SUMMARY
In multicellular organisms, neurons integrate a diverse array of external cues to affect downstream changes in organismal health. Specifically, activation of the endoplasmic reticulum (ER) unfolded protein response (UPRER) in neurons increases lifespan by preventing age-onset loss of ER proteostasis and driving lipid depletion in a cell non-autonomous manner. The mechanism of this communication is dependent on the release of small clear vesicles from neurons. We find dopaminergic neurons are necessary and sufficient for activation of cell non-autonomous UPRER to drive lipid depletion in peripheral tissues, whereas serotonergic neurons are sufficient to drive protein homeostasis in peripheral tissues. These signaling modalities are unique and independent and together coordinate the beneficial effects of neuronal cell non-autonomous ER stress signaling upon health and longevity.
In Brief
Higuchi-Sanabria et al. identify two distinct neuronal populations that are critical in signaling non-autonomous UPRER from neurons to the periphery: serotonergic neurons drive induction of protein changes and dopaminergic neurons drive induction of lipid changes.
Graphical Abstract
INTRODUCTION
Under conditions of stress, multicellular organisms must coordinate an effective response across diverse tissues to maintain organismal homeostasis. For most stress responses, including the endoplasmic reticulum (ER) unfolded protein response (UPRER), there is an age-associated decline in protein folding and the proteotoxic stress response. This can be circumvented by overexpressing the primary transcription factor xbp-1s, which can preserve organismal health and extend lifespan (Frakes and Dillin, 2017; Higuchi-Sanabria et al., 2016; Taylor and Dillin, 2013). In particular, overexpression of spliced xbp-1 (xbp-1s) specifically in the Caenorhabditis elegans nervous system extended lifespan and increased ER stress tolerance in a cell non-autonomous manner by promoting protein homeostasis (Frakes et al., 2020; Taylor and Dillin, 2013) and driving lipid homeostasis (Daniele et al., 2020; Imanikia et al., 2019a). Although the identity of the secreted ER-stress signal (SERSS) from neurons to the periphery required in this paradigm is still unknown, small clear vesicles (SCVs), which could be host to numerous neurotransmitters and biogenic amines, were implicated in neurons, whereas dense core vesicles (DCVs), which are host to numerous neuropeptides, were implicated in glia (Frakes et al., 2020; Taylor and Dillin, 2013).
Downstream of the SERSS, the intestine activates UPRER in an xbp-1s-dependent manner, which contributed to the lifespan extension found in this paradigm. Previously, the beneficial effect of neuronal non-autonomous UPRER induction in the periphery was ascribed to increased protein homeostasis by upregulation of chaperones (Taylor and Dillin, 2013). However, by characterizing the specific changes that occur in the signal receiving cell, namely, the intestine, it was uncovered that this neuronal signal results in significant ER expansion and lipid depletion. These phenomena are chaperone independent and are both necessary and sufficient for the lifespan extension found when xbp-1 s is overexpressed in neurons (Daniele et al., 2020). Thus, neuronal xbp-1 s activates two distinct pathways in the distal tissue: a protein homeostasis arm through chaperone in duction and a metabolic arm that involves ER remodeling and lipid depletion. The depletion of lipids occurs by two mechanisms: (1) upregulation of lysosomal lipases and desaturases, which depletes triglycerides and increases oleic acid levels (Imanikia et al., 2019a); and (2) activation of lipophagy by a conserved RME-1/RAB-10/EHBP-1 complex (Daniele et al., 2020). However, how these two distinct signals are elicited and the neural subtypes involved in these divergent mechanisms are yet to be discovered.
A similar communication from neurons to periphery is also observed in vertebrates. When Xbp1s is overexpressed in Pomc neurons of the hypothalamus of mice, the UPRER is upregulated and has beneficial impacts on metabolic physiology (e.g., improved glucose levels, improved insulin sensitivity, and protection against high-fat diet-induced obesity) (Williams et al., 2014). In this model, Xbp1s increases Pomc neuron activity, which in turn increases energy expenditure by promoting brown adipose tissue (BAT) thermogenesis and browning of white adipose tissue (WAT), which results in an overall decrease in fat mass and body weight, consistent with the findings in C. elegans. The direct conservation of these phenomena downstream of neuronal Xbp1s suggests that mechanistic understanding of the neuronal circuits and cellular changes in response to these signals would serve as a critical foundation for understanding age- and disease-linked declines in ER homeostasis and ER quality control.
Here, we performed screens to identify the specific neurons important for sensing and transmitting UPRER to the periphery. These studies specifically segregate two distinct and independent mechanisms of UPRER through two separate sets of neurons: protein homeostasis through chaperone induction by serotonin signaling and metabolic remodeling through lipid depletion by dopamine signaling. These phenomena are generally discussed as simultaneous and interchangeable responses to UPRER activation. However, we challenge this notion to argue not only that they can be separated but also that unique machinery exist to elicit these divergent responses.
RESULTS
Screening of Neurotransmitters Reveal Serotonin Signaling as Necessary and Sufficient for Chaperone Induction of Non-autonomous UPRER
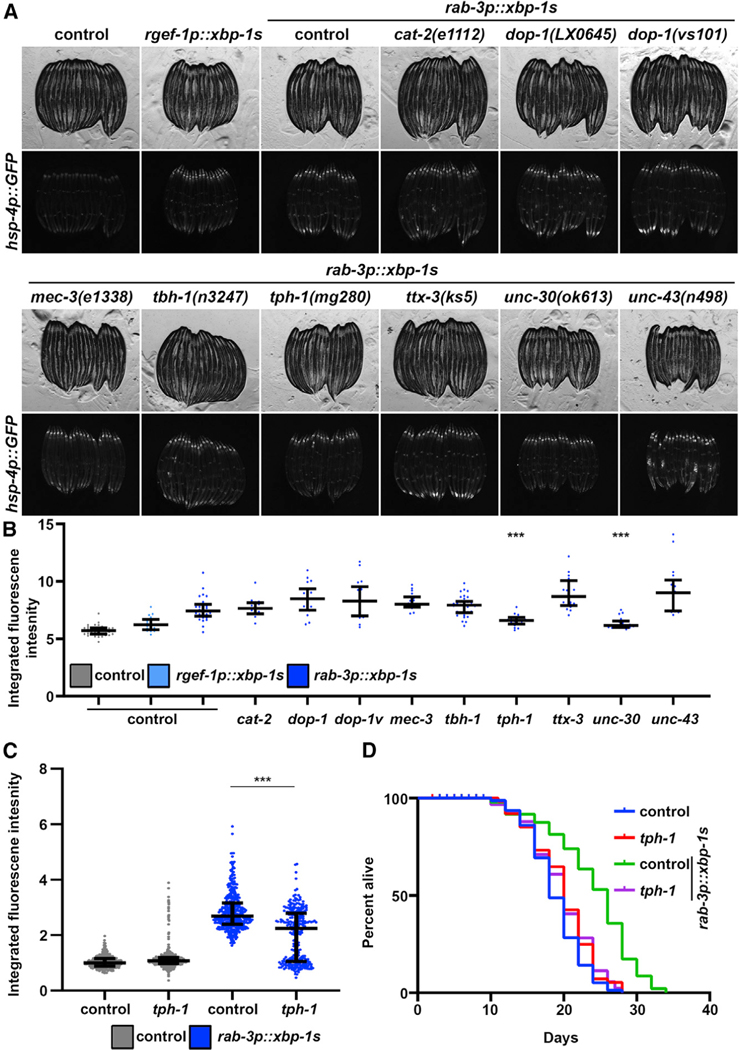
The goal of this study was to identify specific neuronal subtypes involved in the cell non-autonomous transmission of UPRER. Our lab described a requirement for neurotransmitter release by SCVs to promote cell non-autonomous signaling downstream of xbp-1 s (Salio et al., 2006; Taylor and Dillin, 2013). Thus, using high intestinal expression of the putative UPRER reporter hsp-4p::GFP as the readout of cell non-autonomous UPRER induction, we crossed these reporter animals to a panel of mutants for neurotransmitter biosynthesis and function to screen for mutations that prevent non-autonomous UPRER signaling. Specifically, pan-neuronal overexpression of the active, spliced variant of xbp-1, xbp-1s (referred to as neuronal xbp-1s animals), resulted in robust activation of UPRER and induction of the hsp-4p::GFP reporter. Suppressors of non-autonomous UPRER induction can be visualized as a loss of fluorescence using this reporter. Screening a panel of neurotransmitter mutants identified mutations in mec-3, tph-1, and unc-30 as strong suppressors of non-autonomous UPRER induction (Figure 1A).
Figure 1. A Screen for Neurotransmitters Reveals Serotonin as an Essential Signal for Non-autonomous UPRER of Chaperones.
(A) Fluorescent micrographs of day 1 adult hsp-4p::GFP control or neuronal xbp-1s (rgef-1p::GFP or rab-3p::GFP) animals carrying mutations for genes encoding neurotransmitter synthesis or signaling pathways. All images are contrast matched.
(B) Quantification of (A) using ImageJ as described in STAR Methods with control represented in gray and neuronal xbp-1s represented in blue (light blue, rgef-1p::xbp-1s; dark blue, rab-3p::xbp-1s). Lines represent median and interquartile range, with each dot representing a single animal. ***p < 0.001 compared to rab-3p::xbp-1s controls using Student’s t test. dop-1, dop-1(LX0645); dop-1v, dop-1(vs101).
(C) Quantification of hsp-4p::GFP in day 1 adult hsp-4p::GFP control (gray) or neuronal xbp-1s (rab-3p::xbp-1s; blue) animals with and without tph-1(mg280) mutation for serotonin synthesis. Lines represent median and interquartile range, with each dot representing a single animal. Data are representative of three independent trials. ***p < 0.001 using non-parametric Mann-Whitney testing.
(D) Lifespans of control and neuronal xbp-1s (rab-3p::xbp-1s) animals with and without tph-1(mg280) mutation for serotonin synthesis. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
The mec-3 gene encodes a LIM homeobox protein, and mutations in mec-3 specifically affect sensory neurons and decrease glutamatergic signaling (Serrano-Saiz et al., 2013; Xue et al., 1992). tph-1 encodes the tryptophan hydroxylase enzyme, which is a key enzyme for serotonin biosynthesis; animals with tph-1 mutations fail to synthesize serotonin (Sze et al., 2000). unc-30 plays a critical role in GABA synthesis, and mutations in unc-30 have severe defects in GABAergic differentiation, axon guidance, and downstream motor/behavioral phenotypes (Westmoreland et al., 2001). Among these three neuronal circuits, only serotonin signaling has been described to be critical in promoting neurotransmission of stress signals, including the UPR of the mitochondria (UPRMT) (Berendzen et al., 2016) and the heat-shock response (HSR) (Tatum et al., 2015). Furthermore, the loss of tph-1 did not cause gross morphological changes to the organism, such as mec-3 and unc-30 mutations, which cause severe motor defects. Moreover, further quantitative analysis of reporters normalized to animal size revealed significant differences (p < 0.001) for only tph-1 and unc-30 mutations (Figure 1B). Therefore, we sought to further characterize serotonin signaling as a potential “master regulator” of stress signaling and thus playing a similar role in mediating neurotransmission of the UPRER to the periphery. First, we confirmed that the deletion mutant tph-1(mg280) failed to promote non-autonomous UPRER signaling, as measured by hsp-4p::GFP induction across a large population by quantification using a large-particle biosorter (Figure 1C; Daniele et al., 2017). More importantly, mutations in tph-1 fully suppressed the lifespan extension found in neuronal xbp-1s animals, providing strong evidence that serotonin signaling is critical for the beneficial effects of neuronal xbp-1s (Figure 1D).
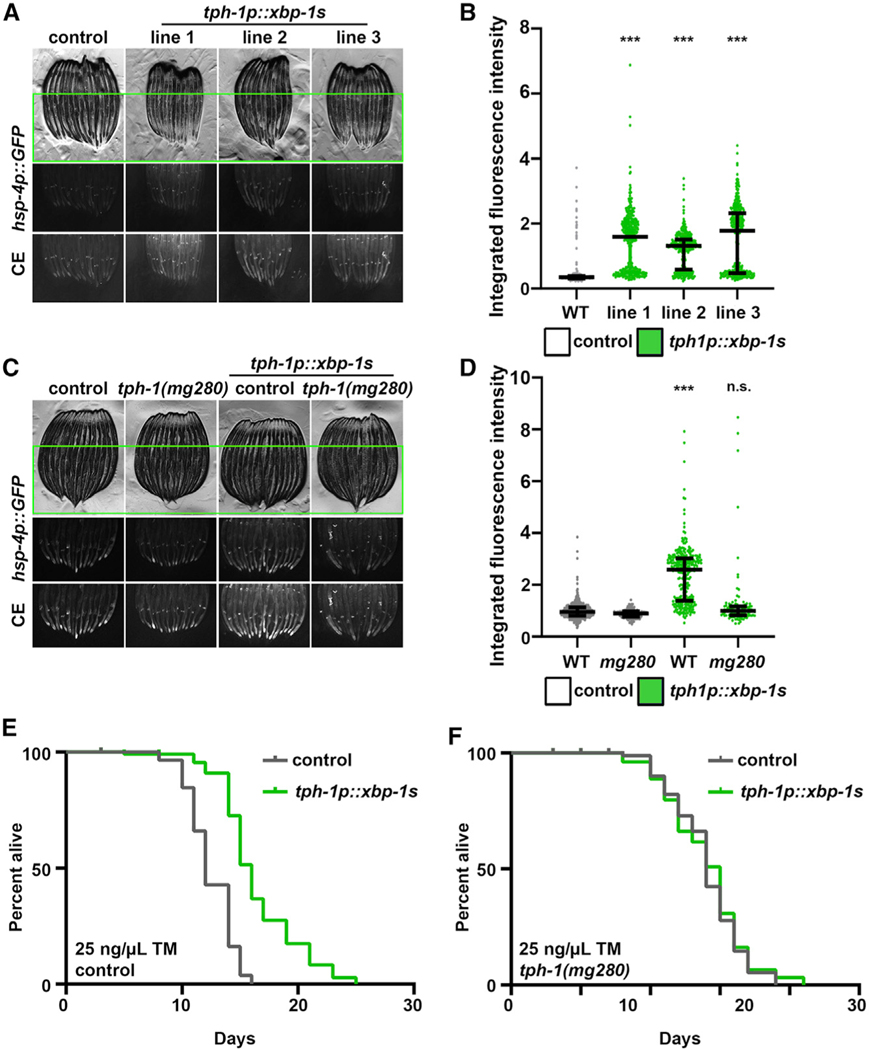
To determine whether serotonergic neurons were sufficient to drive non-autonomous UPRER in peripheral tissue, we created transgenic animals overexpressing xbp-1s specifically in serotonergic neurons (tph-1p::xbp-1s, named serotonergic xbp-1s). Three independent overexpression lines of serotonergic xbp-1s overexpression showed robust induction of the UPRER in the periphery (Figures 2A and 2B). Moreover, the peripheral activation of the UPRER was fully dependent on serotonin production (tph-1), suggesting that serotonergic xbp-1s drives non-autonomous UPRER induction through a similar serotonin-dependent mechanism as pan-neuronal xbp-1s and not through some non-specific, off-target effect of serotonergic xbp-1s expression (Figures 2C and 2D). To determine whether the induction of chaperones in the periphery was beneficial for whole-animal physiology, we performed survival assays upon exposure to the chemical agent tunicamycin, which blocks N-linked glycosylation in the ER to produce protein misfolding stress (Heifetz et al., 1979). Indeed, we found that serotonergic xbp-1s animals have a significant increase in survival under ER stress, which is also fully dependent on serotonin signaling (Figures 2E and 2F). Under basal conditions, serotoninergic xbp-1s animals also exhibited a mild lifespan extension, which is also dependent on serotonin signaling (Figures S1A and S1B). Both the lifespan extension and induction of the UPRER are dependent on xbp-1, as RNAi knockdown of xbp-1 fully suppressed these phenotypes (Figures S1C–S1F). These data suggest that serotonergic xbp-1s is sufficient to drive non-autonomous UPRER induction in the periphery, which has direct implications in organismal physiology and lifespan.
Figure 2. Serotonergic xbp-1s Is Sufficient to Drive Chaperone Induction and ER Stress Resistance.
(A) Fluorescent micrographs of day 1 adult hsp-4p::GFP control or three independent integration lines of serotonergic xbp-1s (tph-1p::xbp-1s) animals. All images are contrast matched. CE, contract enhanced images for clarity; some pixels may be saturated. Only the lower 60% of the worm is shown to remove co-injection marker signal.
(B) Quantification of (A): day 1 adult hsp-4p::GFP control (gray) or serotonergic xbp-1s (tph-1p::xbp-1s; green) animals. Lines represent median and interquartile range, with each dot representing a single animal. Data are representative of three independent trials. ***p < 0.001 using non-parametric Mann-Whitney testing against control. Quantification is applied in the lower 60% of the worm to remove co-injection marker signal.
(C) Fluorescent micrographs of day 1 adult hsp-4p::GFP control or serotonergic xbp-1s (tph-1p::xbp-1s; line 3 was selected for all further analyses) animals with and without tph-1(mg280) mutation for serotonin synthesis. Some pixels may be saturated. Only the lower 60% of the worm is shown to remove co-injection marker signal.
(D) Quantification of (C): day 1 adult hsp-4p::GFP control (gray) or serotonergic xbp-1s (tph-1p::xbp-1s; green) animals. Lines represent median and interquartile range, with each dot representing a single animal. Data are representative of three independent trials. ***p < 0.001 using non-parametric Mann-Whitney testing against matching control. Quantification is applied in the lower 60% of the worm to remove co-injection marker signal.
(E) Tunicamycin (TM) survival assay of control and serotonergic xbp-1s (tph-1p::xbp-1s) animals. Animals were moved to 25 ng/μL TM plates at day 1 of adulthood. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
(F) TM survival assay of control and serotonergic xbp-1s (tph-1p::xbp-1s) animals carrying tph-1(mg280) mutation for serotonin synthesis. Animals were moved to 25 ng/μL TM plates at day 1 of adulthood. Assays for (E) and (F) were performed simultaneously and can be directly compared. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
Pan-neuronal xbp-1s can drive two independent arms of the UPRER in the periphery: a protein homeostasis arm by upregulation of chaperones and a metabolic arm that promotes lipid depletion through the conserved EHBP-1/RME-1/RAB-10 lipophagy machinery (Daniele et al., 2020). To determine whether serotonergic xbp-1s was also sufficient to drive the lipid homeostasis arm of non-autonomous UPRER induction, we used the lipid droplet marker dhs-3p::DHS-3::GFP to measure lipid levels in the intestine (Na et al., 2015; Zhang et al., 2012). We have previously validated that this reporter is a reliable marker that correlates well with lipid levels in worms (Daniele et al., 2020). Interestingly, we found that serotonergic xbp-1s animals exhibit increased lipid levels and thus failed to deplete lipids (Figures S2A and S2). Moreover, unlike pan-neuronal xbp-1s, the lifespan extension of serotonergic xbp-1s is not dependent on ehbp-1, the gene encoding the primary lipophagy component required for lipid depletion (Figure S2C; Daniele et al., 2020). Thus, it is possible that serotonergic xbp-1s animals promote only the protein homeostasis arm of UPRER by chaperones and that a different set of neurons is responsible for promoting the lipid homeostasis arm of UPRER.
Finally, we sought to determine whether increased serotonin signaling itself was sufficient to drive non-autonomous UPRER induction. We used a well-validated mutant of the serotonin reuptake receptor MOD-5. Mutants of mod-5 have been shown to exhibit increased serotonin signaling (Ranganathan et al., 2001). To our surprise, mod-5(n3314) mutant animals failed to induce the hsp-4p::GFP expression (Figures S2D and S2E). Moreover, unlike serotonergic xbp-1s animals, mod-5(n3314) animals have a very mild, non-significant increase in lipid levels (Figure S2F), suggesting that simply increasing serotonin signaling is not sufficient to phenocopy serotonergic xbp-1s. These data suggest that serotonin signaling is necessary, but not sufficient, for transmission of neuronal UPRER to the periphery, and it is possible that other neurotransmitters are involved in the transmission of signals from serotonergic xbp-1s.
Screening of Neurotransmitters Reveal Dopamine Signaling as Necessary and Sufficient for Lipid Depletion Downstream of Non-autonomous UPRER
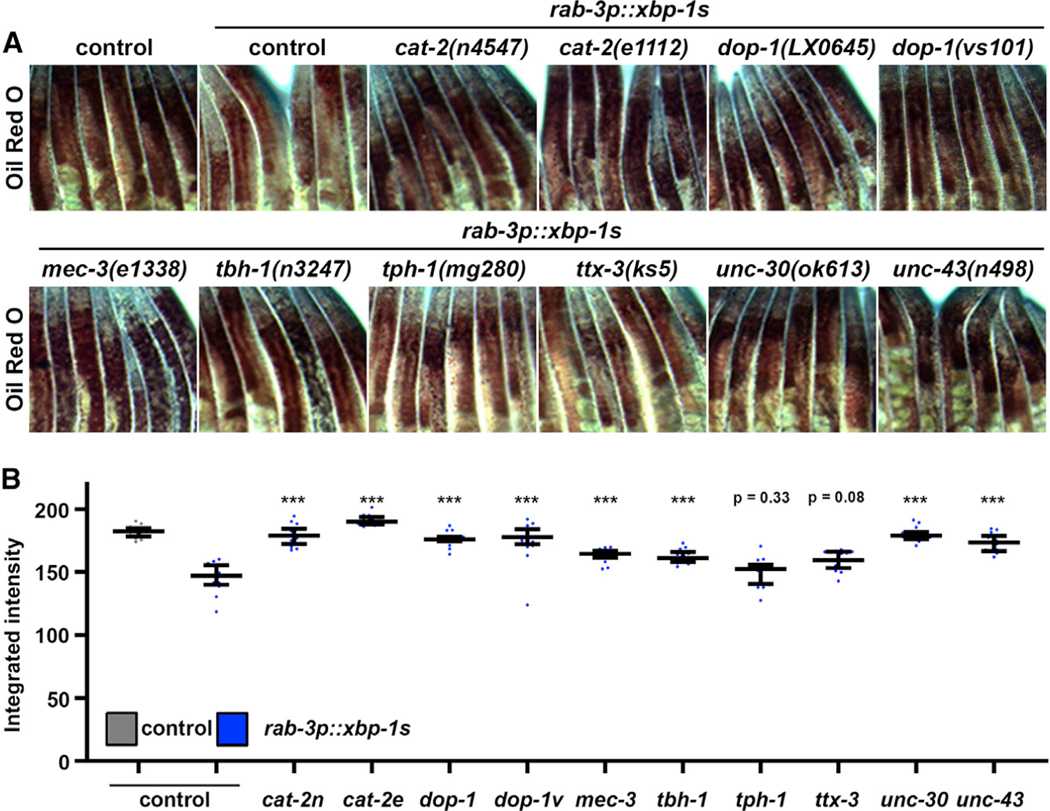
Because serotonergic xbp-1s animals failed to elicit changes to lipid levels, we performed a similar screen of neurotransmitter mutants measuring whole-animal lipid levels using oil red O (Escorcia et al., 2018) in an effort to identify the neurotransmitter involved in lipid-mediated changes downstream of neuronal UPRER. As previously described (Imanikia et al., 2019a), oil red O staining can robustly measure the decrease in lipids found in neuronal xbp-1s animals. Interestingly, most of the mutants tested—with the exception of serotoninergic (tph-1) and thermosensory (ttx-3) mutants—showed suppression of the decreased lipid phenotype in neuronal xbp-1s animals (Figures 3A and 3B). Four separate dopamine mutants exhibited suppression, two each for cat-2 [cat-2(n4547) and cat-2(e1112)] and dop-1 [dop-1(vs101) and dop-1(LX0645). cat-2 encodes for tyrosine hydroxylase, the enzyme responsible for the initial and rate-limiting step in catecholamine synthesis, including dopamine (Lints and Emmons, 1999; Nagatsu et al., 1964). dop-1 encodes one of the dopamine receptors, which is most similar to mammalian D1-like receptor (Chase et al., 2004). Two distinct mutations in cat-2 and dop-1 suppressed the lipid depletion found in neuronal xbp-1 s animals. Moreover, several studies have already implicated dopamine signaling in fat metabolism, and increased dopamine signaling in C. elegans has been shown to decrease fat content through increased β-oxidation (Barros et al., 2014). Animals mutated for dat-1, which encodes the dopamine reuptake receptor, have increased exogenous dopamine levels, and we found that these animals also exhibited a significant decrease in lipids using the DHS-3::GFP marker (Figures S3A and S3B). Therefore, we focused our studies on lipid metabolism on dopaminergic neurons.
Figure 3. A Screen for Neurotransmitters Reveals Dopamine as an Essential Signal for Non-autonomous UPRER of Lipid Metabolism.
(A) Brightfield micrographs of day 1 adult control or neuronal xbp-1 s (rgef-1p::GFP or rab-3p::GFP) animals carrying mutations for genes encoding neurotransmitter synthesis or signaling pathways stained with oil red O. All images are contrast matched and magnified between the pharynx and the most anterior part of the egg sac.
(B) Quantification of (A) using ImageJ as described in STAR Methods with control represented in gray and neuronal xbp-1 s represented in blue (light blue, rgef-1p::xbp-1 s; dark blue, rab-3p::xbp-1 s). Lines represent median and interquartile range, with each dot representing a single animal. ***p < 0.001 compared to rab-3p::xbp-1 s controls using Student’s t test. cat-2n, cat-2(n4547); cat-2e, (cat-2e1112); dop-1, dop-1(LX0645); dop-1v, dop-1(vs101).
To directly test the requirement of dopaminergic neurons in non-autonomous UPRER, we tested the effects of overexpressed xbp-1s specifically in dopaminergic neurons (dat-1p::xbp-1s, referred to as dopaminergic xbp-1s). Intriguingly, these animals exhibited a significant depletion of lipids, which is dependent on dopamine synthesis (Figures 4A and 4B). However, in stark contrast to the pan-neuronal and serotonergic xbp-1s animals, dat-1p::xbp-1s animals completely failed to induce chaperones, as measured by hsp-4p::GFP and qPCR of xbp-1s downstream targets (Figures S3C–S3E). Moreover, dopaminergic xbp-1s was sufficient to extend lifespan in a dopamine- and ehbp-1-dependent manner (Figures 4C–4E). Finally, similar to serotonergic and pan-neuronal xbp-1s, the beneficial effects of dopaminergic xbp-1s was completely dependent on xbp-1 (Figure 4F; Figures S4A and S4B). These data indicate that dopaminergic xbp-1s signals a unique program distinct from serotonergic signaling, which acts through dopamine to mobilize lipids by increasing EHBP-1-mediated lipophagy. Indeed, knockdown of ehbp-1 is sufficient to suppress the lipid depletion found in dopaminergic xbp-1s animals (Figures S4A and S4B). In agreement, the lipid depletion found in increased dopamine signaling through mutation of the dopamine reuptake receptor is also dependent on xbp-1s and ehbp-1 (Figures S4C and S4D). Taken together, our data provide evidence that dopaminergic signaling promotes lipid depletion through XBP-1-mediated activation of EHBP-1, which is sufficient to extend lifespan and is independent of protein chaperone induction.
Figure 4. Overexpression of xbp-1s in Dopaminergic Neurons Drives Lipid Depletion to Extend Lifespan.
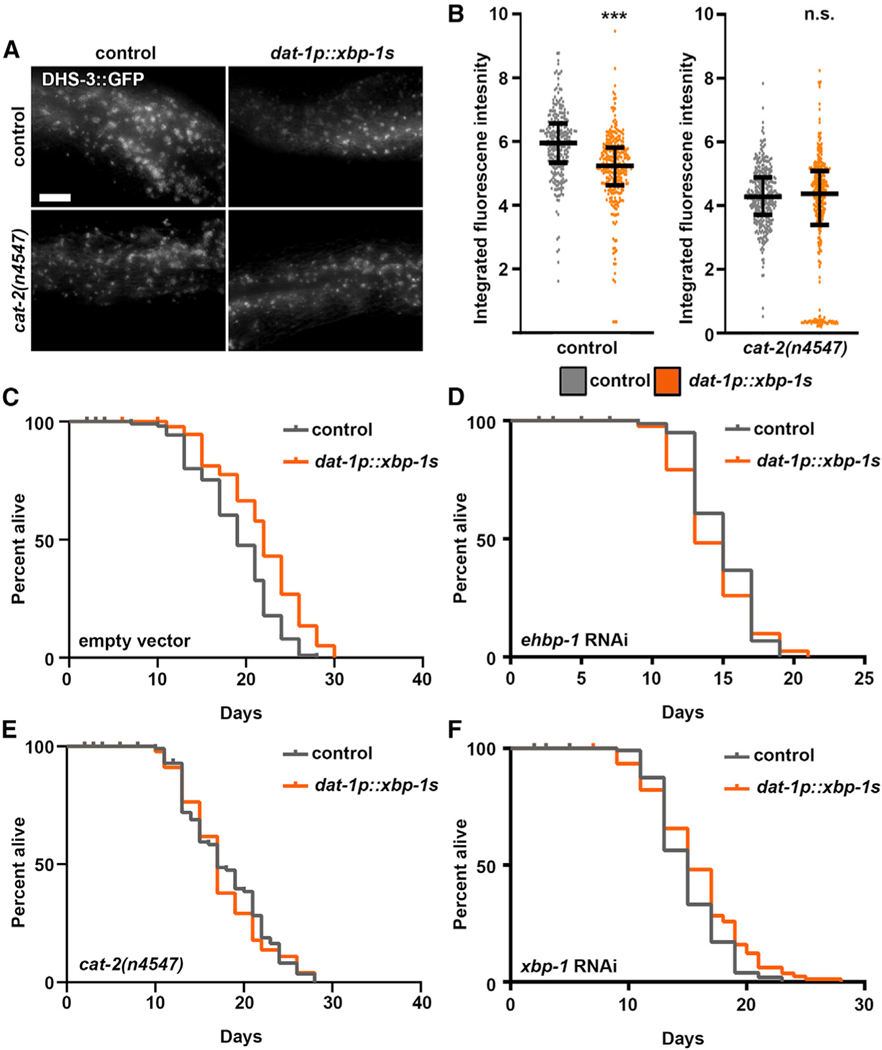
(A) Representative fluorescent micrographs of intestinal lipid droplets (LDs) (by dhs-3p::DHS-3::GFP) in day 2 adult control and dopaminergic xbp-1s (dat-1p::xbp-1s) animals with and without cat-2(n4547) mutation in dopamine synthesis. Imaging was performed equidistant from the vulva to the tail of the worm across all conditions. All images are contrast matched. Scale bar represents 10 μm.
(B) Quantification of dhs-3p::DHS-3::GFP in (a) of control (gray) and dopaminergic xbp-1s (dat-1p::xbp-1s; orange) animals with and without cat-2(n4547) mutation in dopamine synthesis. Lines represent median and interquartile range, with each dot representing a single animal. Data are representative of three independent trials. ***p < 0.001 using non-parametric Mann-Whitney testing against matching control.
(C) Lifespans of control and dopaminergic xbp-1s (dat-1p::xbp-1s) animals grown on empty vector. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
(D) Lifespans of control and dopaminergic xbp-1s (dat-1p::xbp-1s) animals grown on ehbp-1 RNAi from hatch. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
(E) Lifespan measurements of control and dopaminergic xbp-1s (dat-1p::xbp-1s) animals carrying cat-2(n4547) mutation for dopamine synthesis. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
(F) Lifespans of control and dopaminergic xbp-1s (dat-1p::xbp-1s) animals grown on xbp-1 RNAi. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics. Measurements for (D), (E), and (F) were performed simultaneously and can be directly compared.
Serotonergic and Dopaminergic UPRER Signatures Are Unique and Independent
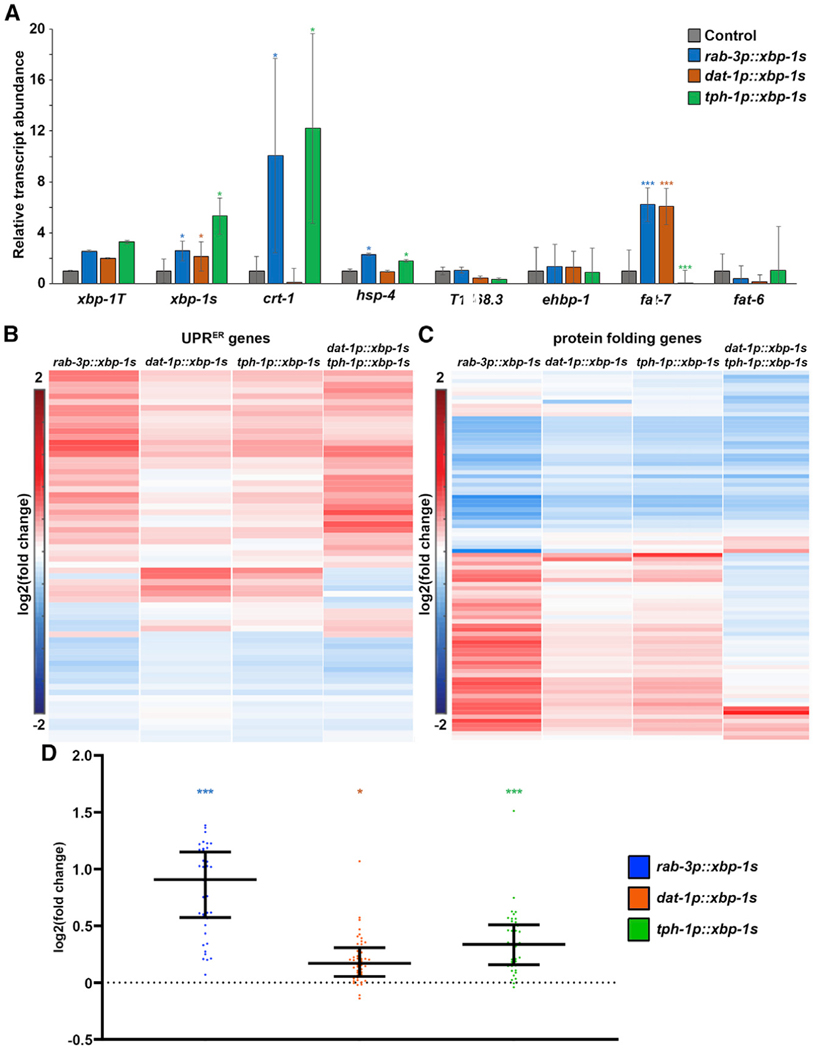
Next, we sought to determine whether serotonergic and dopaminergic UPRER were truly independent of each other. First, we performed transcriptome analysis on serotonergic xbp-1s and dopaminergic xbp-1s animals. As expected, these animals exhibit increased xbp-1s, as measured by qPCR, and overall UPRER signatures similar to pan-neuronal xbp-1s (Figures 5A and 5B). Moreover, consistent with our previous findings, serotonergic xbp-1s significantly increases mRNA transcript abundance of hsp-4, as well as another XBP-1 s target, crt-1, similar to pan-neuronal xbp-1s. Moreover, serotonergic xbp-1s broadly upregulates protein-folding genes and has a signature more similar to pan-neuronal xbp-1s, whereas dopaminergic xbp-1s either minimally affects or completely fails to promote expression of these genes (Figures 5A, 5C, and 5D). Finally, dopaminergic xbp-1s showed increased levels of the desaturase fat-7, similar to pan-neuronal xbp-1s, whereas serotonergic xbp-1s failed to upregulate these transcripts (Figure 5A).
Figure 5. Serotonergic and Dopaminergic xbp-1s Elicit Two Different Transcriptional Programs.
(A) qPCR of transcripts in control (gray), pan-neuronal xbp-1s (blue), dopaminergic xbp-1s (orange), and serotonergic xbp-1s (green) animals grown on empty vector from hatch. RNA was isolated in day 1 adults, and data were collected against a standard curve to calculate relative transcript abundance against control. Data are pooled across three biological replicates and four technical replicates per sample. Data are represented as mean ± standard deviation. *p < 0.05; ***p < 0.001 using Student’s t test.
(B) RNA sequencing (RNA-seq) was performed in control, pan-neuronal xbp-1s, dopaminergic xbp-1s, serotonergic xbp-1s, and dopaminergic/serotonergic double xbp-1s as described in the STAR Methods. Heatmap indicates log2 (fold change) of genes in comparison to control. Here, canonical UPRER target genes are represented as genes that showed decreased expression when ire-1 was mutated (Shen et al., 2005) and/or are part of the Gene Ontology (GO) term 0030968 UPRER using BioMart WormBase Parasite. Warmer colors indicate increased expression, and cooler colors indicate decreased expression. See Table S3 for actual values of log2 (fold change).
(C) Heatmap indicates log2 (fold change) of genes in comparison to control. Here, protein-folding genes are represented as per GO term 0006457 using BioMart WormBase Parasite. Warmer colors indicate increased expression, and cooler colors indicate decreased expression. See Table S4 for actual values of log2 (fold change).
(D) log2 (fold change) of genes are shown for all upregulated genes of pan-neuronal xbp-1s (blue), dopaminergic xbp-1s (orange), and serotonergic xbp-1s (green) of protein-folding genes represented as per GO term 0006457 using BioMart WormBase Parasite (bottom half of heatmap of C). Middle line represents median, and whiskers represent interquartile range. ***p < 0.001; *p < 0.05 using Mann-Whitney testing calculated from normalized reads of each gene compared to wildtype control.
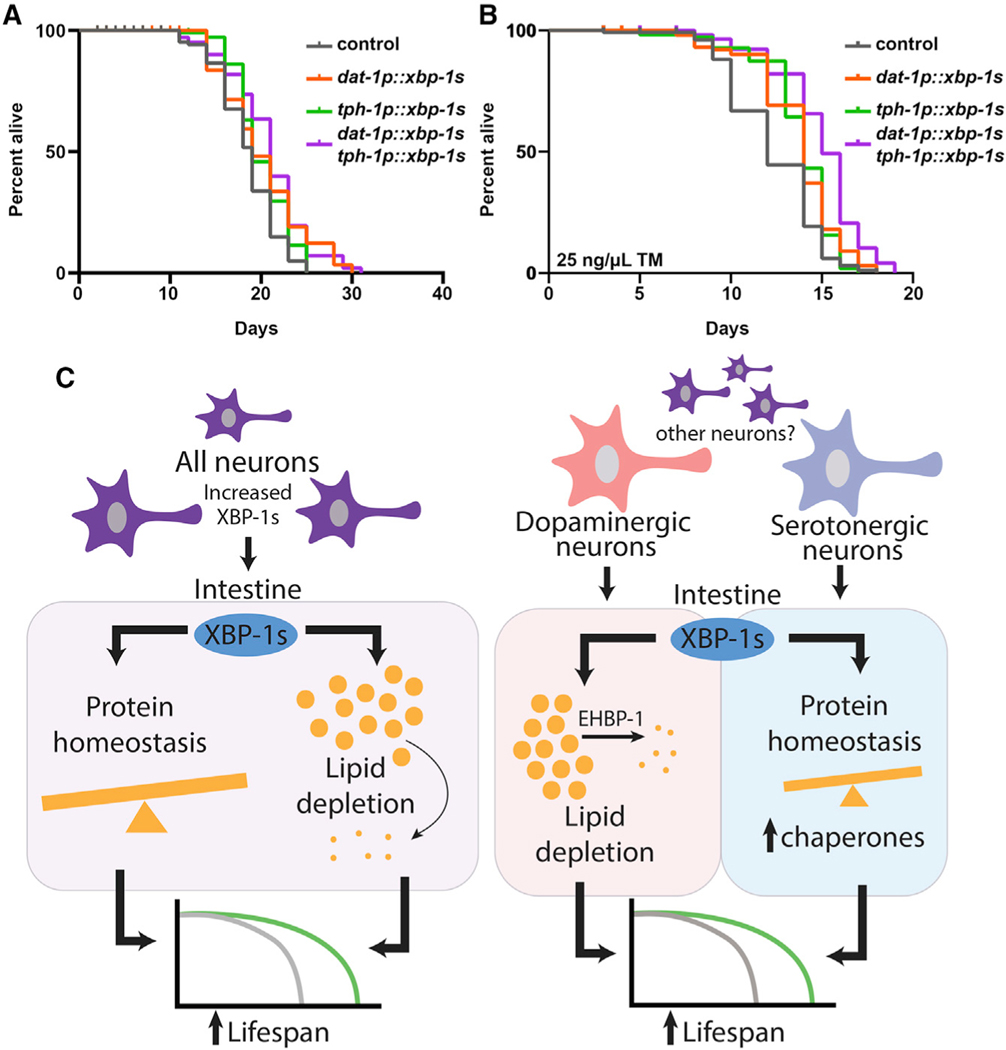
In addition to transcriptional changes, we found that peripheral chaperone induction driven by serotonergic xbp-1s was independent of dopamine production, and mutations in cat-2 had no effect on hsp-4p::GFP induction (Figures S5A and S5B). In addition, the lifespan extension and ER stress resistance found in serotonergic xbp-1s was completely independent of dopamine synthesis (Figures S5C and S5D). Similarly, the lipid depletion found in dopaminergic xbp-1s animals was completely independent of serotonin production, as mutations in tph-1 did not suppress the lipid reduction found in these animals (Figures S6A and S6B). The lifespan extension found in dopaminergic xbp-1s animals was also completely independent of serotonin (Figure S6C). Finally, we found that the lifespan extension and ER stress resistance of dopaminergic and serotonergic xbp-1s are additive (Figures 6A and 6B), providing additional evidence that the two signaling modalities are truly distinct and unique.
Figure 6. Serotonergic and Dopaminergic Neurons Have Independent Roles in Modulating Unique Branches of Non-autonomous UPRER.
(A) Lifespan measurements of control, serotonergic xbp-1s (tph-1p::xbp-1s), dopaminergic xbp-1 s (dat-1p::xbp-1s), and serotonergic xbp-1 s (tph-1p::xbp-1s)/dopaminergic xbp-1s (dat-1p::xbp-1s) animals. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
(B) TM survival assay of control, serotonergic xbp-1s (tph-1p::xbp-1s), dopaminergic xbp-1s (dat-1p::xbp-1s), and serotonergic xbp-1s (tph-1p::xbp-1s)/dopaminergic xbp-1s (dat-1p::xbp-1s) animals. Animals were moved to 25 ng/μL TM plates at day 1 of adulthood. Data are representative of three independent trials. See Table S1–S2 for lifespan statistics.
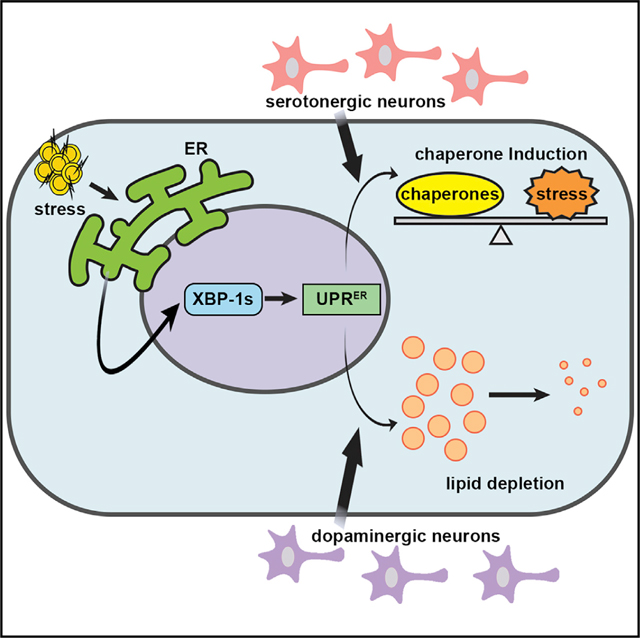
(C) Schematic/model showing pan-neuronal xbp-1s overexpression results in lifespan extension by two independent mechanisms: activation of chaperones to promote protein homeostasis and activation of an EHBP-1-mediated lipophagy machinery to deplete lipids. Serotonergic xbp-1s is sufficient to drive chaperone induction to ER stress resistance and extend lifespan but fails to activate EHBP-1-mediated lipophagy. In contrast, dopaminergic xbp-1s drives depletion of lipids through EHBP-1 but fails to activate chaperones to promote protein homeostasis. These divergent signals are independent and together impart the beneficial impact of pan-neuronal overexpression of xbp-1s on whole-animal physiology.
DISCUSSION
Neuronal transmission of stress signals has been identified in multiple stress response pathways (Durieux et al., 2011; Prahlad et al., 2008; Taylor and Dillin, 2013). For the UPRER, recent work has identified that two distinct signaling modalities exist to affect whole-animal physiology and lifespan: first, chaperone induction drives protein homeostasis; and second, enhanced lipophagy by EHBP-1 drives lipid depletion. These systems were independent of each other and equally important for driving the beneficial effects of neuronal xbp-1s overexpression (Daniele et al., 2020). Here, we found that these paradigms diverge at the source of the neurons that drive the ER stress signal: serotonergic xbp-1s drives non-autonomous activation of chaperones to promote protein homeostasis and dopaminergic xbp-1s drives non-autonomous activation of lipophagy to promote lipid homeostasis. These two divergent signals are independently sufficient to drive beneficial effects on lifespan and together make up at least part of the neuronal circuitry involved in pan-neuronal xbp-1s signaling (Figure 6C). Interestingly, we found that the combination of serotonergic and dopaminergic xbp-1s caused an egg-laying defect, which was more severe than is found in pan-neuronal xbp-1s animals. Moreover, the transcriptional landscape of the combination was significantly different from the pan-neuronal xbp-1s animals. Thus, it is clear that the combination of the two individual signaling paradigms is not completely equivalent to pan-neuronal xbp-1s, despite that they show an additive phenotype for lifespan and resistance to ER stress. This may be due to differences in dosage or the requirement to mobilize other neuronal subtypes to fully phenocopy pan-neuronal xbp-1s.
Because we use RNAi in our experiments, it is possible that xbp-1 RNAi is directly affecting our neuronal xbp-1s expression, despite neurons being refractory to RNAi. Therefore, more stringent tissue-specific studies are required for more conclusive evidence that xbp-1s is required directly in the periphery, but our data strongly suggest this is the case. Thus, it would be interesting to determine how two distinct signals can exist downstream of a single transcription factor, XBP-1s, solely based on which neuron elicits the non-autonomous stress signal. It is possible that based on which neuronal signal is received, peripheral tissues (e.g., the intestine), modify their response by recruiting variable cofactors for xbp-1s to drive a unique transcriptional program. Our work also invites the intriguing possibility of the existence of other neuronal populations involved (e.g., glutaminergic and GABAergic), which either work in concert with—or independent of—the serotonergic and dopaminergic circuits described. Indeed, our preliminary screens correlate glutamatergic and GABAergic neurons in chaperone induction and glutamatergic, GABAergic, and octopaminergic neurons in lipid metabolism. Although these neurons were not further investigated in this study due to their pleiotropic phenotypes, including severe motor defects, it is possible that these circuits play an additional role in signaling ER stress. More importantly, it is possible that one of these circuits—or some other neuronal circuit not studied here—feeds into the serotonergic circuit. We found that although serotonin is necessary for non-autonomous induction of protein homeostasis, it was not sufficient, as increasing serotonin signaling by mutation of the serotonin reuptake receptor had no effect on UPRER. Moreover, the suppression of chaperone induction in pan-neuronal xbp-1s by mutation of serotonin biosynthesis was not complete, suggesting an attractive hypothesis that some other neurotransmitter is responsible for the remainder of the signaling. Finally, serotonergic xbp-1s itself, although sufficient to induce chaperones, created a bimodal population. Taken together, these data clearly indicate the complexity of serotonin-mediated non-autonomous signaling, and further dissection of all neurons and their neurotransmitters is required to properly map the neuronal circuitry involved.
It is still unclear how serotonergic xbp-1s and dopaminergic xbp-1s signal two unique events. One possible reason is that increased xbp-1s in neurons non-specifically promotes the release of neurotransmitters, potentially by increasing release of SCVs, resulting in increased release of serotonin by serotonergic neurons and dopamine by dopaminergic neurons. Ultimately, it is the neurotransmitter itself that drives unique responses, for which dopamine promotes lipid depletion and serotonin—likely with other neurotransmitters—promotes protein homeostasis. Indeed, we found that increased dopamine signaling by mutations of the dopamine reuptake receptor decreased lipid levels in an xbp-1-dependent manner.
Another intriguing phenomenon from this study was the opposing role of serotonergic and dopaminergic signaling on lipid homeostasis. Although dopaminergic xbp-1s promoted lipid depletion, which had direct implications on organismal health, serotonergic xbp-1s resulted in increased levels of lipid droplets. One possible explanation is that downstream of serotonin signaling, XBP-1 s is recruited primarily to the promoters of genes involved in protein homeostasis. This may titrate XBP-1 s away from genes involved in lipid homeostasis, which may result in a global increase in lipid levels. Indeed, we found that fat-7 transcript levels, which increase in both pan-neuronal and dopaminergic xbp-1s, were decreased in serotonergic xbp-1s. Finally, it is interesting that although dopaminergic xbp-1s animals fail to induce chaperones, they exhibit increased resistance to tunicamycin, a drug that causes ER stress by blocking N-linked glycosylation and producing protein misfolding stress. Consistent with these findings, two recent studies showed that promoting lipid homeostasis by oleic acid supplementation or hyperactivation of lysosomal lipases was sufficient to drive protein homeostasis by promoting clearance of aggregation-prone proteins, despite not directly promoting chaperone induction (Imanikia et al., 2019a, 2019b). Thus, it is possible that promoting lipid homeostasis is sufficient to drive protein homeostasis in the absence of chaperones in the periphery. Still to be determined is whether this is due to a causative and direct impact of lipids on protein homeostasis or an indirect effect for which there is a decreased burden on ER, such that quality-control machinery can be mobilized strictly to regulate protein homeostasis.
Overall, the major finding that there exists two distinct and independent mechanisms of UPRER and that they are separable at the source of the neurons involved in signaling these effects is intriguing. Although this phenomenon is described in C. elegans, non-autonomous UPRER transmission is well-described in animal models, such as from the brain to the liver in mice (Brandt et al., 2018; Williams et al., 2014). Moreover, dopamine signaling has direct implications on lipid homeostasis and overall metabolism in higher eukaryotes (Folgueira et al., 2019; Fulton and Alquier, 2019; Leite and Ribeiro, 2020), which invites the attractive hypothesis that dopamine and serotonin signaling may play critical roles in diverging UPRER signals in higher eukaryotes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Please contact Andrew Dillin (dillin@berkely.edu), corresponding author, for any requests.
Materials Availability
All strains are available either at the Caenorhabditis Genome Center (CGC) or by request to the lead contact.
Data and Code Availability
All data is available in the main text or the supplementary materials. Raw sequencing data is available through ArrayExpress Annotaire 2.0, accession number: E-MTAB-9771. All other data can be requested from the lead contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains and maintenance
All C. elegans strains are derivatives of the Bristol N2 strain from Caenorhabditis Genetics Center (CGC) and are listed in Key Resources Table. For standard maintenance of worms, animals are grown on standard nematode growth media (NGM) plates and fed OP50 E. coli B bacteria at 15°C, for a maximum of 25 generations. For all experiments, animals are synchronized using a standard bleaching protocol where carcasses are degraded using a bleach solution (1.8% sodium hypochlorite, 0.375M KOH), followed by 3 washed with M9 solution (22 mM KH2PO4 monobasic, 42.3 mM Na2HPO4, 85.6 mM NaCl, 1 mM MgSO4), followed by L1 arresting synchronization, which is performed by floating eggs in M9 overnight in a 20°C incubator on a rotator for a maximum of 16 hours. L1 arresting was performed to better age-match animals, as some mutants and transgenic animals used in this study exhibit some variability in growth rates. All experiments were performed without L1-arresting at least once to ensure that there were no artifacts produced from L1-arresting. Synchronized animals were plated on RNAi bacteria (NGM + 1 μM IPTG and 100 μg/mL carbenicillin; HT115 E. coli K strain containing pL4440 vector control or pL4440 with RNAi of interest) until the desired stage of adulthood. All RNAi constructs used in this study were isolated from the Vidal library or previously characterized (Daniele et al., 2020).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
|
| ||
| OP50 | CGC | N/A |
| HT115 | CGC | N/A |
| DH5α | Invitrogen | N/A |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| (+)-5-Fluorodeoxyuridine (FUDR) | Spectrum Chemical | F2026-10GMBL |
| Agarose, low melting | Sigma-Aldrich | A9414-10G |
| Alexa Fluor 488 Phalloidin | Life Technologies | A12380 |
| Bacto Peptone | Fisher Scientific | DF0118072 |
| BD Difco granulated agar | VWR | 90000-782 |
| Calcium chloride dihydrate | VWR | 97061-904 |
| Carbenicillin | BioPioneer | C0051-25 |
| Chloroform | Sigma-Aldrich | 34854 |
| Cholesterol | Sigma-Aldrich | 57-88-5 |
| IPTG dioxane free | Denville Scientific | CI8280-4 |
| Isopropanol | Fisher Scientific | AC327272500 |
| LB Broth Miller | Fisher Scientific | BP1426500 |
| Magnesium sulfate heptahydrate | VWR | EM-MX0070-3 |
| Potassium phosphate dibasic | VWR | EM-PX1570-2 |
| Potassium phosphate monobasic | VWR | EM-PX1565-5 |
| Sodium Chloride | EMD Millipore | SX0420-5 |
| Sodium phosphate dibasic | VWR | 71003-472 |
| Tetracycline hydrochloride | Sigma-Aldrich | T7660-5G |
| Trizol | Fisher Scientific | 15596018 |
| Tunicamycin | Millipore | 654380 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| QIAquick PCR Purification Kit | QIAGEN | 28106 |
| QIAprep Spin Miniprep Kit | QIAGEN | 27106 |
|
| ||
| Deposited Data | ||
|
| ||
| RNA-seq | Annotare 2.0 ArrayExpress | E-MTAB-9771 |
|
| ||
| Experimental Models: Organisms/Strains | ||
|
| ||
| C. elegans: Bristol (N2) strain as wild type (WT) | CGC | N2 |
| C. elegans: AGD927: uthIs270 [rab-3p::Xbp-1 s, myo-2p::tdTomato] | Taylor and Dillin, 2013 | N/A |
| C. elegans: AGD928: uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP] | Taylor and Dillin, 2013 | N/A |
| C. elegans: AGD1237: N2, unc-30(ok613) IV; uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP] | This study | N/A |
| C. elegans: AGD1497: N2, uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP];tbh1(n3247) | This study | N/A |
| C. elegans: AGD1498: N2, uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP]; ttx-3(ks5) | This study | N/A |
| C. elegans: AGD1500: N2, uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP]; tph1(mg280) | This study | N/A |
| C. elegans: AGD1526: N2, uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP] V; cat2(e1112) | This study | N/A |
| C. elegans: AGD1767: N2, uthIs460[dat-1p::Xbp-1 s, myo-2p::tdTomato] | This study | N/A |
| C. elegans: AGD1771: N2, uthIs464[rgef-1p::Xbp-1 s, myo-2p::tdTomato] | This study | N/A |
| C. elegans: AGD2048: dhs-3p::dhs-3::GFP; hjSi158 [vha-6p::SEL-1(1-79)::mCherry::HDELlet-858 3′ UTR] | Daniele et al., 2020 | N/A |
| C. elegans: AGD2049: dat-1(ok157), dhs-3p::dhs-3::GFP; hjSi158[vha-6p::SEL-1(1-79)::mCherry::HDELlet-858 3′ UTR] | This study | N/A |
| C. elegans: AGD2064: N2, uthIs460[dat-1p::Xbp-1 s, myo-2p::tdTomato](#Aa), dhs-3p::dhs-3::GFP; hjSi158[vha-6p::SEL-1(1-79)::mCherry::HDELlet-858 3′ UTR] | This study | N/A |
| C. elegans: AGD2065: N2, dhs-3p::dhs-3::GFP; hjSi158 [vha-6p::SEL-1(1-79)::mCherry::HDELlet-858 3′ UTR]; uthIs270 [rab-3p::Xbp-1 s, myo-2p::tdTomato] | Daniele et al., 2020 | N/A |
| C. elegans: AGD2149: N2, uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP]; dop1(vs101) | This study | N/A |
| C. elegans: AGD2207: N2, cat-2(n4547) II; zcls4 [hsp-4p::GFP]V | This study | N/A |
| C. elegans: AGD2209: N2, cat-2(n4547) II; dhs-3p::dhs-3::GFP; hjSi158[vha-6p::SEL-1(1-79)::mCherry::HDELlet-858 3′ UTR] | This study | N/A |
| C. elegans: AGD2147: N2, uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP]; mec-3(e1338)IV | This study | N/A |
| C. elegans: AGD2155: N2, uthIs270[rab-3p::Xbp-1 s, myo-2p::tdTomato]; zcIs4[hsp-4p::GFP]; unc-43(n498)IV | This study | N/A |
| C. elegans: AGD2164: N2, uthIs460[dat-1p::xbp-1 s, myo-2p::tdTomato](#Aa); cat-2(n4547) | This study | N/A |
| C. elegans: AGD2275: N2, uthIs460[dat-1p::Xbp-1 s, myo-2p::tdTomato](#Aa); zcls4(hsp-4p::GFP)V | This study | N/A |
| C. elegans: AGD2474: N2, uthIs460[dat-1p::xbp-1 s, myo-2p::tdTomato](#Aa); ldrIs [dhs-3p::dhs-3::GFP + unc-76(+)]; hjSi158[vha-6p::SEL-1(1-79)::mCherry::HDEL::let-858 3′ UTR]; cat-2(n4547) II | This study | N/A |
| C. elegans: AGD2496: N2, tph-1(mg280)II;; zcls4[hsp-4p::GFP]V | This study | This study |
| C. elegans: AGD2503: N2, uthIs501(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line1 | This study | This study |
| C. elegans: AGD2504: N2, uthIs502(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line2 | This study | This study |
| C. elegans: AGD2505: N2, uthIs503(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line3 | This study | This study |
| C. elegans: AGD2506: N2, uthIs501(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) linel; zcls4[hsp-4p::GFP]V | This study | This study |
| C. elegans: AGD2507: N2, uthIs502(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line2; zcls4[hsp-4p::GFP]V | This study | This study |
| C. elegans: AGD2508: N2, uthIs503(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line3; zcls4[hsp-4p::GFP]V | This study | This study |
| C. elegans: AGD2686: N2, uthIs460[dat-1p::xbp-1 s, myo-2p::tdTomato], ldrIs[dhs-3p::dhs-3::GFP + unc-76(+)]; hjSi158[vha-6p::SEL-1(1-79)::mCherry::HDEL::let-858 3′ UTR]; tph-1(mg280)II | This study | This study |
| C. elegans: AGD2695: N2, uthIs460[dat-1p::xbp-1 s, myo-2p::tdTomato]; tph-1(mg280) II | This study | This study |
| C. elegans: AGD2713: N2, uthIs503(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line3; uthIs460[dat-1p::xbp-1 s, myo-2p::tdTomato] | This study | This study |
| C. elegans: AGD2714: N2, uthIs503(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line 3; ldrIs[dhs-3p::dhs-3::GFP + unc-76(+)]; hjSi158 [vha-6p::SEL-1(1-79)::mCherry::HDEL::let-858 3′ UTR]” | This study | This study |
| C. elegans: AGD2715: N2, uthIs503(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) line3; cat-2(n4547) | This study | This study |
| C. elegans: AGD2716: N2, uthIs503(tph-1p::xbp-1 s::unc-54 UTR; myo-2p::GFP) 57E4; tph-1(mg280) | This study | This study |
| C. elegans: AGD2967: N2, mod-5(n3314)I, zcls4[hsp-4p::GFP]V | This study | This study |
| C. elegans: AGD2968: N2, mod-5(n3314)I, ldrIs [dhs-3p::dhs-3::GFP + unc-76(+)]; hjSi158[vha-6p::SEL-1(1-79)::mCherry::HDEL::let-858 3′ UTR] | This study | This study |
| C. elegans: MT15620: tph-1(mg280) | CGC | GR1321 |
| C. elegans: MT15620: cat-2(n4547) | CGC | MT15620 |
| C. elegans: RM2702: dat-1(ok157) | CGC | RM2702 |
| C. elegans: SJ4005: zcls4(hsp-4p::GFP)V | Taylor and Dillin, 2013 | SJ4005 |
|
| ||
| Oligonucleotides | ||
|
| ||
| RHS427 xbp-1T qPCR F | cacctccatcaacaacaacat | Forward primer to qPCR xbp-1 total |
| RHS428 xbp-1T qPCR R | aaccgtctgctccttcctcaa | Reverse primer to qPCR xbp-1 total |
| RHS429 xbp-1 s qPCR F | cgtgcctttgaatcagcagtg | Forward primer to qPCR xbp-1 spliced |
| RHS430 xbp-1 s qPCR R | cgaggtgtccatcttcttgtt | Reverse primer to qPCR xbp-1 spliced |
| RHS433 crt-1 qPCR F | atgacgagatggacggagaat | Forward primer to qPCR crt-1 |
| RHS434 crt-1 qPCR R | ctgacttgacctgccacaaat | Reverse primer to qPCR crt-1 |
| RHS435 T14G8.3 qPCF F | gccagtggagccaaaagcaaa | Forward primer to qPCR T14G8.3 |
| RHS436 T14G8.3 qPCR R | ccaagcggttcatagcctctt | Reverse primer to qPCR T14G8.3 |
| RHS473 hsp4 qPCR V2 F | GAATCAACCCTGACGAAGCAG | Forward primer to qPCR hsp-4 |
| RHS474 hsp4 qPCR V2 R | CCTCCGACAGTCTCAATACCC | Reverse primer to qPCR hsp-4 |
| RHS475 ehbp-1 qPCR F | GATCCGTGAAACAGAAGTTGG | forward primer to qPCR ehbp-1 |
| RHS476 ehbp-1 qPCR R | CCCAATTGGTCAATTTCCGAA | reverse primer to qpCR ehbp-1 |
| RHS682 fat-6 qPCR F | TCTACCAGCTCATCTTCGAGGC | forward primer to qPCR fat-6 |
| RHS683 fat-6 qPCR R | GATCACGAG CCCATTCGATGAC | reverse primer to qPCR fat-6 |
| RHS684 fat-7 qPCR F | GGAAGGAGACAGCATTCATTGCG | forward primer to qPCR fat-7 |
| RHS685 fat-7 qPCR R | GTCTTGTGGGAATGTGTGGTGG | reverse primer to qPCR fat-7 |
| RHS529 tphlp-xbpis F | AGGTCGACTCTAGAGGATCC cgcgaattgcggccgacata |
forward primer to PCR tph-1p |
| RHS530 tph1p-xbp1s R | CGTTTTGGATAGTTGCTCAT atgattgaagagagcaatgctac |
reverse primer to PCR tph-1p |
| RHS531 tph1p-xbp1s F2 | gcattgctctcttcaatcatATGAGCAACTATC CAAAACGTATTTA |
Forward primer to PCR xbp-1 s::UTR + vector from pAF18 (3) |
| RHS532 tph1p-xbp1s R2 | tatgtcggccgcaattcgcgGGATCCTCTAG AGTCGACCTG |
reverse primer to PCR xbp-1 s::UTR + vector from pAF18 |
| VR1 | CTGCAGaatgtttctagtcgtttttg | Forward primer to PCR dat-1p from gDNA |
| VR2 | tttttatgggttttggtaggCCCGGG | Reverse primer to PCR dat-1p from gDNA |
| VR3 | CCCGGGATGAGCAACTATCCAAAACG | Forward primer to PCR xbp-1 s::UTR + vector from pAF18 (3) |
| VR4 | TGCAGGCATGCAAGCTTCTGCAG | Reverse primer to PCR xbp-1 s::UTR + vector from pAF18 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pRHS55: tph-1p::xbp-1 scDNA::unc-54 3′ UTR | This study | N/A |
| pVR1: dat-1p::xbp-1 s cDNA::unc-54 3′ UTR | This study | N/A |
| pRT5: pAD1 rgef-1p::xbp-1 s cDNA::unc-54 3′ UTR | Taylor and Dillin, 2013 | N/A |
| pEK1 : myo-2p::GFP::unc-54 3′UTR | This study | N/A |
| pEK2myo-2p::tdtomato::unc-52 3′ UTR | This study | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| Zen 2 Blue Edition | Zeiss | N/A - download available from Zeiss website |
| LASX | Leica | N/A - download available from Leica website |
Transgene integration was performed by injecting N2 worms with pVR1 (dat-1p::xbp-1 s) or pRHS55 (tph-1p::xbp-1 s) at 25 ng/μL, pEK2 (myo-2p::tdtomato) for pVR1 or pEK1 (myo-2p::GFP) for pRHS55 at 2.5 ng/μL and 100 ng/μL of pD64 vehicle as filler DNA. Worms positive for fluorescent pharynx were selected to identify for stable arrays. Integration was performed by gamma irradiation. Briefly, L4 worms were irradiated with 4000–4400 rems of radiation and integrants were identified by selecting animals that maintained fluorescent pharynxes at 100% frequency in the F3 generation. Multiple independent lines were isolated, backcrossed to N2 animals 8x to eliminate mutations, and animals with the most similar phenotypes to the array animals were used for further experimentation.
pVR1 (cloned into pre-existing pRT5 plasmid using PstI/XmaI restriction cloning, replacing rgef-1p for dat-1p) dat-1p
aatgtttctagtcgtttttgtattttaaagcacattcccaattctgaatacttttctgaaatccatgaaatggaacttgaatccagttttcactaaaacgacctcatacactttctctcgtatcctcaaaatatctatgacattatcattagcttcgctagtttcatttctttcaaatattatgcattcttaaattccgatacccgcgtgcaaaagtgctctattgagcaactttgggatcatatgtacacaccaatgcccttttcccaaatcttttcctgtccttttctctaaaaacaataaatccatgcctattccagtatgacccctttgaagcagatataatcgcacaaacatatacacatagctcggataaatgtagaaaaagaagaaaagaagtataagtagatagatgctttccggcaattatccaccgcaccgtagtcttcaccaactgagactgcgtcgttaggagacgccgacatgattcagaagcagaatttggaagaaaaacgacgatgatattgaggctggcacacatacaccggaatattcgacatgccaccacatctagattccaaggcaatctctacctcttcccattctttcggtttttttgttctgacaagaaaagtggatagctacgggctcaatgagctgattttatttttaaatatcttaaaactatactagattcatgtgttttcaggtccatattccaaattagtcgaaaagctgatcccgctacggtttactcgaatctcaacaatttttagcc
xbp-1 s
ATGAGCAACTATCCAAAACGTATTTATGTGCTCCCAGCACGCCACGTGGCAGCGCCACAGCCTCAGAGAATGGCTCCCAAGCGTGCACTTCCAACAGAACAAGTTGTCGCACAACTTCTTGGCGATGATATGGGACCATCTGGGCCACGCAAAAGAGAACGACTGAATCATTTGAGTCAGGAGGAGAAAATGGATCGTCGGAAACTTAAAAATCGAGTCGCAGCCCAAAATGCTAGAGACAAAAAGAAGGAAAGATCAGCAAAGATCGAGGATGTGATGCGCGATCTGGTGGAGGAGAACCGCCGGCTCCGCGCTGAAAACGAACGTCTTCGCCGTCAAAATAAAAATCTTATGAACCAGCAGAACGAGTCCGTCATGTATATGGAAGAGAACAACGAAAACTTGATGAACAGCAATGATGCATGCATCTACCAGAACGTCGTCTACGAAGAAGAAGTCGTCGGTGAGGTTGCACCAGTTGTCGTCGTCGGAGGAGAGGATCGCCGTGCCTTTGAATCAGCAGTGGGAACAGGCCCGATCCACCTCCATCAACAACAACATCAGCAACCAACTCCGTCGTATGGATTCCAAGAAGAACAACACAATCAGTGTGGATATGTATCTAACTATCATCTCGATTCTATGCAACCACATGGATCGCAACAAGAAGATGGACACCTCGAACAAATCCTCGAACATCTCAAGAGCCCAAGCGGAGAGTTCGATCGATTCGTTGCTGGCTACATTGAGGAAGGAGCAGACGGTTATGCAGCGTCTTGTTCAAGCGGATCCATGTACACATCTTCAGAAACGCGTGAAACACTTTCGCCGAATTCCCTAGCCATGTCCCCGTCGATGAGCAGCTCGAGCACTGACTGGGATGATGAGCTTTTGGGATGTGGAACCGAAACTGGAACTGGAACCGACGAGCTGCTTACCGA CCCCGGAAACTGGAACTTTGAAACTTTCGACGAAAATTCAATCGACCTAAATTTCTTCCAAAATTAA
unc-54 3′ UTR
CATCTCGCGCCCGTGCCTCTGACTTCTAAGTCCAATTACTCTTCAACATCCCTACATGCTCTTTCTCCCTGTGCTCCCACCCCCTATTTTTGTTATTATCAAAAAACTTCTCTTAATTTCTTTGTTTTTTAGCTTCTTTTAAGTCACCTCTAACAATGAAATTGTGTAGATTCAAAAATAGAATTAATTCGTAATAAAAAGTCGAAAAAAATTGTGCTCCCTCCCCCCATTAATAATAATTCTATCCCAAAATCTACACAATGTTCTGTGTACACTTCTTATGTTTTTTACTTCTGATAAATTTTTTTGAAACATCATAGAAAAAACCGCACACAAAATACCTTATCATATGTTACGTTTCAGTTTATGACCGCAATTTTTATTTCTTCGCACGTCTGGGCCTCTCATGACGTCAAATCATGCTCATCGTGAAAAAGTTTTGGAGTATTTTTGGAATTTTTCAATCAAGTGAAAGTTTATGAAATTAATTTTCCTGCTTTTGCTTTTTGGGGTTTCCCCTATTGTTTGTCAAGATTTCGAGGACGGCGTTTTTCTTGCTAAAATCACAAGTATTGATGAGCACGATGCAAGAAAGATCGGAAGAAGGTTTGGGTTTGAGG CTCAGTGGAAG
pRHS55 (cloned into pre-existing pRT5 plasmid using gibson assembly to replace rgef-1p with tph-1p) tph-1p
cgcgaattgcggccgacatattgaaagaatgaaatataaattacgttttttgaataaaaaattttaaaatggtaactagaatataaacgttcaaatttagaacacgtattaaacataaatttgattgaaaaatttttgaaatcaaattcaatttcttcgaaaaacttgaataaccaaaaaaaaagtttttgaaaacatttgttcaaatattgtagatattgtagaaattggttttcacacagctttgggtattcagttttcgtgttccaacaagaaaataatgaaatcttcaagagatgaaatttggatttttacaaaaacacggtctgtaacttggctgaaaatgaaaataattaataaaattgataaaatattgcaaaatatttcacagttactattcaacattattttgctattaaaattcaaaagatgttgatttatgactagtacatcagaacagttttcaaaaattttgatggtttttgggagaatgccttttactctgtgtaactcaaaaaataattggcaattgtttgtaataaaaagtatttcaaaagaaagaaaaattgtttcttatgattttatcagttgatcattcgttttctagacatgtatttttctagatatgctagtttgaatctacacaatttagtgagtttgaatttagattccagcgatatgcaatgattttgaagttaatgagtatttgcctgtttttactaggtcattccaaaaatttcaactgcagaactcaacactgaaccttaaggtaaagcgaaatctgaacttttttttgctagtttgagtttttattttaagagatcgaaaaattttccttgaactttgaaacatgacaattcaaacttttcaaatgtagtaagctccgatgcgttcccgttcattattcttcttcaataaattcgaaatctgacatcattctcatcttttcccatcatcacaagccgtgggctcatttattctcccacggaaaccatgacagcaaaaataaatagagtggcgccttattcgactcatttcgtttttttttctccggatattagattgtgtggcaggcggctccattgtatattacgtgccgaattccagaagcaccacgccatcggatatctaaaagaggaggtgtctttgtttgcgcataataaaacaatcaatcaacacagcaaagacccctctcaacctcatttcatgattttctttggtttttaggtagcattgctctcttcaatcat
xbp-1 s
ATGAGCAACTATCCAAAACGTATTTATGTGCTCCCAGCACGCCACGTGGCAGCGCCACAGCCTCAGAGAATGGCTCCCAAGCGTGCACTTCCAACAGAACAAGTTGTCGCACAACTTCTTGGCGATGATATGGGACCATCTGGGCCACGCAAAAGAGAACGACTGAATCATTTGAGTCAGGAGGAGAAAATGGATCGTCGGAAACTTAAAAATCGAGTCGCAGCCCAAAATGCTAGAGACAAAAAGAAGGAAAGATCAGCAAAGATCGAGGATGTGATGCGCGATCTGGTGGAGGAGAACCGCCGGCTCCGCGCTGAAAACGAACGTCTTCGCCGTCAAAATAAAAATCTTATGAACCAGCAGAACGAGTCCGTCATGTATATGGAAGAGAACAACGAAAACTTGATGAACAGCAATGATGCATGCATCTACCAGAACGTCGTCTACGAAGAAGAAGTCGTCGGTGAGGTTGCACCAGTTGTCGTCGTCGGAGGAGAGGATCGCCGTGCCTTTGAATCAGCAGTGGGAACAGGCCCGATCCACCTCCATCAACAACAACATCAGCAACCAACTCCGTCGTATGGATTCCAAGAAGAACAACACAATCAGTGTGGATATGTATCTAACTATCATCTCGATTCTATGCAACCACATGGATCGCAACAAGAAGATGGACACCTCGAACAAATCCTCGAACATCTCAAGAGCCCAAGCGGAGAGTTCGATCGATTCGTTGCTGGCTACATTGAGGAAGGAGCAGACGGTTATGCAGCGTCTTGTTCAAGCGGATCCATGTACACATCTTCAGAAACGCGTGAAACACTTTCGCCGAATTCCCTAGCCATGTCCCCGTCGATGAGCAGCTCGAGCACTGACTGGGATGATGAGCTTTTGGGATGTGGAACCGAAACTGGAACTGGAACCGACGAGCTGCTTACC GACCCCGGAAACTGGAACTTTGAAACTTTCGACGAAAATTCAATCGACCTAAATTTCTTCCAAAATTAA
unc-54 3′ UTR
CATCTCGCGCCCGTGCCTCTGACTTCTAAGTCCAATTACTCTTCAACATCCCTACATGCTCTTTCTCCCTGTGCTCCCACCCCCTATTTTTGTTATTATCAAAAAACTTCTCTTAATTTCTTTGTTTTTTAGCTTCTTTTAAGTCACCTCTAACAATGAAATTGTGTAGATTCAAAAATAGAATTAATTCGTAATAAAAAGTCGAAAAAAATTGTGCTCCCTCCCCCCATTAATAATAATTCTATCCCAAAATCTACACAATGTTCTGTGTACACTTCTTATGTTTTTTACTTCTGATAAATTTTTTTGAAACATCATAGAAAAAACCGCACACAAAATACCTTATCATATGTTACGTTTCAGTTTATGACCGCAATTTTTATTTCTTCGCACGTCTGGGCCTCTCATGACGTCAAATCATGCTCATCGTGAAAAAGTTTTGGAGTATTTTTGGAATTTTTCAATCAAGTGAAAGTTTATGAAATTAATTTTCCTGCTTTTGCTTTTTGGGGTTTCCCCTATTGTTTGTCAAGATTTCGAGGACGGCGTTTTTCTTGCTAAAATCACAAGTATTGATGAGCACGATGCAAGAAAGATCGGAAGAAGGTTTGGGTTTGAGGCTCAGTGGAAG
METHOD DETAILS
Stereomicroscopy for fluorescent transcriptional reporters
Fluorescent animals were grown on standard RNAi plates as described above until day 1 of adulthood. Animals were moved onto standard NGM plates containing 5 μL of 100 mM sodium azide to paralyze worms. Paralyzed worms were lined up and imaged immediately on a Leica M250FA automated fluorescent stereomicroscope equipped with a Leica DFC3000G camera, standard GFP filter, and driven by LAS-X software.
Lifespan and survival analysis
Lifespan measurements were performed on solid NGM plates with RNAi bacteria. Worms were synchronized as described above, and adult animals were moved away from their progeny daily onto fresh RNAi plates until no progeny were visible (~7–10 days). Animals were then scored every 1–2 days for death until all animals are scored. Animals with bagging, vulval explosion, or other ageunrelated deaths were censored and removed from quantification. For tunicamycin survival assays, animals are moved to plates containing 25 ng/μL tunicamycin at day 1 of adulthood. Animals are not proved for the duration of the experiment, as no progeny survive at this concentration of tunicamycin, and drug efficacy was not visibly reduced during the duration of the experiment. Animals are scored daily for tunicamycin survival for better dynamic range.
Oil Red O Staining
Staining of lipids using Oil Red O was performed as previously described (18). Briefly, day 1 adult animals were washed off plates with PBST (0.02% Tween 20) and washed three times in PBST to remove bacteria. Animals were fixed in 40% isopropanol for 3 minutes at room temperature, with proper mixing on a rotator. Animals were then stained with Oil Red O at a final concentration of 1% w/v in 60% isopropanol (Oil Red O solution was prepared by dissolving Oil Red O at 2% w/v in 100% isopropanol, then diluting into water to 60% and allowing this solution to mix by rocking on a nutator for 2 hours at room temperature, then filtering Oil Red O through a 0.2 μm cellulose sterile syringe filter. 600 μL was added to a 100 μL suspension of worms for a final concentration of 1% w/v). Staining was performed for 2 hours on a rotator at 20°C, then animals were washed by removing Oil Red O solution up to 100 μL, then adding in 600 μL of PBST and rotating for another 30 minutes at 20°C. Animals were then imaged on a Leica M250FA automated fluorescent stereomicroscope equipped with a Leica DFC310 FX color camera and driven by LAS-X software.
qPCR
Worms were grown on standard NGM plates with empty vector bacteria on solid agar from hatch at 20°C until D1. Worms were collected by washing off with M9. M9 was subsequently aspirated, replaced with trizol, and worms were freeze/thawed 3x with liquid nitrogen. After the final thaw, chloroform was added at a 1:5 ratio with chloroform:trizol volume for aqueous separation of RNA, which was performed via centrifugation in heavy gel phase-lock tubes. The aqueous phase was transferred into isopropanol, then RNA purification was performed using a QIAGEN RNeasy Mini Kit (74106) as per manufacturer’s directions. 2 μg of RNA was used for cDNA synthesis using the QIAGEN QuantiTect Reverse Transcriptase kit (205314) as per manufacturer’s directions. qPCR analysis was performed using a general standard curve protocol using SYBR-green. Four technical replicates were performed for three independent biological replicates per sample. All primers are listed in Key Resources Table.
RNA-sequencing
Animals were synchronized and grown to D1 on EV RNAi plates. ~2,000 animals were harvested using M9. M9 was subsequently aspirated, replaced with trizol, and worms were freeze/thawed 3x with liquid nitrogen. After the final thaw, chloroform was added at a 1:5 ratio of chloroform:trizol for aqueous separation of RNA using heavy gel phase-lock tubes (VWR, 10847–802). The aqueous phase was mixed with isopropanol and RNA purification was performed using a QIAGEN RNeasy Mini Kit (74106) as per manufacturer’s directions. Library preparation, RNA-sequencing, and analysis was performed by Genewiz. Briefly, library preparation was performed with Poly A selection and HiSeq Sequencing using NEBNext Ultra RNA Library Prep Kit for Illumina following manufacturer’s instructions (NEB, Ipswich, MA, USA). Sequencing libraries were clustered onto 1 lane of a flowcell, loaded on Illumina HiSeq instrument (4000 or equivalent) and samples were sequenced using 2×150bp Paired End configuration. Base calling was conducted on HiSeq Control Software. Raw reads were trimmed to remove adapters and nucleotides with poor quality using Trimmomatic v.0.36, then mapped to the reference genome on ENSEMBL using STAR aligner v.1.5.2, counting genes that fell within exon regions. DESeq2 was used for differential expression analysis across groups of samples (three biological replicates per sample). p values were generated using the Wald test and adjusted using the Benjamini-Hochberg procedure.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical information is presented in every figure legend, including sample size, number of replicates, and statistical analysis used. Detailed protocols for quantification and analysis are also provided below.
Stereomicroscopy
Quantification of screening data of hsp-4p::GFP animals were performed on ImageJ. Integrated fluorescent intensity measurements were made along the bottom portion of the animal starting at the lower spermathecal to the tail. Data is represented as integrated intensity normalized to the area of the region of interest drawn.
Oil Red O
Quantification was performed on ImageJ. Images were first stacked to RGB (Image = > Color = > Stack to RGB), then converted to a 32 bit black and white image (Image = > Type = > 32 bit). Images were then inverted, so that Oil Red O staining was recognized as the brightest points. Integrated density was measured across the anterior and posterior portions of the worm, ignoring the region where eggs were present. Anterior and posterior sections were first normalized to the area of the region of interest (ROI), then averaged to calculate integrated Oil Red O values.
Large particle biosorter analysis
For large-scale quantification of fluorescent animals, a Union Biometrica complex object parameter analysis sorter (COPAS) was used (Bar-Ziv et al., 2020). Fluorescent animals were washed off plates using M9, and run through the COPAS biosort using a 488 nm light source. Integrated fluorescent intensity was normalized to the time of flight of the animal, then normalized to the extinction to correct for both worm length and worm thickness. For experiments including transgenic animals with myo-2p::GFP co-injection markers, signal is only collected in the bottom 60% of animals to remove signal from the head. All biosorter data is represented dot-line plot, where the middle line represents median and the two outer lines represent interquartile range (the 25th and 75th percentile), and each dot represents a single animal. All experiments are performed at least three independent times, and statistics are calculated using Prism7 software and non-parametric Mann-Whitney testing.
Lifespan and survival
All survival data is represented as a Kaplan-Meier curve, and statistics are calculated using Log-Rank testing using Prism7 software. All experiments are performed at least three times, with all experiments performed by at least two independent researchers, with all conditions blinded each time
qPCR
Standard Student’s t test was used for qPCR data.
RNA-seq
A proprietary pipeline was used by Genewiz to calculate p values provided in Tables S3 and S4. p values were generated using the Wald test and adjusted using the Benjamini-Hochberg procedure.
Supplementary Material
Highlights.
Non-autonomous signaling of UPRER is mediated by serotonergic and dopaminergic neurons
Serotonergic neurons drive protein homeostasis to extend lifespan
Dopaminergic neurons drive lipid homeostasis to extend lifespan
ACKNOWLEDGMENTS
We are grateful to all the members of the Dillin lab for intellectual and technical support, especially Melissa Sanchez and Anel Esquivel. This work was supported by grant 1K99AG065200-01A1 through the National Institute on Aging (NIA) to R.H.-S., the Bert Lubin Scholarship Fund through CHORI to M.d.l.R.R., and 4R01AG042679-04 through the NIA and the Howard Hughes Medical Institute to A.D. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108489.
REFERENCES
- Barros A.G.d.A., Bridi JC, de Souza BR, de Castro Júnior, de Lima Torres KC, Malard L, Jorio A, de Miranda DM, Ashrafi K, and Romano-Silva MA (2014). Dopamine signaling regulates fat content through β-oxidation in Caenorhabditis elegans. PLoS One 9, e85874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Ziv R, Frakes AE, Higuchi-Sanabria R, Bolas T, Frankino PA, Gildea HK, Metcalf MG, and Dillin A. (2020). Measurements of Physiological Stress Responses in C. Elegans. Journal of visualized experiments : JoVE. 10.3791/61001. [DOI] [PMC free article] [PubMed]
- Berendzen KM, Durieux J, Shao L-W, Tian Y, Kim H-E, Wolff S, Liu Y, and Dillin A. (2016). Neuroendocrine Coordination of Mitochondrial Stress Signaling and Proteostasis. Cell 166, 1553–1563.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt C, Nolte H, Henschke S, Engstrӧm Ruud L, Awazawa M, Morgan DA, Gabel P, Sprenger H-G, Hess ME, Günther S, et al. (2018). Food Perception Primes Hepatic ER Homeostasis via Melanocortin-Dependent Control of mTOR Activation. Cell 175, 1321–1335.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase DL, Pepper JS, and Koelle MR (2004). Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat. Neurosci 7, 1096–1103. [DOI] [PubMed] [Google Scholar]
- Daniele JR, Esping DJ, Garcia G, Parsons LS, Arriaga EA, and Dillin A. (2017). “High-Throughput Characterization of Region-Specific Mitochondrial Function and Morphology”. Sci. Rep 7, 6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele JR, Higuchi-Sanabria R, Durieux J, Monshietehadi S, Ramachandran V, Tronnes SU, Kelet N, Sanchez M, Metcalf MG, Garcia G, et al. (2020). UPRER promotes lipophagy independent of chaperones to extend life span. Sci. Adv 6, eaaz1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, and Dillin A. (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escorcia W, Ruter DL, Nhan J, and Curran SP (2018). Quantification of Lipid Abundance and Evaluation of Lipid Distribution in Caenorhabditis elegans by Nile Red and Oil Red O Staining . J. Vis. Exp, 57352. [DOI] [PMC free article] [PubMed]
- Folgueira C, Beiroa D, Porteiro B, Duquenne M, Puighermanal E, Fondevila MF, Barja-Fernández S, Gallego R, Hernández-Bautista R, Castelao C, et al. (2019). Hypothalamic dopamine signaling regulates brown fat thermogenesis. Nat. Metab 1, 811–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes AE, and Dillin A. (2017). The UPRER: Sensor and Coordinator of Organismal Homeostasis. Mol. Cell 66, 761–771. [DOI] [PubMed] [Google Scholar]
- Frakes AE, Metcalf MG, Tronnes SU, Bar-Ziv R, Durieux J, Gildea HK, Kandahari N, Monshietehadi S, and Dillin A. (2020). Four glial cells regulate ER stress resistance and longevity via neuropeptide signaling in C. elegans. Science 367, 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton S, and Alquier T. (2019). Lipid signalling in the mesolimbic dopamine pathway. Neuropsychopharmacology 44, 221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz A, Keenan RW, and Elbein AD (1979). Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry 18, 2186–2192. [DOI] [PubMed] [Google Scholar]
- Higuchi-Sanabria R, Charalel JK, Viana MP, Garcia EJ, Sing CN, Koenigsberg A, Swayne TC, Vevea JD, Boldogh IR, Rafelski SM, and Pon LA (2016). Mitochondrial anchorage and fusion contribute to mitochondrial inheritance and quality control in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 27, 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanikia S, Sheng M, Castro C, Griffin JL, and Taylor RC (2019a). XBP-1 Remodels Lipid Metabolism to Extend Longevity. Cell Rep. 28, 581–589.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanikia S, Özbey NP, Krueger C, Casanueva MO, and Taylor RC (2019b). Neuronal XBP-1 Activates Intestinal Lysosomes to Improve Proteostasis in C. elegans. Curr. Biol 29, 2322–2338.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite F, and Ribeiro L. (2020). Dopaminergic Pathways in Obesity-Associated Inflammation. J. Neuroimmune Pharmacol 15, 93–113. [DOI] [PubMed] [Google Scholar]
- Lints R, and Emmons SW (1999). Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFbeta family signaling pathway and a Hox gene. Development 126, 5819–5831. [DOI] [PubMed] [Google Scholar]
- Na H, Zhang P, Chen Y, Zhu X, Liu Y, Liu Y, Xie K, Xu N, Yang F, Yu Y, et al. (2015). Identification of lipid droplet structure-like/resident proteins in Caenorhabditis elegans. Biochim. Biophys. Acta 1853, 2481–2491. [DOI] [PubMed] [Google Scholar]
- Nagatsu T, Levitt M, and Udenfriend S. (1964). Tyrosine hydroxylase. The initial step in norepinephrine biosynthesis. J. Biol. Chem 239, 2910–2917. [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, and Morimoto RI (2008). Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R, Sawin ER, Trent C, and Horvitz HR (2001). Mutations in the Caenorhabditis elegans serotonin reuptake transporter MOD-5 reveal serotonin-dependent and -independent activities of fluoxetine. J. Neurosci 21, 5871–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salio C, Lossi L, Ferrini F, and Merighi A. (2006). Neuropeptides as synaptic transmitters. Cell Tissue Res. 326, 583–598. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, and Hobert O. (2013). Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, and Kaufman RJ (2005). Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS genetics 1, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Victor M, Loer C, Shi Y, and Ruvkun G. (2000). Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560–564. [DOI] [PubMed] [Google Scholar]
- Tatum MC, Ooi FK, Chikka MR, Chauve L, Martinez-Velazquez LA, Steinbusch HWM, Morimoto RI, and Prahlad V. (2015). Neuronal serotonin release triggers the heat shock response in C. elegans in the absence of temperature increase. Curr. Biol 25, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, and Dillin A. (2013). XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153, 1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland JJ, McEwen J, Moore BA, Jin Y, and Condie BG (2001). Conserved function of Caenorhabditis elegans UNC-30 and mouse Pitx2 in controlling GABAergic neuron differentiation. J. Neurosci 21, 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KW, Liu T, Kong X, Fukuda M, Deng Y, Berglund ED, Deng Z, Gao Y, Liu T, Sohn J-W, et al. (2014). Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab. 20, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Finney M, Ruvkun G, and Chalfie M. (1992). Regulation of the mec-3 gene by the C.elegans homeoproteins UNC-86 and MEC-3. EMBO J. 11, 4969–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Na H, Liu Z, Zhang S, Xue P, Chen Y, Pu J, Peng G, Huang X, Yang F, et al. (2012). Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol. Cell. Proteomics 11, 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials. Raw sequencing data is available through ArrayExpress Annotaire 2.0, accession number: E-MTAB-9771. All other data can be requested from the lead contact.