Abstract
Amyloid cross‐seeding and amyloid inhibition are two different research subjects being studied separately for different pathological purposes, in which amyloid cross‐seeding targets to study the co‐aggregation of different amyloid proteins and potential molecular links between different neurodegenerative diseases, while amyloid inhibition aims to design different molecules for preventing amyloid aggregation. While both amyloid cross‐seeding and amyloid inhibition are critical for better understanding the pathological causes of different neurodegenerative diseases including Parkinson disease (PD) and Type 2 diabetes (T2D), less efforts have been made to reconcile the two phenomena. Herein, we proposed a new preventive strategy to demonstrate (a) the cross‐seeding of octapeptide TKEQVTNV from α‐synuclein (associated with PD) with hIAPP (associated with T2D) and (b) the cross‐seeding‐promoted hIAPP fibrillization and cross‐seeding‐reduced hIAPP toxicity. Collective results confirmed that TKEQVTNV can indeed cross‐seed with hIAPP monomers and oligomers, not protofibrils, to form β‐structure‐rich fibrils and to accelerate hIAPP fibrillization. Moreover, such cross‐seeding‐induced promotion effect by TKEQVTNV also rescued the pancreatic cells from hIAPP‐induced cytotoxicity by increasing cell viability and reducing cell apoptosis simultaneously. This work provides a new angle to discover amyloid fragments and use them as amyloid modulators (inhibitors or promotors) to interfere with amyloid aggregation of other amyloid proteins, as well as sequence/structure basis to explore the amyloid cross‐seeding between different amyloid proteins that may help explain a potential molecular talk between different neurodegenerative diseases.
Keywords: amyloid aggregation, amyloid cross‐seeding, hIAPP, Parkinson disease, Type 2 diabetes, α‐synuclein
Abbreviations
- AD
Alzheimer disease
- AFM
atomic force microscopy
- Aβ
amyloid‐β
- BBB
blood–brain barrier
- CD
circular dichroism spectroscopy
- DMSO
dimethyl sulfoxide
- HFIP
1,1,1,3,3,3‐hexafluoro‐2‐propanol
- hIAPP
human islet amyloid polypeptide
- LDH
lactate dehydrogenase
- MTT
3‐(4,5‐dimethylthiazole‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- NAC
nonamyloid component
- PD
Parkinson disease
- T2D
Type 2 diabetes
- ThT
thioflavin T
- α‐syn
α‐synuclein
1. INTRODUCTION
Pathological accumulation of misfolded and aggregated proteins is associated with many neurodegenerative diseases, for example, the aggregation of amyloid‐β (Aβ), α‐synuclein (α‐syn), and human islet amyloid polypeptide (hIAPP) is associated with Alzheimer disease (AD), Parkinson disease (PD), and Type 2 diabetes (T2D), respectively. 1 , 2 , 3 , 4 Among these neurodegenerative diseases, T2D has shown to have the highest incidence rate worldwide (1 out of 3 adults), resulting in ~1.5 million deaths anuually. 5 , 6 While the exact pathophysiological causes of T2D still remain unclear, significant efforts and progresses have been made to develop different preventive strategies and components for targeting the production, aggregation, and clearance of hIAPP, which is considered to play an vital role in β‐cell dysfunction and death. 7 , 8 Particularly, numerous research works have developed different types of amyloid inhibitors against hIAPP aggregation, including small molecules, 9 , 10 , 11 , 12 , 13 polymers, 14 , 15 nanoparticles, 16 , 17 and peptides. 18 , 19 These amyloid hIAPP inhibitors achieve their inhibition functions via different ways 8 , 20 by (a) delaying or preventing the misfolding of hIAPP into β‐strand structures, (b) remodeling hIAPP aggregation pathways away from key neurotoxic species, (c) forming less toxic or nontoxic hIAPP‐inhibitor complexes, (d) disintegrating toxic species, and (e) attenuating the hIAPP‐induced cell toxicity. Almost all of these inhibitors are effective to inhibit hIAPP aggregation, but none of them have achieved to clinical success for T2D treatment.
Alternatively, it is more fundamental and mechanistic importance for exploring new strategies for modulating hIAPP fibrillization and toxicity. First, given a well‐known fact that prefibrillar soluble oligomers of hIAPP and other amyloid proteins are the key neurotoxic species, another strategy is to design or discover compounds to promote hIAPP aggregation by quickly bypassing toxic hIAPP oligomers formed at an intermediate stage, instead of inhibiting hIAPP aggregation. Second, in comparison with widely used organic compounds as hIAPP inhibitors, 21 peptide‐based inhibitors of hIAPP are less explored, but provide a promising alternative because of their high biocompatibility and biodegradability, less cytotoxicity, weak immunogenicity, high binding affinity and specificity to targets, and ease of synthesis and modification. 22 In the past decade, peptide‐based amyloid inhibitors (not only limited to hIAPP) were mostly derived from the amyloidogenic fragments of their parent amyloid proteins with or without slight sequence modifications. 23 As specific to peptide inhibitors of hIAPP, a number of short peptides of SNNFGA (hIAPP20–25), GAILSS (hIAPP24–29), NYGAILSS (modified hIAPP22–29), NFGAILPP (modified hIAPP22‐29), and FLPNF (modified hIAPP11–15) have been reported to inhibit hIAPP aggregation in vitro. 24 , 25 , 26 Similarly, KLVFFA (Aβ16‐21), GGVVIA(Aβ37‐42) selected from Aβ showed highly sequence‐specific inhibitory activity against Aβ1–42 aggregation and cytotoxicity. 27 Central NAC (nonamyloid component) region (α‐syn61–95) 28 and AVVT (α‐syn69–72) 29 from α‐synuclein and peptide E59–71 from β2‐microglobulin 30 were also demonstrated their strong influence with the self‐assembly of their parent amyloid proteins. Hypothetically, since these amyloid fragments are selected from their parent sequences, it is not surprising that they exhibit high tendency to interact with their parent proteins so as to impose their inhibition effects on amyloid aggregations.
More importantly, recent studies have reported the co‐existence and co‐interactions of some different amyloid proteins in blood and tissues including α‐syn and hIAPP, 31 Aβ and hIAPP, 32 Aβ and α‐syn, 33 Aβ and tau, 34 Aβ and transthyretin, 35 hIAPP, and insulin. 36 Such the cross‐sequence interactions between different amyloid proteins are known as amyloid cross‐seeding (“proteinopathies”), which may be correlated with the co‐occurrence of different neurodegenerative diseases in the same patients. 37 Amyloid cross‐seeding not only indicates the presence of one type of amyloid proteins will trigger the pathological event of another protein misfolding disease (PMD)‐associated amyloid proteins, but also provides a potential new design strategy of amyloid modulators, that is, amyloid fragments from one amyloid protein could cross‐interact with another full‐length amyloid protein to interfere with its aggregation via cross‐seeding interactions.
Our previous works have demonstrated (a) the existence of cross‐seeding between different full‐length amyloid proteins and between different full‐length amyloid proteins and protein fragments (e.g., Aβ and hIAPP, 32 Aβ and GNNQQNY, hIAPP and GNNQQNY, 38 hIAPP and rIAPP, 39 ) and (b) the use of short protein fragments to modulate the aggregation and toxicity of their parent amyloid proteins. 24 , 25 , 26 , 27 , 28 , 29 , 30 Inspired by these works, here we are the first to (a) explore and identify the existence of the cross‐seeding between an octapeptide TKEQVTNV derived from the NAC region of α‐synuclein (associated with PD) and full‐length hIAPP (associated with T2D) (Figure 1) and (b) discover TKEQVTNV as amyloid promotor to interact with hIAPP monomers and oligomers, not protofibrils, to accelerate hIAPP fibrillization and thus to reduce hIAPP‐induced cell toxicity. Selection of TKEQVTNV from α‐synuclein 40 , 41 and hIAPP as our study system is attributed to a close clinical correlation between T2D and PD. Accumulating clinical data have shown that T2D, PD, and AD are known to promote each other's incidence in the same patients. 42 Evidently, T2D patients have ~28%–85% higher risk for developing PD in different studied regions. 43 , 44 , 45 , 46 From a pathological viewpoint, T2D and PD also share common symptoms of insulin resistance, 47 excess oxidative stress, 48 and chronic inflammation, 49 thus providing additional evidence for a close relationship between T2D and PD. While the mechanistic link between T2D and PD is still unclear, it is intuitive to hypothesize that molecular cross‐talk between T2D and PD is likely attributed to direct interactions between hIAPP and α‐synuclein. 50 , 51 For instance, several studies have shown that hIAPP promoted α‐synuclein fibrillization, 31 and vice versa, 52 further implying that both hIAPP and α‐synuclein can be templates for facilitating fibril growth via cross‐seeding between hIAPP and α‐synuclein. Given the cross‐seeding between hIAPP and α‐synuclein and its potential implication for a pathological link between T2D and PD, here through the step‐by‐step tests using a combination of biophysical, imaging, cellular techniques, we demonstrated the cross‐seeding between TKEQVTNV and hIAPP, which not only accelerated hIAPP fibrillization in a dose dependent manner, but also protected cells from hIAPP‐induced toxicity. This work provides not only a new strategy for discovering heterogeneous amyloid proteins or fragments as modulators of another amyloid proteins, but also the better fundamental understanding of amyloid cross‐seeding (Figure 1).
FIGURE 1.
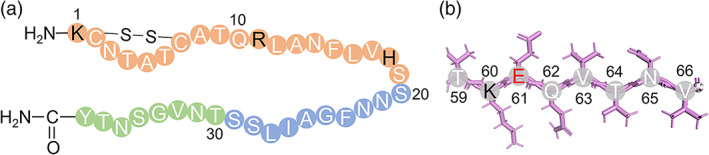
(a) A full‐length hIAPP1–37 and (b) fragmented peptide 59TKEQVTNV66 from α‐synuclein59–66. Color ID: N‐Terminal residues (orange circles), C‐Terminal residues (green circles), hydrophobic core (blue circles), positively charged residues (black letters), and negatively charged residues (red letters)
2. METHODS AND MATERIALS
2.1. Materials
Human amylin (hIAPP1–37, ≥ 95%) was purchased from CPC Scientific (Sunnyvale, CA). Synthetic TKEQVTNV was obtained from Genscript (Piscataway, NJ). 1,1,1,3,3,3‐hexafluoro‐2‐propanol (HFIP, ≥99.9%), dimethyl sulfoxide (DMSO, ≥99.9%), and thioflavin T (ThT, 98%) were purchased from Sigma Aldrich (St. Louis, MO). All other chemicals were of the highest grade available.
2.2. Peptide purification and preparation
Both hIAPP and TKEQVTNV were obtained in a lyophilized form and stored at −20°C once they arrived. To obtain homogenous monomeric peptide, both peptide powder was reconstituted in HFIP at 1 mg/ml concentration and incubated at the ambient temperature for 2 hr, then sonicated in ice bath for 30 min before centrifuged at 14,000 rpm and 4°C for 30 min. Finally, supernatant was aliquoted according to experimental setting and stored at −80°C. Before each experiment, the sub‐packaged peptides were lyophilized using freeze dryer for more than 30 min, then predissolved in 10 mM NaOH, sonicated for 10 s and further diluted in different buffer to a final concentration of 25 μM.
2.3. ThT fluorescence assay
Two millimolar ThT stock solution was obtained by dissolving ThT powder into Milli‐Q water and stored in dark place at room temperature. Samples were prepared on ice by mixing hIAPP‐10 mM Tris buffer with TKEQVTNV in different molar ratios (1:0–1:4) to achieve a final test volume of 200 μl. After transferring the samples to 96‐well plate in the plate reader, 1 μl 2 mM ThT was quickly added to each well before the aggregation was initiated at 37°C, then fluorescence intensity data were recorded consistently at 30 min intervals for 24 hr. The kinetic top‐read mode of a SpectraMax M3 microplate reader (Molecular Devices, CA) with excitation at 450 nm and emission at the range of 470 to 500 nm was used to monitor the ThT fluorescence.
2.4. Circular dichroism spectroscopy
The secondary structures of peptides were examined by far‐UV circular dichroism (CD) spectroscopy with a J‐1500 spectropolarimeter (Jasco Inc., Japan) using a continuous scanning mode at room temperature. hIAPP‐10 mM PBS buffer with and without TKEQVTNV incubated at 37°C for 0, 4, 8, 12, and 24 hr. At each time point, 150 μl sample solution was placed into a 1 mm optical path length CD cuvette and scanned between 190 and 250 nm with a step size of 0.5 nm and 50 nm/min scan rate. All received spectra were analyzed by subtracting PBS buffer baseline to remove background influence. The secondary structural contents were determined by using the Beta Structure Selection (BeStSel) algorithm 53 (http://bestsel.elte.hu/).
2.5. Atomic force microscopy
The morphology changes of hIAPP mediated by TKEQVTNV was observed by using Nanoscope III multimode atomic force microscopy (AFM) with an Extender eletroncis module (Veeco Corp, Santa Barbara, CA) in a ScanAsyst Mode. A 20 μl sample used in CD experiment was taken for AFM measurement at different time points in order to correlate morphological changes with their conformational change.
Twenty microliter samples at 6, 12, and 24 hr were dropped on a piece of cleaved mica for 5 min, rinsed three times with Mill‐Q water to totally remove additional salt and dried with an air stream before stored in a sealed container. All images were recorded at the 512 × 512 pixel resolution at a typical scan rate of 1.0 Hz and with the vertical tip oscillation frequency of 45–95 kHz. Representative AFM images were obtained by scanning six different locations on the mica surface.
2.6. Cell culture
Rat insulinoma cells RIN‐m5F (ATCC® CRL‐11605TM, Manassas, VA) were chosen as model pancreatic β‐cells to evaluate hIAPP‐mediated cytotoxicity, and cultured in sterile‐filtered RPMI‐1640 medium (Sigma Aldrich, St. Louis, MO) mixed with 10% (v/v) FBS (VWR, Radnor, PA) and 1% (v/v) penicillin/streptomycin (Sigma Aldrich) in a T75 flask at 37°C and 5% CO2 in a humidified incubator. After the cells attached 80% surface area, they were then harvested by using 0.25 mg/ml Trypsin/EDTA solution (Sigma Aldrich) and seeded in 96‐well plate (2 × 104 per well).
2.7. MTT reduction assay
Cellular metabolic activity as an indicator of cell viability was measured by MTT reduction assay. Oxidoreductase enzymes released from mitochondrion reduced the MTT to formazan and therefore exhibited colored solution to quantify the cell viability. 54 96‐well plate with cells were incubated for 24 hr to allow them attached. Then replace with fresh cell culture medium with hIAPP/TKEQVTNV/hIAPP‐TKEQVTNV solutions and cultured for another 24 hr at 37°C and 5% CO2 in a humidified incubator. For seeding cell assay, during the same 24 hr incubation, TKEQVTNV was added into cell‐hIAPP solution at 1, 5, and 11 hr. In MTT experiment, cell culture was replaced with 0.5 mg/ml MTT fresh media followed by incubated for 4 hr. Then the culture medium was discarded and replaced by 150 μl DMSO per well to dissolve the formazan crystals. Finally, absorbance was recorded at 540 nm using SpectraMax M3 microplate reader. Each sample repeated three times and cell viability were calculated in comparison with untreated cells. Each sample was counted in triplicate and reported as mean ± SD.
2.8. Lactate dehydrogenase cytotoxicity assay
The amyloid‐induced cytotoxicity was measured by lactate dehydrogenase (LDH) assay. Soluble enzyme LDH found in cytoplasm would release into cell medium and therefore act as an indicator of irreversible cell death due to cell membrane damage. 55 LDH assay was performed using CytoSelect™ LDH Cytotoxicity Assay Kit (Cell Biolabs, San Diego, USA). In short, after 24 hr cell‐drug incubation, 10 μl sterile water or Triton X‐100 solution was added to each well as positive and negative control, respectively. After 10 min incubation at room temperature, 90 μl cell medium of each well was transferred to a clean 96‐well plate before adding 10 μl LDH cytotoxicity assay reagent per well. Last, plate was incubated for another 30 min and read OD using 450 nm. Relative cytotoxicity percentage were proportional normalized by the difference of positive and negative control. Each sample was counted in triplicate and reported as mean ± SD.
2.9. LIVE/DEAD viability/cytotoxicity assay
LIVE/DEAD cell assay was further used to visualize the amyloid‐mediated cell viability and cytotoxicity. hIAPP incubated with and without TKEQVTNV for 24 hr was stained using a LIVE/DEAD® Viability/Cytotoxicity Kit (L3224, Invitrogen) and imaged by fluorescence microscope (Olympus IX81) to visualize the live and dead insulinoma cells. The representative images of the live and dead cells were chosen from three different locations.
3. RESULTS AND DISCUSSION
3.1. TKEQVTNV cross‐seeds with hIAPP1 –37 to promote amyloid aggregation
To probe the cross‐seeding behavior between TKEQVTNV and hIAPP, we first performed the self‐ and hetero‐aggregation process of TKEQVTNV and hIAPP using thioflavin T (ThT) assay. Figure 2a shows amyloid fibril formation at different molar ratios of hIAPP:TKEQVTNV from 1:1 to 1:4. As a control, the self‐aggregation of 25 μM hIAPP showed a typical nucleation‐polymerization kinetics of amyloid fibrillation, including a 0.5 hr lag phase (nuclei formation), 0.5–11 hr exponential growth phase (protofibril formation), and 11–24 hr saturation phase (mature fibril formation), thus reaching a final ThT plateau of ~340 a.u. Differently, pure TKEQVTNV did not produce any ThT signal at a high concentration of 100 μM, thus ruling out the possibility of TKEQVTNV self‐aggregation for influencing the cross‐seeding results between TKEQVTNV and hIAPP.
FIGURE 2.
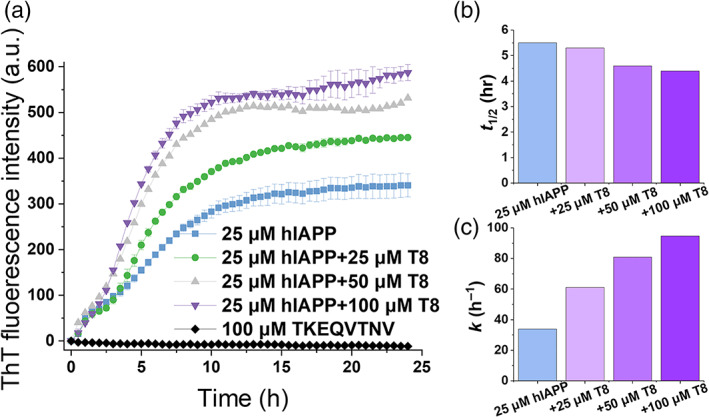
Aggregation kinetics of amyloid cross‐seeding between TKEQVTNV and hIAPP using ThT assay. (a) Cross‐seeding aggregation kinetics of hIAPP (25 μM) and TKEQVTNV at different molar ratios of hIAPP/TKEQVTNV (1:1, 1:2, and 1:4). Error bars represent the standard deviation of three replicate experiments. ThT kinetic parameters of (b) fibrillization half‐life t 1/2 and (c) fibrillization rate constant k at t 1/2
For comparison, when incubating hIAPP (25 μM) with equimolar nonamylogenic TKEQVTNV (25 μM) at the same incubation conditions, TKEQVTNV clearly promoted hIAPP aggregation as evidenced by (a) the higher maximal ThT intensity (I max) in the final equilibrium phase, (b) the steeper slope in the growth phase, and (c) the less time to reach half of I max. Consistently, the presence of the other two hIAPP:TKEQVTNV ratios of 1:2 and 1:4 also led to similar increase of ThT signals in both growth and equilibrium phases, confirming that TKEQVTNV can indeed cross‐seed with hIAPP to promote hIAPP aggregation in a dose‐dependent manner. Specifically, as hIAPP/TKEQVTNV molar ratio increased from 1:1 to 1:4, I max was increased by 30.9% at 1:1, 56.2% at 1:2, and 72.6% at 1:4, respectively. Additionally, the fibrillization half‐life (t 1/2) was reduced from 5.5 to 4.4 hr (Figure 2b) and the corresponding rate constant (k) of hIAPP fibrillization was dramatically increased by 80.8%–180.1% (Figure 2c). It is interesting to note that for all cross‐seeding cases, they had similar ThT curves at the first 2 hr, implying the common cross‐seeding pathway between hIAPP and TKEQVTNV. In other words, at the beginning of cross‐seeding hIAPP nucleus are likely formed, then hIAPP seeds could serve as templates to recruit TKEQVTNV monomers to form heterologous aggregates, therefore producing the higher ThT signals in the final equilibrium stage. Additionally, increase of TKEQVTNV concentration from 50 to 100 μM did not produce the higher ThT aggregation curve, indicating that TKEQVTNV has reached the saturated concentration to cross‐seed with hIAPP to promote hIAPP fibrillization.
3.2. TKEQVTNV modifies the morphologies and secondary structures of hIAPP1 –37
To better understand the cross‐seeding results from ThT curves, AFM, and CD were used to characterize the morphological and conformational changes of hIAPP and TKEQVTNV. Figure 3 shows a series of topographical AFM images of hIAPP (25 μM) in the absence and presence of TKEQVTNV (25–100 μM) as a function of incubation time at 37°C. At a first glance, both homo‐ and cross‐seeding aggregation can form amyloid fibrils during 24 hr, but they exhibited different fibril formation kinetics. As a control, pure hIAPP (25 μM) produced a few protofibrils of different lengths of 300–1,500 nm at 6 hr that were sparsely distributed in the scanning area. After 12 hr, longer and thicker mature fibrils with average heights of 14–15 nm were observed and continued to grow into denser fibrils with average heights of 17–18 nm. As compared to pure hIAPP, cross‐seeding samples of hIAPP‐TKEQVTNV had much faster aggregation kinetics as evidenced by more and denser fibrils formed at every time point of 6, 12, and 24 hr. To be detailed, at 6 hr, co‐incubation of hIAPP and TKEQVTNV significantly accelerated the aggregation process by forming more protofibrils that were strongly aggregated and entangled with each other. Such cross‐seeding‐induced promotion effect became more pronounced at the longer incubation time. Moreover, for all cross‐seeding samples at three different molar ratios, fibrils formed at 24 hr enabled to nearly cover the whole scanning area with average heights of 20–35 nm, in sharp contrast to the scattered thin hIAPP fibrils alone. Additionally, hIAPP‐TKEQVTNV samples (hairy, short fibrils) displayed different morphologies from pure hIAPP samples (thin, long fibrils), such morphological differences between homo‐ and cross‐seeding samples become more obvious at the low concentration of TKEQVTNV (25–50 μM). AFM results were generally in line with ThT results to support that TKEQVTNV cross‐seeds with hIAPP to promote hIAPP fibrillization in a concentration‐dependent manner.
FIGURE 3.
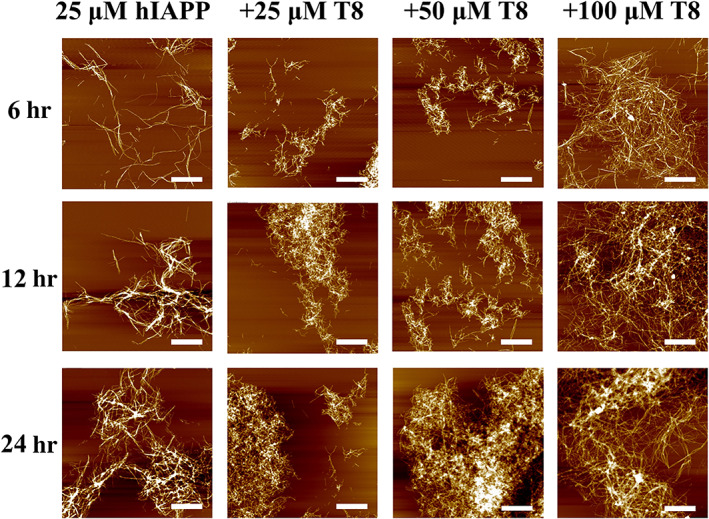
AFM images for pure hIAPP peptides (25 μM) in the absence (first column) and presence (second to fourth column with T8) of TKEQVTNV at different molar ratios of hIAPP/TKEQVTNV (1:1, 1:2, and 1:4) at 6, 12, and 24 hr. Scale bars are 1 μm
To gain further insights into cross‐seeding between hIAPP and TKEQVTNV, we monitored the secondary structural changes of cross‐seeding samples using CD. Use of 24 hr timeline allowed to cover all of aggregation states from monomeric to mature fibrillar hIAPP. First, as shown in Figure 4a, freshly prepared hIAPP (25 μM) exhibited the structural transition from random coil (a representative negative peak at ~198 nm) at 0 hr, to α‐helix conformation (negative peak at ~208 and ~222 nm) at 8 hr, finally to characteristic cross β‐sheet structure (a negative peak at ~215 nm at the expense of disappearance of ~198 nm peak) at 24 hr. For comparison, pure TKEQVTNV always maintained its random coil structure during 24 hr incubation (Figure S1a). Second, co‐incubation of hIAPP with different ratios of TKEQVTNV (1:1–1:4) exhibited similar conformational transition from random coils to β‐rich structures (Figure S1b), as evidenced by the co‐occurrence of the peak decrease at ~198 nm and the peak increase at ~215 nm. Third, to better qualify the extent of structural transition of hIAPP as induced by TKEQVTNV, we analyzed the original CD spectra by subtracting pure TKEQVTNV signals from cross‐seeding signals. CD spectra in Figure 4b showed that cross‐seeding of TKEQVTNV with hIAPP at 1:4 ratio exhibited the faster conformational transition than pure hIAPP in three aspects: (a) in the early aggregation stage of 0–4 hr, hIAPP‐TKEQVTNV sample showed an obvious decline in peak 198 nm, while hIAPP remained almost unchanged for this peak; (b) in the growth stage of 8–12 hr, cross‐seeding samples showed nearly all the β‐sheet structure with a tiny peak at 198 nm. However, at the same time point, pure hIAPP showed a smaller β‐sheet peak and an obvious random coil peak; (c) in the final equilibrium stage of 24 hr, cross‐seeding samples indeed promoted the β‐sheet formation by 7.6% at the expense of random coil and α‐helix.
FIGURE 4.
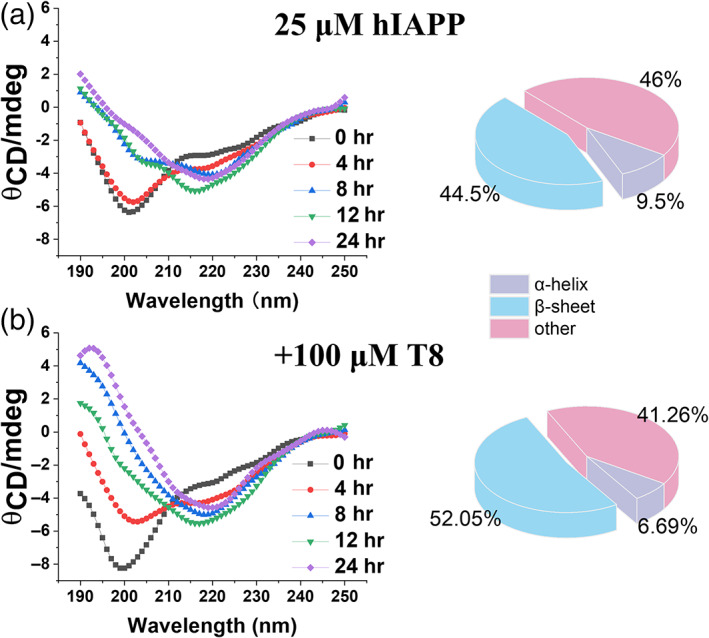
Time‐dependent circular dichroism (CD) spectra to monitor the secondary structure change of (a) pure hIAPP (25 μM) and (b) cross‐seeding samples of hIAPP‐TKEQVTNV (25 μM:100 μM) during 24 hr aggregation
3.3. TKEQVTNV cross‐seeds with different hIAPP1 –37 seeds to modulate the cross‐seeding pathways
Upon demonstrating (a) the cross‐seeding TKEQVTNV with freshly prepared hIAPP monomers and (b) the cross‐seeding‐induced acceleration of hIAPP fibrilization, it is more interesting and important to examine whether TKEQVTNV could still cross‐seed with the larger sizes of hIAPP seeds, if so, whether the cross‐seeding could also promote hIAPP fibrillation. To test this goal, we designed different cross‐seeding tests by adding nonamyloidogenic TKEQVTNV (100 μM) to hIAPP (25 μM) seeds, which were preformed at 1, 5, and 11 hr encompassing the lag, growth, and early equilibrium phases. As shown in Figure 5a, when TKEQVTNV was added to a preformed hIAPP seed solution obtained at 1 hr, ThT signal (red curve) experienced an instantaneous and significant increase as compared to that of pure hIAPP. This growth process lasted for almost ~10 hr and finally reached to a stable plateau of ~1,094, which was ~45% higher than that of pure hIAPP aggregation (black curve). In the second case of cross‐seeding TKEQVTNV with 5 hr‐seeded hIAPP, similar increase of ThT signals was also observed (blue curve), that is, upon adding TKEQVTNV to 5 hr‐seeded hIAPP solution, ThT aggregation curve suddenly increased immediately, followed by a quick approach to a stable plateau of ~890 after 2 hr growth time, showing ~18% increase of final fibrils. The first two cases confirm that (a) TKEQVTNV can cross‐seed with smaller hIAPP seeds formed at the early lag and growth phases, (b) such cross‐seeding also promoted hIAPP fibrillization, and (c) cross‐seeding‐induced promote effect became less efficient when TKEQVTNV tended to cross‐seed with the larger hIAPP seeds. In the third cross‐seeding case, when adding TKEQVTNV to preformed hIAPP fibrils from 11 hr incubation, cross‐seeding nearly disappeared as indicated by no any ThT signal increase, instead the two overlapped ThT curves between mixed TKEQVTNV‐hIAPP (green curve) and pure hIAPP (black curve).
FIGURE 5.
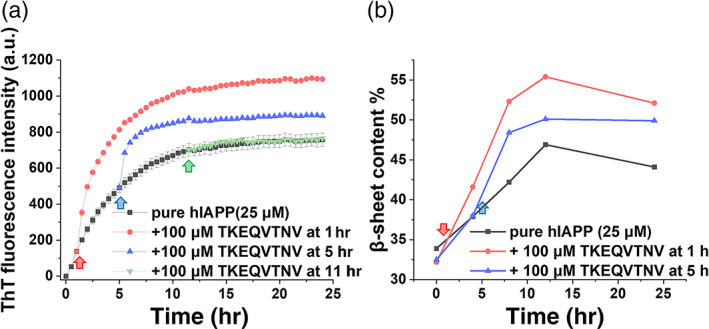
Cross‐seeding of TKEQVTNV with different sizes of hIAPP seeds. (a) Time‐dependent ThT fluorescence curves and (b) β‐structure contents upon adding TKEQVTNV (100 μM) to hIAPP (25 μM) seeds preformed at the lag (1 hr), growth (5 hr), or equilibrium (11 hr) phases
In parallel, we also performed CD to quantify the secondary structure changes for three cross‐seeding cases by adding TKEQVTNV (100 μM) to 25 μM hIAPP seeds preformed at different time points. β‐sheet contents in Figure 5b, as calculated from original CD curves in Figure S2, confirmed the different cross‐seeding efficiency between TKEQVTNV and hIAPP species. Specifically, after adding TKEQVTNV to 1 hr hIAPP seeds, β‐sheet content immediately increased to 42% at 4 hr and continuously increased to 52% at 24 hr. Similarly, the cross‐seeding of TKEQVTNV with 5 hr hIAPP seeds also led to a continuous increase of β‐sheet content to 50% finally. For comparison, the overall β‐sheet content of the three systems was in a decrease order of TKEQVTNV‐1 hr hIAPP seeds > TKEQVTNV‐5 hr hIAPP seeds > pure hIAPP. Collectively, TKEQVTNV cross‐seeds with different hIAPP seeds exhibited different cross‐seeding efficiency, suggesting the existence of cross‐seeding barriers that could be attributed to several factors. TKEQVTNV has a high tendency to interact with hIAPP monomers, probably because they have similar random coils for easily entangling with each other. Once hIAPP seeds are formed, they will recruit TKEQVTNV to adopt similar β‐structure to form hybrid hIAPP‐TKEQVTNV aggregates. The nonamyloidogenic nature of TKEQVTNV makes this cross‐seeding process more energetically expensive to overcome the higher cross‐seeding barriers for achieving structural transition and aggregation. When hIAPP protofibrils are formed, additional energy barriers arise from incompatible hydrophobicity and lack of reacting sites between hydrophobic hIAPP fibrils and hydrophilic TKEQVTNV.
3.4. Cross‐seeding of TKEQVTNV with hIAPP1 –37 alleviates amyloid‐mediated cytotoxicity
Different from amyloid inhibitors that have been well studied to prevent amyloid aggregation and amyloid‐induced cytotoxicity, amyloid promoters have recently been found to show protection capacity against amyloid cytotoxicity by quickly bypassing toxic oligomeric aggregates. 13 As described above, since TKEQVTNV significantly accelerated hIAPP fibrillization, it is possible for TKEQVTNV‐induced cross‐seeding to alleviate hIAPP‐associated cytotoxicity. To test this hypothesis, we applied 3‐(4,5‐dimethylthiazole‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assay for cell viability and lactate dehydrogenase (LDH) assay for cell apoptosis to examine the potential protection role of TKEQVTNV in pancreatic cells using the RIN‐m5F cell line incubated for 24 hr. As a control, we first set untreated RIN‐m5F cells as a baseline by assigning the results to 100% cell viability and 0% cytotoxicity. Pure TKEQVTNV presented nearly no cytotoxicity to cells as indicated by 105%–113% cell viability (Figure 6a) and 0.27%–0.71% cytotoxicity (Figure 6b) during 24 hr cell culture, indicating that TKEQVTNV at any concentration between 25 and 125 μM has no toxicity to cells and does not affect the normal growth of cells. In sharp contrast, pure hIAPP (25 μM) was significantly toxic to pancreatic cells, as evidenced by 47% of cell viability and 49% of cell apoptosis. In parallel, when introducing TKEQVTNV into hIAPP‐cultured cell solutions at a 1:1–1:4 ratio for 24 hr, cell viability/cell apoptosis was 49.9%/46.9%, 51.9%/44.6%, and 51.4%/42.6%, showing a concentration‐dependent protection effect on hIAPP‐induced cytotoxicity. Seeing is believing, to more intuitively understand the influence of cross‐seeding between TKEQVTNV and hIAPP on pancreatic cells, we further conducted live/dead cell assay to directly visualize cell viability. Figure 6c consistently showed that (a) TKEQVTNV alone exhibited no toxicity to cells, as observed by the large proportion of live cells (green stains) as compared to dead ones (red stains); (b) co‐incubation of TKEQVTNV with hIAPP led to much less dead cells as compared with pure hIAPP system, again confirming the protection ability of TKEQVTNV from hIAPP‐induced cell toxicity.
FIGURE 6.
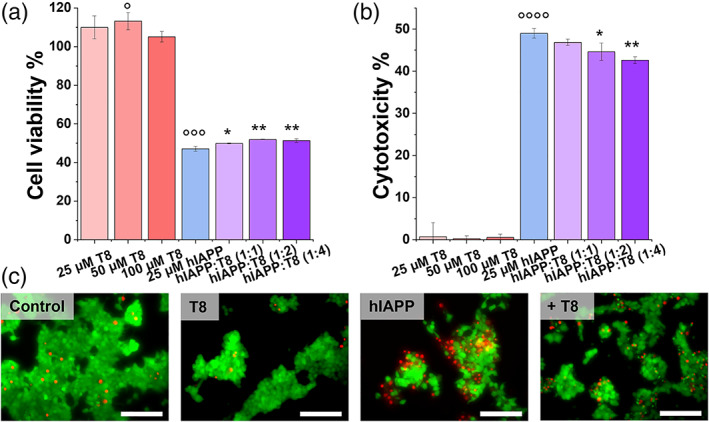
Protection role of cross‐seeding of TKEQVTNV with freshly prepared hIAPP1–37 monomers in amyloid‐mediated cytotoxicity by (a) MTT assay for cell viability and (b) LDH assay for cell cytotoxicity when the incubation of hIAPP (25 μM) with and without TKEQVTNV at molar ratios of 1:1, 1:2, and 1:4 for 24 hr. Data were normalized by the pure cell group. The t‐test was used for data analysis (n = 3) for cells treated with TKEQVTNV or hIAPP alone relative to control. (°°°p <.005, °°°°p <.001), as well as cells treated with mixed TKEQVTNV‐hIAPP relative to cells treated with hIAPP alone (*p <.05; **p <.01). (c) Representative fluorescence microscopy images of cells treated with 25 μM hIAPP in the absence or presence of 100 μM TKEQVTNV for 24 hr. Untreated cells were set as control for comparison. Red and green fluorescence represent the dead and live cells, respectively. Scale bars are 100 μm
Instead of treating cells with the freshly prepared TKEQVTNV and hIAPP in cell medium, we also designed a new set of seeding experiments to examine whether TKEQVTNV (100 μM) could also protect cells from the preformed hIAPP seeds (25 μM) at 1, 5, and 11 hr. As shown in Figure 7, at the first glance, whenever TKEQVTNV was added into the hIAPP‐seeded cell mediums, it can protect cells from toxic hIAPP seeds. Specifically, when incubating TKEQVTNV with 1, 5, and 11 hr hIAPP seeds, the resultant cell viability/cytotoxicity was 51.2%/35.0%, 50.6%/36.2%, and 48.7%/37.7%, respectively, showing a similar cell protection effect. Of note, it appears that TKEQVTNV is more effective to protect cells from hIAPP‐seeds‐induced cell toxicity than from hIAPP monomer‐induced cell toxicity.
FIGURE 7.
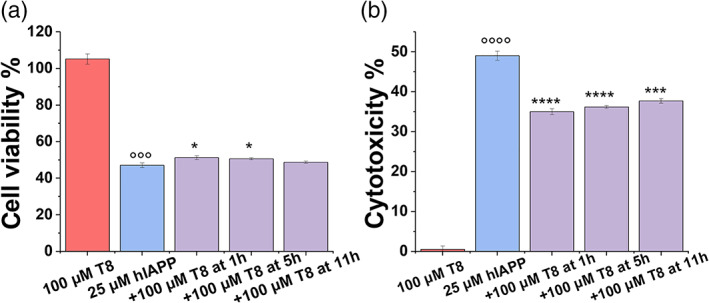
Protection role of cross‐seeding of TKEQVTNV with different hIAPP1–37 seeds in amyloid‐mediated cytotoxicity by (a) MTT assay for cell viability and (b) LDH assay for cell cytotoxicity when adding TKEQVTNV (100 μM) to 1, 5, and 11 hr‐seeded hIAPP (25 μM) solution for 24 hr. Data were normalized by the pure cell group. The t‐test was used for data analysis (n = 3) for cells treated with TKEQVTNV or hIAPP alone relative to control. (°°°p <.005, °°°°p <.001), as well as cells treated with mixed TKEQVTNV‐hIAPP relative to cells treated with hIAPP alone (*p <.05; **p <.01, ***p <.005, ****p <.001)
While some drugs have been approved to alleviate T2D symptoms (i.e., blood sugar level), there is still no cure for T2D due to the complex T2D pathologies and unclear T2D mechanisms. Current prevailing “amyloid cascade hypothesis” postulates that the misfolding and aggregation of intrinsically disordered hIAPP into highly ordered, β‐structure‐rich species (namely amyloids) is considered as a central pathogenic cause of T2D. The hIAPP‐mediated toxicity mechanisms involve different pathways to induce β‐cell death, including the formation of excessive reactive oxygen species, the increase of end axoplasmic reticulum stress, the initiation of inflammatory response, and membrane disruption. To this end, different types of modulators (including inhibitors and promotors) were developed to regulate the production, aggregation, and clearance of hIAPP and ameliorate hIAPP‐induced islet‐β cell dysfunction, cell membrane disruption, and glucose intolerance. 13 , 21 , 56 However, most of them fail the large‐scale clinic trials 37 due to their (a) poor solubility to be dissolved in human body fluids (blood and spinal fluid) for drug transport; (b) poor blood–brain barrier (BBB) permeability to deliver drugs to target cells/organs; (c) low binding affinity and selectivity to capture hIAPP efficiently and accurately; and (d) inability to address multiple pathogenic pathways involving other PMDs. Recently, “amyloid cross‐seeding hypothesis” have also observed (a) the co‐occurrence of different PMDs in the same individuals and (b) the increased risk for developing different PMDs if a PMD is presented. This implies the existence of certain pathological links, despite still unknown, between different PMDs. It is intuitive to hypothesize that molecular cross‐talk between different PMDs is likely attributed to direct interactions between their corresponding disease‐causative proteins, leading to the co‐aggregation of different amyloid proteins. 37
Here, we proposed a strategy to reconcile amyloid cross‐seeding and amyloid prevention in a binary system of α‐synuclein fragmental peptide (TKEQVTNV) and hIAPP. Specifically, TKEQVTNV can indeed cross‐seed with hIAPP aggregates of different sizes to promote hIAPP fibrillization, regulate structural conversion toward β‐sheet structures, and rescue hIAPP‐mediated cell death. However, cross‐seeding efficiency of TKEQVTNV‐hIAPP depends on hIAPP concentrations and its aggregation states, indicating the existence of cross‐species barriers along the folding pathways of different amyloid proteins. As a result, the difference in cell protection effects could be attributed to different cross‐seeding efficiencies between TKEQVTNV and different hIAPP seeds. It is generally accepted that during the hIAPP fibrillization process, oligomeric hIAPP aggregates induce their toxicity mainly through (a) calcium dyshomeostasis and (b) membrane leakage. 57 As shown in Figure 8a, hIAPP oligomers have the ability to interact with cell membranes to form calcium permeable transition pores in the mitochondria and to disrupt cell membranes, both resulting in calcium hemostasis responsible for cell death. When TKEQVTNV was added to monomeric hIAPP solution, TKEQVTNV acted as amyloid promoter to accelerate hIAPP fibrillization by quickly bypassing the most toxic oligomer‐rich stage, which in turn reduced cell toxicity to some extents (Figure 8b). Such TKEQVTNV‐induced hIAPP fibrillization was also confirmed by ThT (Figure 2) and CD (Figure 4) results as discussed above. To better understand a more complete picture of cross‐seeding scenario between TKEQVTNV and hIAPP, the seeding experiments have shown that TKEQVTNV had the lower cross‐seeding efficiency to facilitate hIAPP fibrillation via hIAPP seeds than via hIAPP monomers (Figure 8c). As a result, while less hIAPP seeds or oligomers are converted into hIAPP fibrils by TKEQVTNV, TKEQVTNV‐hIAPP seed complexes are less toxic than hIAPP seeds with relatively inert ability to interact with cell membrane, thus still resulting in the comparable, but different, cell protection effect to the TKEQVTNV‐hIAPP monomer cases.
FIGURE 8.
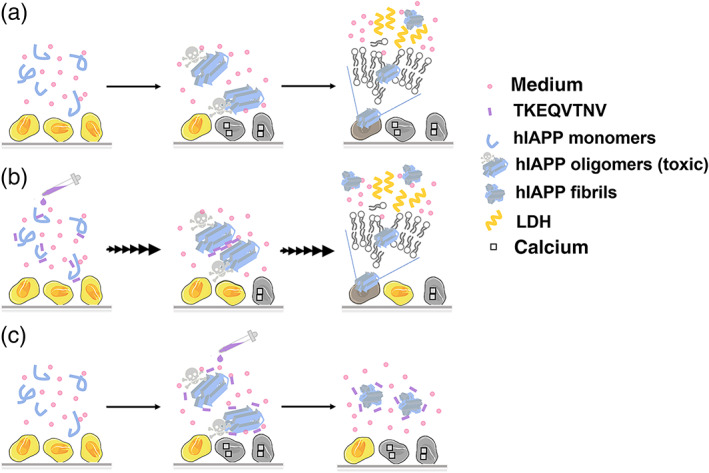
Hypothetical cross‐seeding‐induced cell toxicity/protection models for (a) pure hIAPP system, (b) TKEQVTNV‐hIAPP monomers system, and (c) TKEQVTNV‐hIAPP seeds system
4. CONCLUSIONS
Given a clinical and pathological correlation between T2D and PD, collective aggregation data from ThT assays, CD spectra, and AFM images confirmed that TKEQVTNV can interact with hIAPP monomers at different molar ratios and hIAPP oligomers at the lag and early growth phases to promote β‐structure formation by 8% and hIAPP fibrillization by up to 73%. But, TKEQVTNV was unable to accelerate fibrillar hIAPP aggregation. Fundamentally, different cross‐seeding efficiencies between TKEQVTNV and different hIAPP aggregates also indicate the existence of co‐aggregation barriers along the folding and aggregation pathways. Further in vitro MTT and LDH assays showed that the nonamyloidogenic TKEQVTNV also protected pancreatic cells from both hIAPP monomer‐ and hIAPP oligomer‐induced toxicity by increasing cell viability from 47% to 52% and decreasing cell apoptosis from 49% to 38%. Reduction of hIAPP cytotoxicity by TKEQVTNV is likely attributed to different detoxification pathways by quickly bypassing oligomer‐forming stage and forming less toxic and inert hIAPP‐TKEQVTNV complexes. Based on our “like‐interacts‐like” hypothesis that the cross‐seeding between any two amyloid proteins/fragments are likely driven by their common β‐structures and the subsequent β‐structure‐to‐β‐structure interactions, some NAC fragments of α‐synuclein including E61−V66, A69−Q79, and I88−V95 possess similar self‐assembly ability to form β‐structures, 40 thus they are expected to strongly interact with hIAPP aggregates via (a) conformationally similar β‐structure interactions; (b) opposite charged residues (E, K, R) through electrostatic attraction; and (c) hydrophilic residues of N, T, Q, S to form hydrogen bonding networks. Further cross‐seeding studies of these α‐synuclein fragments with hIAPP are required to examine our speculation. More importantly, different from well‐studied amyloid inhibition, cross‐seeding‐induced amyloid promotion provides a different angle to develop the peptide‐based prevention strategies/molecules and to further reveal the fundamentals of amyloidosis.
CONFLICT OF INTEREST
We declare no conflict of interest in this manuscript.
AUTHOR CONTRIBUTIONS
Yijing Tang: Conceptualization (equal); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); writing – original draft (equal). Dong Zhang: Conceptualization (supporting); formal analysis (supporting); methodology (supporting); validation (equal); writing – original draft (supporting). Yonglan Liu: Data curation (supporting); formal analysis (supporting); software (equal); writing – original draft (supporting). Yanxian Zhang: Formal analysis (supporting); investigation (supporting); validation (supporting); writing – original draft (supporting). Yifan Zhou: Data curation (supporting); methodology (supporting); validation (supporting). Yung Chang: Conceptualization (equal); methodology (supporting); resources (supporting). Bowen Zheng: Formal analysis (supporting); methodology (supporting); validation (supporting). Alice Xu: Formal analysis (supporting); methodology (supporting); validation (supporting). Jie Zheng: Conceptualization (lead); funding acquisition (lead); investigation (equal); project administration (lead); supervision (lead); writing – original draft (lead).
Supporting information
FIGURE S1 Time‐dependent circular dichroism (CD) spectra to monitor the secondary structure change of (a) pure TKEQVTNV of 25–100 μM and (b) cross‐seeding samples of hIAPP‐TKEQVTNV at different molar ratios of 1:1, 1:2, and 1:8 at 0 and 24 hr.
FIGURE S2 Time‐dependent circular dichroism (CD) spectra to monitor the secondary structure change of co‐incubating samples of TKEQVTNV (100 μM) and hIAPP (25 μM) seeds formed at different time points of 1 and 5 hr.
ACKNOWLEDGMENTS
The authors trained three K12 students of Keven Gong from Hudson Middle School, Bowen Zheng from Copley High School, and Alice Xu from Hudson High School via this project. The authors thank financial support from NSF‐CBET‐2107619 and Faculty Summer Research Fellowship from The University of Akron.
Tang Y, Zhang D, Liu Y, Zhang Y, Zhou Y, Chang Y, et al. A new strategy to reconcile amyloid cross‐seeding and amyloid prevention in a binary system of α‐synuclein fragmental peptide and hIAPP . Protein Science. 2022;31:485–497. 10.1002/pro.4247
Funding information National Science Foundation (US), Grant/Award Number: 2107619; The University of Akron
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. [DOI] [PubMed] [Google Scholar]
- 2. Mukherjee A, Morales‐Scheihing D, Butler PC, Soto C. Type 2 diabetes as a protein misfolding disease. Trends Mol Med. 2015;21:439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu Q, Lee J‐Y, Luo Y. Nanoparticles targeting hepatic stellate cells for the treatment of liver fibrosis. Eng Sci. 2019;6:12–21. [Google Scholar]
- 4. Musunuri B, Shetty S, Shetty DK, et al. Acute‐on‐chronic liver failure mortality prediction using an artificial neural network. Eng Sci. 2021;15:187–196. [Google Scholar]
- 5. Vos T, Allen C, Arora M, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang H, Naghavi M, Allen C, et al. Global, regional, and national life expectancy, all‐cause mortality, and cause‐specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1459–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Westermark P, Andersson A, Westermark GT. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol Rev. 2011;91:795–826. [DOI] [PubMed] [Google Scholar]
- 8. Konarkowska B, Aitken JF, Kistler J, Zhang S, Cooper GJ. The aggregation potential of human amylin determines its cytotoxicity towards islet β‐cells. FEBS J. 2006;273:3614–3624. [DOI] [PubMed] [Google Scholar]
- 9. Meng F, Abedini A, Plesner A, Verchere CB, Raleigh DP. The flavanol (−)‐epigallocatechin 3‐gallate inhibits amyloid formation by islet amyloid polypeptide, disaggregates amyloid fibrils, and protects cultured cells against IAPP‐induced toxicity. Biochemistry. 2010;49:8127–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ren B, Liu Y, Zhang Y, et al. Genistein: A dual inhibitor of both amyloid β and human islet amylin peptides. ACS Chem Nerosci. 2018;9:1215–1224. [DOI] [PubMed] [Google Scholar]
- 11. Ren B, Liu Y, Zhang Y, et al. Tanshinones inhibit hIAPP aggregation, disaggregate preformed hIAPP fibrils, and protect cultured cells. J Mater Chem B. 2018;6:56–67. [DOI] [PubMed] [Google Scholar]
- 12. Bahramikia S, Yazdanparast R. Inhibition of human islet amyloid polypeptide or amylin aggregation by two manganese‐salen derivatives. Eur J Pharmacol. 2013;707:17–25. [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Zhang D, Tang Y, et al. Aromadendrin: A dual amyloid promoter to accelerate fibrillization and reduce cytotoxicity of both amyloid‐β and hIAPP. Mater Advan. 2020;1:1241–1252. [Google Scholar]
- 14. Xin Y, Wang X, Luo L, Meng F. Conformation‐dependent manipulation of human islet amyloid polypeptide fibrillation by shiitake‐derived lentinan. ACS Appl Mater Interf. 2018;10:31069–31079. [DOI] [PubMed] [Google Scholar]
- 15. Meng Q‐Y, Wang H, Cui Z‐B, Yu W‐G, Lu X‐Z. Chitosan oligosaccharides attenuate amyloid formation of hIAPP and protect pancreatic β‐cells from cytotoxicity. Molecules. 2020;25:1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang L, Zhu S, Lu T, et al. The effects of a series of carbon dots on fibrillation and cytotoxicity of human islet amyloid polypeptide. J Mater Chem B. 2016;4:4913–4921. [DOI] [PubMed] [Google Scholar]
- 17. Yousaf M, Huang H, Li P, Wang C, Yang Y. Fluorine functionalized graphene quantum dots as inhibitor against hIAPP amyloid aggregation. ACS Chem Nerosci. 2017;8:1368–1377. [DOI] [PubMed] [Google Scholar]
- 18. Mishra A, Misra A, Vaishnavi TS, et al. Conformationally restricted short peptides inhibit human islet amyloid polypeptide (hIAPP) fibrillization. Chem Commun. 2013;49:2688–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L, Lei L, Li Y, Wang L, Li F. A hIAPP‐derived all‐d‐amino‐acid inhibits hIAPP fibrillation efficiently at membrane surface by targeting α‐helical oligomeric intermediates. FEBS Lett. 2014;588:884–891. [DOI] [PubMed] [Google Scholar]
- 20. Aitken JF, Loomes KM, Scott DW, et al. Tetracycline treatment retards the onset and slows the progression of diabetes in human amylin/islet amyloid polypeptide transgenic mice. Diabetes. 2010;59:161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang Y, Zhang D, Zhang Y, et al. Introduction and fundamentals of human islet amyloid polypeptide inhibitors. ACS Appl Bio Mater. 2020;3:8286–8308. [DOI] [PubMed] [Google Scholar]
- 22. Danho W, Swistok J, Khan W, et al. Opportunities and challenges of developing peptide drugs in the pharmaceutical industry. Peptides for youth. New York, USA: Springer, 2009; p. 467–469. [DOI] [PubMed] [Google Scholar]
- 23. Saini RK, Goyal D, Goyal B. Targeting human islet amyloid polypeptide aggregation and toxicity in type 2 diabetes: An overview of peptide‐based inhibitors. Chem Res Toxicol. 2020;33:2719–2738. [DOI] [PubMed] [Google Scholar]
- 24. Shi Y, Lv W, Jiao A, Zhang C, Zhang J. A novel pentapeptide inhibitor reduces amyloid deposit formation by direct interaction with hIAPP. Int J Endocrinol. 2019;2019:9062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin J, Jiao A, Lv W, et al. Pentapeptide protects INS‐1 cells from hIAPP‐mediated apoptosis by enhancing autophagy through mTOR pathway. Front Pharmacol. 2019;10:896–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scrocchi LA, Chen Y, Waschuk S, et al. Design of peptide‐based inhibitors of human islet amyloid polypeptide fibrillogenesis. J Mol Biol. 2002;318:697–706. [DOI] [PubMed] [Google Scholar]
- 27. Lu J, Cao Q, Wang C, et al. Structure‐based peptide inhibitor design of amyloid‐β aggregation. Front Mol Neurosci. 2019;12:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jensen PH, Højrup P, Hager H, et al. Binding of a β to α‐and β‐synucleins: Identification of segments in α‐synuclein/NAC precursor that bind a β and NAC. Biochem J. 1997;323:539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. El‐Agnaf OM, Paleologou KE, Greer B, et al. A strategy for designing inhibitors of α‐synuclein aggregation and toxicity as a novel treatment for Parkinson's disease and related disorders. FASEB J. 2004;18:1315–1317. [DOI] [PubMed] [Google Scholar]
- 30. Jones S, Manning J, Kad NM, Radford SE. Amyloid‐forming peptides from β2‐microglobulin—Insights into the mechanism of fibril formation in vitro. J Mol Biol. 2003;325:249–257. [DOI] [PubMed] [Google Scholar]
- 31. Horvath I, Wittung‐Stafshede P. Cross‐talk between amyloidogenic proteins in type‐2 diabetes and Parkinson's disease. Proc Natl Acad Sci U S A. 2016;113:12473–12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu R, Zhang M, Chen H, Jiang B, Zheng J. Cross‐seeding interaction between β‐amyloid and human islet amyloid polypeptide. ACS Chem Nerosci. 2015;6:1759–1768. [DOI] [PubMed] [Google Scholar]
- 33. Mandal PK, Pettegrew JW, Masliah E, Hamilton RL, Mandal R. Interaction between Aβ peptide and α synuclein: Molecular mechanisms in overlapping pathology of Alzheimer's and Parkinson's in dementia with Lewy body disease. Neurochem Res. 2006;31:1153–1162. [DOI] [PubMed] [Google Scholar]
- 34. Stancu IC, Vasconcelos B, Terwel D, Dewachter I. Models of β‐amyloid induced tau‐pathology: The long and “folded” road to understand the mechanism. Mol Neurodegen. 2014;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X, Zhang X, Ladiwala ARA, et al. Mechanisms of transthyretin inhibition of β‐amyloid aggregation in vitro. J Neurosci. 2013;33:19423–19433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu P, Zhang S, Chen MS, et al. Co‐assembly of human islet amyloid polypeptide (hIAPP)/insulin. Chem Commun. 2012;48:191–193. [DOI] [PubMed] [Google Scholar]
- 37. Ren B, Zhang Y, Zhang M, et al. Fundamentals of cross‐seeding of amyloid proteins: An introduction. J Mater Chem B. 2019;7:7267–7282. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Zhang M, Liu Y, et al. Dual amyloid cross‐seeding reveals steric zipper‐facilitated fibrillization and pathological links between protein misfolding diseases. J Mater Chem B. 2021;9:3300–3316. [DOI] [PubMed] [Google Scholar]
- 39. Hu R, Zhang M, Patel K, et al. Cross‐sequence interactions between human and rat islet amyloid polypeptides. Langmuir. 2014;30:5193–5201. [DOI] [PubMed] [Google Scholar]
- 40. Atsmon‐Raz Y, Miller Y. A proposed atomic structure of the self‐assembly of the non‐amyloid‐β component of human α‐synuclein as derived by computational tools. J Phys Chem B. 2015;119:10005–10015. [DOI] [PubMed] [Google Scholar]
- 41. Pollock‐Gagolashvili M, Miller Y. Two distinct polymorphic folding states of self‐assembly of the non‐amyloid‐β component differ in the arrangement of the residues. ACS Chem Nerosci. 2017;8:2613–2617. [DOI] [PubMed] [Google Scholar]
- 42. Atsmon‐Raz Y, Miller Y. The non amyloid‐β component (NAC) of human α‐synuclein oligomers induces the formation of new Aβ oligomers: Insight into the molecular interactions that link Parkinson's disease and Alzheimer's disease. Biophys J. 2016;110:529a. [DOI] [PubMed] [Google Scholar]
- 43. Xu Q, Park Y, Huang X, et al. Diabetes and risk of Parkinson's disease. Diabetes Care. 2011;34:910–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yue X, Li H, Yan H, Zhang P, Chang L, Li T. Risk of Parkinson disease in diabetes mellitus: An updated meta‐analysis of population‐based cohort studies. Medicine. 2016;95:e3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu G, Jousilahti P, Bidel S, Antikainen R, Tuomilehto J. Type 2 diabetes and the risk of Parkinson's disease. Diabetes Care. 2007;30:842–847. [DOI] [PubMed] [Google Scholar]
- 46. Wahlqvist ML, Lee M‐S, Hsu C‐C, Chuang S‐Y, Lee J‐T, Tsai H‐N. Metformin‐inclusive sulfonylurea therapy reduces the risk of Parkinson's disease occurring with type 2 diabetes in a Taiwanese population cohort. Parkinsonism Relat Disord. 2012;18:753–758. [DOI] [PubMed] [Google Scholar]
- 47. Hogg E, Athreya K, Basile C, Tan EE, Kaminski J, Tagliati M. High prevalence of undiagnosed insulin resistance in non‐diabetic subjects with Parkinson's disease. J Parkinsons Dis. 2018;8:259–265. [DOI] [PubMed] [Google Scholar]
- 48. Hwang O. Role of oxidative stress in Parkinson's disease. Exper Neurobiol. 2013;22:11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Herrero M‐T, Estrada C, Maatouk L, Vyas S. Inflammation in Parkinson's disease: Role of glucocorticoids. Front Neuroanat. 2015;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheong JL, de Pablo‐Fernandez E, Foltynie T, Noyce AJ. The association between type 2 diabetes mellitus and Parkinson's disease. J Parkinsons Dis. 2020;10:775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Atsmon‐Raz Y, Miller Y. Molecular mechanisms of the bindings between non‐amyloid β component oligomers and amylin oligomers. J Phys Chem B. 2016;120:10649–10659. [DOI] [PubMed] [Google Scholar]
- 52. Mucibabic M, Steneberg P, Lidh E, et al. α‐Synuclein promotes IAPP fibril formation in vitro and β‐cell amyloid formation in vivo in mice. Sci Rep. 2020;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Micsonai A, Wien F, Kernya L, et al. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci U S A. 2015;112:E3095–E3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berridge MV, Tan AS. Characterization of the cellular reduction of 3‐(4, 5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyltetrazolium bromide (MTT): Subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch Biochem Biophys. 1993;303:474–482. [DOI] [PubMed] [Google Scholar]
- 55. Fotakis G, Timbrell JA. In vitro cytotoxicity assays: Comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol Lett. 2006;160:171–177. [DOI] [PubMed] [Google Scholar]
- 56. Rodríguez‐Comas J, Moreno‐Vedia J, Obach M, et al. Alpha1‐antitrypsin ameliorates islet amyloid‐induced glucose intolerance and β‐cell dysfunction. Mol Metabol. 2020;37:100984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harris JR, Marles‐Wright J. Macromolecular protein complexes II: Structure and function. Cham, Switzerland: Springer, 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Time‐dependent circular dichroism (CD) spectra to monitor the secondary structure change of (a) pure TKEQVTNV of 25–100 μM and (b) cross‐seeding samples of hIAPP‐TKEQVTNV at different molar ratios of 1:1, 1:2, and 1:8 at 0 and 24 hr.
FIGURE S2 Time‐dependent circular dichroism (CD) spectra to monitor the secondary structure change of co‐incubating samples of TKEQVTNV (100 μM) and hIAPP (25 μM) seeds formed at different time points of 1 and 5 hr.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.


