single cases are illustrated in Figs. 4 and 5, showing the hippocampual segmentation on RAW brain images by method with arrows pointing to the erroneous segmented voxels. Fig. 4 presents one single case in which RAVEL results in a more accurate hippocampal segmentation than RAW. The RAW brain image, without any segmentation on, is depicted in Fig. 4a. Figure 4b shows that the segmentation based on RAW images (in yellow) has extraneous segmented voxels over the CSF and adjacent white matter areas, when compared to segmentation based on RAVEL (in red). The orange area shows the overlap of the two segmentations and the remained yellow and red areas are for RAW and RAVEL segmentations, respectively. Figure 5a depicts the RAW brain image for the second case. Fig. 5b presents this case in which RAVEL (in red) results in a less accurate hippocampal segmentation than RAW (in yellow), by not capturing the entire hippocampus, pointed by arrows. The overlapped area of these two segmentations is in orange.
Fig. 4.
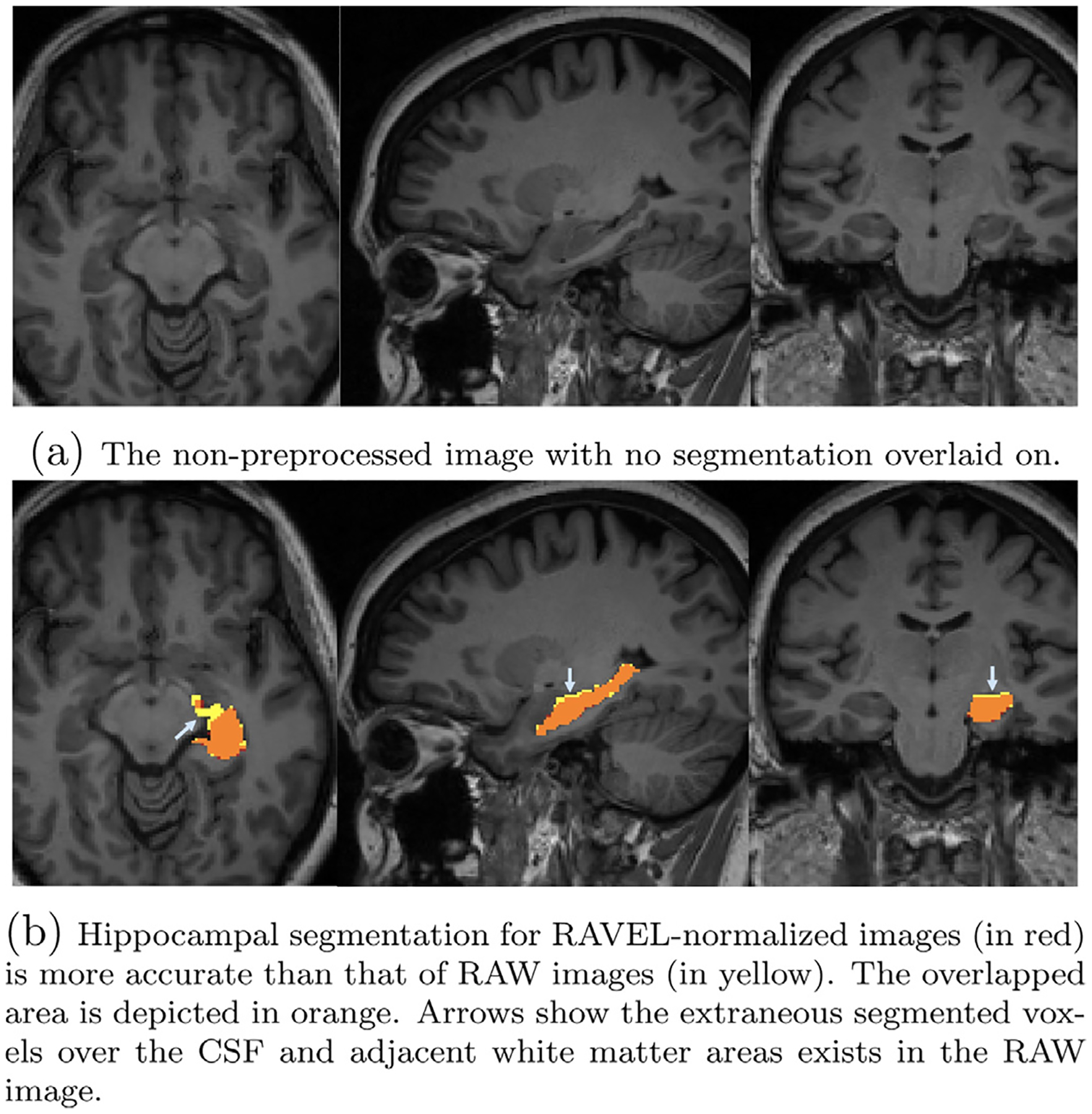
Axial, sagittal, and coronal slices of a single subject MRI with overlaid hippocampal segmentations generated by FS for RAW and RAVEL-normalized images. Initially referenced in Section 3.2.1. For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
Fig. 5.
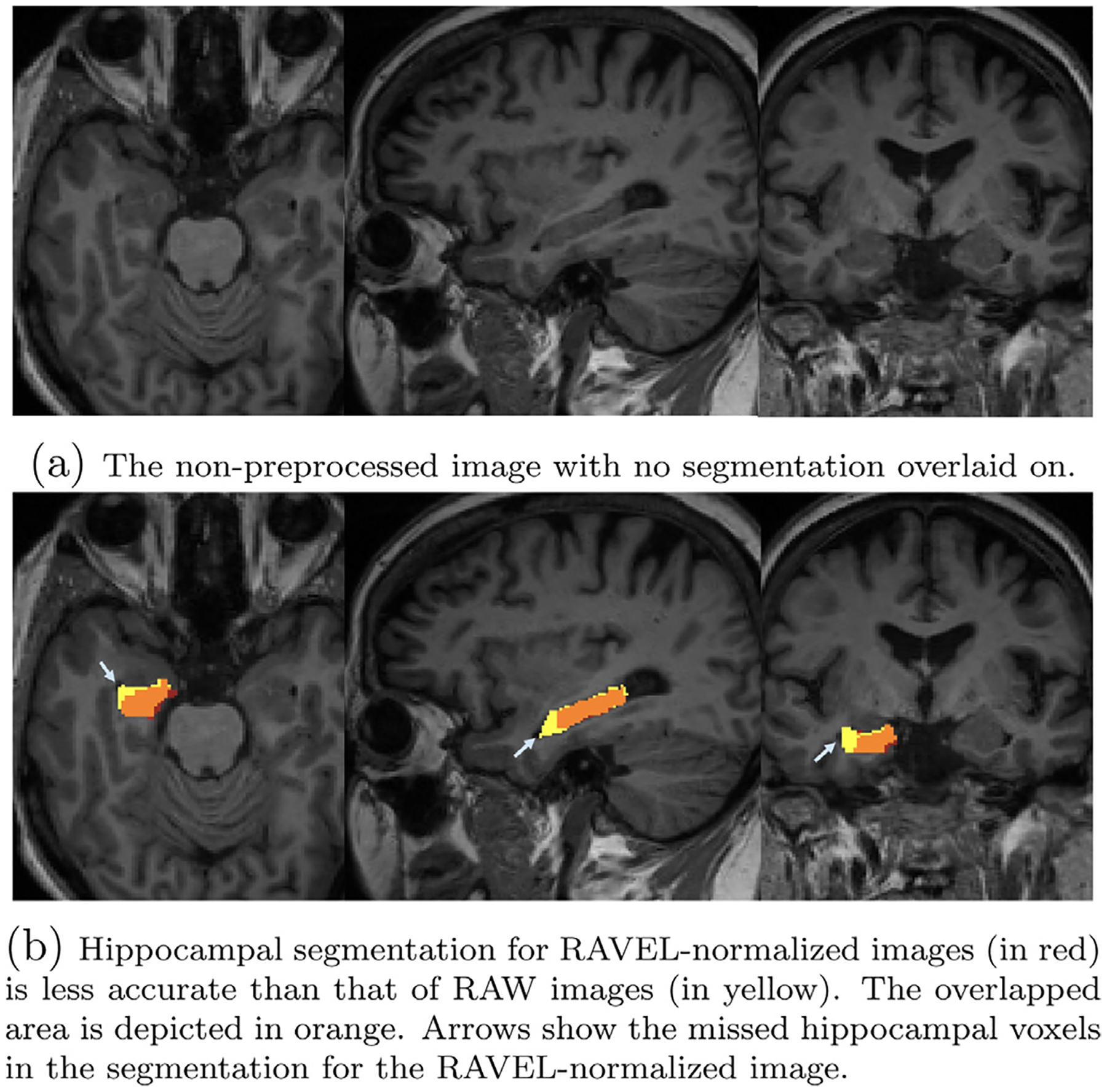
Axial, sagittal, and coronal slices of two single subjects MRI with overlaid hippocampal segmentations generated by FS for RAW and RAVEL-normalized images. Initially referenced in Section 3.2.1. For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
3.2.2. Harmonization result
For most of the derived imaging measures reported in Table 1, RAVEL decreased bias (13 decreases versus 7 increases) and increased variability (SD) of differences (6 decreases versus 14 increases), when compared to the measures from the RAW data in column 1. The comparison has been done based on absolute values of bias. The number of measures with statistically significant bias decreased from 11 for RAW to 6 for RAVEL and the RAVEL-normalized images resulted in change of RMSDs as variances (11 decreases versus 9 increases), when compared to RAW. Fig. 6 presents these total number of changes (decrease, increase, and no change) in (a) bias, (b) variation (SD) of cross-scanner differences, and (c) variance (RMSD), in addition to (d) the number of statistically significant biases over all 20 summary measures.
Fig. 6.
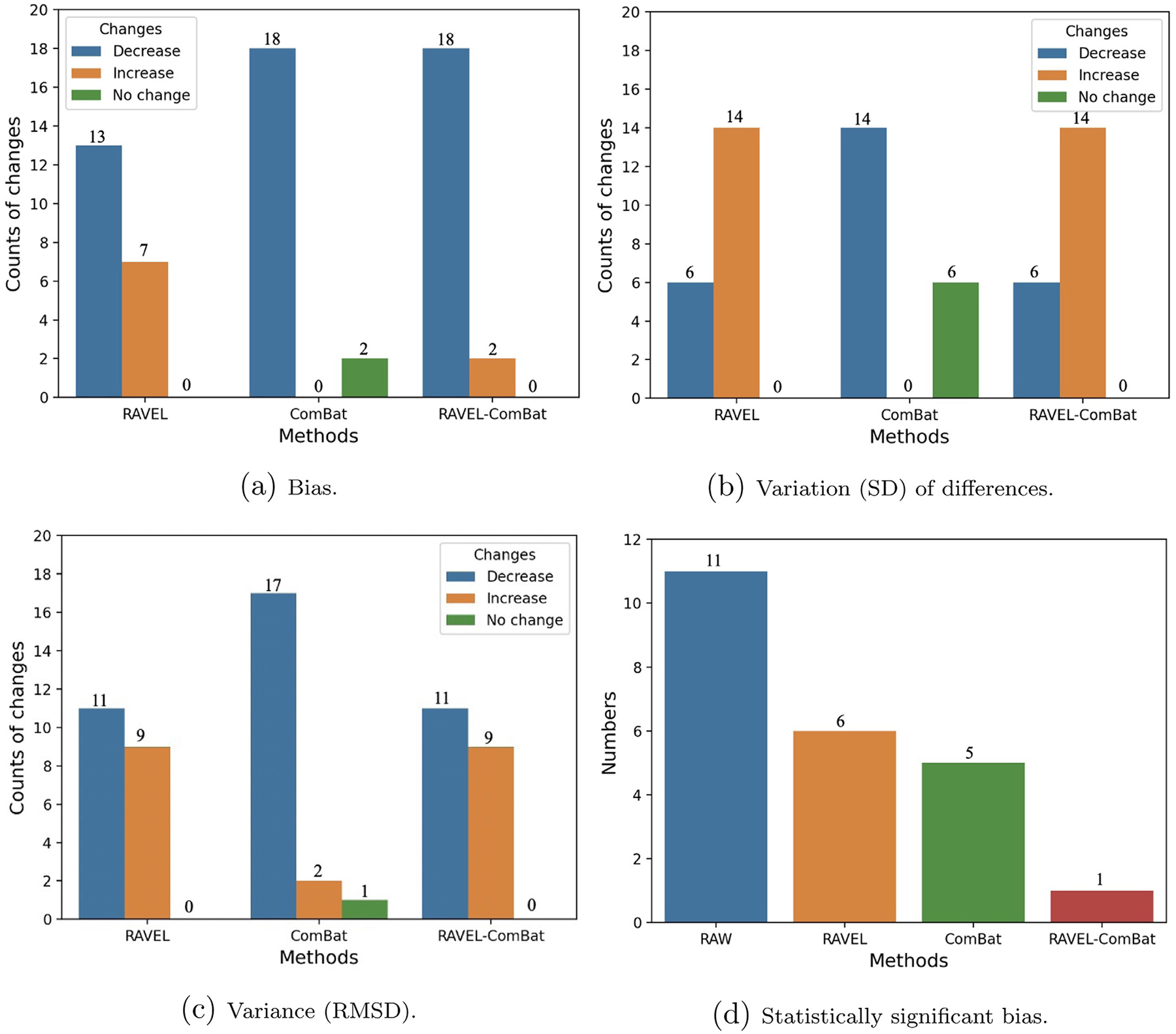
Bar plots showing number of regional summary measures with changes (classified as decrease, increase, and no change) in (a) cross-scanner bias, (b) variation (SD), and (c) variance (RMSD) for tested methods compared to RAW. Part (d) shows the number of regional summary measures with statistically significant cross-scanner bias for each method. Statistical measures were calculated over 20 FS-derived summary quantities (listed in Section 2.6). For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
Columns fourth and fifth in Table 1 show the mean (SD) of cross-scanner differences and RMSD values for all summary measures extracted from the RAVEL-normalized images. These results are complemented by the statistically significant biases (highlighted values) and increased RMSDs (values in bold). This table is accompanied by Fig. 7, visualizing the results of this table for the summary measures, including volumes respectively of inferior temporal and middle temporal for left and right hemispheres, as well as cortical thickness of entorhinal and inferior parietal in left hemisphere. In Fig. 7, the cross-scanner differences of all subjects were depicted as a line plot for each method. The smoother line plots indicate methods which resulted in lower variation (SD) of cross-scanner differences. The line plots closer to x-axis depict methods which resulted in smaller variances (cross-scanner differences are closer to zero).
Fig. 7.
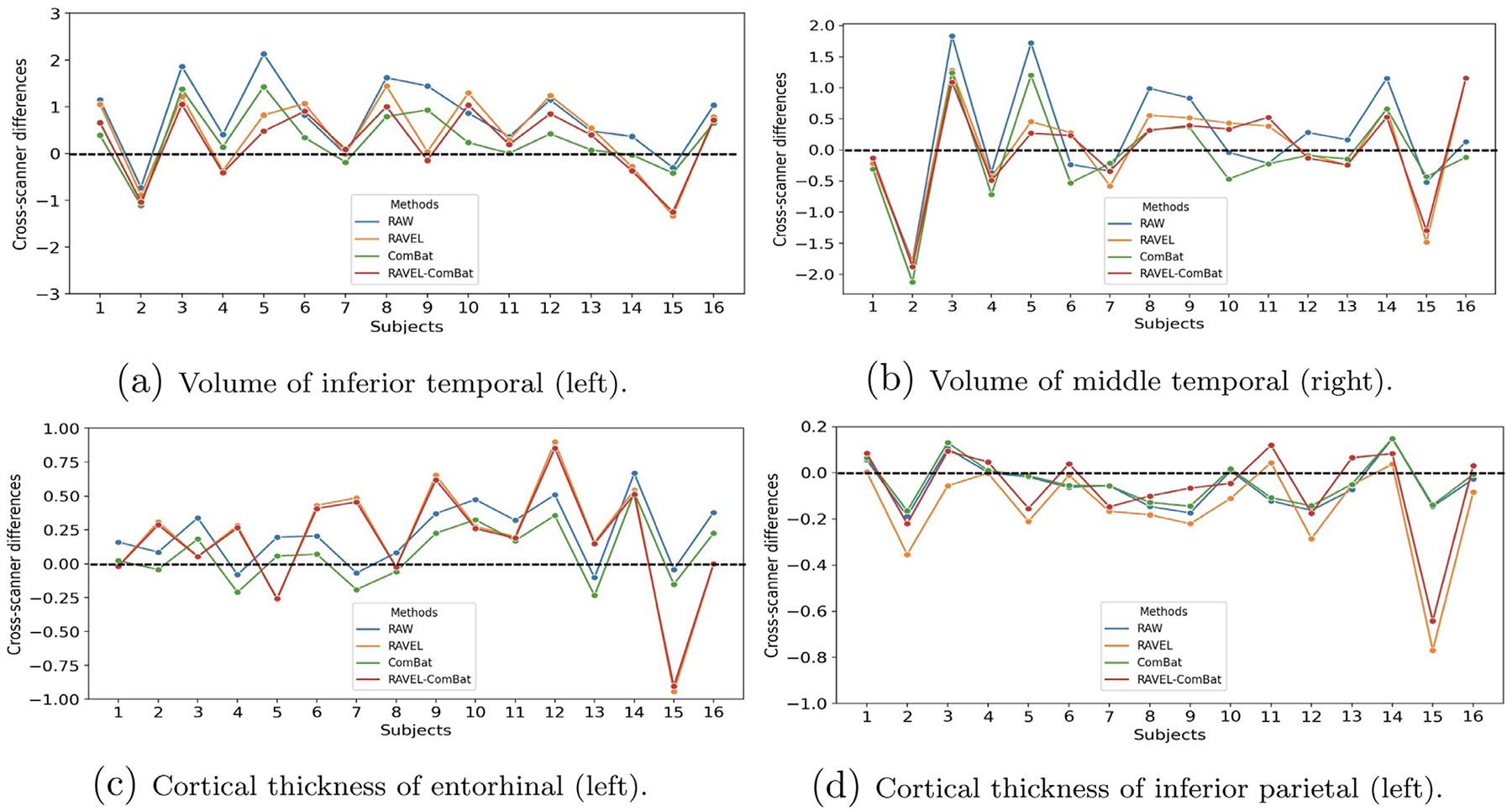
Line plots depicting cross-scanner differences (Siemens 3T - GE 1.5T) for all subjects and methods. The plots depicted for 4 summary measures which were also reported in Table 1. The plotted differences were in millimeter (mm) and cubic centimeter (cm)3 for cortical thicknesses and volumes, respectively. A smoother line plot indicates a lower SD of differences and a plot closer to x-axis (zero differences) shows lower variance. The line plots showed that in (a) all three methods succeed in harmonization, in (b) all methods increased variance, in (c) and (d) RAVEL and RAVEL-ComBat increased variance while ComBat succeed in harmonization. For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
Based on the results in Table 1, the volume of inferior temporal (left hemisphere) is one of the measures that RAVEL harmonized by decreasing bias, SD of differences, and variance, resulting in no statistically significant differences of bias. These results were supported in Fig. 7a where the line plot for RAVEL is smoother than RAW and closer to x-axis. However, the results in Table 1 showed that RAVEL resulted in increased variance for the rest of the 3 summary measures. Such pattern could also be observed in Fig. 7b, c, and d, as the plots for RAVEL deviated from x-axis due to increased peaks when compared to the plots for RAW.
Based on our observations, RAVEL had some potential harmonization effect on our data by showing decrease in either bias, variance, or number of summary measures with statistically significant bias. However, this effect does not seem to be consistent among (1) summary measures (increased bias and variance for some summary measures), and (2) subjects (increased SD of differences for some summary measures, for example Fig. 7b, c, and d).
3.3. ComBat
The results for ComBat-harmonized measures in Table 1 showed that ComBat decreased bias and SD of cross-scanner differences for most of the measures. Considering absolute values of bias, these statistics were 18 and 14 decreases for bias and SD of differences, respectively, while no changes have been seen for the rest of measures. These results were supported by the decreased number of statistically significant biases (11 for RAW decreased to 5 for ComBat) and RMSD values (17 decreases, 2 increases, and 1 no change). These statistics were depicted in Fig. 6.
Based on the results in Table 1, ComBat successfully harmonized volume of inferior temporal and cortical thickness of entorhinal (both for left hemisphere), by decreasing bias, SD of differences, and variance as well as removing statistical significance of bias. These results were supported by the corresponding line plots of ComBat in Figs. 7a and c, where they were similar to RAW but smoother and closer to x-axis. However, the results for cortical thickness of inferior parietal (left hemisphere) did not change noticeably (ComBat almost overlapped RAW in Fig. 7d) and the volume of middle temporal (right) still retained the increase in variance (Fig. 7b).
Based on our observations, ComBat had potential harmonization effect on our data by showing decrease in bias, variance, SD of differences, or number of summary measures with statistically significant bias. This effect seems to be more consistent across summary measures and subjects which makes ComBat to be preferred over RAVEL for the task of regional harmonization.
3.4. RAVEL-ComBat pipeline
Results of comparing RAVEL-ComBat to RAW in Table 1 showed that this method decreased bias for most of the summary measures (18 decreases versus 2 no changes), when the absolute values of bias were compared. The number of summary measures with statistically significant bias decreased from 11 for RAW to 1 for this pipeline. However, RAVEL-ComBat almost increased the SD of differences (6 decreases versus 14 increases) as well as the variance for almost half of the summary measures (11 decreases versus 9 increases). These results were summarized in Fig. 6.
Results in Table 1 showed that RAVEL-ComBat followed almost similar pattern with RAVEL in harmonization of the 4 selected summary measures. RAVEL-ComBat successfully harmonized volume of inferior temporal (left), while increased the variance for the other 3 measures. These results were also visualized in Fig. 7 where all the line plots for RAVEL-ComBat closely followed RAVEL’s. However, comparing these two methods, minor improvements have been observed for RAVEL-ComBat which were (1) decreasing the number of biases and (2) resulting smaller increases in RMSD values (the number of changes in variance are similar between the methods). Such differences could be seen for cortical thickness of inferior parietal (left) in Fig. 7d.
Even though RAVEL-ComBat was improved by ComBat due to its decreased number of biases, this method was still more similar to RAVEL in terms of regional harmonization. Although the number of statistically significant biases decreased noticeably using RAVEL-ComBat, the harmonized measures still suffer from increased SD of differences and variance when compared to RAW and ComBat. In conclusion, ComBat would be preferred over RAVEL and RAVEL-ComBat as these two methods are inconsistent among subjects and summary measures.
4. Discussion
Unwanted technical variability in multi-site clinical trials and observational studies is an increasingly common problem and mainly introduced as two types of variability: (1) intensity unit effects, and (2) scanner effects. Scanner effects can be removed by harmonization techniques categorized into image level harmonization and regional level harmonization methods. The image level harmonization techniques for MRs were largely categorized in literature (Dewey et al., 2019, 2020; Zuo et al., 2021) into either unsupervised methods which use unpaired data, or supervised methods which use paired data.
A group of unsupervised methods approached harmonization as the image-to-image (I2I) synthesis problem, in which the images of one scanner are synthesized to be more similar to images of the other scanner, called target s scanner. However, if the synthesis process is left unconstrained on preserving the brain structure, it may result in unintentional anatomical modification of images during harmonization. In response, several approaches harmonized images at contrast level, aiming to preserve the structural information of them (Dewey et al., 2020; Zuo et al., 2021). These methods disentangled images into structural and contrast components using a cross-modality paired data. Even though this data is not paired across scanners, it still made these approaches situational. Another group of harmonization methods approached I2I translation using generative adversarial networks (GAN). Modanwal et al. (2020) used a CycleGAN to constraint on anatomical information of the images and Liu et al. (2021) preserved the structural information of images by adapting them to the style (contrast information) of target images. While the former is fundamentally limited to two scanners, the latter required an arbitrarily chosen target scanner.
Among the two supervised methods, DeepHarmony (Dewey et al., 2019), an I2I synthesis method used two U-Net networks for synthesizing the images of the two scanners that it can harmonize. While DeepHarmony is limited to two scanners, mica (Wrobel et al., 2020) is a multi-scanner harmonization method aimed to modify voxels of images to have the cumulative distribution function (CDF) of their corresponding voxels in the target scanner. This method though, required an arbitrarily chosen target scanner. Even though the requirement of paired data (either across modalities or scanners) made many of the current harmonization methods situational, the experimental benefits from the paired images are quite informative at these early stages.
In this study, we focused on regional level harmonization of summary measures pertinent to AD. We hypothesized that the pipeline of intensity normalization method using RAVEL, and harmonization method using ComBat, would result in better removal of unwanted variability and consequently would improve regional level harmonization. Accordingly, we collected a paired cohort of 16 healthy elderly study participants scanned on two different MRI scanners, GE 1.5T and Siemens 3T. We assumed that technical variability manifests as within-subject differences for summary measures and reducing these differences would achieve regional level harmonization appearing as lowered bias and variance.
Consistent with previous reports, our results showed that RAVEL further normalized the White-Stripe-normalized images, specifically CSF and GM areas (Fortin et al., 2016). Moreover, RAVEL preserved the anatomical information of images. For example, it preserved the segmentation accuracy for the hippocampus. These results are consistent with our previous findings from the group’s previously reported results for a multi-site Down Syndrome study (Minhas et al., 2020). Regarding regional harmonization, RAVEL, ComBat, and RAVEL-ComBat effectively harmonized the 1.5T and 3T MRI summary measures in this study, in that all techniques reduced the number of statistically significant biases across the regional cortical thicknesses and volumes examined. ComBat, however, demonstrated a more consistent harmonization effect across subjects and summary measures as compared to RAVEL and RAVEL-ComBat. Based on the results on our data, ComBat would be preferred to RAVEL and RAVEL-ComBat for the task of regional harmonization to avoid the inconsistency across subjects and summary measures that were observed with the other two pipelines.
Despite demonstrating an overall reduction in the number of statistically significant biases between FreeSurfer outcome measures from 1.5T and 3T MRI scans (Table 1 and Fig. 6d), the application of RAVEL introduced a significant difference in the left inferior parietal cortical thickness and increased RMSD across multiple regional cortical thickness and volume measures (Table 1). There are multiple possibilities as to why RAVEL introduced unwanted differences and variability. The quality of FreeSurfer segmentations, and therefore outcome measures, is dependent on the GM-WM contrast in the T1 MR image, with increased contrast resulting in more accurate FreeSurfer segmentations. To examine the effects of RAVEL on GM-WM contrast for the 1.5T and 3T scans, we calculated the area under the receiver operating characteristic (AU-ROC) for classification of voxel intensity values as GM relative to WM. For this, we first extracted the histograms of GM and WM using the tissue mask in the EveTemplate package (Oishi et al., 2009). We then looked at the classification of GM voxels from WM as the problem of estimating the separation of their histograms. For classifying the GM and WM voxels, we set the voxel intensity thresholds from one end of the union of histograms to the other. Every threshold position generated a point on the AUC curve. A complete separation of histogram would result in AUROC = 1 and completely overlapped histograms would give AUROC = 0.5. AUROC values across subjects for 1.5T and 3T scans before and after RAVEL are shown in Fig. 8. RAW 3T scans consistently had better GM-WM contrast than RAW 1.5T scans (mean(SD) RAW 3T AUROC: 0.849(0.028); mean(SD) RAW 1.5T AUROC: 0.812(0.033)). The application of RAVEL reduced the mean of absolute differences between 3T and 1.5T AUROC values from 0.037 ± 0.021 to 0.017 ± 0.018. As such, just as RAVEL effectively normalized MRI voxel intensity distributions across scanners, it also normalized GM-WM contrasts. However, in doing so, RAVEL reduced GM-WM contrast, on average, across 3T MRIs (mean(SD) RAW 3T AUROC: 0.849(0.028); mean(SD) RAVEL 3T AUROC: 0.840(0.018)). This reduction in contrast may have reduced quantitative accuracy and increased variability in cortical thickness and volume FreeSurfer measures.
Fig. 8.
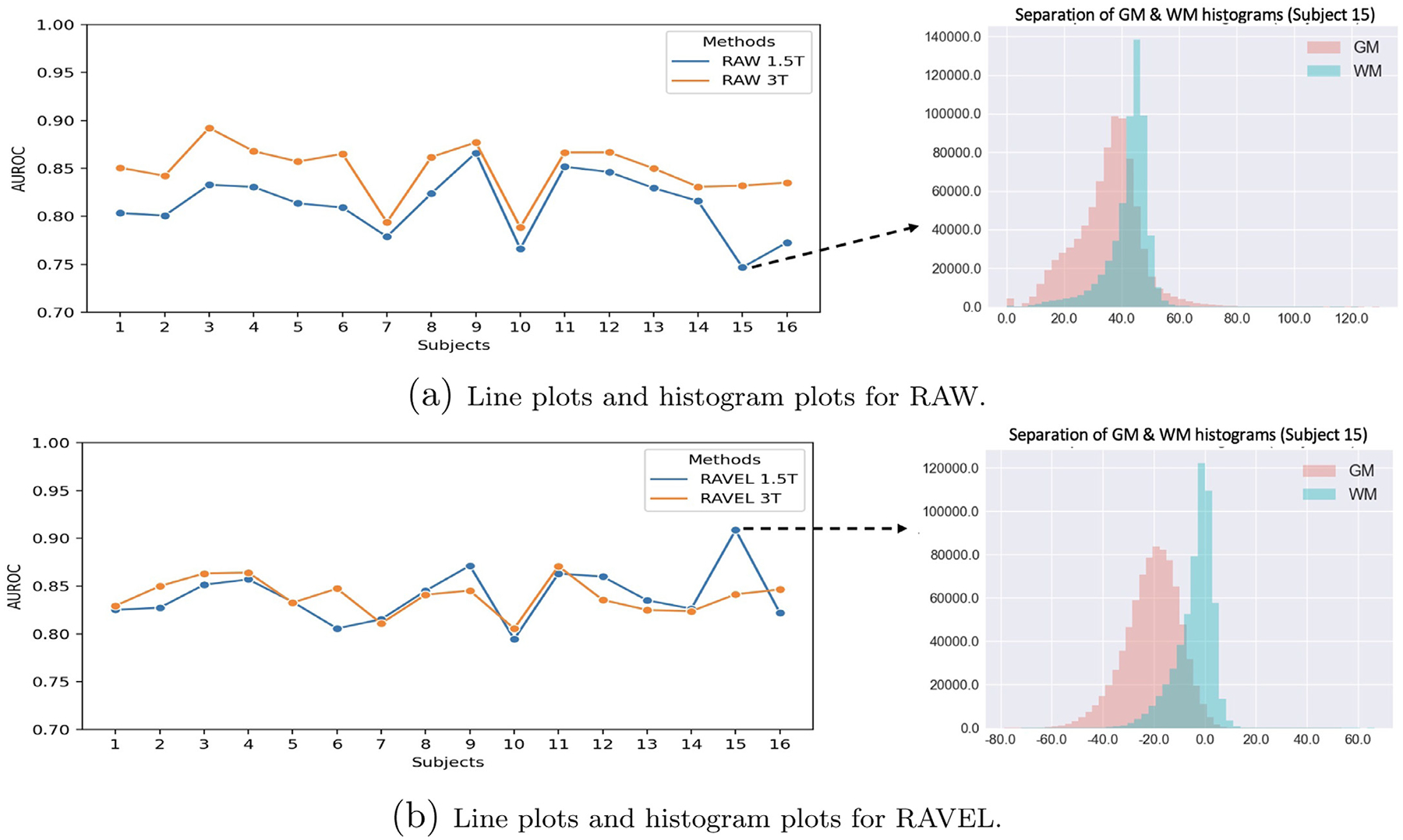
AUROC for classification of voxel intensity values as GM relative to WM. The AUROCs were estimated as the separation of the GM and WM histograms. On the left, the line plots depict the AUROC of the classification for all subjects. On the right, the plots show the overlap/separation of the histograms of tissues for one single subject. The two plots were depicted separately for each scanner (GE 1.5T and Siemens 3T), for RAW (a) and RAVEL (b). For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
Differences in motion artifacts may have also affected and possibly confounded harmonization of FreeSurfer outcome measures via RAVEL. Previous studies have demonstrated that motion artifacts, including blurring, ghosting, and ringing, reduce FreeSurfer measures of regional GM cortical thickness and volume (Alexander-Bloch et al., 2016; Backhausen et al., 2016). The effect of motion artifacts on RAVEL is not well understood, and characterizing it is beyond the scope of this study. Nevertheless, motion artifacts may introduce variability in CSF regions, from which the unwanted scanner-associated variation component is estimated for RAVEL. In this study, motion artifacts, or the lack thereof, were not consistent across 1.5T and 3T MRI acquisitions. Significant motion artifacts were observed in the frontal cortex of the 1.5T scan but not 3T scan for a single subject, as shown in Fig. 9.
Fig. 9.
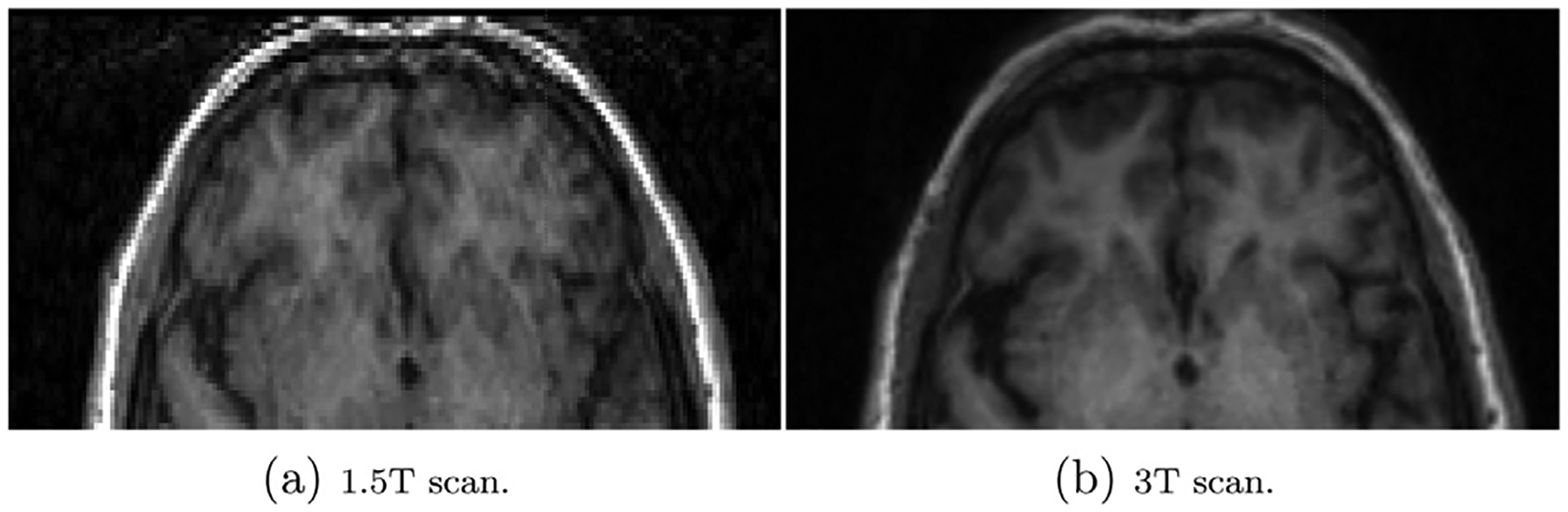
Inconsistent motion artifact across scanners for a single subject in our data. More significant motion artifacts were observed in the frontal cortex of 1.5T scan (a) relative to the 3T scan (b).
Further investigation into ComBat led to some additional insight with respect to effects of preprocessing before applying ComBat. In this study we applied the preprocessing steps recommended for RAVEL to our images for all four methods (RAW, RAVEL, ComBat, and RAVEL-ComBat) in order to avoid confounding the comparisons with inconsistent pipeline. However, the choice of preprocessing could affect the results of RAW and ComBat, which do not necessarily need any pre-processing before the segmentation with FreeSurfer. We investigated this issue by skipping the preprocessing step in the process of preparing RAW and ComBat-harmonized images and generated two new sets of data. We then compared RAW and ComBat with their corresponding new data using paired t-test. Our results showed that preprocessing was a source of variability in our data which resulted in statistically significant different values for summary measures within each scanner: RAW (1.5T: 9 measures, 3T: 10 measures) and ComBat (1.5T: 9 measures, 3T: 7 measures). This significant effect of the preprocessing step on results should be considered in studies when ComBat is used for the purpose of data harmonization and not for method comparison. The details of the experiments were reported in Supplementary File 2 and Supplementary Tables 3 and 4. Moreover, ComBat could be modified for handling the dependence for within-subject scans which is the case for our paired cohort. Thus, we added the subjects as a fixed effect to ComBat which resulted in a non-significant F-test when tested versus the original ComBat. We also handled the dependence by adapting the longitudinal Combat (Beer et al., 2020): we ran the model without the inclusion of time but, we included a random intercept. We presented these results in Supplementary Table 5.
Our study adds to the literature in at least three unique ways. First, we provided a thorough investigation of the existing RAVEL in a paired cohort. Second, we reported unique results of application on ComBat harmonization before and after performing RAVEL. Finally, we performed a neuroradiological rating of the hippocampus that can be used to assess AD progression. Additionally, we provide some insights on the effects of RAVEL intensity normalization on the GM-WM image contrast. Our study does have some limitations too. We only evaluated one cohort. In addition, we only assessed two methods: RAVEL and ComBat. As future work, we will study the generalizability of our results to other cohorts and will add more non-situational harmonization methods, such as mica (Wrobel et al., 2020), to our method comparison. We will also investigate the disadvantages observed with RAVEL in this study, including 1) increased variability across subjects and imaging measures, and 2) the unimproved segmentation accuracy of important regions of brain, such as hippocampus. These two aspects will benefit the neuroimaging community with better methods of normalization and accordingly harmonization.
Supplementary Material
Acknowledgment
This work was supported by the National Institutes of Health/NIA grants: R01 AG063752 (to D.L. Tudorascu) and RF1 AG025516 (to W.E. Klunk), in addition to the University of Pittsburgh ADRC Grant: P30 AG066468 (partial support for S.J. Hwang in form of an internal pilot). We thank Dr. James Wilson for the useful discussion.
Footnotes
Declaration of Competing Interest
The authors declare that they have no competing interests.
Credit authorship contribution statement
Mahbaneh Eshaghzadeh Torbati: Conceptualization, Methodology, Writing – original draft. Davneet S. Minhas: Methodology, Writing – review & editing. Ghasan Ahmad: Writing – review & editing. Erin E. O’Connor: Investigation, Writing – review & editing. John Muschelli: Writing – review & editing. Charles M. Laymon: Writing – review & editing. Zixi Yang: Writing – review & editing. Ann D. Cohen: Writing – review & editing. Howard J. Aizenstein: Funding acquisition, Writing – review & editing. William E. Klunk: Funding acquisition, Writing – review & editing. Bradley T. Christian: Writing – review & editing. Seong Jae Hwang: Methodology, Writing – review & editing. Ciprian M. Crainiceanu: Methodology, Writing – review & editing. Dana L. Tudorascu: Funding acquisition, Methodology, Conceptualization, Supervision, Writing – review & editing.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at 10.1016/j.neuroimage.2021.118703.
2 Used the public code from https://github.com/ncullen93/neuroCombat
References
- Alexander-Bloch A, Clasen L, Stockman M, Ronan L, Lalonde F, Giedd J, Raznahan A, 2016. Subtle in-scanner motion biases automated measurement of brain anatomy from in vivo MRI. Hum. Brain Mapp 37 (7), 2385–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal 12 (1), 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhausen LL, Herting MM, Buse J, Roessner V, Smolka MN, Vetter NC, 2016. Quality control of structural MRI images applied using freesurfer-a hands-on workflow to rate motion artifacts. Front. Neurosci 10, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer JC, Tustison NJ, Cook PA, Davatzikos C, Sheline YI, Shinohara RT, Linn KA, Initiative ADN, et al. , 2020. Longitudinal combat: a method for harmonizing longitudinal multi-scanner imaging data. Neuroimage 220, 117129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AA, Beer JC, Tustison NJ, Cook PA, Shinohara RT, Shou H, Initiative ADN, et al. , 2019. Removal of scanner effects in covariance improves multivariate pattern analysis in neuroimaging data. bioRxiv 858415. [Google Scholar]
- Dewey BE, Zhao C, Reinhold JC, Carass A, Fitzgerald KC, Sotirchos ES, Saidha S, Oh J, Pham DL, Calabresi PA, et al. , 2019. DeepHarmony: a deep learning approach to contrast harmonization across scanner changes. Magn. Reson. Imaging 64, 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey BE, Zuo L, Carass A, He Y, Liu Y, Mowry EM, Newsome S, Oh J, Calabresi PA, Prince JL, 2020. A disentangled latent space for cross-site MRI harmonization. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 720–729. [Google Scholar]
- Ellis KA, Bush AI, Darby D, De Fazio D, Foster J, Hudson P, Lautenschlager NT, Lenzo N, Martins RN, Maruff P, et al. , 2009. The Australian imaging, biomarkers and lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease. Int. Psychogeriatr 21 (4), 672–687. [DOI] [PubMed] [Google Scholar]
- Fischl B, 2012. FreeSurfer. Neuroimage 62 (2), 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J-P, Cullen N, Sheline YI, Taylor WD, Aselcioglu I, Cook PA, Adams P, Cooper C, Fava M, McGrath PJ, et al. , 2018. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 167, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J-P, Parker D, Tunç B, Watanabe T, Elliott MA, Ruparel K, Roalf DR, Satterthwaite TD, Gur RC, Gur RE, et al. , 2017. Harmonization of multi-site diffusion tensor imaging data. Neuroimage 161, 149–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J-P, Sweeney EM, Muschelli J, Crainiceanu CM, Shinohara RT, Initiative ADN, et al. , 2016. Removing inter-subject technical variability in magnetic resonance imaging studies. Neuroimage 132, 198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM, 2010. The clinical use of structural MRI in alzheimer disease. Nat. Rev. Neurol 6 (2), 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen R, Bouvy WH, Mendrik AM, Viergever MA, Biessels GJ, De Bresser J, 2016. Robustness of automated methods for brain volume measurements across different MRI field strengths. PLoS ONE 11 (10), e0165719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellier P, 2003. Consistent intensity correction of MR images. In: Proceedings 2003 International Conference on Image Processing (Cat. No. 03CH37429), vol. 1. IEEE, pp. I–1109. [Google Scholar]
- Jäger F, Deuerling-Zheng Y, Frericks B, Wacker F, Hornegger J, 2006. A new method for MRI intensity standardization with application to lesion detection in the brain. Vision modeling and visualization, vol. 2006. Citeseer. 296–276 [Google Scholar]
- Johnson WE, Li C, Rabinovic A, 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8 (1), 118–127. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Marizzoni M, Sala-Llonch R, Bosch B, Bartrés-Faz D, Arnold J, Benninghoff J, Wiltfang J, Roccatagliata L, Nobili F, et al. , 2013. Brain morphometry reproducibility in multi-center 3 T MRI studies: a comparison of cross-sectional and longitudinal segmentations. Neuroimage 83, 472–484. [DOI] [PubMed] [Google Scholar]
- Karayumak SC, Bouix S, Ning L, James A, Crow T, Shenton M, Kubicki M, Rathi Y, 2019. Retrospective harmonization of multi-site diffusion MRI data acquired with different acquisition parameters. Neuroimage 184, 180–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, 1999. Array of hope. Nat. Genet 21 (1), 3–4. [DOI] [PubMed] [Google Scholar]
- Leek JT, Storey JD, 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3 (9), e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Clarkson MJ, Bartlett JW, Clegg S, Jack CR Jr, Weiner MW, Fox NC, Ourselin S, Initiative ADN, et al. , 2010. Robust atrophy rate measurement in Alzheimer’s disease using multi-site serial MRI: tissue-specific intensity normalization and parameter selection. Neuroimage 50 (2), 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Maiti P, Thomopoulos SI, Zhu A, Chai Y, Kim H, Jahanshad N, 2021. Style transfer using generative adversarial networks for multi-site MRI harmonization. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madabhushi A, Udupa JK, 2006. New methods of MR image intensity standardization via generalized scale. Med. Phys 33 (9), 3426–3434. [DOI] [PubMed] [Google Scholar]
- Minhas DS, Yang Z, Muschelli J, Laymon CM, Mettenburg JM, Zammit MD, Johnson S, Mathis CA, Cohen AD, Handen BL, et al. , 2020. Statistical methods for processing neuroimaging data from two different sites with a down syndrome population application. In: International Conference on Information Processing and Management of Uncertainty in Knowledge-Based Systems. Springer, pp. 367–379. [Google Scholar]
- Modanwal G, Vellal A, Buda M, Mazurowski MA, 2020. MRI image harmonization using cycle-consistent generative adversarial network. In: Medical Imaging 2020: Computer-Aided Diagnosis, vol. 11314. International Society for Optics and Photonics, p. 1131413. [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L, 2005. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s disease neuroimaging initiative (ADNI). Alzheimer’s Dementia 1 (1), 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielson DM, Pereira F, Zheng CY, Migineishvili N, Lee JA, Thomas AG, Bandettini PA, 2018. Detecting and harmonizing scanner differences in the ABCD study-annual release 1.0. BioRxiv 309260. [Google Scholar]
- Ning L, Bonet-Carne E, Grussu F, Sepehrband F, Kaden E, Veraart J, Blumberg SB, Khoo CS, Palombo M, Kokkinos I, et al. , 2020. Cross-scanner and cross-protocol multi-shell diffusion MRI data harmonization: algorithms and results. Neuroimage 221, 117128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyúl LG, Udupa JK, 1999. On standardizing the MR image intensity scale. Magn. Reson. Med 42 (6), 1072–1081. [DOI] [PubMed] [Google Scholar]
- Nyúl LG, Udupa JK, Zhang X, 2000. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging 19 (2), 143–150. [DOI] [PubMed] [Google Scholar]
- Oishi K, Faria A, Jiang H, Li X, Akhter K, Zhang J, Hsu JT, Miller MI, van Zijl PCM, Albert M, et al. , 2009. Atlas-based whole brain white matter analysis using large deformation diffeomorphic metric mapping: application to normal elderly and Alzheimer’s disease participants. Neuroimage 46 (2), 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis M, 2014. Neuroimaging in parkinson disease: from research setting to clinical practice. Nat. Rev. Neurol 10 (12), 708. [DOI] [PubMed] [Google Scholar]
- Pomponio R, Erus G, Habes M, Doshi J, Srinivasan D, Mamourian E, Bashyam V, Nasrallah IM, Satterthwaite TD, Fan Y, et al. , 2020. Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. Neuroimage 208, 116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team., 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- Schnack HG, van Haren NEM, Brouwer RM, van Baal GCM, Picchioni M, Weisbrod M, Sauer H, Cannon TD, Huttunen M, Lepage C, et al. , 2010. Mapping reliability in multicenter MRI: voxel-based morphometry and cortical thickness. Hum. Brain Mapp 31 (12), 1967–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DL, Tagge I, Powers K, Ahn S, Bakshi R, Calabresi PA, Todd Constable R, Grinstead J, Henry RG, Nair G, et al. , 2019. Multisite reliability and repeatability of an advanced brain MRI protocol. J. Magn. Reson. Imaging 50 (3), 878–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz CG, Gunter JL, Wiste HJ, Przybelski SA, Weigand SD, Ward CP, Senjem ML, Vemuri P, Murray ME, Dickson DW, et al. , 2016. A large-scale comparison of cortical thickness and volume methods for measuring Alzheimer’s disease severity. Neuroimage Clin. 11, 802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Xiao Y, Subbanna N, Francis S, Arnold DL, Collins DL, Arbel T, 2011. Evaluating intensity normalization on MRIs of human brain with multiple sclerosis. Med. Image Anal 15 (2), 267–282. [DOI] [PubMed] [Google Scholar]
- Shinohara RT, Crainiceanu CM, Caffo BS, Gaitán MI, Reich DS, 2011. Population-wide principal component-based quantification of blood–brain-barrier dynamics in multiple sclerosis. Neuroimage 57 (4), 1430–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara RT, Oh J, Nair G, Calabresi PA, Davatzikos C, Doshi J, Henry RG, Kim G, Linn KA, Papinutto N, et al. , 2017. Volumetric analysis from a harmonized multisite brain MRI study of a single subject with multiple sclerosis. Am. J. Neuroradiol 38 (8), 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA, Jarso S, Pham DL, Reich DS, Crainiceanu CM, 2014. Australian imaging biomarkers lifestyle flagship study of ageing, and Alzheimer’s disease neuroimaging initiative. statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 6 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA, Jarso S, Pham DL, Reich DS, Crainiceanu CM, et al. , 2014. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 6, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney EM, Shinohara RT, Shea CD, Reich DS, Crainiceanu CM, 2013. Automatic lesion incidence estimation and detection in multiple sclerosis using multisequence longitudinal MRI. Am. J. Neuroradiol 34 (1), 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: improved n3 bias correction. IEEE Trans. Med. Imaging 29 (6), 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachinger C, Rieckmann A, Pölsterl S, Detect and correct bias in multi-site neuroimaging datasets, 2020. arXiv preprint arXiv:2002.05049 [DOI] [PubMed] [Google Scholar]
- Weisenfeld NL, Warfteld SK, 2004. Normalization of joint image-intensity statistics in MRI using the Kullback-Leibler divergence. In: 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821) IEEE, pp. 101–104. [Google Scholar]
- Wrobel J, Martin ML, Bakshi R, Calabresi PA, Elliot M, Roalf D, Gur RC, Gur RE, Henry RG, Nair G, et al. , 2020. Intensity warping for multisite MRI harmonization. Neuroimage 223, 117242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Linn KA, Cook PA, Phillips ML, McInnis M, Fava M, Trivedi MH, Weissman MM, Shinohara RT, Sheline YI, 2018. Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Hum. Brain Mapp 39 (11), 4213–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Dewey BE, Carass A, Liu Y, He Y, Calabresi PA, Prince JL, 2021. Information-based disentangled representation learning for unsupervised MR harmonization. In: International Conference on Information Processing in Medical Imaging. Springer, pp. 346–359 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


