Abstract
Genetic polymorphisms at the IFNL4 loci are known to influence the clinical outcome of several different infectious diseases. Best described is the association between the IFNL4 genotype and hepatitis C virus clearance. However, an influence of the IFNL4 genotype on the adaptive immune system was suggested by several studies but never investigated in humans. In this cross-sectional study, we have genotyped 201 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-positive participants for 3 IFNL4 polymorphisms (rs368234815, rs12979860, and rs117648444) and stratified them according to the IFNλ4 activity. Based on this stratification, we investigated the association between the IFNL4 genotype and the antibody as well as the CD8+ T cell response in the acute phase of the SARS-CoV-2 infection. We observed no differences in the genotype distribution compared with a Danish reference cohort or the 1,000 Genome Project, and we were not able to link the IFNL4 genotype to changes in either the antibody or CD8+ T cell responses of these patients.
Keywords: Interferon lambda-4, COVID-19, SARS-CoV-2, genetics, antibody, T cell response
Introduction
Interferon lambda 4 (IFNλ4) is a recently identified member of the IFNλ family. Humans possess 4 genes belonging to the IFNλ family: IFNL1, IFNL2, IFNL3, and IFNL4. The first 3 members (IFNL1-3) share a high degree of similarity and were identified by 2 independent groups in 2003 (Kotenko and others 2003; Sheppard and others 2003) as a novel family of genes encoding virally induced IFNs. In the following decade, genome-wide association studies (GWAS) linked the clearance of hepatitis C virus (HCV) to genetic variation within the type III IFN loci (Ge and others 2009; Suppiah and others 2009; Tanaka and others 2009; Thomas and others 2009), and this subsequently led to the discovery of IFNλ4 (Prokunina-Olsson and others 2013).
Upon identification of IFNλ4, the investigators also identified a dinucleotide variant (rs368234815, TT/ΔG) located in the first exon of IFNλ4. Rs368234815 ΔG is the ancestral allele and generates the full-length IFNλ4 protein, whereas the rs368234815 TT allele leads to a frameshift and therefore aborts the expression of IFNλ4. The ΔG/TT variation is associated with spontaneous HCV clearance as well as with the response to treatment (Prokunina-Olsson and others 2013). This variant is also in high linkage disequilibrium (LD) with the initially discovered and still often clinical denoted GWAS marker (rs12979860, C/T). Surprisingly, patients harboring the functional IFNL4 have a lower HCV clearance rate than that of patients who have a nonfunctional IFNλ4 (Prokunina-Olsson and others 2013).
While the functional IFNL4 is unfavorable in terms of HCV clearance, the functional version of this gene is associated with lower levels of liver inflammation and fibrosis in HCV-infected patients (Bochud and others 2012; Eslam and others 2017; Mohlenberg and others 2019), as well as in patients with nonalcoholic fatty liver disease (Eslam and others 2015; Petta and others 2017), proving an advantage for patients. The causal role of the IFNλ4 protein in lower HCV clearance rates is further supported by the finding of a genetic variant (rs117648444, G/A) resulting in a single amino acid substitution of a proline to a serine at position 70 (IFNλ4 P70S) in functional IFNλ4, which substantially affects the antiviral activity of IFNλ4 (Terczynska-Dyla and others 2014).
HCV patients harboring the impaired IFNλ4 S70 variant display lower IFN-stimulated gene expression levels, but better treatment response rates and better spontaneous clearance rates than those patients carrying the fully active IFNλ4 P70 variant (Terczynska-Dyla and others 2014). Thus, it is clear that the disease course of HCV is affected by the IFNL4 genotype.
Recently, we demonstrated that treatment with the recombinant murine IFNλ2 protein specifically enhanced the generation of IgG1 and IgA antibodies in a mouse model of influenza A virus infection (Ye and others 2019). Furthermore, it has been suggested that IFNλ4 could shape the adaptive immune response in humans since the genetic findings surrounding IFNL4 are mainly in relatively complex diseases (Larrubia and others 2014; Sutti and Albano 2020).
Another indication came recently, when the ability to produce IFNL4 has been associated with higher antibody levels against HCV (Waldenström and others 2021). Until now this hypothesis has been difficult to study since IFNλ4 is absent in rodents, however, a unique chance occurred with the emergence of the novel coronavirus designated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in December 2019 (Wang and others 2020; Zhou and others 2020; Zhu and others 2020) and becoming a pandemic in the spring of 2020 (WHO 2020).
We can safely assume that patients are infected with the SARS-CoV-2 virus for the first time, and thus, we can study the primary response of the adaptive immune system. Therefore, we seized the chance to investigate the potential link between the IFNL4 genotype and the adaptive immune response raised toward the SARS-CoV-2. We got access to patients diagnosed with SARS-CoV-2 infection and genotyped 3 polymorphisms within the IFNL4 gene. In this study, we stratify the patients based on the functional activity of IFNλ4 and investigate the relationship with the antibody and CD8+ T cell response.
Materials and Methods
Study design
The study was conducted on 203 participants from the CoroNAT cohort (Nielsen and others 2021; Vibholm and others 2021), collected at the Department of Infectious Diseases at Aarhus University Hospital, Denmark, from April 3 to July 9 2020. The study was approved by The National Health Ethics Committee (case number 1-10-72-76-20) and the Danish Data Protection Agency. Patient flow diagram is shown in Supplementary Fig. S1. Demographic and clinical data on participants were collected to determine if certain parameters were correlated to the IFNL4 genotype (Table 1).
Table 1.
Cohort Characteristics
| Characteristics | Total cohort |
|---|---|
| Patients, n | 201 |
| Female sex, n (%) | 110 (54.7) |
| Age, years, average (range) | 47 (20–79) |
| Body mass index, kg/m2, median (range) | 25.4 (18.8–47.8) |
| Duration of COVID-19 symptoms, days, median (range) | 12 (0–47) |
| COVID-19 disease severity, n (%) | |
| Outpatient | 167 (83.1) |
| Hospitalized | 34 (16.9) |
| Smoking, n (%) | |
| Never | 133 (65.5) |
| Current | 9 (4.4) |
| Previous | 61 (30.0) |
| Race, n (%) | |
| Asian | 2 (1.0) |
| Black or African European | — |
| White or Caucasian | 197 (98.0) |
| Other | 2 (1.0) |
| Haplotype | |
| No IFNλ4 | 87 (43.3) |
| IFNλ4-S70 | 25 (12.4) |
| IFNλ4-P70 | 89 (44.3) |
All participants were assigned a COVID-19 severity group depending on their course of disease. Group 1 consisted of asymptomatic or moderately sick participants able to recover at home. Group 2 comprises all severely ill hospitalized participants, regardless of intensive care unit admission or oxygen supplementation. Haplotype is determined on the basis of genotyping of rs368234815, rs12979860, and rs117648444 using a KASP assay.
Blood samples were collected at a minimum of 14 days after full recovery (no ongoing COVID-19 symptoms, except loss of sense of smell/taste, and cognitive deficits, which are symptoms equivalent to long-COVID-19) and at a maximum of 12 weeks after a first positive SARS-CoV-2 Real Time-PCR. Participants were allocated into 2 groups according to the severity of COVID-19 based on the following criteria on COVID-19 severity: (1) Outpatients (able to stay at home either with no or some limitation to their daily activities), and (2) Hospitalized [both nonintensive care unit (ICU) and ICU admission]. Each participant provided written informed consent before any study activities.
DNA extraction and genotyping
Genomic DNA was purified from cryopreserved peripheral blood mononuclear cell using the DNeasy Blood and Tissue Kit (No. 69504; Qiagen) following their specification. Genotyping was performed using a competitive allele-specific PCR assay designed and optimized by LGC following their protocol on a Roche LC480-Series instrument. The following polymorphisms were genotyped: rs368234815 (ΔG/TT) (±IFNλ4), rs12979860 (C/T) (GWAS marker), and rs117648444 (G/A) (IFNλ4-P70/S70) using primer sets provided by LGC (Supplementary Fig. S2). Genotyping was performed blinded to clinical phenotypes.
Neutralization assay
The SARS-CoV-2 neutralization capacity of plasma from participants from the CoroNAT cohort was assessed through infection of Vero76 cmyc hTMPRSS2 cells, with VSV*ΔG(luc)-SARS-2-S pseudovirus particles as described in Nielsen and others (2021). Neutralization curves were plotted with 3 parameter nonlinear fits from which IC50 values were calculated.
Total Ig, IgM, and IgA detection
Serum levels of anti-SARS-CoV-2 antibodies were detected by semiquantitative ELISA on participants from the CoroNAT cohort as described in Vibholm and others (2021).
Dextramer staining by flow cytometry
The dextramer stains were performed on the HLA-A2-positive patient samples from the CoroNAT cohort as described in Vibholm and others (2021) and their location visualized in Fig. 5 of Nielsen and others (2021). PBMCs were incubated at room temperature for 30 min with the following SARS-CoV-2 dextramers (all from Immundex): A*0201/TLACFVLAAV-PE (Cat. No. WB3848-PE), A*0201/GMSRIGMEV-FITC (Cat. No. WB5751-FITC), A*0201/LLLDRLNQL-APC (Cat. No. WB5762-APC), A*0201/ILLNKHIDA-PE (Cat. No. WB5848-PE), A*0201/RLNEVAKNL-FITC (Cat. No. WB5750-FITC), A*0201/YLQPRTFLL-APC (Cat. No. WB5824-APC), A*0201/VLNDILSRL-PE (Cat. No. WB5823-PE), A*0201/NLNESLIDL-FITC (Cat. No. WB5850-FITC), A*0201/FIAGLIAIV-APC (Cat. No. WB5825-APC), A*0201/LLLNCLWSV-PE (Cat. No. WB3513-PE), or positive/negative control dextramers: A*0201/NLVPMVATV-PE (Cat. No. WB2132-PE, Pos. Control, CMV), A*0201/NLVPMVATV-FITC (Cat. No. WB2132-FITC, Pos. Control, CMV), A*0201/NLVPMVATV-APC (Cat. No. WB2132-APC, Pos. Control, CMV), A*0201/Neg. Control-PE (Cat. No. WB2666-PE), A*0201/Neg. Control-FITC (Cat. No. WB2666-FITC), A*0201/Neg. Control-APC (Cat. No. WB2666-APC). Cells were washed and stained with viability dye (Zombie Violet, Cat. No. 423114; Biolegend) and CD8 (Clone RPA-T8, Cat. No. 563795; BD) and acquired on a 5-laser Fortessa flow cytometer.
Statistical analyses
Graphs and data analyses were performed using GraphPad Prism 7.0 and StataIC 16.1. The 3 polymorphisms were analyzed both separately and as haplotypes. The LD metrics (D′ and r2) between polymorphisms were analyzed and plotted using Haploview 4.2 (Barrett and others 2005).
Testing for a normal distribution when the data were divided according to the genotype or haplotype was performed by an unpaired Student's-T-test or Mann–Whitney test as appropriate. P < 0.05 was interpreted as statistically significant. P values are indicated as follows: n.s. = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001, and **** = P < 0.0001.
Results
The participants
Two hundred one SARS-CoV-2-positive patients were included from a Danish COVID-19 cohort collected at Aarhus University hospital; the CoroNAT cohort (Nielsen and others 2021; Vibholm and others 2021). Clinical characteristics of participants are shown in Table 1, and the patient flow diagram is shown in Supplementary Fig. S1. In our study, the participants were fairly evenly divided between male and female, with 110 (54.7%) participants being female. The average age was 47 years and the median body mass index was 25.4 kg/m2. Participants experienced a range of symptoms, from asymptomatic to hospitalization and ICU admission, with a median duration of hospitalization of 12 days. The participants were divided into 2 groups based on hospitalization status as a marker for disease severity. The majority of the participants were nonhospitalized (ie, outpatients, 83.1%). Furthermore, the majority of the cohort was nonsmokers and of white origin.
Genetics of IFNL4
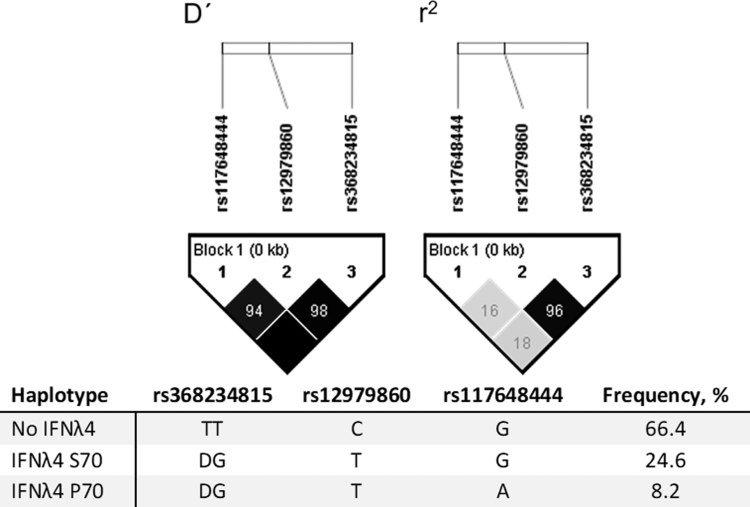
Participants were genotyped for the following polymorphisms: rs368234815 (ΔG/TT) (±IFNλ4), rs12979860 (C/T) (GWAS marker), and rs117648444 (G/A) (IFNλ4-P70/S70) (Supplementary Table S1), from which the allele frequency was calculated (Supplementary Table S2). Participants were allocated into 3 groups according to the haplotype of IFNL4 based on the genotyping: No IFNλ4; rs368234815 TT/rs12979860 C, IFNλ4-P70; rs368234815 ΔG/rs12979860 T/rs117648444 G, and IFNλ4-S70; rs368234815 ΔG/rs12979860 T/rs117648444 A (Table 1). The participants were assigned to their IFNL4 haplotype using a dominant model: participant heterozygote for the IFNλ4 determining variant [rs368234815 (ΔG/TT)] was considered IFNλ4 positive. Eighty-seven participants (43.3%) belonged to the No IFNλ4 group, 25 participants (12.4%) belonged to the IFNλ4-S70 group, and 89 (44.3%) participants belonged to the IFNλ4-P70 group.
Furthermore, we observed a strong LD between rs368234815 and rs12979860 (D′ = 0.98 and r2 = 0.96) (Fig. 1). This observation is comparable with what is detected in the European population of the 1,000 Genome Project (Supplementary Fig. S3). The genotype and allele frequencies of the 3 SNPs were comparable with the European population of the 1,000 Genome Project (Supplementary Table S2).
FIG. 1.
Haploview LD plots of rs368234815, rs12979860, and rs117648444 for the participants. The plots show the LD metrics generated with Haploview. For D′ the color depicts the LOD; black: LOD ≥2 and gray <2, and the number is D′. For r2 the color and number depict r2; shades of gray: 0 < r2 < 1 and black: r2 = 1. If the value is 100, it is for simplicity not written. LD, linkage disequilibrium; LOD, logarithm of the odds.
Antibody response
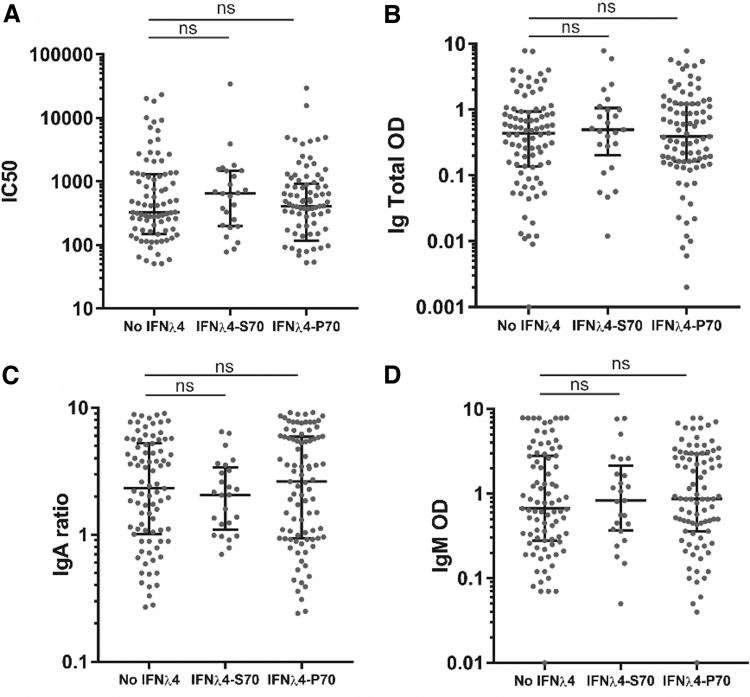
The participants had their antibody response measured 4 weeks after recovery (Nielsen and others 2021) and we stratified the results according to the IFNL4 haplotype (Fig. 2). The first parameter we analyzed was the antibody neutralization potency measured by IC50. The IC50 was extrapolated based on the functional neutralization capacity of total plasma antibodies in vitro measured using VSV pseudotyped with the SARS-CoV-2 spike protein. Antibody neutralizing potency was evaluated by serial dilutions of participant plasma, yielding infectivity titration curves for each of the participants leading to extrapolation of the IC50 values. When allocating the IC50 values to the IFNL4 haplotypes, we observed no significant difference between the different haplotypes (Fig. 2A).
FIG. 2.
Antibody response toward the SARS-CoV-2 spike assigned to the IFNλ4 haplotype. (A) IC50 values calculated from neutralization curves. (B) Blank-corrected chemiluminescent signal of total Ig against SARS-CoV-2 spike measured by ELISA. Total Ig is shown as OD (1:100). (C) Blank-corrected chemiluminescent signal of IgA against SARS-CoV-2 spike measured by ELISA. IgA is shown as ratio against standard. (D) Blank-corrected chemiluminescent signal of IgM against SARS-CoV-2 spike measured by ELISA. IgM is shown as OD (1:11). Error bars show median and interquartile range. Statistical comparison by Mann–Whitney U test. ns = P > 0.05, n = 201. Ig, immunoglobulin; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The next parameter we analyzed was the total immunoglobulin (Ig) levels of serum anti-SARS-CoV-2 antibodies detected by ELISA. Again, allocation to the IFNL4 haplotypes proved to be nonsignificant between the different haplotypes (Fig. 2B). The SARS-CoV-2-specific serum IgA and IgM were also measured on the participants by ELISA. To investigate any differences between the effectivity of seroconversion in the participants, we also allocated the IgA and IgM measurements to the IFNL4 haplotypes (Fig. 2C, D, respectively). Again, we observed no significant differences between the IFNL4 haplotypes.
We observed the same nonsignificant results when we allocated the antibody response to either the presence or absence of functional IFNλ4 or the individual polymorphisms: rs368234815 (ΔG/TT) (±IFNλ4), rs12979860 (C/T) (GWAS marker), or rs117648444 (G/A) (IFNλ4-P70/S70) (Supplementary Fig. S4). In summary, the haplotype as well as the genotype of IFNL4 did not impact the antibody response in our analysis.
CD8+ T cell response
Samples collected 4 weeks after recovery from the 105 HLA-A2-positive participants were subjected to a dextramer staining flow cytometry analysis to assess the CD8+ T cell responses toward 9 different CD8+ T cell epitopes. The epitopes were covering different parts of the SARS-CoV-2 genome (Nielsen and others 2021). Again, we stratified the participants according to the IFNL4 genotype. The first analysis we performed was to determine whether the CD8+ T cells from the participants were able to bind to the dextramers based on the IFNL4 haplotype (responders) or not (nonresponders) (Fig. 3A).
FIG. 3.
SARS-CoV-2 responsive CD8+ T cells assigned to IFNλ4 haplotype. (A) Bar chart showing whether participants responded to the investigated epitopes or not, n = 201. (B) CD8+ T cell response shown as the cumulative number of SARS-CoV-2 epitopes targeted by the responsive participants, n = 106. (C) Distribution of the cumulative CD8+ T cell responses in participants, n = 105. Ten percent of the participants had no detectable CD8+ T cell epitope response, and are not shown on the graph, but were included in statistical tests. Error bars show median and interquartile range. Statistical comparison by Mann–Whitney U test. ns = P > 0.05.
There was a slight tendency toward the functional IFNL4 haplotypes (IFNλ4-S70 and IFNλ4-P70) having a higher number of responsive participants compared with nonresponsive participants, however, this tendency is nonsignificant. Since we observed a tendency toward a higher responsiveness in the functional IFNL4 haplotypes, we investigated the participants with CD8+ T cells able to bind the dextamers (responders) in more detail. We allocated the number of positive epitopes for each responsive participant according to the IFNL4 haplotype (Fig. 3B). However, we detected no significant differences in the number of epitopes recognized between the groups. We also compared the strength of the CD8+ T cell response, defined as the cumulative frequencies of all dextramer responses, across the IFNλ4 haplotypes, but the distributions were again nonsignificant (Fig. 3C).
We observed similar nonsignificant associations when we assigned the CD8+ T cell response to either the presence or absence of functional IFNλ4 or the individual polymorphisms: rs368234815 (ΔG/TT) (±IFNλ4), rs12979860 (C/T) (GWAS marker), or rs117648444 (G/A) (IFNλ4-P70/S70) (Supplementary Fig. S5). In conclusion, the CD8+ T cell response does not seem to be influenced by the IFNL4 haplotype as well as genotype, even though there is a tendency that participants with functional IFNλ4 might have a better response. This will, however, require further investigation.
Discussion
The COVID-19 pandemic offered a unique opportunity to study the impact of IFNL4 genetics on the primary response of the adaptive immune system. This study set out to characterize the link between the IFNL4 genotype and the antibody, as well as the CD8+ T cell response toward an acute SARS-CoV-2 infection. First, we performed a genetic analysis showing a strong LD between rs368234815 and rs12979860 as expected within a northern European cohort (Prokunina-Olsson 2019).
Next, we analyzed the impact of IFNL4 genetics on the adaptive immune response by stratifying the participants according to IFNL4 haplotype and performing a comparison of different antibody and T cell response parameters. We did not observe any differences in any of the analyzed antibody or T cell parameters (Figs. 2, 3, and Supplementary Figs. S4–S5). We analyzed the antibody response in the entire cohort, but the T cell response only in a proportion of the cohort due to sample quality and scarcity. A different subset of participants might possibly lead to differential results, however, we do not believe this to be the situation.
In the original study surrounding the CoroNAT cohort, we conclude that the SARS-CoV-2 infections lead to the production of effective neutralizing antibodies (Nielsen and others 2021). Furthermore, this study shows that the neutralization capacity is increased with disease severity. It is possible that the disease severity has a greater impact on the adaptive immune response compared with the IFNL4 haplotype.
Our findings do not show any strong linkage between IFNL4 and the adaptive immune response during the acute phase of COVID-19. This is in accordance with the GWAS performed on COVID-19 patients, which have found no association with the IFNL4 locus (Severe Covid-19 GWAS group 2020; Oh and others 2020; Pairo-Castineira and others 2021). However, our study includes fewer participants in comparison with the GWAS associating IFNL4 with HCV (Ge and others 2009; Suppiah and others 2009; Tanaka and others 2009; Thomas and others 2009; Waldenström and others 2021).
In addition, the functional IFNλ4 is protective toward inflammation (Eslam and others 2017; Mohlenberg and others 2019) and it is known that host-mediated lung inflammation drives the mortality in COVID-19 patients (Dorward and others 2021). This implies that further studies investigating other clinical outcomes might be needed. Furthermore, emerging data from the COVID-19 pandemic suggest there could be substantial long-term effect following SARS-CoV-2 infection leading to “long COVID-19” or “postacute COVID-19 syndrome” in some patients (George and others 2020; Nalbandian and others 2021). This group of patients share some characteristics with chronically infected patients. Genetic analyses of patients with long-term effects of COVID-19 might be more likely to produce clear results. Thus, the relationship between IFNL4 genetics and COVID-19 still remains to be investigated in more depth, and further genetic studies are needed.
Supplementary Material
Authors' Contributions
M.M., I.M., L.K.V., S.F.N., M.H.S., R.O., M.K., J.D.G. O.S.S., T.R.O., M.T., and R.H. contributed to data collection, data analysis, and data interpretation. M.M. and R.H. contributed to the study design and wrote the article draft. M.M. performed the literature search, and created figures and tables. The final version of this article was reviewed and approved by all the authors.
Disclaimer
The content of this publication and the opinions expressed reflect those of the individual authors solely and do not necessarily reflect the views or policies of the Department of Health and Human Services, the National Institutes of Health, the National Cancer Institute, or the Food and Drug Administration, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author Disclosure Statement
T.R.O'B. is a coinventor on patents for the IFN-λ4 protein that are held by the U.S. National Cancer Institute The other authors report no conflict of interest.
Funding Information
The funding source had no role in the study design, collection, analysis, and interpretation of data, writing of the article, or the decision to submit the article for publication. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics.
Supplementary Material
References
- 2020. Genomewide association study of severe Covid-19 with respiratory failure. Severe Covid-19 GWAS Group. N Engl J Med 383(16):1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. [DOI] [PubMed] [Google Scholar]
- Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, Goossens N, Kuske L, Mullhaupt B, Gerlach T, Heim MH, Moradpour D, Cerny A, Malinverni R, Regenass S, Dollenmaier G, Hirsch H, Martinetti G, Gorgiewski M, Bourliere M, Poynard T, Theodorou I, Abel L, Pol S, Dufour JF, Negro F. 2012. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology 55(2):384–394. [DOI] [PubMed] [Google Scholar]
- Dorward DA, Russell CD, Um IH, Elshani M, Armstrong SD, Penrice-Randal R, Millar T, Lerpiniere CEB, Tagliavini G, Hartley CS, Randle NP, Gachanja NN, Potey PMD, Dong X, Anderson AM, Campbell VL, Duguid AJ, Qsous WA, BouHaidar R, Baillie JK, Dhaliwal K, Wallace WA, Bellamy COC, Prost S, Smith C, Hiscox JA, Harrison DJ, Lucas CD. 2021. Tissue-specific immunopathology in fatal COVID-19. Am J Respir Crit Care Med 203(2):192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslam M, Hashem AM, Leung R, Romero-Gomez M, Berg T, Dore GJ, Chan HL, Irving WL, Sheridan D, Abate ML, Adams LA, Mangia A, Weltman M, Bugianesi E, Spengler U, Shaker O, Fischer J, Mollison L, Cheng W, Powell E, Nattermann J, Riordan S, McLeod D, Armstrong NJ, Douglas MW, Liddle C, Booth DR, George J, Ahlenstiel G. 2015. Interferon-lambda rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat Commun 6:6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslam M, McLeod D, Kelaeng KS, Mangia A, Berg T, Thabet K, Irving WL, Dore GJ, Sheridan D, Gronbaek H, Abate ML, Hartmann R, Bugianesi E, Spengler U, Rojas A, Booth DR, Weltman M, Mollison L, Cheng W, Riordan S, Mahajan H, Fischer J, Nattermann J, Douglas MW, Liddle C, Powell E, Romero-Gomez M, George J. 2017. IFN-lambda3, not IFN-lambda4, likely mediates IFNL3-IFNL4 haplotype-dependent hepatic inflammation and fibrosis. Nat Genet. May; 49(5):795–800. DOI: 10.1038/ng.3836 [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461(7262):399–401. [DOI] [PubMed] [Google Scholar]
- George PM, Wells AU, Jenkins RG. 2020. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med 8(8):807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 4(1):69–77. [DOI] [PubMed] [Google Scholar]
- Larrubia JR, Moreno-Cubero E, Lokhande MU, García-Garzón S, Lázaro A, Miquel J, Perna C, Sanz-de-Villalobos E. 2014. Adaptive immune response during hepatitis C virus infection. World J Gastroenterol 20(13):3418–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlenberg M, Terczynska-Dyla E, Thomsen KL, George J, Eslam M, Gronbaek H, Hartmann R. 2019. The role of IFN in the development of NAFLD and NASH. Cytokine 124:154519. [DOI] [PubMed] [Google Scholar]
- Nalbandian A, Sehgal K, Gupta A, Madhavan MV, McGroder C, Stevens JS, Cook JR, Nordvig AS, Shalev D, Sehrawat TS, Ahluwalia N, Bikdeli B, Dietz D, Der-Nigoghossian C, Liyanage-Don N, Rosner GF, Bernstein EJ, Mohan S, Beckley AA, Seres DS, Choueiri TK, Uriel N, Ausiello JC, Accili D, Freedberg DE, Baldwin M, Schwartz A, Brodie D, Garcia CK, Elkind MSV, Connors JM, Bilezikian JP, Landry DW, Wan EY. 2021. Post-acute COVID-19 syndrome. Nat Med 27:601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SS, Vibholm LK, Monrad I, Olesen R, Frattari GS, Pahus MH, Højen JF, Gunst JD, Erikstrup C, Holleufer A, Hartmann R, Østergaard L, Søgaard OS, Schleimann MH, Tolstrup M. 2021. SARS-CoV-2 elicits robust adaptive immune responses regardless of disease severity. EBioMedicine 68:103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JH, Tannenbaum A, Deasy JO. 2020. Identification of biological correlates associated with respiratory failure in COVID-19. BMC Med Genomics 13(1):186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, Walker S, Parkinson N, Fourman MH, Russell CD, Furniss J, Richmond A, Gountouna E, Wrobel N, Harrison D, Wang B, Wu Y, Meynert A, Griffiths F, Oosthuyzen W, Kousathanas A, Moutsianas L, Yang Z, Zhai R, Zheng C, Grimes G, Beale R, Millar J, Shih B, Keating S, Zechner M, Haley C, Porteous DJ, Hayward C, Yang J, Knight J, Summers C, Shankar-Hari M, Klenerman P, Turtle L, Ho A, Moore SC, Hinds C, Horby P, Nichol A, Maslove D, Ling L, McAuley D, Montgomery H, Walsh T, Pereira AC, Renieri A, Millar J, Nichol A, Walsh T, Shankar-Hari M, Ponting C, Meikle J, Finernan P, McMaster E, Law A, Baillie JK, Paterson T, Wackett T, Armstrong R, Clark R, Coutts A, Donnelly L, Gilchrist T, Hafezi K, Macgillivray L, Maclean A, McCafferty S, Morrice K, Weaver J, Boz C, Golightly A, Ward M, Mal H, Szoor-McElhinney H, Brown A, Hendry R, Stenhouse A, Cullum L, Law D, Law S, Law R, Swets M, Day N, Taneski F, Duncan E, Parkinson N, Collier D, Wood S, Zak A, Borra C, Matharu M, May P, Alldis Z, Mitchelmore O, Bowles R, Easthope A, Bibi F, Lancoma-Malcolm I, Gurasashvili J, Pheby J, Shiel J, Bolton M, Patel M, Taylor M, Zongo O, Ebano P, Harding P, Astin-Chamberlain R, Choudhury Y, Cox A, Kallon D, Burton M, Hall R, Blowes S, Prime Z, Biddle J, Prysyazhna O, Newman T, Tierney C, Kassam J, Shankar-Hari M, Ostermann M, Campos S, Bociek A, Lim R, Grau N, Jones TO, Whitton C, Marotti M, Arbane G, Bonner S, Hugill K, Reid J, Welters I, Waugh V, Williams K, Shaw D, Roman JF, Martinez ML, Johnson E, Waite A, Johnston B, Hamilton D, Mulla S, McPhail M, Smith J, Baillie JK, Barclay L, Hope D, McCulloch C, McQuillan L, Clark S, Singleton J, Priestley K, Rea N, Callaghan M, Campbell R, Andrew G, Marshall L, McKechnie S, Hutton P, Bashyal A, Davidson N, Summers C, Polgarova P, Stroud K, Pathan N, Elston K, Agrawal S, Battle C, Newey L, Rees T, Harford R, Brinkworth E, Williams M, Murphy C, White I, Croft M, Bandla N, Gellamucho M, Tomlinson J, Turner H, Davies M, Quinn A, Hussain I, Thompson C, Parker H, Bradley R, Griffiths R, Scriven J, Nilsson A, Bates M, Dasgin J, Gill J, Puxty A, Cathcart S, Salutous D, Turner L, Duffy K, Puxty K, Joseph A, Herdman-Grant R, Simms R, Swain A, Naranjo A, Crowe R, Sollesta K, Loveridge A, Baptista D, Morino E, Davey M, Golden D, Jones J, Moreno Cuesta J, Haldeos A, Bakthavatsalam D, Vincent R, Elhassan M, Xavier K, Ganesan A, Purohit D, Abdelrazik M, Morgan J, Akeroyd L, Bano S, Lawton T, Warren D, Bromley M, Sellick K, Gurr L, Wilkinson B, Nagarajan V, Szedlak P, Cupitt J, Stoddard E, Benham L, Preston S, Laha S, Slawson N, Bradshaw Z, Brown J, Caswell M, Melling S, Bamford P, Faulkner M, Cawley K, Jeffrey H, London E, Sainsbury H, Nagra I, Nasir F, Dunmore C, Jones R, Abraheem A, Al-Moasseb M, Girach R, Padden G, Egan J, Brantwood C, Alexander P, Bradley-Potts J, Allen S, Felton T, Manna S, Farnell-Ward S, Leaver S, Queiroz J, Maccacari E, The Gen OI, Gen OC, Gen Oc-i, Central m, laboratory t, Data analysis t, Barts Health Nhs Trust LUK, Guy's, St Thomas' Hospital LUK, James Cook University Hospital MUK, The Royal Liverpool University Hospital LUK, King's College Hospital LUK, Royal Infirmary of Edinburgh EUK, John Radcliffe Hospital OUK, Addenbrooke's Hospital CUK, Morriston Hospital SUK, Ashford, St Peter's Hospital SUK, Royal Stoke University Hospital SUK, Queen Elizabeth Hospital BUK, Glasgow Royal Infirmary GUK, Kingston Hospital SUK, The Tunbridge Wells H, Maidstone Hospital KUK, North Middlesex University Hospital Nhs Trust LUK, Bradford Royal Infirmary BUK, Blackpool Victoria Hospital BUK, Countess of Chester Hospital CUK, Wythenshawe Hospital MUK, St George's Hospital LUK. 2021. Genetic mechanisms of critical illness in COVID-19. Nature 591(7848):92–98. [DOI] [PubMed] [Google Scholar]
- Petta S, Valenti L, Tuttolomondo A, Dongiovanni P, Pipitone RM, Camma C, Cabibi D, Di Marco V, Fracanzani AL, Badiali S, Nobili V, Fargion S, Grimaudo S, Craxi A. 2017. Interferon Lambda 4 rs368234815 TT>deltaG variant is associated with liver damage in patients with nonalcoholic fatty liver disease. Hepatology 66(6):1885–1893. [DOI] [PubMed] [Google Scholar]
- Prokunina-Olsson L. 2019. Genetics of the human Interferon Lambda region. J Interferon Cytokine Res 39(10):599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45(2):164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J, Ostrander C, Dong D, Shin J, Presnell S, Fox B, Haldeman B, Cooper E, Taft D, Gilbert T, Grant FJ, Tackett M, Krivan W, McKnight G, Clegg C, Foster D, Klucher KM. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol 4(1):63–68. [DOI] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, Riordan S, Sheridan D, Smedile A, Fragomeli V, Muller T, Bahlo M, Stewart GJ, Booth DR, George J. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41(10):1100–1104. [DOI] [PubMed] [Google Scholar]
- Sutti S, Albano E. 2020. Adaptive immunity: an emerging player in the progression of NAFLD. Nat Rev Gastroenterol Hepatol 17(2):81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. 2009. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41(10):1105–1109. [DOI] [PubMed] [Google Scholar]
- Terczynska-Dyla E, Bibert S, Duong FH, Krol I, Jorgensen S, Collinet E, Kutalik Z, Aubert V, Cerny A, Kaiser L, Malinverni R, Mangia A, Moradpour D, Mullhaupt B, Negro F, Santoro R, Semela D, Semmo N, Heim MH, Bochud PY, Hartmann R. 2014. Reduced IFNlambda4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun 5:5699. [DOI] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461(7265):798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibholm LK, Nielsen SSF, Pahus MH, Frattari GS, Olesen R, Andersen R, Monrad I, Andersen AHF, Thomsen MM, Konrad CV, Andersen SD, Højen JF, Gunst JD, Østergaard L, Søgaard OS, Schleimann MH, Tolstrup M. 2021. SARS-CoV-2 persistence is associated with antigen-specific CD8 T-cell responses. EBioMedicine 64:103230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenström J, Hellstrand K, Westin J, Nilsson S, Christensen P, Färkkilä M, Mørch K, Langeland N, Norkrans G, Lagging M. 2021. Presence of interferon-λ 4, male gender, absent/mild steatosis and low viral load augment antibody levels to hepatitis C virus. Scand J Gastroenterol 56:849–854. [DOI] [PubMed] [Google Scholar]
- Wang C, Horby PW, Hayden FG, Gao GF. 2020. A novel coronavirus outbreak of global health concern. Lancet 395(10223):470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2020. Coronavirus disease 2019 (COVID-19). Situation Report 51. World Health Organization, Iris, 1-9 https://apps.who.int/iris/handle/10665/331475. [Google Scholar]
- Ye L, Schnepf D, Becker J, Ebert K, Tanriver Y, Bernasconi V, Gad H, Hartmann R, Lycke N, Staeheli P. 2019. Interferon-λ enhances adaptive mucosal immunity by boosting release of thymic stromal lymphopoietin. Nat Immunol 20:593–601. [DOI] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382(8):727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.