Abstract
Color patterns are often linked to the behavioral and morphological characteristics of an animal, contributing to the effectiveness of such patterns as antipredatory strategies. Species‐rich adaptive radiations, such as the freshwater fish family Cichlidae, provide an exciting opportunity to study trait correlations at a macroevolutionary scale. Cichlids are also well known for their diversity and repeated evolution of color patterns and body morphology. To study the evolutionary dynamics between color patterns and body morphology, we used an extensive dataset of 461 species. A phylogenetic supertree of these species shows that stripe patterns evolved ~70 times independently and were lost again ~30 times. Moreover, stripe patterns show strong signs of correlated evolution with body elongation, suggesting that the stripes’ effectiveness as antipredatory strategy might differ depending on the body shape. Using pedigree‐based analyses, we show that stripes and body elongation segregate independently, indicating that the two traits are not genetically linked. Their correlation in nature is therefore likely maintained by correlational selection. Lastly, by performing a mate preference assay using a striped CRISPR‐Cas9 mutant of a nonstriped species, we show that females do not differentiate between striped CRISPR mutant males and nonstriped wild‐type males, suggesting that these patterns might be less important for species recognition and mate choice. In summary, our study suggests that the massive rates of repeated evolution of stripe patterns are shaped by correlational selection with body elongation, but not by sexual selection.
Keywords: body morphology, convergence, CRISPR‐Cas9 cichlid, motion dazzle, pigmentation, trait correlation
Across the ~1200 cichlid species of the East African rift lakes, melanic horizontal stripes have evolved numerous times. By applying comparative analyses, hybrid crosses, and a behavioral experiment using a CRISPR/Cas9 knockout mutant, our study aims at understanding the ecological function of these horizontal stripe patterns. We suggest that the massive rates of repeated evolution of stripe patterns are shaped by correlational selection with body elongation, but not by sexual selection.
1. INTRODUCTION
Across the more than 1200 cichlid species of the East African rift lakes, melanic horizontal stripes have evolved numerous times and are a prime example of convergent evolution (Kratochwil et al., 2018). Different functions of stripes have been proposed including camouflage (Cott, 1940; Longley, 1916; Stevens & Merilaita, 2009; Thayer, 1918) and social signaling (Barlow, 2008; Seehausen et al., 1999). The repeated evolution of stripes suggests that they evolved by natural selection and constitute adaptations to similar ecological niches (Harvey & Pagel, 1991) with shared selection pressures (Arendt & Reznick, 2008; Losos, 2011; McGhee, 2011; Ord & Summers, 2015). Alternatively, it has been hypothesized that the evolution of coloration in cichlids is driven by sexual selection (i.e., nuptial coloration) (Allender et al., 2003; Couldridge & Alexander, 2002; Knight & Turner, 1999; Maan et al., 2004). Yet, a comparative study of East African cichlid fishes has shown that strength of sexual selection has no detectable effect on stripe pattern evolution (Seehausen et al., 1999), and to the best of our knowledge, an influence of stripe patterns for mate choice or preference has never been formally tested. From an ecological viewpoint, the evolution of stripes has been mainly linked to shoaling and piscivorous feeding, suggesting that these trait combinations might be effective strategies to decrease predation risk or to avoid being seen by prey (Seehausen et al., 1999).
Cryptic color patterns (e.g., spots, blotches, vertical bars) allow individuals to blend in with the background and reduce the probability of detection by predators, but they may no longer be advantageous when the prey moves (Cott, 1940; Thayer, 1918) because motion‐sensitive visual circuits of a stationary predator can easily detect movement against a stationary background. Conversely, horizontal stripes that might be considered being conspicuous in many environments may become beneficial when the individual moves (Allen et al., 2013; Brodie, 1992; Jackson et al., 1976). This special type of camouflage is called motion dazzle as it prevents successful capture during motion by causing predators to misjudge the direction or speed of prey movement. Several studies showed that such motion dazzle patterns might be involved in hampering a predator's ability to intercept a moving prey (Allen et al., 2013; Brodie, 1992; von Helversen et al., 2013; Jackson et al., 1976; Kelley & Kelley, 2014; Rojas et al., 2014). It is generally accepted that motion dazzle patterns are advantageous for mobile species that are highly detectable against the stationary background, while cryptic pigmentation patterns are advantageous for less‐mobile species that rely on camouflage to reduce detection (Halperin et al., 2016). A beneficial trait correlation of stripe patterns with body length was first shown for several reptile species (Allen et al., 2013; Murali & Kodandaramaiah, 2017). This trait correlation redirects predator attacks to the tail and thereby reduces the probability of individuals being captured (von Helversen et al., 2013) suggesting that the effectiveness of dazzle patterns depends on body shape (Murali & Kodandaramaiah, 2017).
In fish, body elongation is a major axis of body shape divergence (Astudillo‐Clavijo et al., 2015; Clabaut et al., 2007; Claverie & Wainwright, 2014; Kautt et al., 2020; Muschick et al., 2012) and has implications for a fish's susceptibility to predators (Chivers et al., 2008; Price et al., 2015), swimming performance (Rouleau et al., 2010), and attractiveness to mates (Head et al., 2013). Thereby, a combination of stripes and an elongated body shape could represent an adaptation to predator–prey interactions, for example, high swimming performance while impairing a predators’ perception of its prey through motion dazzle.
Understanding how correlations of traits (e.g., body shape and color patterns) evolve, neutrally or under different types of selection, and identifying the underlying genetic correlations can contribute to the understanding of the evolutionary effects of trait correlations (Dingemanse et al., 2012; Lande & Arnold, 1983; Sih et al., 2004). Current models of rapid divergence emphasize the importance of linkage and pleiotropy in adaptation and speciation (Noor & Bennett, 2009; Seehausen et al., 2014; Via, 2012). Hence, horizontal stripes could have evolved together with other phenotypic traits such as body shape due to a number of adaptive or nonadaptive reasons like genetic constraints such as linkage, in which two or more loci each affecting different traits are genetically linked and therefore tend to be inherited together. Pleiotropy, in which a single locus causally affects two or more traits (McKinnon & Pierotti, 2010; Saltz et al., 2017), could lead to a similar outcome. In general, trait correlations caused by pleiotropy are not expected to break down simply through neutral processes (Jones et al., 2003). On the other hand, trait correlations caused by genetic linkage and built up by correlational selection are normally expected to erode eventually through recombination, unless (correlational) selection is strong and persistent. This causes trait correlations that are generated by genetic linkage to be transient, and it was argued that they contribute little to evolutionary change (Saltz et al., 2017).
Cichlids are ideal for the investigation of trait correlations both from an evolutionary (looking across whole phylogenies) and a genetic viewpoint, since many species can be hybridized and most often even produce fertile offspring. Such hybrid crosses for example permitted the identification of the genetic basis of horizontal stripe patterns in East African cichlids. A single gene, agouti‐related peptide 2 (agrp2), was identified to facilitate the repeated evolution of stripes (Henning et al., 2014; Kratochwil et al., 2018). High expression of the “stripe inhibitor” gene agrp2 prevents stripe pattern formation, and low expression permits their appearance (Kratochwil et al., 2018, 2019). Agouti family genes have been shown to control both color pattern divergence (Henning et al., 2014; Kratochwil et al., 2018) and growth, obesity, and energy metabolisms (Duhl et al., 1994; Song & Cone, 2007). Therefore, this locus is of particular interest to investigate trait correlations, since agrp2 might act pleiotropically.
Knowledge of the genetic basis of stripes, a potentially pleiotropic function of the underlying gene(s), and the availability of large comparative datasets allow us now to take a further step toward understanding the ecological function of horizontal stripe patterns. Moreover, technological advances in genome editing methods introduce the potential to solely examine the effect of coloration traits in a behavioral experiment. By applying novel CRISPR/Cas9 tools, we can effectively remove stripe patterns and thus examine the role of sexual selection in response to the altered color pattern.
Here, we investigate (1) whether there is a correlation between body elongation and stripe patterns using comparative analyses, (2) whether the two traits are genetically constrained by linkage or pleiotropy for which we analyze two sets of hybrid crosses, and (3) whether there are signatures of selection on these traits. (4) Moreover, we explore whether stripes potentially play a role in sexual selection by using a classic two‐choice behavioral experiment. To do so, we let females of a nonstriped species choose between a nonstriped wild‐type male and a striped agrp2 knockout mutant of the same species which was generated using the CRISPR/Cas9 genome editing technique.
2. METHODS
2.1. Data collection
We compiled a large photographic dataset of 461 species from all major East African cichlid lineages. Images were compiled from various online image databases and textbooks (listed in Table S2). In striped species, stripe patterns are usually present in both sexes but they are sometimes covered by the male's nuptial coloration. We therefore scored a species as striped when either the male or the female possessed a stripe pattern and measured the elongation index of the striped individual. However, only a small percentage of species shows stripes in only one sex and, if this is the case, it mostly affects how clear the pattern is.
For many species, there is only a single photograph available. All images were required to be in lateral view so that standard length and body elongation could be measured reliably. We further analyzed individuals for the presence of stripe pattern in three different ways (Figure S2): binary (0/no stripes, 1/stripes), categorical (0/no stripes, 1/spotted, 2/partially striped, 3/fully striped, 4/oblique stripe; only used for Table S1), and continuous (percentage of the midlateral stripe region covered by melanin). To calculate the elongation index, we measured the standard length, from the tip of the snout to the base of the caudal fin, of each fish as well as body depth, maximum distance between the most anterior part of the pelvic fin and dorsal fin. Since most photographs did not have a scale bar, the measures of standard length and body depth do not account for absolute size. The elongation index was then calculated as the standard length divided by body depth. For all measurements, we used the software Fiji (Schindelin et al., 2012). The raw data are available in the electronic (Table S1).
To examine the evolution of stripes in cichlids, we built a supertree combining trees from several different studies (Dunz & Schliewen, 2013; Hulsey et al., 2017, 2018, 2019; Hulsey, Zheng, et al., 2018; Irisarri et al., 2018; Kratochwil et al., 2018; Malinsky et al., 2018; McGee et al., 2016; Meier et al., 2017). Because it facilitated including the most species of cichlids into a single tree, we first generated a phylogeny of cichlid species using the mitochondrial nd2 gene (NCBI accession numbers listed in Table S3) as this is the most comprehensive dataset available at this time. For this, we used the evolutionary model GTR + Gamma + Inv. The codon positions were examined as separate partitions. For tree building, we used BEAST v1.10.1 (Suchard et al., 2018) that generates trees with branch lengths in units of time. The recovered set of nd2 based phylogenetic hypotheses was highly similar to previous results for this gene (Salzburger et al., 2005; Wagner et al., 2012).
After examining the results with the nd2 gene tree alone, we combined this tree with ten other published phylogenies of African cichlids that incorporated information from the nuclear genome (Dunz & Schliewen, 2013; Hulsey et al., 2019; Hulsey, Holzman, et al., 2018; Hulsey, Zheng, et al., 2017, 2018; Irisarri et al., 2018; Kratochwil et al., 2018; Malinsky et al., 2018; McGee et al., 2016; Meier et al., 2017). Consensus phylogenies from these studies were combined using the “mrp.supertree” function with the “pratchet” method as implemented in the R program “phytools” (Revell, 2012) which estimates the MRP (matrix representation parsimony) supertree from a set of input trees. We used the “optim.parsimony” method as implemented in the R package “phangorn” (Schliep, 2011). The function “optim.parsimony” tries to find the maximum parsimony tree nearest neighbor interchange rearrangements as well as subtree pruning and regrafting.
Our final dataset, which covered phylogenetic as well as phenotypic information, included 461 species of East African cichlids (Data S1). To infer transitions from having no stripe to gaining stripes, q01, as well as from having stripes to losing them, q10, we imported a set of trees into the program Mesquite and then used the “Summarize State Changes Over Trees” function.
2.2. Comparative analyses
To determine whether there was an association between the presence of horizontal stripes and body elongation, we performed phylogenetic ANOVAs across the African cichlid supertree using “phytools” (Revell, 2012). We performed phylogenetic ANOVAs running simulations (nsim = 1000) with three stripe phenotypes as grouping factors: 1. presence of both midlateral and dorsolateral stripes versus species lacking either of these stripes. 2. Dorsolateral stripe versus species with no stripe. 3. Midlateral stripe versus species with no stripe.
To determine whether there was macroevolutionary evidence of stabilizing selection on body elongation in association with the presence or absence of stripes, we also fit a series of Brownian motion (BM) and Ornstein–Uhlenbeck (OU) models to body elongation evolution using the package “OUwie” (Beaulieu et al., 2012).
Prior to the evolutionary model fitting, we generated stochastic character maps (simmaps) using the function “make.simmap” implemented in the R package “phytools” (Revell, 2012). For each input tree simulated, we conducted a single simmap simulation using the “sym” transition model that treats the transition between striped and nonstriped as equal. We chose the “sym” function because the range of inferred transitions between striped and nonstriped overlapped, and we did not want to bias the results.
The support for more parameter‐rich models over models with fewer parameters was assessed using AIC scores. We asked first whether there was more support for a model containing a single optimum value, α, as compared to a simple model of BM for body elongation evolution in the cichlids examined. This first OU model did not incorporate any differences between striped and nonstriped species. So, we next asked whether there was support for an OU process with different optima associated with the presence and absence of stripes. Support for this two‐optima model over a model with a single optimum would further support the hypothesis that stripes are evolutionarily associated with a difference in body elongation. Finally, we asked whether there was support for different selective pulls toward the morphological optimum for nonstriped, α 1, and striped, α 2, cichlid lineages.
2.3. Experimental crosses
All data on obtaining experimental crosses and quantification of stripes are described in Kratochwil et al. (2018). Here, we phenotyped F2 individuals from a Lake Malawi cross between a striped species (Pseudotropheus cyaneorhabdos) and a nonstriped species (Chindongo demasoni; previously: Pseudotropheus demasoni) and a second cross of Lake Victoria cichlids again consisting of a striped species (Haplochromis sauvagei) and a nonstriped species (Pundamilia nyererei). For all fishes, we estimated the elongation index and three scores to quantify the number and/or continuity of stripes (binary, categorical, continuous). We checked for normal distribution of data using the Shapiro–Wilk test of normality and decided to use a nonparametric test to calculate trait correlation. We used Kendall's tau‐b correlation coefficient to test whether body elongation and stripes (continuous measurement) are correlated. Additionally, we compared the striped and nonstriped individuals (based on binary scoring) using a Wilcoxon test.
2.4. Mate preference experiments
To test for a role of stripe patterns in species recognition and sexual selection, we tested ten P. nyererei females for a preference in a classical two‐choice experimental setup. Females were allowed to choose between two males of the same species: a nonstriped wild‐type male and a striped sibling CRISPR‐Cas9 agrp2 mutant (Kratochwil et al., 2018) of the same size. The mutant stimulus male differed genetically only from its sibling stimulus male in that the agrp2 had a loss of function mutation induced by CRISPR‐Cas9. The two male individuals (the same mutant and the same wild‐type male were used with all females) used for the experiment have successfully sired offspring before entering the experiment. The experimental setup consisted of three separate tanks placed next to each other to reduce the test to visual cues only because the mutation in agrp2 rendered the stripe pattern phenotype of the mutant male. It was shown that species‐isolating female choice is likely based primarily on such visual information (Jordan et al., 2003).
A single female was placed in the middle, while the left and right tanks each contained one stimulus male. The central tank was delineated into three areas (the left choice area in front of the left male, the neutral area in the center, and the right choice area in front of the right male). The aquarium was lit by one daylight neon lamp situated next to the camera, while the room was maintained in the dark to avoid reflections. The room temperature was constantly kept at 25°C under a 12:12‐h light:dark cycle.
All fish were acclimatized overnight (minimum 12 h) and in groups with other fish to reduce stress levels. After acclimatization, all other fish were carefully removed from the setup, and we allowed another acclimatization of 20 min. We then recorded females’ positions continuously with a digital video camera positioned above the setup for 40 min. Then, we switched the two stimuli males to correct for female side preference and filmed the female again for 40 min. For tracking, we used the R package “trackR” (https://github.com/swarm‐lab/trackR). From this, we calculated a preference index as the amount of time spent in front of male A divided by the total amount of time spent in the two choice areas. A female was assumed to prefer male A over male B when the preference index (amount of time spent in front of male A divided by the total amount of time spent in the two choice areas) was above 50%. All calculations were performed using R 3.0 software (R Development Core Team, 2019).
3. RESULTS
3.1. Horizontal stripes evolved repeatedly across East African radiations
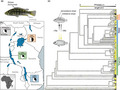
Of the 461 species in our dataset, 33% (152 species) show either a midlateral or a dorsolateral stripe (Figure 1a,c). In total, 32% (149 species) show only a midlateral stripe and 26% (119 species) show a dorsolateral stripe (Figure 1b). There are some species that only have a midlateral but no dorsolateral stripe (N = 32), but only three species show exclusively a dorsolateral but no midlateral stripe. Ancestral state reconstruction of the presence of horizontal stripes (binary scoring) revealed that stripes evolved on average 73 (range: 64–82) times independently and were lost on average 28 (range: 19–37) times (Figure 1c). Based on these, many frequent gains and losses the ancestral state cannot be reliably determined (PP = 0.5).
FIGURE 1.
Repeated evolution of horizontal stripes across East African cichlid radiations. (a) Picture of a female Haplochromis chilotes showing both stripe (black) and bar patterns (grey). (b) Map of East Africa highlighting Lakes Victoria, Tanganyika, and Malawi that encompass the three main radiations. Pie charts give the percentage of striped species (overall ~30%, in gray). The percentage of striped species was inferred from the presented data (461 species). Color codes show the major radiations of Lakes Tanganyika (green), Malawi (blue), and Victoria (orange). (c) Phylogenetic supertree of 461 East African cichlids with likelihood reconstructions of ancestral states (yellow–white–black gradient with yellow indicating a nonstriped state and black a striped state) of the stripe phenotype that evolved ~70 times independently. White branches moving away from the tips of the tree represent the inherent uncertainty in our ancestral state reconstructions
3.2. Stabilizing selection on body elongation is acting stronger in striped species
To determine whether there is macroevolutionary evidence of stabilizing selection on body elongation in association with horizontal stripes, we fit a series of Brownian motion (BM) and Ornstein–Uhlenbeck (OU) models to the evolution of body elongation (Beaulieu et al., 2012). These comparative analyses revealed that stabilizing selection is supported for body elongation in association with stripe patterns. The OUMA model (Table 1) is the most supported model. While this model supports a different mean (optima) between striped and nonstriped species, it supports greater stabilizing selection (α) of body elongation in striped as compared to nonstriped species. This suggests that stabilizing selection on body elongation is acting more strongly on striped species than on nonstriped species and striped species have a higher optimum elongation index (Table 1). Also, there is support for a different body elongation optimum, or mean value, for the striped species versus the nonstriped species which is consistent with the phylogenetic ANOVA results (Figure 2).
TABLE 1.
Results of different models of trait evolution
| AIC | Optima | σ | α | ||||
|---|---|---|---|---|---|---|---|
| No stripe | Stripe | No stripe | Stripe | No stripe | Stripe | ||
| BM1 | 1149.77 | 3.28 | 3.28 | 44.04 | 44.04 | NA | NA |
| OU1 | 897.99 | 3.19 | 3.19 | 83.40 | 83.40 | 87.09 | 87.09 |
| OUM | 895.11 | 3.06 | 3.35 | 85.07 | 85.07 | 91.11 | 91.11 |
| OUMA | 876.98 | 3.06 | 3.31 | 84.66 | 84.66 | 91.90 | 92.24 |
Shown are different Akaike's information criteria (AIC) values for different Brownian motion (BM) and Ornstein–Uhlenbeck (OU) models to test for macroevolutionary evidence of stabilizing selection on body elongation in association with horizontal stripes. Different elongation optima for striped and nonstriped species are reported for the different models; σ gives the rate of divergence and α is a parameter of stabilizing selection.
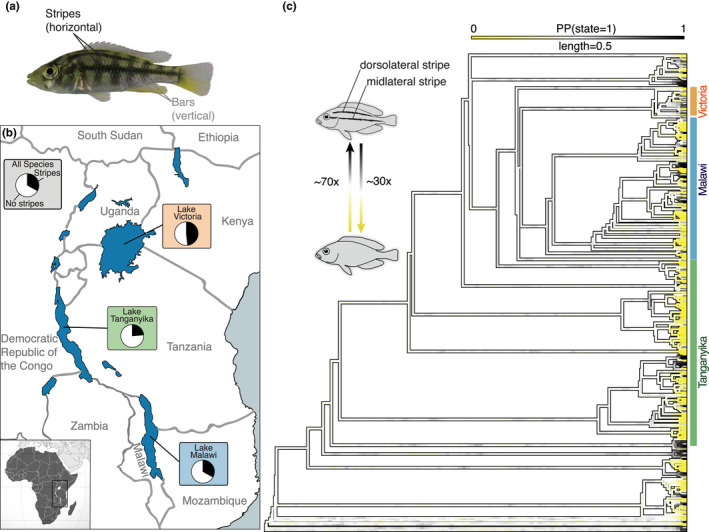
FIGURE 2.
Striped fishes show an elongated body shape across cichlid radiations of the East African Great Lakes. (a) While both Lake Malawi (blue) and Lake Tanganyika (green) cichlids show a substantially elongated body shape if they have any horizontal stripes (phylogenetic ANOVA), this pattern is not supported in Lake Victoria cichlids (orange; phylogenetic ANOVA; p = .199). (b) There is no evidence for genetic linkage of horizontal stripes and body elongation since the elongation index, measured in F 2 offspring from two hybrid crosses, is not significantly correlated with a stripe phenotype. Importantly, the two parental species of the Lake Malawi (i and j) and Lake Victoria (k and l) hybrid crosses show different elongation indices which is given below the species' names. Photographs show striped species endemic to Lake Malawi (c and d), Lake Victoria (e and f), and Lake Tanganyika (g and h)
3.3. Horizontal stripes are largely associated with body elongation
To test whether horizontal stripes are associated with body shape, that is, elongation, across East African radiations, we calculated the elongation index as the standard length divided by body depth and employed a phylogenetic ANOVA. The results show that both the dorsolateral and midlateral stripe individually are significantly associated with body elongation (phylogenetic ANOVA; p = .018 in the case of just the dorsolateral stripe and phylogenetic ANOVA; p = .012 with the midlateral stripe, Table 1 and Figure 2). Showing any stripe pattern (either dorsolateral or a midlateral stripe) is significantly associated with body elongation as well (phylogenetic ANOVA; p = .009). While both cichlids from the older Lake Malawi and Lake Tanganyika cichlid radiations show an association between a substantially elongated body shape and horizontal stripes (phylogenetic ANOVA; p = .011 in Lake Malawi cichlids and phylogenetic ANOVA; p = .004 in Lake Tanganyika cichlids), this association is not statistically supported in Lake Victoria cichlids (phylogenetic ANOVA; p = .199).
To investigate whether stripes and body elongation are genetically linked traits and therefore inherited together, we analyzed two independent hybrid crosses from Lake Victoria and Lake Malawi for a correlation between body elongation and stripes. One might have expected to find the same correlation of stripes with body elongation in F2 offspring of those crosses (see Section 2.3). However, there was no correlation between body elongation and presence of stripes in the crosses from Lake Malawi (Kendall rank correlation; r = −.0024, p = .96) and Lake Victoria (Kendall rank correlation; r = .022, p = .73). A comparison of elongation indices between the striped and nonstriped F2 offspring showed that elongation does not differ significantly between individuals of the Lake Malawi cross (Wilcoxon test; p = .5406) as well as the Lake Victoria cross (Wilcoxon test; p = .7604). Hence, we conclude that, these two traits are inherited independently and are neither linked genetically (linked genes in the same genetic region controlling both traits) nor linked through pleiotropy (same gene controlling both traits).
3.4. No evidence for a role of sexual selection acting on stripes
To test for a role of horizontal stripes in species recognition and/or mate preference, we performed a two‐choice experiment with females of the nonstriped species P. nyererei measuring the time the female spends in the vicinity of the wild‐type or the CRISPR‐Cas9 phenotypically choice male. Females could choose between two stimulus males of the same size: a striped CRISPR‐Cas9 knockout (Kratochwil et al., 2018) and a wild‐type sibling showing no stripes from the same species. From this, we calculated a preference index as the amount of time spent in front of male A divided by the total amount of time spent in the two choice areas (see Section 2). The mean preference index across trials (N = 10) is 0.46 which is suggesting that P. nyererei females do not visually differentiate between the nonstriped wild‐type P. nyererei and the striped mutant P. nyererei (or at least do not prefer one over the other) since they spent approximately the same time with both stimulus males.
4. DISCUSSION
Here, we reconstructed the number of times horizontal stripes evolved as well as their phenotypic associations in a robust phylogenetic framework by generating a supertree encompassing 461 East African cichlid species. This supertree consists of several published phylogenies based on mitochondrial and nuclear data. Our results enhance our understanding of the association of stripes with the evolution of body shape and mate choice in African cichlids.
Comparative analyses showed that stripes are present in one third of East African cichlid species and evolved many times independently with stripes having been gained ~70 times and having been lost again ~30 times. While the exact number of transitions might be influenced by gene flow and lack of treelike structure within the Lake Malawi and Victoria radiations, we can conclude that stripes evolved many times independently and more frequently than one might have expected.
Moreover, stripes are not confined to specific clades in the molecular phylogeny (Figure 1c) reflecting the high degree of evolutionary lability of stripes. Based on our phylogeny, we were not able to estimate the ancestral state of African cichlids, but based on previous work suggests that the frequent loss of stripes is explained by the “stripe inhibitor” gene agrp2 (Kratochwil et al., 2018), it seems more parsimonious that stripe presence is the ancestral state of East African cichlids (although it was likely not as exaggerated as in extant striped cichlids from the Great Lakes) and that the independent loss of stripes was caused by independent mutations affecting agrp2 expression (Kratochwil, 2019; Kratochwil et al., 2018; Urban et al., 2021) and possibly other genes (Gerwin et al., 2021).
Exploring the selective forces which have led to the repeated gains and losses of stripe patterns, comparative analyses revealed that the presence of stripe patterns is significantly correlated with body elongation in most East African cichlids (Figure 2). While the mean elongation index is slightly higher in striped Lake Victoria cichlids (3.134) than in nonstriped species (3.047), an association between stripes and an elongated body shape is not supported statistically. One possible explanation is the relatively small sample size of Lake Victoria cichlids (N = 29) compared to Lake Malawi (N = 137) and Lake Tanganyika cichlids (N = 174) in which this association was significant. Also, as evident from Figure 2, the Lake Tanganyika radiation, the oldest lake, shows the most variation, followed by the Lake Malawi radiation and the youngest radiation of Lake Victoria. Further analyses revealed that the trait correlation between stripe pattern and body elongation is subject to stabilizing selection that is acting more strongly on striped species than on nonstriped species. Notably, different elongation optima are supported for striped species than for nonstriped species (Table 1). In reptiles, a combination of body length and stripes reduces the probability with which moving prey is captured by affecting the predators’ perception of speed (von Helversen et al., 2013). This motion dazzle effect of stripes is a form of defensive color pattern suggested to prevent successful capture during motion by causing predators to misjudge the direction or speed of prey movement. It has been hypothesized to play a role in motion camouflage in a variety of animals such as snakes (Brodie, 1989, 1992, 1993; Creer, 2005; Jackson et al., 1976) and lizards (Murali et al., 2018), and many studies have demonstrated its effect in humans (Hogan et al., 2016, 2017; Stevens et al., 2011). While in fish, body elongation has key implications for fitness, such as susceptibility to predators (Chivers et al., 2008; Price et al., 2015) and swimming performance (Rouleau et al., 2010), stripe patterns in cichlids were previously associated with shoaling behavior as well as a piscivorous feeding mode (Seehausen et al., 1999). The evolution of vertical bar patterns, on the other hand, was associated with structurally complex habitats, such as rocky substrates and vegetation. Vertically barred snakes were shown to be more secretive and rely on crypsis or aggression as their primary mechanism of defense (Jackson et al., 1976). While vertical bars and horizontal stripes can be easily obtained by hybridization (Gerwin et al., 2021) in nature, they rarely occur in the same species, which indicates that vertical bars and horizontal stripes might have different ecological roles.
We propose that the association of body elongation and stripes in cichlids could similarly reduce the probability with which individuals are captured by predators (von Helversen et al., 2013) by creating a motion dazzle pattern (Murali & Kodandaramaiah, 2017) as it has been shown for reptiles (Allen et al., 2013; Murali & Kodandaramaiah, 2017). However, such a motion dazzle pattern could also be beneficial for predatory species where it could cause the prey to misjudge the predator's speed which is suggested by the association of stripes with a piscivorous feeding mode. Such a type of morphological integration has been suggested to play a role in the rapid evolution of a number of traits in cichlids and other adaptively diverging groups (Albertson et al., 2005; Hulsey, Machado‐Schiaffino, et al., 2017; Husemann et al., 2017) suggesting an important role of either pleiotropy or genetic linkage (Saltz et al., 2017). However, body elongation and stripes segregate in F2 offspring of the two hybrid crosses (Figure 2b). While in each hybrid crosses, the major effect “stripe suppressor gene” argp2 separates striped cichlids from nonstriped cichlids (Henning et al., 2014; Kratochwil et al., 2018), a correlation with body elongation was not supported. This result implies that body elongation and horizontal stripes do not have a shared genetic basis or shared major effect loci and are thus not correlated due to genetic linkage or pleiotropy of agrp2. The correlation of the traits might therefore rather be caused by selection pressures on both body shape and color patterns. Hence, the trait correlation of body elongation and stripes is not morphologically integrated, nor do they represent genetic modularity, but evolve due to strong correlational selection which favors combinations of traits that work in concert (Brodie et al., 1995).
In cichlids, color patterns were shown to be important in species recognition (Couldridge & Alexander, 2002; Seehausen & van Alphen, 1998; Seehausen et al., 1998) and it was proposed that disruptive sexual selection is one of the main contributors to the rapid rate of speciation seen in cichlids (Maan et al., 2004). Such premating barriers are considered essential for reproductive isolation in closely related species, such as cichlids (Butlin, 1987; Coyne & Orr, 2004; Rometsch et al., 2020). However, several recent studies have shown that species divergence of reef fishes is frequently first characterized by the evolution of different color patterns, especially among breeding males (Allen et al., 2015; Hench et al., 2019; Rocha & Bowen, 2008; Victor & Randall, 2010, 2014). Yet, when testing for a role of sexual selection (Figure 3), we found that females did not differentiate between a striped and a nonstriped male. This result fits the observation that in many species, both males and females have stripes and that therefore the stripe patterns do not appear to matter in terms of mate choice. Other sensory modalities (e.g., olfaction or sound) might matter (more) in terms of mate choice but were not tested here.
FIGURE 3.
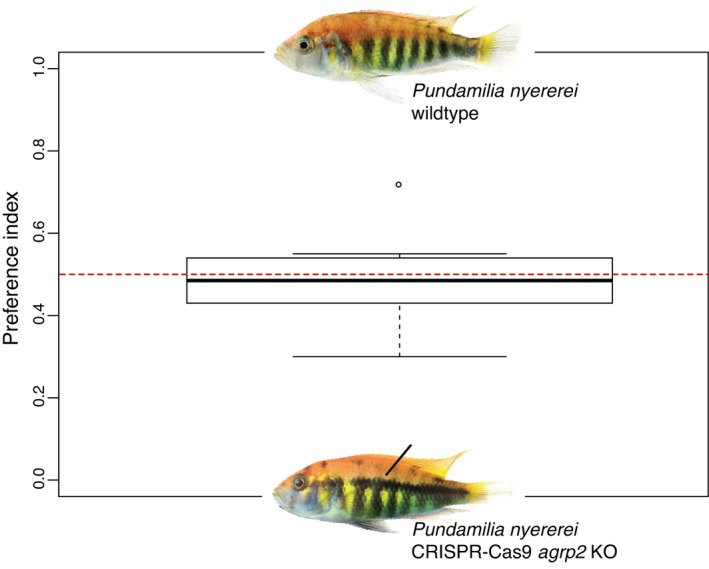
Horizontal stripes may not be relevant for species recognition and mate preference. The preference index was calculated as the amount of time spent in front of male A divided by the total amount of time spent in the two choice areas. The dashed red line indicates 0.5 which means no deviation from random choice. A value >0.5 would indicate a preference for the wild‐type Pundamilia nyererei, while a value <0.5 indicates a preference for the striped agrp2 knockout
The importance of the males’ nuptial coloration was highlighted in several studies on mate preference in cichlids (e.g., Maan et al., 2004; Seehausen, 1997; Seehausen & van Alphen, 1998) because it is often based on pigments such as carotenoids (red to yellow pigment) which most animals cannot synthesize de novo and thus have to be incorporated by diet. Thereby red/yellow coloration can have an important role in honest signaling of male quality and play an important part in sexual selection (Evans & Norris, 1996; Seehausen & van Alphen, 1998). Melanin, on the other hand, which is the pigment responsible for stripe patterns, can be synthesized de novo (Braasch et al., 2007) and thus may be a less informative indicator of male quality.
In summary, our results suggest that the correlated evolution between horizontal stripes and an elongated body shape is not the consequence of genetic linkage or pleiotropy, but most likely of correlational selection or a correlated response to selection. Since different elongation optima are supported for striped and nonstriped species, this trait correlation could result in an optimized optical illusion pattern. Such a pattern could potentially represent an adaptation to predator–prey interactions. Additionally, by taking advantage of a genetically modified striped CRISPR/Cas9 fish, we show that stripe patterns likely have no influence on mate preference. Therefore we suggest that the repeated evolution of stripe patterns in African cichlids has been predominantly driven by ecological selection.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
Sabine Urban: Formal analysis (lead); Visualization (lead); Writing – original draft (lead). Jan Gerwin: Data curation (equal); Writing – review & editing (supporting). C. Darrin Hulsey: Conceptualization (equal); Formal analysis (supporting); Funding acquisition (equal); Supervision (supporting); Writing – review & editing (supporting). Axel Meyer: Conceptualization (equal); Funding acquisition (equal); Writing – review & editing (supporting). Claudius F. Kratochwil: Conceptualization (equal); Funding acquisition (equal); Supervision (lead); Writing – review & editing (supporting).
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the International Max Planck Research School for Organismal Biology (to S.U.), a grant by the Hector Fellow Academy (to A.M.), the Baden‐Württemberg Foundation, and the Deutsche Forschungsgemeinschaft (DFG, KR 4670/2‐1 and KR 4670/4‐1 to C.F.K. as well as ME 1725/21‐1 to A.M. and C.D.H). The authors thank the staff of the animal care facility of the University of Konstanz for their valuable help. Open Access funding enabled and organized by Projekt DEAL. WOA Institution: N/A Blended DEAL.
Urban, S. , Gerwin, J. , Hulsey, C. D. , Meyer, A. , & Kratochwil, C. F. (2022). The repeated evolution of stripe patterns is correlated with body morphology in the adaptive radiations of East African cichlid fishes. Ecology and Evolution, 12, e8568. 10.1002/ece3.8568
Contributor Information
Axel Meyer, Email: axel.meyer@uni-konstanz.de.
Claudius F. Kratochwil, Email: claudius.kratochwil@helsinki.fi.
DATA AVAILABILITY STATEMENT
Data are provided in manuscript and supplementary material.
REFERENCES
- Albertson, R. C. , Streelman, J. T. , Kocher, T. D. , & Yelick, P. C. (2005). Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proceedings of the National Academy of Sciences of the United States of America, 102, 16287–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, G. R. , Erdmann, M. V. , & Dailami, M. (2015). Cirrhilabrus marinda, a new species of wrasse (Pisces: Labridae) from eastern Indonesia, Papua New Guinea, and Vanuatu. Journal of the Ocean Science Foundation, 15, 1–13. [Google Scholar]
- Allen, W. L. , Baddeley, R. , Scott‐Samuel, N. E. , & Cuthill, I. C. (2013). The evolution and function of pattern diversity in snakes. Behavioral Ecology, 24, 1237–1250. [Google Scholar]
- Allender, C. J. , Seehausen, O. , Knight, M. E. , Turner, G. F. , & Maclean, N. (2003). Divergent selection during speciation of Lake Malawi cichlid fishes inferred from parallel radiations in nuptial coloration. Proceedings of the National Academy of Sciences of the United States of America, 100, 14074–14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt, J. , & Reznick, D. (2008). Convergence and parallelism reconsidered: What have we learned about the genetics of adaptation? Trends in Ecology & Evolution, 23, 26–32. [DOI] [PubMed] [Google Scholar]
- Astudillo‐Clavijo, V. , Arbour, J. H. , & Lopez‐Fernandez, H. (2015). Selection towards different adaptive optima drove the early diversification of locomotor phenotypes in the radiation of Neotropical geophagine cichlids. BMC Evolutionary Biology, 15, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, G. (2008). The cichlid fishes: Nature's grand experiment in evolution. Basic Books. [Google Scholar]
- Beaulieu, J. M. , Jhwueng, D. C. , Boettiger, C. , & O'Meara, B. C. (2012). Modeling stabilizing selection: Expanding the Ornstein‐Uhlenbeck model of adaptive evolution. Evolution, 66, 2369–2383. [DOI] [PubMed] [Google Scholar]
- Braasch, I. , Schartl, M. , & Volff, J. N. (2007). Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evolutionary Biology, 7, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie, E. D. 3rd . (1989). Genetic correlations between morphology and antipredator behaviour in natural populations of the garter snake Thamnophis ordinoides . Nature, 342, 542–543. 10.1038/342542a0 [DOI] [PubMed] [Google Scholar]
- Brodie, E. D. 3rd . (1992). Correlational selection for color pattern and antipredator behavior in the garter snake Thamnophis ordinoides . Evolution, 46, 1284–1298. [DOI] [PubMed] [Google Scholar]
- Brodie, E. D. (1993). Consistency of individual differences in anti‐predator behaviour and colour pattern in the garter snake, Thamnophis ordinoides . Animal Behaviour, 45, 851–861. [Google Scholar]
- Brodie, E. D. 3rd , Moore, A. J. , & Janzen, F. J. (1995). Visualizing and quantifying natural selection. Trends in Ecology & Evolution, 10, 313–318. [DOI] [PubMed] [Google Scholar]
- Butlin, R. (1987). Speciation by reinforcement. Trends in Ecology & Evolution, 2, 8–13. [DOI] [PubMed] [Google Scholar]
- Chivers, D. P. , Zhao, X. , Brown, G. E. , Marchant, T. A. , & Ferrari, M. C. O. (2008). Predator‐induced changes in morphology of a prey fish: The effects of food level and temporal frequency of predation risk. Evolutionary Ecology, 22, 561–574. 10.1007/s10682-007-9182-8 [DOI] [Google Scholar]
- Clabaut, C. , Bunje, P. M. , Salzburger, W. , & Meyer, A. (2007). Geometric morphometric analyses provide evidence for the adaptive character of the Tanganyikan cichlid fish radiations. Evolution, 61, 560–578. [DOI] [PubMed] [Google Scholar]
- Claverie, T. , & Wainwright, P. C. (2014). A morphospace for reef fishes: Elongation is the dominant axis of body shape evolution. PLoS One, 9, e112732. 10.1371/journal.pone.0112732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cott, H. B. (1940). Adaptive coloration in animals. Methuen & Co., Ltd. [Google Scholar]
- Couldridge, V. C. K. , & Alexander, G. J. (2002). Color patterns and species recognition in four closely related species of Lake Malawi cichlid. Behavioral Ecology, 13, 59–64. [Google Scholar]
- Coyne, J. A. , & Orr, H. A. (2004). Speciation. Sinauer Associates Sunderland. [Google Scholar]
- Creer, D. A. (2005). Correlations between ontogenetic change in color pattern and antipredator behavior in the racer, Coluber constrictor . Ethology, 111, 287–300. [Google Scholar]
- Dingemanse, N. J. , Barber, I. , Wright, J. , & Brommer, J. E. (2012). Quantitative genetics of behavioural reaction norms: Genetic correlations between personality and behavioural plasticity vary across stickleback populations. Journal of Evolutionary Biology, 25, 485–496. [DOI] [PubMed] [Google Scholar]
- Duhl, D. M. , Vrieling, H. , Miller, K. A. , Wolff, G. L. , & Barsh, G. S. (1994). Neomorphic agouti mutations in obese yellow mice. Nature Genetics, 8, 59–65. [DOI] [PubMed] [Google Scholar]
- Dunz, A. R. , & Schliewen, U. K. (2013). Molecular phylogeny and revised classification of the haplotilapiine cichlid fishes formerly referred to as “Tilapia”. Molecular Phylogenetics and Evolution, 68, 64–80. [DOI] [PubMed] [Google Scholar]
- Evans, M. R. , & Norris, K. (1996). The importance of carotenoids in signaling during aggressive interactions between male firemouth cichlids (Cichlasoma meeki). Behavioral Ecology, 7, 1–6. [Google Scholar]
- Gerwin, J. , Urban, S. , Meyer, A. , & Kratochwil, C. F. (2021). Of bars and stripes: A Malawi cichlid hybrid cross provides insights into genetic modularity and evolution of modifier loci underlying colour pattern diversification. Molecular Ecology, 30(19), 4789–4803. [DOI] [PubMed] [Google Scholar]
- Halperin, T. , Carmel, L. , Hawlena, D. , & McGraw, K. (2016). Movement correlates of lizards’ dorsal pigmentation patterns. Functional Ecology, 31, 370–376. [Google Scholar]
- Harvey, P. H. , & Pagel, M. D. (1991). The comparative method in evolutionary biology. Oxford University Press. [Google Scholar]
- Head, M. L. , Kozak, G. M. , & Boughman, J. W. (2013). Female mate preferences for male body size and shape promote sexual isolation in threespine sticklebacks. Ecology and Evolution, 3, 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hench, K. , Vargas, M. , Höppner, M. P. , McMillan, W. O. , & Puebla, O. (2019). Inter‐chromosomal coupling between vision and pigmentation genes during genomic divergence. Nature Ecology & Evolution, 3, 657–667. [DOI] [PubMed] [Google Scholar]
- Henning, F. , Lee, H. J. , Franchini, P. , & Meyer, A. (2014). Genetic mapping of horizontal stripes in Lake Victoria cichlid fishes: Benefits and pitfalls of using RAD markers for dense linkage mapping. Molecular Ecology, 23, 5224–5240. 10.1111/mec.12860 [DOI] [PubMed] [Google Scholar]
- Hogan, B. G. , Cuthill, I. C. , & Scott‐Samuel, N. E. (2017). Dazzle camouflage and the confusion effect: The influence of varying speed on target tracking. Animal Behavior, 123, 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, B. G. , Scott‐Samuel, N. E. , & Cuthill, I. C. (2016). Contrast, contours and the confusion effect in dazzle camouflage. Royal Society Open Science, 3, 160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsey, C. D. , Alfaro, M. E. , Zheng, J. , Meyer, A. , & Holzman, R. (2019). Pleiotropic jaw morphology links the evolution of mechanical modularity and functional feeding convergence in Lake Malawi cichlids. Proceedings of the Royal Society B: Biological Sciences, 286, 20182358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsey, C. D. , Holzman, R. , & Meyer, A. (2018). Dissecting a potential spandrel of adaptive radiation: Body depth and pectoral fin ecomorphology coevolve in Lake Malawi cichlid fishes. Ecology and Evolution, 8, 11945–11953. 10.1002/ece3.4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsey, C. D. , Machado‐Schiaffino, G. , Keicher, L. , Ellis‐Soto, D. , Henning, F. , & Meyer, A. (2017). The integrated genomic Architecture and evolution of dental divergence in East African Cichlid Fishes (Haplochromis chilotes × H. nyererei). G3 (Bethesda), 7, 3195–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsey, C. D. , Zheng, J. , Faircloth, B. C. , Meyer, A. , & Alfaro, M. E. (2017). Phylogenomic analysis of Lake Malawi cichlid fishes: Further evidence that the three‐stage model of diversification does not fit. Molecular Phylogenetics and Evolution, 114, 40–48. [DOI] [PubMed] [Google Scholar]
- Hulsey, C. D. , Zheng, J. , Holzman, R. , Alfaro, M. E. , Olave, M. , & Meyer, A. (2018). Phylogenomics of a putatively convergent novelty: Did hypertrophied lips evolve once or repeatedly in Lake Malawi cichlid fishes? BMC Evolutionary Biology, 18, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husemann, M. , Tobler, M. , McCauley, C. , Ding, B. , & Danley, P. D. (2017). Body shape differences in a pair of closely related Malawi cichlids and their hybrids: Effects of genetic variation, phenotypic plasticity, and transgressive segregation. Ecology and Evolution, 7, 4336–4346. 10.1002/ece3.2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irisarri, I. , Singh, P. , Koblmuller, S. , Torres‐Dowdall, J. , Henning, F. , Franchini, P. , Fischer, C. , Lemmon, A. R. , Lemmon, E. M. , Thallinger, G. G. , Sturmbauer, C. , & Meyer, A. (2018). Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nature Communications, 9, 3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J. F. , Ingram, W. , & Campbell, H. W. (1976). The dorsal pigmentation pattern of snakes as an antipredator strategy: A multivariate approach. The American Naturalist, 110, 1029–1053. [Google Scholar]
- Jones, A. G. , Arnold, S. J. , & Burger, R. (2003). Stability of the G‐matrix in a population experiencing pleiotropic mutation, stabilizing selection, and genetic drift. Evolution, 57, 1747–1760. [DOI] [PubMed] [Google Scholar]
- Jordan, R. , Kellogg, K. , Juanes, F. , Stauffer, J. , & Montgomery, W. L. (2003). Evaluation of female mate choice cues in a group of Lake Malawi Mbuna (Cichlidae). Copeia, 2003, 181–186. [Google Scholar]
- Kautt, A. F. , Kratochwil, C. F. , Nater, A. , Machado‐Schiaffino, G. , Olave, M. , Henning, F. , Torres‐Dowdall, J. , Harer, A. , Hulsey, C. D. , Franchini, P. , Pippel, M. , Myers, E. W. , & Meyer, A. (2020). Contrasting signatures of genomic divergence during sympatric speciation. Nature, 588, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L. A. , & Kelley, J. L. (2014). Perceptual biases and animal illusions: a response to comments on Kelley and Kelley. Behavioral Ecology, 25(3), 468–469. 10.1093/beheco/aru040 [DOI] [Google Scholar]
- Knight, M. , & Turner, G. (1999). Reproductive isolation among closely related Lake Malawi cichlids: Can males recognize conspecific females by visual cues? Animal Behaviour, 58, 761–768. 10.1006/anbe.1999.1206 [DOI] [PubMed] [Google Scholar]
- Kratochwil, C. F. (2019). Molecular mechanisms of convergent color pattern evolution. Zoology (Jena), 134, 66–68. [DOI] [PubMed] [Google Scholar]
- Kratochwil, C. F. , Liang, Y. , Gerwin, J. , Woltering, J. M. , Urban, S. , Henning, F. , Machado‐Schiaffino, G. , Hulsey, C. D. , & Meyer, A. (2018). Agouti‐related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science, 362, 457–460. [DOI] [PubMed] [Google Scholar]
- Kratochwil, C. F. , Liang, Y. , Urban, S. , Torres‐Dowdall, J. , & Meyer, A. (2019). Evolutionary dynamics of structural variation at a key locus for color pattern diversification in cichlid fishes. Genome Biology and Evolution, 11, 3452–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R. , & Arnold, S. J. (1983). The measurement of selection on correlated characters. Evolution, 37, 1210–1226. [DOI] [PubMed] [Google Scholar]
- Longley, W. H. (1916). Observations upon tropical fishes and inferences from their adaptive coloration. Proceedings of the National Academy of Sciences of the United States of America, 2, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos, J. B. (2011). Convergence, adaptation, and constraint. Evolution, 65, 1827–1840. [DOI] [PubMed] [Google Scholar]
- Maan, M. E. , Seehausen, O. , Soderberg, L. , Johnson, L. , Ripmeester, E. A. , Mrosso, H. D. , Taylor, M. I. , van Dooren, T. J. , & van Alphen, J. J. (2004). Intraspecific sexual selection on a speciation trait, male coloration, in the Lake Victoria cichlid Pundamilia nyererei . Proceedings of the Royal Society B: Biological Sciences, 271, 2445–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky, M. , Svardal, H. , Tyers, A. M. , Miska, E. A. , Genner, M. J. , Turner, G. F. , & Durbin, R. (2018). Whole‐genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nature Ecology & Evolution, 2, 1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee, M. D. , Neches, R. Y. , & Seehausen, O. (2016). Evaluating genomic divergence and parallelism in replicate ecomorphs from young and old cichlid adaptive radiations. Molecular Ecology, 25, 260–268. [DOI] [PubMed] [Google Scholar]
- McGhee, G. R. (2011). Convergent evolution: Limited forms most beautiful. MIT Press. [Google Scholar]
- McKinnon, J. S. , & Pierotti, M. E. (2010). Colour polymorphism and correlated characters: Genetic mechanisms and evolution. Molecular Ecology, 19, 5101–5125. [DOI] [PubMed] [Google Scholar]
- Meier, J. I. , Marques, D. A. , Mwaiko, S. , Wagner, C. E. , Excoffier, L. , & Seehausen, O. (2017). Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nature Communications, 8, 14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali, G. , & Kodandaramaiah, U. (2017). Body size and evolution of motion dazzle coloration in lizards. Behavioral Ecology, 29, 79–86. [Google Scholar]
- Murali, G. , Merilaita, S. , & Kodandaramaiah, U. (2018). Grab my tail: Evolution of dazzle stripes and colourful tails in lizards. Journal of Evolutionary Biology, 31, 1675–1688. [DOI] [PubMed] [Google Scholar]
- Muschick, M. , Indermaur, A. , & Salzburger, W. (2012). Convergent evolution within an adaptive radiation of cichlid fishes. Current Biology, 22, 2362–2368. [DOI] [PubMed] [Google Scholar]
- Noor, M. A. F. , & Bennett, S. M. (2009). Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity, 103(6), 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ord, T. J. , & Summers, T. C. (2015). Repeated evolution and the impact of evolutionary history on adaptation. BMC Evolutionary Biology, 15, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, S. A. , Friedman, S. T. , & Wainwright, P. C. (2015). How predation shaped fish: The impact of fin spines on body form evolution across teleosts. Proceedings of the Royal Society B: Biological Sciences, 282, 20151428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3, 217–223. [Google Scholar]
- Rocha, L. , & Bowen, B. (2008). Speciation in coral‐reef fishes. Journal of Fish Biology, 72, 1101–1121. [Google Scholar]
- Rojas, B. , Devillechabrolle, J. , & Endler, J. A. (2014). Paradox lost: Variable colour‐pattern geometry is associated with differences in movement in aposematic frogs. Biology Letters, 10, 20140193. 10.1098/rsbl.2014.0193 [DOI] [Google Scholar]
- Rometsch, S. J. , Torres‐Dowdall, J. , & Meyer, A. (2020). Evolutionary dynamics of pre‐ and postzygotic reproductive isolation in cichlid fishes. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 375, 20190535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau, S. , Glémet, H. , & Magnan, P. (2010). Effects of morphology on swimming performance in wild and laboratory crosses of brook trout ecotypes. Functional Ecology, 24, 310–321. [Google Scholar]
- Saltz, J. B. , Hessel, F. C. , & Kelly, M. W. (2017). Trait correlations in the genomics era. Trends in Ecology & Evolution, 32, 279–290. [DOI] [PubMed] [Google Scholar]
- Salzburger, W. , Mack, T. , Verheyen, E. , & Meyer, A. (2005). Out of Tanganyika: Genesis, explosive speciation, key‐innovations and phylogeography of the haplochromine cichlid fishes. BMC Evolutionary Biology, 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. , Tinevez, J. Y. , White, D. J. , Hartenstein, V. , Eliceiri, K. , Tomancak, P. , & Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep, K. P. (2011). phangorn: Phylogenetic analysis in R. Bioinformatics, 27, 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen, O. (1997). Distribution of and reproductive isolation among color morphs of a rock‐dwelling Lake Victoria cichlid (Haplochromis nyererei) (vol 5, pg 195, 1996). Ecology of Freshwater Fish, 6, 57. 10.1111/j.1600-0633.1996.tb00133.x [DOI] [Google Scholar]
- Seehausen, O. , Butlin, R. K. , Keller, I. , Wagner, C. E. , Boughman, J. W. , Hohenlohe, P. A. , Peichel, C. L. , Saetre, G. P. , Bank, C. , Brannstrom, A. , Brelsford, A. , Clarkson, C. S. , Eroukhmanoff, F. , Feder, J. L. , Fischer, M. C. , Foote, A. D. , Franchini, P. , Jiggins, C. D. , Jones, F. C. , … Widmer, A. (2014). Genomics and the origin of species. Nature Reviews Genetics, 15, 176–192. [DOI] [PubMed] [Google Scholar]
- Seehausen, O. , Mayhew, P. J. , & van Alphen, J. J. M. (1999). Evolution of colour patterns in East African cichlid fish. Journal of Evolutionary Biology, 12, 514–534. [Google Scholar]
- Seehausen, O. , & van Alphen, J. J. M. (1998). The effect of male coloration on female mate choice in closely related Lake Victoria cichlids (Haplochromis nyererei complex). Behavioral Ecology and Sociobiology, 42, 1–8. [Google Scholar]
- Seehausen, O. , Witte, F. , Alphen, J. J. M. V. , & Bouton, N. (1998). Direct mate choice maintains diversity among sympatric cichlids in Lake Victoria. Journal of Fish Biology, 53, 37–55. [Google Scholar]
- Sih, A. , Bell, A. M. , Johnson, J. C. , & Ziemba, R. E. (2004). Behavioral syndromes: An integrative overview. The Quarterly Review of Biology, 79, 241–277. [DOI] [PubMed] [Google Scholar]
- Song, Y. , & Cone, R. D. (2007). Creation of a genetic model of obesity in a teleost. The FASEB Journal, 21, 2042–2049. [DOI] [PubMed] [Google Scholar]
- Stevens, M. , & Merilaita, S. (2009). Defining disruptive coloration and distinguishing its functions. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 364, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, M. , Searle, W. T. , Seymour, J. E. , Marshall, K. L. , & Ruxton, G. D. (2011). Motion dazzle and camouflage as distinct anti‐predator defenses. BMC Biology, 9, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard, M. A. , Lemey, P. , Baele, G. , Ayres, D. L. , Drummond, A. J. , & Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evolution, 4, vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer, G. H. (1918). Concealing‐coloration in the animal kingdom: An exposition of the laws of disguise through color and pattern: Being a summary of Abbott H. Thayer's discoveries. Macmillan Company. [Google Scholar]
- Urban, S. , Nater, A. , Meyer, A. , & Kratochwil, C. F. (2021). Different sources of allelic variation drove repeated color pattern divergence in cichlid fishes. Molecular Biology and Evolution, 38, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via, S. (2012). Divergence hitchhiking and the spread of genomic isolation during ecological speciation‐with‐gene‐flow. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 367, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor, B. C. , & Randall, J. E. (2010). Gramma dejongi, a New Basslet (Perciformes: Grammatidae) from Cuba, a Sympatric Sibling Species of G. loreto . Zoological Studies, 49, 865–871. [Google Scholar]
- Victor, B. C. , & Randall, J. E. (2014). Pseudojuloides edwardi, n. sp. (Perciformes: Labridae): An example of evolution of male‐display phenotype outpacing divergence in mitochondrial genotype. Journal of the Ocean Science Foundation, 11, 1–12. [Google Scholar]
- von Helversen, B. , Schooler, L. J. , & Czienskowski, U. (2013). Are stripes beneficial? Dazzle camouflage influences perceived speed and hit rates. PLoS One, 8, e61173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, C. E. , Harmon, L. J. , & Seehausen, O. (2012). Ecological opportunity and sexual selection together predict adaptive radiation. Nature, 487, 366–369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Data are provided in manuscript and supplementary material.


