Abstract
We have previously shown that the Kunitz-type serine protease inhibitor Spint1a, also named Hai1a, is required in the zebrafish embryonic epidermis to restrict the activity of the type II transmembrane serine protease (TTSP) Matriptase1a / St14a, thereby ensuring epidermal homeostasis. A closely related Kunitz-type inhibitor is Spint2/Hai2, which in mammals plays multiple developmental roles that are either redundant or non-redundant with those of Spint1. However, the molecular bases for these non-redundancies are not fully understood. Here, we study spint2 during zebrafish development. It is co-expressed with spint1a in multiple embryonic epithelia, including the outer / peridermal layer of the epidermis. However, unlike spint1a, spint2 expression is absent from the basal epidermal layer but present in hatching gland cells. Hatching gland cells derive from the mesendodermal prechordal plate, from where they undergo a thus far undescribed transit into, and coordinated sheet migration within, the interspace between the outer and basal layer of the epidermis to reach their final destination on the yolk sac. Hatching gland cells usually survive their degranulation that drives embryo hatching but die several days later. In spint2 mutants, cohesion among hatching gland cells and their collective intra-epidermal migration are disturbed, leading to a discontinuous organization of the gland. In addition, cells undergo precocious cell death before degranulation, so that embryos fail to hatch. Chimera analyses show that Spint2 is required in hatching gland cells, but not in the overlying periderm, their potential migration and adhesion substrate. Spint2 acts independently of all tested Matriptases, Prostasins and other described Spint1 and Spint2 mediators. However, it displays a tight genetic interaction with and acts at least partly via the cell-cell adhesion protein E-cadherin, promoting both hatching gland cell cohesiveness and survival, in line with formerly reported effects of E-cadherin during morphogenesis and cell death suppression. In contrast, no such genetic interaction was observed between Spint2 and the cell-cell adhesion molecule EpCAM, which instead interacts with Spint1a. Our data shed new light onto the mechanisms of hatching gland morphogenesis and hatching gland cell survival. In addition, they reveal developmental roles of Spint2 that are strikingly different from those of Spint1, most likely due to differences in the expression patterns and relevant target proteins.
Keywords: Spint2, hatching gland, morphogenesis, collective cell migration, cohesion, cell death, E-cadherin, zebrafish
Introduction
Membrane-anchored extracellular serine proteases, such as the type II transmembrane proteases Matriptase (also named Supressor of tumorgenicity 14 / St14) and Hepsin, and the GPI-anchored protease Prostasin/Prss8, play crucial roles during numerous developmental processes and the manifestation of various diseases, especially epithelial-derived cancers (carcinomas). They modify their substrate proteins, including growth factors and protease-activated receptors, pericellularly to activate pathways important especially in epithelial homeostasis (Tanabe and List, 2017). St14, for example, is required for the development of the epidermal barrier (List et al., 2006; Netzel-Arnett et al., 2006), and dysregulated high activity has been implicated with numerous types of carcinogenesis, identifying it as a potential oncogene (Szabo et al., 2008; Tanabe and List, 2017). The activity of these membrane-anchored extracellular proteases is regulated at multiple levels. A central negative regulatory role is played by the cognate Kunitz-type transmembrane serine protease inhibitors Spint1, also named Hai1 (Hepatocyte growth factor activator inhibitor-1) (Shimomura et al., 1997) and Spint2, also called Placental Bikunin and Hai2 (Kawaguchi et al., 1997; Marlor et al., 1997). Loss of Spint1 or Spint2 inhibitory function results in the activation of numerous oncogenic downstream pathways, identifying them as potential tumor suppressors (Kataoka et al., 2018). Thus, low expression levels and hypermethylation of the Spint2 locus is linked to poor prognosis in several carcinomas characterized by elevated cell proliferation, motility, and invasiveness (Roversi et al., 2018). Of note, the relevant Spint2 target proteases mediating these effects differ between different carcinomas and have for instance been identified as Matriptase in some (Tsai et al., 2014), and cell surface-associated plasmin in other cases (Wu et al., 2019). In the latter case, this results in the induction of epithelial-mesenchymal transitions characterized by increased Vimentin and Slug expression and decreased levels of the cell-cell adhesion protein E-cadherin (Wu et al., 2019). In striking contrast, however, other carcinomas are characterized by increased, rather than decreased Spint2 expression, implying a tumor-promoting, rather than tumor-suppressive function (Graumann et al., 2019; Yamamoto et al., 2018).
Spint1 and Spint2 also play important roles during development. Mammalian Spint1 and Spint2 are largely co-expressed and share crucial functions but can also act independently of each other. In mouse, global loss-of-function mutations in either of them lead to strong and St14-dependent impairments of placental labyrinth development (Fan et al., 2007; Szabo et al., 2009; Szabo et al., 2007; Tanaka et al., 2005), pointing to non-redundant roles of Spint1 and Spint2, which nevertheless are both mediated via Matriptase. In addition, Spint2 mutant mice show St14-independent defects in neural tube closure that are not shared by Spint1 mutants, pointing to Spint2-specific functions via additional, not yet identified Spint2 targets (Szabo et al., 2009).
Furthermore, conditional loss-of-function studies have been performed in mice. Keratinocyte-specific inactivation of mouse Spint1 leads to St14-dependent disruption of epidermal integrity (Kawaguchi et al., 2015; Nagaike et al., 2008), similar to the epidermal defects observed in global spint1a mutants in zebrafish (Armistead et al., 2020; Carney et al., 2007; Schepis et al., 2018). Similarly, mouse Spint2 mutants, in which the early requirements of Spint2 for placental development have been genetically circumvented or compensated, display strongly disturbed integrity of the intestinal epithelium (Kawaguchi et al., 2019; Szabo et al., 2019), recapitulating Congenital sodium diarrhea (CSA) / tufting enteropathy (CTE) caused by hypomorphic SPINT2 mutations in human (Faller et al., 2014; Heinz-Erian et al., 2009; Holt-Danborg et al., 2019; Salomon et al., 2014). In this case, the up-regulation of other membrane-tagged proteases in addition to Matriptase, leading to the loss of the epithelial cell-cell adhesion molecule EpCAM, was identified as the underlying pathomechanism (Kawaguchi et al., 2019; Szabo et al., 2019; Wu et al., 2017), consistent with the identification of EPCAM itself as another CTE disease gene in human (Salomon et al., 2014; Sivagnanam et al., 2008).
Taken together, despite their similar inhibitory potentials towards the same serine proteases and co-expression in many tissues, the in vivo roles of Spint1 and Spint2 differ in many cases and are far from fully understood. Therefore, as formerly done for spint1, we have generated and analyzed global spint2 mutants in the zebrafish. In contrast to Spint1 in mouse and zebrafish, zebrafish Spint2 is dispensable for epidermal development, but, like Spint2 in mouse, is required for late intestinal integrity. In addition, it is required for proper morphogenesis and functionality of the hatching gland, an organ absent in amniotes. Hatching gland cells are of mesendodermal origin and derive from the prechordal plate (Melby et al., 1996; Shih and Fraser, 1996), while its later morphogenesis had been little studied thus far. We show that, starting at early segmentation stages, the hatching gland primordium translocates from the interior of the embryo into the interspace between the outer and basal layer of the epidermis, followed by a cell intercalation-driven transition from a multi- to a mono-layered organization and the cohesive migration of the resulting mono-layered sheet into the epidermis of the yolk sac. Spint2 is required for persistent cohesion and collective migration of this monolayer of hatching gland cells, as well as for their survival and the eventual hatching of the embryo. Tests with multiple known substrates of mammalian Spint1 and Spint2 all yielded negative results, pointing to the involvement of a yet unknown protease in mediating the effects caused by the loss of Spint2 function. However, Spint2 displays a strong genetic interaction with E-cadherin, consistent with its formerly described, ST14-independent working mechanism in lung cancer cells (Wu et al., 2019), whereas it does not interact with EpCAM, as would have been expected from its reported working mechanism in mammalian intestinal cells and the etiology of CTE.
Materials and Methods
Zebrafish lines
Wild-type fish were used from intercrosses of TL and EK. The following transgenic lines were used: Tg(Ola.Actb:Hsa.hras-egfp)vu119 (Cooper et al., 2005), Tg(krt4:GFP)gz7 (Gong et al., 2002), Tg(krt4:tomatocaax)fr48Tg (Richardson et al., 2016; Teixeira Rosa et al., 2019), −1.8gsc:GFPml1Tg (Doitsidou et al., 2002), Tg(gsc:Cre)fr44Tg (Pogoda et al., 2018) and Tg(ubi:Zebrabow-M)a13 (Pan et al., 2013). For Tg(bact:tdtomato-caax)fr51Tg, plasmid bact:tdtomato-caax was generated using the Tol2 kit (Kwan et al., 2007) and injected into wild-type embryos together with tol2 mRNA, followed by standard screening procedures to identify appropriate founders for transgenic line establishment.
Generation and genotyping of st14c TALEN mutant.
TALENs against exon3 of zmp:0000001114 (st14c) were designed using the TAL Effector Nucleotide Targeter 2.0 web-tool from Cornell University (tal-nt.cac.cornell.edu). Target sequence of the forward strand (Left TALEN) was 5’-TAGAACCAAAGAACT-3’ (position 2-16 of exon3), the second (Right) TALEN was constructed against the sequence stretch 5’-GAGCTGTGTTTCTCAATG-3’ (position 32-49 of exon3, reverse strand). The 16bp spacer region harbors an MboII restriction side to be used for later genotyping. TALEN assembly of the RVD-containing repeats was conducted using the Golden Gate approach (Cermak et al., 2011). Once assembled the RVD were cloned into pCS2TAL3DD to generate the Left TALEN gene and into pCS2TAL3RR to generate the right TALEN gene (Dahlem et al., 2012). 5’-capped mRNA was generated by in vitro transcription of pCS2TAL3DD and pCS2TAL3RR TALEN plasmid templates via the SP6 mMessage mMachine kit from Ambion after linearization of the plasmids with NotI. Lastly, around 100pg of Left and Right TALEN mRNA were injected together into the zygote. Embryos were raised and their progeny screened for germline transmission by PCR using the primers 5’-GAATAGCTATGTACAGTTCCTC-3‘ and 5’-CGCTGCTTGTTGGAATTGTG-3‘ followed by MboII digestion. A 20bp deletion was identified at position 193 of the coding region resulting in a frame shift at aa 65 followed by a truncation, which was used to establish line st14cfr50.
Generation and genotyping of spint2 Crispr/Cas mutant
The guide RNA AGATCGCGGAGACCACCAAA targeting exon1 of spint2 was designed using the webtool at http://crispr.mit.edu/. The specific Alt-R® CRISPR-Cas9 crRNA and the universal Alt-R® TM tracrRNA were obtained from IDT, Belgium, and annealed in equal amounts of 30 μM each in Nuclease-Free Duplex Buffer (IDT) by heating to 95°C for 5 min and subsequent cooling to room temperature. 9 μM of complexed RNA was injected together with 150 ng/μl of Cas9 mRNA generated via the SP6 mMessage mMachine kit from Ambion from plasmid pCS2-Cas9 (Addgene, (Gagnon et al., 2014) in Danieaux buffer into the zygote. Embryos were raised and their progeny screened for germline transmission by PCR using the primers 5’-CGCGTGTATCTGGGACC-3‘ and 5’-TCGTTTAAGAAAGCCTACATGACA-3‘ followed by a T7 endonuclease assay (NEB) to detect indels and to establish line spint2fr49 with a 31bp deletion in exon 1(see Results). Genotyping was performed using the same primers followed by BslI digestion, which results in a cleaved wild-type PCR product and non-cleaved mutant product.
Morpholinos
The following morpholinos were obtained from GeneTools (Philomath, CA), diluted in Danieau buffer to the indicated concentration, and 1.5 nl were injected into 1-cell stage embryos according to standard protocols.
| Gene | Morpholino sequence | Concentration | Reference |
|---|---|---|---|
| c-met | ATAGTGAATTGTCATCTTTGTTCCT | 0.2 mM | (Haines et al., 2004) |
| cdh1 | ATCCCACAGTTGTTACACAAGCCAT | 0.0033 mM | (Montero et al., 2005) |
| epcam | AGGCAACTAAAACCTTCATTGGTGAG | 0.2 mM | (Slanchev et al., 2009) |
| mstr1ra | AACAAGGTCTTTGGGCTGATGAACA | 0.2 mM | (Carney et al., 2007) |
| mstr1rb | CTAAATGAGTGGCCCAATGGACCAT | 0.2 mM | (Carney et al., 2007) |
| par2b | GTAGCTCTCGGACACCGCCATATTC | 0.2 mM | (Armistead et al., 2020) |
| plasminogen | GAACTCCTTTGTGTACCTCCATGTC | 0.2 mM | (Carney et al., 2007) |
| prss60.1 | AACCTCCACATCTTCACCAGCTATC | 0.2 mM | this study |
| prss60.2 (1. ATG) | GGCACAAGTCAATCTCCACATCTTC | 0.2 mM | this study |
| prss60.2 (2. ATG) | AAGTGGCACAAGTCAATCTCCACAT | 0.2 mM | this study |
| prss60.3 | TGAGAAGAACTCTTACCATTCAACT | 0.2 mM | this study |
| spint1a | ACCCTGAGTAGAGCCAGAGTCATCC | 0.05 mM (full) 0.01 mM (low) |
(Carney et al., 2007) |
| spint2 (ATG) | CCTTTTTACCTGTCCCAGCAGCTGT | 0.07 mM (full) 0.0033 mM (low) |
this study |
| spint2 (splice) | ATTTCTGGCTTTGTTTACCTGTGGT | 0.2 mM | this study |
| spint2 5mm control | CCTTTTTACCTGTCCCAGCAGCTGT | 0.07 mM | this study |
| st14c (1. ATG) | ATCCAAAGGCACCAAACAGACGAGC | 0.2 mM | this study |
| st14c (2. ATG) | GCATACGCGGCAGAACTCATGTTTC | 0.2 mM | this study |
| standard control | CCTCTTACCTCAGTTACAATTTATA | 0.07 mM | GeneTools |
| zgc:100868 | TCTACAGGTTTTACAGCTGACTGCC | 0.2 mM | this study |
| zgc:92313 | CTACATGAGGTGAGCCGCCCCATTC | 0.2 mM | this study |
Plasmid generation and injection
To label hatching gland cells with LifeAct-Ruby, a plasmid was generated using the Tol2 kit (Kwan et al., 2007), with the described gsc promoter in the p5E- (Pogoda et al., 2018) and the LifeAct-Ruby sequence (Yoo et al., 2010) in the pME vector. The plasmid was injected into the zygote of wild-type embryos to label hatching gland cells in a mosaic fashion. For rescue experiments, the spint2 or cdh1 cDNA was cloned into the BamHI/EcoRI site of pSGH2 (Bajoghli et al., 2004), which harbors a bicistronic heat-inducible promoter to express eGFP together with spint2 or cdh1. The spint2 cDNA lacks 5’UTR sequences targeted by the spint2 ATG MO, making the plasmid-encoded transcripts insensitive to MO-mediated silencing. The plasmids were injected into one-cell stage tg(bact:tdtomato-caax)fr50tg embryos in conjunction with 0.07 mM spint2 morpholino. Embryos were heat-shocked at 14 −15 hpf for 45 min by incubation at 40°C, and analyzed at 24 hpf.
Inhibitor treatments
LY294002 (TocrisBioscience, Bristol, UK), SB431542, and PD168393 (Sigma-Aldrich, Germany) were dissolved in DMSO, and solutions were further diluted in E3 embryo medium to concentrations of 25 μM LY294002, 75 μM SB431542, and 10 μM PD168393. Embryos were incubated in inhibitor solution starting from 10 hours post fertilization (hpf) (LY29400 and PD168393) or 3 hpf (SB431542).
Whole-mount in situ hybridization (WISH)
Embryos were fixed in 4% paraformaldehyde (PFA) and dehydrated/rehydrated through a methanol series. Whole-mount in situ hybridization was performed as previously described (Hammerschmidt et al., 1996). Probes were synthesized from linearized cDNA clones, using the Roche digoxygenin RNA synthesis kit. cDNA clones were as described for ctslb (Haines et al., 2004), clone IMAGp998L1718140Q (imaGenes) for spint2, and other cDNA clones were generated by amplifying cDNA sequences by PCR and ligating them into pGemT-easy (Promega). Combined colorimetric WISH and immunostainings were performed as described (Carney et al., 2007).
Sectioning
Durcupan sections (following combined colorimetric WISH and immunostainings) were performed as previously described (Carney et al., 2010), cryosections were generated as previously described (Westcot et al., 2015), followed by immunofluorescent staining or H&E staining. Ultrathin sections were generated and processed, and transmission electron microscopy (TEM) was performed as described (Feitosa et al., 2012).
Immunostainings
Immunostainings were performed essentially as previously described (Hammerschmidt et al., 1996). Larvae were fixed in EAF (40% ethanol, 5% acetic acid, 4% formaldehyde in PBS) for whole mount immunostainings with mouse anti Cdh1 (BD Biosciences, 610188, 1:200), or 4% PFA (paraformaldehyde) in PBS for immunostainings using the following primary antibodies: mouse anti Cdh1 (for cryosections), mouse anti-GFP (Invitrogen; A10262, 1:300), chicken anti-GFP (Invitrogen, A10262, 1:500), and mouse anti-tp63 BC4A4 (Zytomend, 1:200). Secondary antibodies were anti-mouseCy3, anti-mouseAlexa488, and anti-chickenAlexa488 (all Invitrogen, 1:500).
TUNEL staining
Dying cells were visualized by TUNEL staining using the in situ cell death detection kit (Roche) following the manufacturer’s instructions.
Microscopy
Images were taken using a Zeiss Confocal (LSM710 META), an AxioImager microscope (Zeiss) equipped with an Apotome, using the AxioVision software (Zeiss) or a Leica M165FC stereo microscope with DFC425C camera and the Leica Application Suite V3.8 software and processed using the ImageJ software and Adobe Photoshop. Cell shapes of hatching gland cells were determined by the shape descriptor tool in ImageJ, calculating circularity by 4π × Area/Perimeter2, and solidity by Area/Convex area.
Cell migration analysis
Embryos were mounted in 1.5% methyl cellulose and fluorescent images were taken in 5 min intervals using 5 μm Z stacks and a 10x objective on the Axioimager microscope (Zeiss) equipped with an Apotome. Maximum intensity projections were generated and cells were tracked using the Axiovision software to determine speed and migration path, and ImageJ was used to determine the angle. Loss of neighbors was manually determined using MIP time series.
Cell transplantations
Polster cells from Tg(Ola.Actb:Hsa.hras-egfp)vu119 transgenic donor embryos either uninjected or injected with spint2 MO were transplanted at 10 hpf into the tissue below the head region of spint2 morphant or wild-type recipients in which the endogenous mesendoderm development had been suppressed by treatment with SB431542. Chimeric recipients were raised in E3 embryo medium and imaged at 24 hpf.
Statistics
Quantitative experiments were repeated at least three times, reaching similar results. Mean values and standard deviations of all individual specimens (biological samples; n) from one representative or all independent experiments (N) are presented, as specified in the respective figure legends. Statistical analysis was performed using Graph Pad Prism software. For comparison of multiple groups, one-way ANOVA with post-hoc Tukey’s test, for comparison of two groups, an unpaired two-tailed Student’s t-test, was used to determine significance and obtained p-values are mentioned in the respective figure legends.
Results
spint2 is expressed in several epithelia and hatching gland cells of zebrafish embryos
Our former analyses had identified two zebrafish spint1 paralogs (Carney et al., 2007). In contrast, performing database searches via blast and conserved synteny analyses, we identified a single spint2-like locus in the zebrafish genome, located on LG 15, in line with the existence of two spint1 paralogs, but only one spint2 gene in the pufferfish Takifugu rubripes (Figure 1A,B). On the protein level, the predicted sequence of zebrafish Spint2 is 29.8, 32.2, 32.1 and 38.6 % identical to Spint2 from human, mouse, the clawed frog Xenopus laevis and Takifugu rubripes, respectively (Figure 1A,C). In mammals, two Spint2 splicing isoforms have been described, with the longer - like mammalian and zebrafish Spint1 - containing two, and the shorter only one Kunitz-type domain (Roversi et al., 2018). In contrast, despite exhaustive RT-PCR experiments, we only identified one isoform of the zebrafish Spint2 protein, which contains three Kunitz-type domains. However, like mammalian Spint2, it lacks the low-density lipoprotein (LDL)-receptor class A domain interspacing the Kunitz-type domains and the polycystic kidney disease (PKD)-like domain of Spint1 proteins, while its N-terminal cysteine-rich domain, which is absent in mammalian Spint2, can be regarded as a truncated “motif at N-terminus with eight-cysteines” (MANEC-type domain; Hong et al., 2015) characteristic for Spint1 proteins (Figure 1C). Thus, zebrafish Spint2 seems to share features with both mammalian Spint1 and Spint2. Of note, however, it shows the exact same protein structure like its Fugu and Xenopus orthologues, which are also composed of a truncated MANEC-type domain and three Kunitz-type domains.
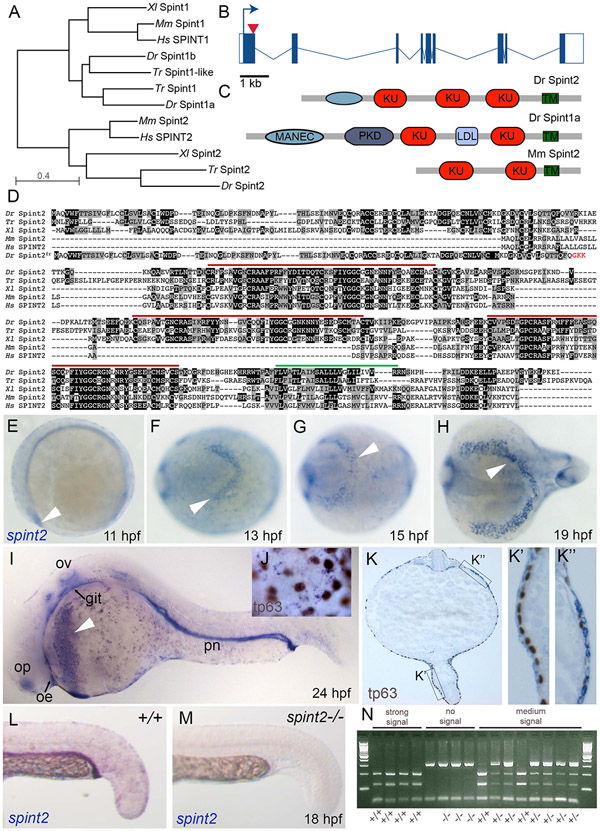
Figure 1: The zebrafish genome contains one ortholog of the Kunitz-type inhibitor spint2, which is expressed in multiple epithelia as well as hatching gland cells.
A. Phylogenetic tree of protein sequences of zebrafish (Dr), pufferfish (Takifugu rubripes, Tr), clawed frog (Xenopus laevis, Xl), mouse (Mm), and human (Hs) Spint1 and Spint2 proteins. The tree was created using COBALT and the Neigbor Joining method on the NCBI webpage. B. Schematic representation of zebrafish spint2 gene structure. Exons are indicated by rectangles, with coding regions in solid blue, introns are indicated with lines. The arrow head indicates the position targeted by the Crispr guide RNA. C. Schematic representation of Zebrafish Spint2 and Spint1a as well as mouse Spint2 protein domains. KU, Kunitz-type domain; PKD, polycystic kidney disease-like domain; MANEC, motif at N-terminus with eight-cysteines; TM, transmembrane domain. D. Multiple alignment of zebrafish (Dr), pufferfish (Takifugu rubripes, Tr), clawed frog (Xenopus laevis, Xl), mouse (Mm), and human (Hs) Spint2 protein sequences by Clustal Omega. Identical amino acid (aa) residues are indicated by black boxes, similar aa residues by grey boxes. Red lines indicate Kunitz-type domains, green line the transmembrane domain. E.-I. Whole mount in situ hybridization (WISH) with a spint2 probe shows expression in the epidermis and hatching gland cells throughout somitogenesis (E-H) and additional expression in the olfactory epithelium (olf), otic vesicle (ot), and pronephric duct (pd) at 24 hpf (I). J-K. WISH for spint2 at 24 hpf followed by tp63 immunostaining shows expression of spint2 in peridermal cells but not in tp63-positive basal cells in whole mounts (J) and cross sections (K, K’) as well as expression in hatching gland cells (K”). L,M. spint2 WISH in spint2+/+ and −/− embryos at 24 hpf. spint2 signal is not detectable in spint2fr49/fr49 embryos. N. Ethidumbromide agarose gel with genotyping results of embryos obtained from an incross of spint2+/fr49 parents after they had been stained via spint2 mRNA WISH. The obtained staining intensity of the individual embryos is indicated.
Performing whole mount in situ hybridization (WISH), we found zebrafish spint2 to be expressed in the hatching gland cells of segmentation stage zebrafish embryos (11-19 hours post fertilization (hpf); Figure 1E-H) and at 24 hpf (Figure 1I,K”), in line with the formerly described hatching gland expression of spint2 in Xenopus embryos (Gawantka et al., 1998). At 24 hpf, zebrafish spint2 is additionally expressed in the epidermis (Figure 1I,J,K) and several other epithelia including the olfactory epithelium, the otic vesicle, the pronephric duct and, at rather low levels, the oral and intestinal epithelium (Figure 1I; for intestine, see also Figure 2). In the zebrafish embryo, the epidermis is a bi-layered epithelium, consisting of basal keratinocytes of ectodermal origin as well as peridermal cells deriving from the enveloping layer (EVL). Co-staining with an antibody against tp63, which is specific for basal keratinocytes, revealed that the expression of spint2 in the epidermis is excluded from tp63-positive basal keratinocytes, indicating that spint2 is solely expressed in the outer peridermal cell layer (Figure 1J,K’). This is different from spint1a and spint1b, which are expressed in both basal keratinocytes and peridermal cells (Carney et al., 2007; Schepis et al., 2018).
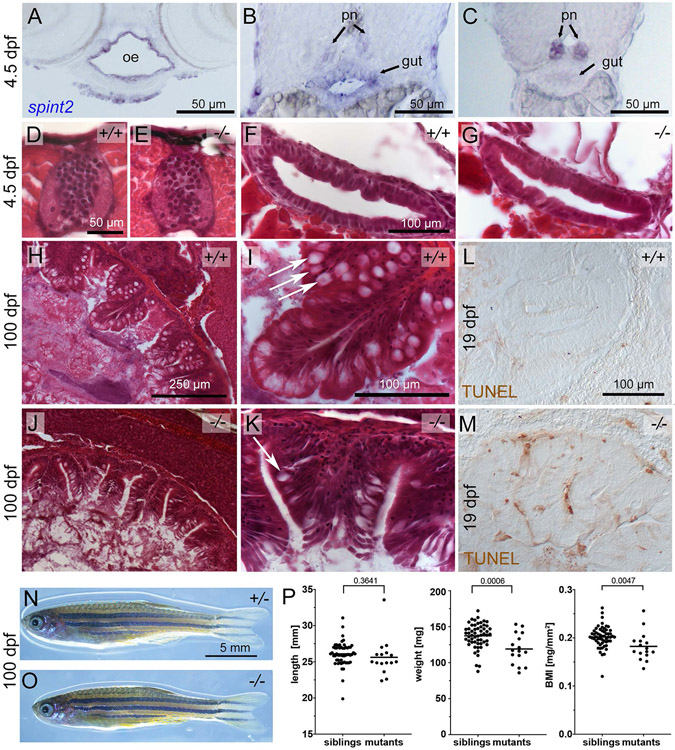
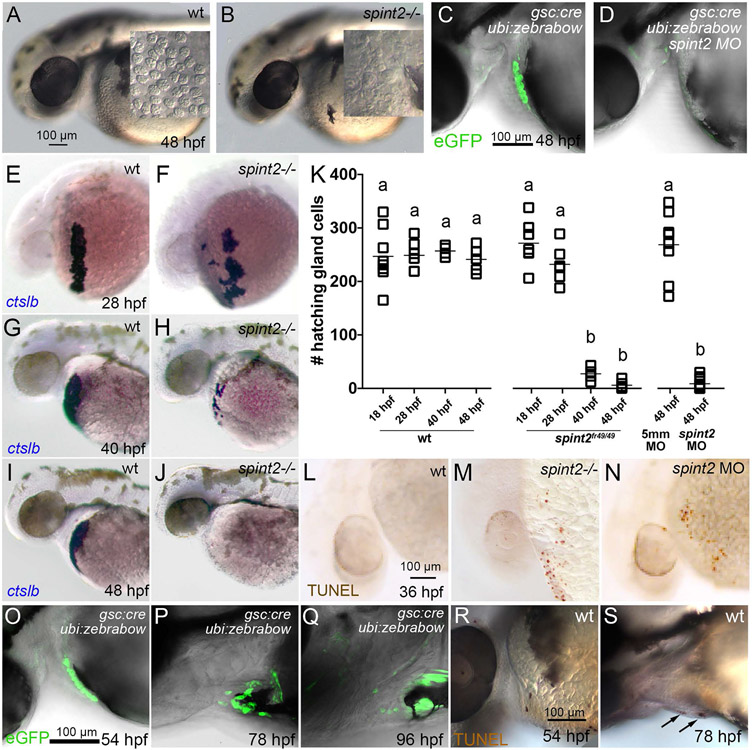
Figure 2: spint2 mutants display a postembryonic intestinal phenotype.
A-C. Cross sections of a 4.5 dpf embryo following spint2 WISH. spint2 expression is strongly detected in the oral epithelium (oe), moderately in the anterior gut (B), and decreasing towards more posterior regions, in contrast to strong spint2_signal in the pronephric ducts (pn) (C). D-G. H&E staining of cross sections of 4.5 dpf spint2 siblings (D, F) and mutants (E, G) showing normal spinal cord morphology (D, E) and gut epithelium (F, G). H-K. H&E staining of cross sections of the rostral intestine of an adult (100 dpf) wild-type sibling (H-I) and a spint2 mutant (J-K). The mutant intestine contains fewer goblet cells (arrows) and a disrupted epithelial integrity. L,M. TUNEL staining of cross sections of the rostral intestine of an 19 dpf juvenile wild-type sibling (L) and a spint2 mutant (M). N,O. Images of an adult (100 dpf) wild-type sibling (N) and a spint2 mutant (O) male raised together. The mutant is of slightly smaller size. P. Quantification of length in mm, weight in mg, and BMI in mg/mm2 of spint2 sibling and mutant males raised together. Dots indicate individual fish, lines represent the mean, and p values were determined by a Student’s t-test. Four experiments / families; 72 individuals.
Zebrafish spint2 is required for intestinal integrity
We next sought to investigate the role of spint2 in zebrafish development by loss-of-function studies. We used two antisense morpholino oligonucleotides (MOs), a translation-blocking MO and a splice-blocking MO at the exon 1 - intron 1/2 border (Figure 1B). Additionally, we applied Crispr/Cas9 technology to generate a stable mutant line with a 31 bp deletion in exon 1 of the spint2 gene (spint2fr49). This deletion causes a frame-shift and a premature stop codon of the new open reading frame after three triplets (Figure 1B,D), resulting in a C-terminally truncated protein of only 109 aa that lacks all Kunitz domains and the transmembrane domain (Figure 1D). Performing spint2 WISH on 24 hpf progeny of an incross of spint2+/− parents and genotyping individuals after photography, we found that homozygous mutants lack spint2 staining (Figure 1L-N), pointing to nonsense-mediated RNA decay of the mutated transcripts. Together, these data suggest that the spint2fr49 allele represents a functional null / amorph.
In contrast to full Spint2 mouse mutants (Szabo et al., 2009), which display early lethality because of compromised placental development, zebrafish spint2 morphants and mutants do not show gross early morphological defects, allowing us – after manual dechorionation of the embryos (see below) - to study its potential roles during later developmental processes.
In contrast to the neural tube closure defects reported for mouse Spint2 mutants, which remain even after concomitant genetic inactivation of St14 (Szabo et al., 2009), zebrafish spint2 mutants show no neurulation defects (data not shown). Furthermore, according to quantifications of multiple sections from multiple embryos, spinal cord dimensions and cell numbers are unaltered in spint2fr49/fr49 mutants during later embryogenesis (Figure 2D,E; and data not shown). This suggests that in contrast to mouse, Spint2 is dispensable for zebrafish neurulation, possibly due to differences in the modes of neurulation (neural tube versus neural keel formation) in mammals versus fish (Schmidt et al., 2013).
In light of the Congenital tufting enteropathy (CTE) phenotype of hypomorphic SPINT2 human patients and mouse mutants after genetic circumvention of the early Spint2 requirement (see Introduction), we also studied the gastrointestinal system of spint2 zebrafish mutants. Sectioning of wild-type embryos at 4.5 days post fertilization (dpf) after spint2 WISH revealed spint2 expression in oral as well as gastrointestinal epithelia (Figure 2A-C). However, we could not find obvious alterations in the histology of the intestinal epithelium in 4 days old spint2fr49/fr49 mutant embryos, approximately 1 day before the onset of external food uptake (Figure 2F,G) (n=5/5). In contrast, in adult (100 dpf) fish mutants, and consistent with the mammalian data, the organization and integrity of intestinal crypts is markedly compromised, with increased desquamation and reduced numbers of goblet cells (Figure 2H-K) (n=4/4). Furthermore, already at 19 dpf, more TUNEL positive cells were observed in intestinal crypts of mutant juveniles (Figure 2L,M) (n=5/5), pointing to intestinal atrophy, very similar to the defects reported for mouse Spint2 mutants (Kawaguchi et al., 2019; Szabo et al., 2019). However, compared to the massive wasting of the mouse mutants (Kawaguchi et al., 2019; Szabo et al., 2019), zebrafish spint2 mutants are of the same size as their siblings, with only moderately reduced body weights and body mass indices (Figure 2N-P). This suggests that in fish, compromised gastrointestinal integrity has less profound physiological consequences than in mammals, in line with former findings for other zebrafish gut mutants (Yang et al., 2009). Future studies have to reveal whether in addition to the gastrointestinal system, Spint2 is also required for maintained integrity of other spint2-expressing epithelia, such as for instance in the pronephros.
Zebrafish spint2 is required for hatching gland morphogenesis
In addition to compromised gut integrity, zebrafish spint2 mutants also display an earlier phenotype that is – for obvious reasons – not shared by the mammalian mutants, affecting the hatching gland of the embryos. As a consequence, in contrast to their siblings, which hatch between 48 and 72 hpf, zebrafish spint2 mutant and morphant embryos fail to hatch (Figure 3A) and, unless manually dechorionated, subsequently die within the chorion.
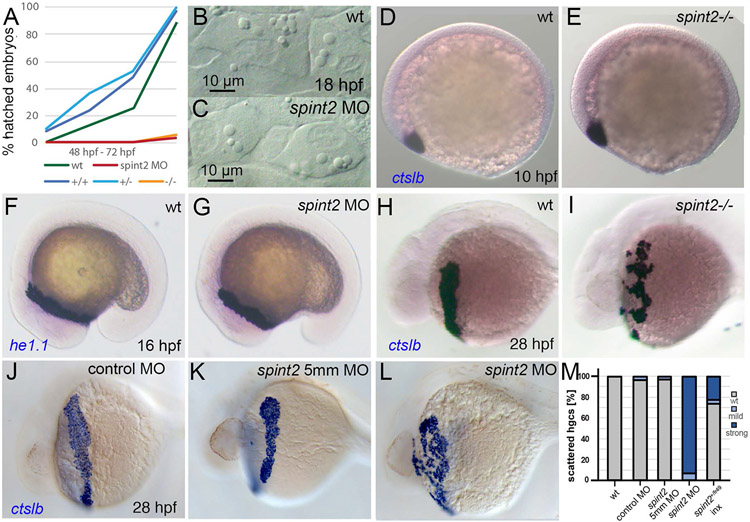
Figure 3: Loss of spint2 results does not affect hatching gland cell specification, but leads to compromised hatching gland morphogenesis.
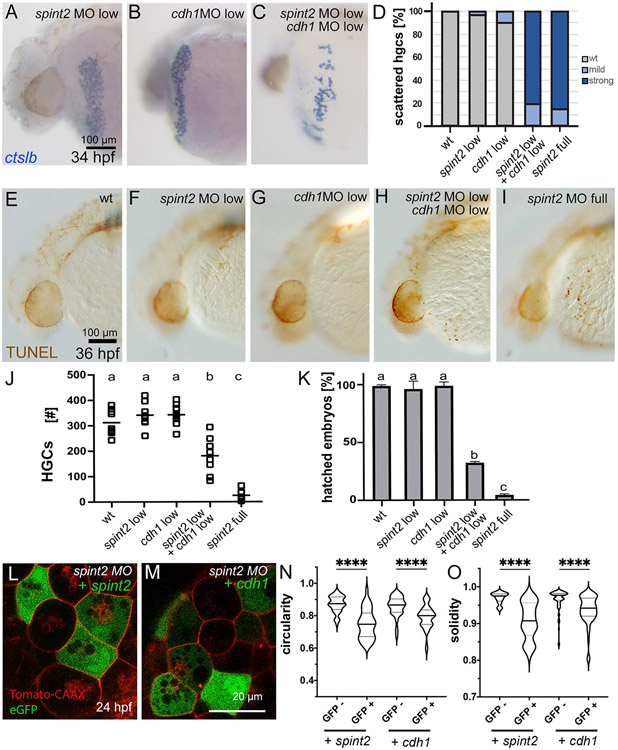
A. Percentage of hatched embryos during 48 – 72 hpf, n=24-50. B-C. DIC images of hatching gland cells displaying characteristic granules in wild type (B) and spint2 MO (C) at 18 hpf. D-G. WISH of hatching gland marker genes shows unaltered expression of ctslb in wild type (D) and spint2 mutant (E) at end of gastrulation (10 hpf), and of he1.1 in wild type (F) and spint2 morphant (G) at 16 hpf. H-L. WISH of ctslb at 28 hpf showing hatching gland cells that are organized in a belt-like structure in wild type sibling (H), standard control- (J) or 5mm control morpholino-injected (K) embryo, but disorganized and scattered in spint2 mutant (I) and spint2 morphant (L). M. Quantification of the hatching gland cell phenotype of mutants and morphants shown in H – L. N=3 independent experiments, n=53-73 embryos.
Hatching is facilitated by the secretion of chorion-degrading enzymes by the so-called hatching gland cells. Hatching gland cells are mesendodermal derivates located in the epidermis on the yolk sac (Inohaya et al., 1997). They derive from the prechordal plate / polster and strongly express the specification marker cathepsin Lb (ctslb, formerly also called hgg1) (Schier et al., 1997) already at this early stage. In spint2 mutants and morphants, ctslb expression patterns and levels are as in wild-type siblings at the end of gastulation (10 hpf; Figure 3D,E for mutant; morphant not shown) and during somitogenesis (16 hpf) (data not shown). Moreover, at 16 hpf, hatching gland cells of spint2 mutants and morphants display normal expression of he1.1, coding for hatching enzyme (Inohaya et al., 1997; Sano et al., 2008) (Figure 3F,G for morphant; mutant not shown). Furthermore, mutants and morphants have normally developed intracellular granules, characteristic vesicular structures filled with hatching enzymes (Fig. 3B,C for morphant; mutant not shown). However, at 28 hpf, differences in the morphology of the hatching glands have become apparent. In wild-type embryos, hatching gland cells are located in a belt-like stripe on the yolk sac, whereas in spint2 mutants (Figure 3H,I,M; n=25/25) and morphants (Figure 3J-M; n=64/64), hatching gland cells appear more scattered and disorganized, pointing to an essential role of Spint2 during hatching gland morphogenesis.
Hatching gland cells display radial cell-cell intercalations and collective cell migration between the two layers of the embryonic epidermis
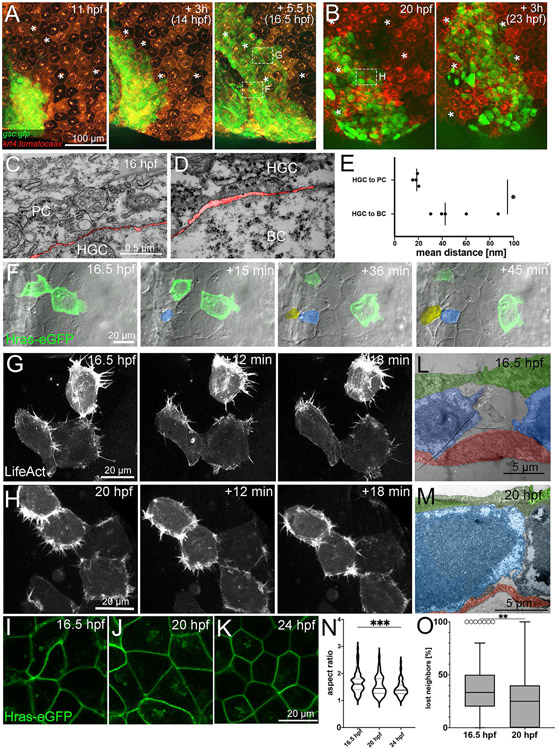
By the end of gastrulation (10 hpf), hatching gland cells have accumulated at the front of developing head, forming a structure named the polster (Melby et al., 1996; Shih and Fraser, 1996), from where they subsequently move onto the yolk sac (Inohaya et al., 1997). Thus, the aforementioned alterations in the spatial organization of hatching gland cells in spint2 deficient embryos that appear between 16 and 28 hpf (see above; Figure 3F-M) could result from defects during such later steps of hatching gland morphogenesis. Whereas the behavior of their progenitor cells during prechordal plate formation is well understood (Montero et al., 2005; Montero and Heisenberg, 2004; Ulrich et al., 2005), the cellular mechanisms underlying the subsequent steps of hatching gland morphogenesis had not been studied in detail as yet. As a first step to do so, we performed a time-series analysis of sections through wild-type embryos stained with the hatching gland cell marker ctslb, and counterstained with a tp63 antibody to label basal keratinocytes (Lee and Kimelman, 2002) or with a GFP antibody to label Tg(krt4:egfp)-expressing surface keratinocytes / peridermal cells (Gong et al., 2002). At the end of gastrulation (10 hpf), the polster consists of a multi-layered mass of hatching gland precursor cells located below the basal keratinocytes in anterior-most positions of the embryonic axis (Figure 4A). During early segmentation stages (14-16 hpf), this cell mass undergoes a displacement through the basal keratinocyte layer to become located above, rather than below it, while still maintaining its multi-layered organization (Figure 4B,C). Shortly later (18 hpf; Figure 4D), however, hatching gland cells most distant from the head have acquired a monolayer organization, while at 24 hpf, the entire hatching gland anlage consists of a single, but correspondingly larger cell layer located between the basal and the outer layer of the epidermis (Figure 4E,G). A similar rearrangement to fewer but larger layers occurs during epiboly, a morphogenetic movement of gastrulation, which has been shown to be largely driven by uni-directional radial cell intercalations (Kane et al., 2005; Warga and Kimmel, 1990). Indeed, similar cell arrangements indicative of intercalations were observed in transmission electron micrographs (TEM) of hatching gland cells in the multi-to-mono-layer-transition zone (Figure 4J,K). Furthermore, actual intercalation events of hatching gland cells from lower to upper layers, but not vice versa, were recorded in time-lapse videos of live embryos via DIC microscopy (Figure 4L, Supplementary Movie 1), identifying uni-lateral cell intercalations as one of the cellular mechanisms underlying hatching gland morphogenesis (Figure 4M).
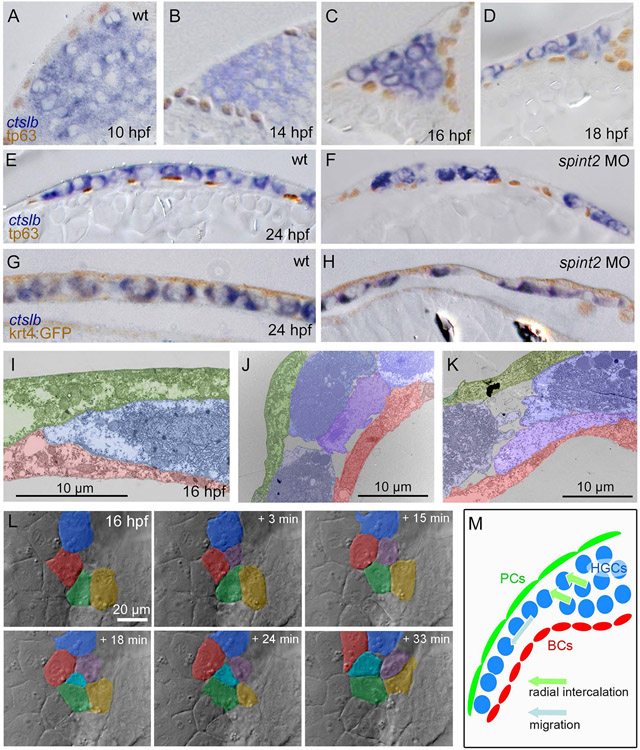
Figure 4: Hatching gland cells undergo intercalations driving the transition from a multi-layered cluster to a single-layered sheet and become localized between the peridermal and basal keratinocyte layer of the epidermis.
A-F. Sections of embryos stained for ctslb mRNA and tp63 protein. Hatching gland precursor cells are organized in a multi-layered cluster that is located below basal keratinocytes at 10 hpf (A) but above basal keratinocytes at 14 hpf (B) and 16 hpf (C). At 18 hpf, the multi-layered cluster has started to rearrange to a mono-layered organization, which is most advanced at the anterior side of the anlage, where hatching gland cells have started to spread onto the yolk sac (D). At 24 hpf, hatching gland cells are organized in single layer that is a continuous sheet in wild type (E) but has gaps in spint2 morphant (F). G-H. Sections of embryos transgenic for the peridermal marker Tg(krt4:egfp) stained for ctslb mRNA and eGFP protein at 24 hpf. Hatching gland cells are located below the peridermal cells in wild-type (G) and morphant (H) embryos. I-K. TEM sections of wild-type embryos at 16 hpf showing hatching gland cells (HGC, false-colored in blue) in between peridermal cells (PC, false-colored in green) and basal keratinocytes (BC, false-colored in red) at the leading edge of the sheet (I) and at the transition from a bi- to the mono-layered state (J,K). L. Stills from a 33 min DIC time-lapse recording of the top layer of hatching gland cells next to the head region, starting at 16 hpf. False-coloring of a subset of hatching gland cells reveals cells intercalating from below. M. Schematic illustrating intercalation and migration events of hatching gland cells around 16 hpf. Hatching gland cells (HGCs) are marked in blue, peridermal cells (PCs) in green, and basal cells (BCs) in red.
In addition, hatching gland cells appear to undergo active cell migrations (Figure 4M). Time-lapse recordings of Tg(−1.8gsc:gfp) (Doitsidou et al., 2002) and Tg(krt4:tomatocaax) (Richardson et al., 2016; Teixeira Rosa et al., 2019) double transgenic embryos with labelled hatching gland and peridermal cells, respectively, revealed that hatching gland cells change positions relative to the periderm during all stages of hatching gland morphogenesis, thus, when still organized in a multi-layered cell mass, as well as during the multi-to-mono-layer transition and as a mono-layered cell sheet (Figure 5A,B; Supplementary Movies 2,3). In TEM images, we found contacts of hatching gland cells to overlaying peridermal cells to be markedly closer than to the underlying basal cells (Figure 5C-E). This suggests that hatching gland cells might primarily use the inner surface of peridermal cells as their migration substrate, similar to the behavior of their progenitors, the prechordal plate cells, which during gastrulation undergo active cell migration on the inner surface of the overlaying ectodermal cells (Montero and Heisenberg, 2004).
Figure 5: Hatching gland cells undergo active cell migration along peridermal cells.
A,B. Fluorescent stills from a time-lapse recording of an embryo transgenic for Tg(−1.8gsc:gfp) and Tg(krt4:tdtomatocaax), starting at 11 hpf (A) and 20 hpf (B). * indicate identical peridermal cells at each time point. Boxed regions in images at 16.5 hpf and 20 hpf indicate regions for which subsequent time-lapse recordings are presented in (F,G,H) at higher magnifications. C,D. TEM sections of contact zones between hatching gland cell (HGC) and peridermal cell (PC) (C) and between HGC and basal keratinocyte (BC) (D) at 16 hpf. The interspace between cells is false-colored in red. E. Quantification of the spaces between hatching gland cells and peridermal and basal cells, respectively. Each dot represents one cell-cell contact. N=2 embryos, n= 4-5 cells. * indicates p value of 0.0213 determined via a Student’s t test. F. Bright-field and fluorescent overlay stills from a time-lapse recording (Supplementary movie 4) of multi-layered zone as indicated in third image of (A), in this case of a wild-type embryo with two transplanted GFP-positive cells transgenic for Tg(Ola.Actb:Hsa.hras-egfp), starting at 16.5 hpf. Cells intercalating from below are false-colored in yellow and blue. G,H. Fluorescent stills from time-lapse recordings (Supplementary movies 5 and 6) of HGCs in mono-layered zone as indicated in third image of A (G) and first image of B (H) of wild-type embryos injected with a 1.8gsc:LifeAct-Ruby plasmid labeling active actin cytoskeletal rearrangements, starting at 16.5 hpf (G) or 20 hpf (H). L,M. TEM sections through HGCs (false-colored in blue) between peridermal cells (PC, false-colored in green) and basal keratinocytes (BC, false-colored in red) at 16.5 hpf (L) or 20 hpf (M). I-K. Confocal images of cell membranes of HGCs of embryos transgenic for Tg(Ola.Actb:Hsa.hras-egfp) to label cell membranes at 16.5 hpf (I), 20 hpf (J), or 24 hpf (K). N. Quantification of aspect ratio of HGCs shown in I-K in violin blot; N=5-9 embryos, n=39-72 cells; *** indicates p value of 0.0001 determined via one-way ANOVA and Tukey’s post hoc test, non-significant differences are not indicated. O. Tukey box and whiskers plot displaying the percentages of neighbors of HGCs that were lost during a tracking period of 50 min in embryos transgenic for Tg(−1.8gsc:gfp), starting at 16.5 hpf or 20 hpf (see also Figure 6A and I). N=3 embryos each, n= 28-57. ** indicates p value of 0.0013 determined via Student’s t test.
Time-lapse recordings of mosaic embryos generated via cell transplantations or plasmid injections, with individually labelled hatching gland cells either in the intercalation zone (with membrane-bound GFP; Figure 5F, Supplementary Movie 4) or in the mono-layered zone (with LifeAct-Ruby; Figure 5G, Supplementary Movie 5), further revealed prominent protrusive activities and cytoskeletal rearrangements within hatching gland cells, indicative of active cell migration. At the edge of the intercalation zone, two neighboring hatching gland cells become separated by several cell diameters within 45 minutes, with much higher protrusive activity of the anterior-wards departing cell, indicative of active cell migration. In addition, this cell seems to be further “pushed” forward by cells intercalating in its back from lower layers, thereby further contributing to the separation of the two former neighbors (Figure 5F and Supplementary Movie 4). But also in the mono-layered part of the gland anlage, where all cells move with similar speeds, cells display high protrusive activity and a dynamic dissociation and re-association behavior, as particularly evident upon visualization of the dynamic actin cytoskeleton (Figure 5G, Supplementary Movie 5), while membrane labelling reveals that despite these dynamics, cells have polygonal shapes and are tightly attached to each other, pointing to epithelial-like properties (Figure 5I).
We also compared the characteristics of hatching gland cells during such early stages of monolayer / sheet migration (16.5 hpf) with later stages of sheet migration (19-20 hpf) and when migration has been completed and the definitive, mono-layered gland is formed (24 hpf). Strikingly, the combination of protrusive cellular activity (Figure 5G,H, Supplementary Movie 5,6) and epithelial characteristics / polygonal cell shapes (Figure 5I-K) persists throughout all stages. However, the tissue becomes progressively more compact (Figure 5H, Supplementary Movie 6), with fewer and smaller gaps between cells (Figure 5L,M). Furthermore, cells become less elongated (Figure 5N) and dynamic re-arrangements within the cell sheet decrease. To address the latter, we tracked neighboring hatching gland cells for 50 min, using the Tg(−1.8gsc:gfp) line (see also below; Figure 6A). Already at early migration stages (16.5 hpf + 50 min), each individual hatching gland cell only loses an average of 35.7 +/−26.2% of its cellular neighbors during the 50 min interval, while at later migration stages, this number has even further dropped (26.1 +/− 22.1%; Figure 5O), pointing to a progressively less dynamic, but yet truly collective sheet migration.
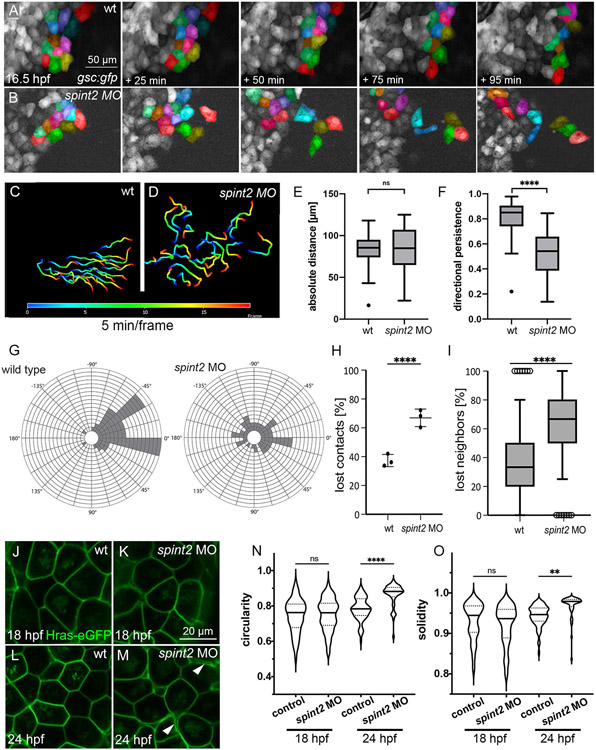
Figure 6: spint2 is required for cohesiveness and collective migration of mono-layered hatching gland cells.
A-B. Fluorescent stills of time-lapse recordings of embryos transgenic for Tg(−1.8gsc:gfp) starting at 16.5 hpf, showing less-organized migration routes of individual hatching gland cells in spint2 morphants (B) compared to wild type (A). A subset of cells is false-colored. C-D. Migration routes of 20 individual hatching gland cells tracked for 90 min starting at 16.5 hpf in a wild-type embryo (C) and spint2 morphant embryo (D), determined using the Axiovision software. The color code indicates the frame number from 0 (16.5 hpf; dark blue) to 20 (18 hpf; red). E,F. Quantification of the absolute migration distance (E) and directional persistence determined by the ratio of the linear displacement to the absolute migration distance (F) of hatching gland cells tracked in C and D; Tukey box and whiskers plot; N=3 embryos, n= 20 cells; p=0.7271 (ns) or <0.0001 (****) (Student’s t-test). G. Rose blots displaying the angle of the migration directionality of individual cells tracked in C and D relative to the average angle of all cells. Each field indicates one cell. H. Quantification of the number of lost cell-cell contacts of neighboring cells tracked for 50 min in wild-type and spint2 morphant embryos starting at 16.5 hpf; N=3, n=72-255, p=0.0023 (Student’s t-test); each dot represents the average percentage of lost contacts in one embryo, error bars represent the standard deviation. I. Tukey box and whiskers plot representing the percentage of neighbors of a cell that were lost during a tracking period of 50 min starting at 16.5 hpf in wild-type (also shown in Figure 5O) and spint2 morphant embryos; N=3 each, n=28-64; **** indicates p value < 0.0001 determined via Student’s t test. J-M. Live confocal images of hatching gland cells of embryos transgenic for Tg(Ola.Actb:Hsa.hras-egfp) at 18 hpf (J,K) or 24 hpf (L,M). Hatching gland cells display a polygonal shape in wild-type and early spint2 morphants (J,K,L) but are rounded up in 24 hpf spint2 morphants (M) Arrowheads point to detachments between adjacent HGCs. N,O. Quantification of the cell shape of HGCs shown in J-M. Violin plots show that the circularity (perfect circular shape =1; elongated shapes <1) as well as the solidity (perfect convex shape =1; gonal shapes <1) indices are not different in wild-type and spint2 morphants at 18 hpf but increased (more circular and less polygonal) in spint2 morphants at 24 hpf; N=5-12 embryos, n=51-94 cells; ** indicates p value of 0.008, **** indicates p value < 0.0001 determined via one-way ANOVA and Tukey’s post hoc test.
Together, this indicates that hatching gland cells within the monolayer zone of the hatching gland anlage are rather cohesive, a hallmark of many tissues or cell sheets undergoing collective cell migrations, including the zebrafish hatching gland primordium during late gastrula stages (Montero and Heisenberg, 2004). In conclusion, in addition to radial cell intercalations in the multi-to-mono-layer-transition zone, active and collective cell migration in the resulting, epithelial-like monolayer contributes to the progressive movement of the hatching gland anlage between the two layers of the epidermis away from the head region to its destination on the yolk sac (Figure 4M).
spint2 is required for cohesiveness and collective migration of mono-layered hatching gland cells
In spint2 mutants, early stages of hatching gland morphogenesis, including the intercalation-driven transition from the multi-layered to mono-layered organization, occurs normally (data not shown). However, after this transition, hatching gland cells of spint2 mutants and morphants are not organized in a continuous sheet like in wild-type siblings and embryos injected with standard control or spint2 5-mismatch control morpholinos, but appear more scattered and often more distant from each other (see Figure 3H-M for whole mount image at 28 hpf and Figure 4E-H for sections at 24 hpf), suggesting that cohesion among migrating hatching gland cells might be compromised. As a first step to test this notion, we performed time-lapse recordings of hatching gland cells in the monolayer zone of Tg(−1.8gsc:gfp) transgenic embryos (Doitsidou et al., 2002) as described above (Figure 5O), now comparing spint2 morphants or spint2fr49/49 mutants with their wild-type siblings from 16.5 through 18 hpf (Figure 6A,B, Supplementary Movies 7,8,9). Tracking of randomly picked individual hatching gland cells of such time-lapse recordings (Figure 6C,D) revealed that the absolute migration distances (and thus the migration velocities) are not altered in spint2 morphants, indicating that their migration potential is not compromised (Figure 6E). However, the directional persistence, as determined by the ratio of the linear displacement to the total trajectory length, is significantly reduced in spint2 morphants, with a much higher standard deviation (Figure 6F). The latter already suggests that spint2 morphant hatching gland cells migrate in a less coordinated manner. This becomes even more apparent when determining the migration direction of individual cells relative to the average direction of the entire cell population. Thus, in wild-type embryos more than 75% of the migrating hatching gland cells remain in a 90° interval, compared to only approximately 40% in spint2 morphants (Figure 6G).
Furthermore, directly addressing the collective migration behavior of hatching gland cells by tracking neighboring cells of 16.5 hpf embryos for 50 min, we found that in wild-type embryos, 37.3+/−4.2% of contacts between neighboring cells are lost during this time frame, compared to 66.9+/−6.1% in spint2 morphants (Figure 6A,B,H). Furthermore, individual hatching gland cells of spint2 morphants in average lose 65.1 +/− 25.7% of their cellular neighbors during this time window, compared to 35.7 +/− 26.2% in wild types (Figure 6I), indicating that their collective migration is severely compromised.
Despite these alterations in collective cell migration, hatching gland cells of spint2 morphants at 18 hpf still have a polygonal shape as in wild-type controls (Figure 6J,K,N,O). However, whereas in wild types, these polygonal cell shape persists even throughout stages when collective cell migration has largely ceased (24 hpf), hatching gland cells of spint2 morphants have rounded up and many have lost contacts to neighboring cells by 24 hpf (Figure 6L-O). Together, this points to reduced cohesiveness and/or increased dynamics among hatching gland cells as the likely cellular basis of the aberrant hatching gland morphogenesis, even though a reflection of such cohesion defects by altered cell shapes of hatching gland cells only becomes obvious several hours later.
spint2 is required for a longer survival of hatching gland cells
But why do spint2 mutants and morphants fail to hatch? Simply because of the aberrant morphogenesis of the hatching gland? In formerly described mutants lacking the Krüppel-like transcription factor Klf17 (formerly known as Klf4), failed hatching is due to failed differentiation of hatching gland cells, reflected by the absence of ctslb expression from earliest stages onwards (Gardiner et al., 2005; Suzuki et al., 2019). In contrast, hatching gland cells of spint2 morphants and mutants display unaltered expression levels of ctslb and other differentiation markers (see above; Figure 3B-L), indicating that failure of hatching is not due to a compromised specification and differentiation of hatching gland cells. However, whereas in live wild-type embryos at 48 hpf, hatching gland cells full with granules are clearly visible on the yolk sac (Figure 7A), no such cells are detectable in spint2fr49/fr49 mutants (Figure 7B). Lineage-tracing experiments expressing Cre recombinase under the control of the gsc promoter in combination with the Tg(ubi:Zebrabow-M)a131 reporter line to label hatching gland precursor cells at late gastrula stages revealed normal numbers of descendants during the first 24 hours of development (data not shown), but their complete absence in spint2 morphants at 48 hpf (Figure 7C,D). Performing a time series of ctslb WISH analyses, we found no significant differences in the number of ctslb-positive hatching gland cells in spint2 morphant or mutant embryos compared to their wild-type siblings at 18 hpf (18-somite stage, 247+/−52, n=8, compared to 271+/−42, n=7) and at 28 hpf (248+/−28 compared to 232+/−35) (Figure 7E,F,K). However, this changes during the second day of development. Whereas in wild-type or 5-mismatch control morpholino-injected embryos, cell numbers stay constant until shortly before hatching (257+/−10 at 40 hpf; 241+/−24 and 267+/−62 at 48 hpf), only 27+/−13 cells are present in spint2 mutants at 40 hpf, and 6+/−6 or 9+/−10 cells at 48 hpf in mutants and morphants (Figure 7G-K). TUNEL staining at 36 hpf further revealed dying hatching gland cells on the yolk sac in mutant and morphant but not in wild-type embryos (Figure 7L-N). This indicates that Spint2-deficient hatching gland cells, although initially properly specified, die before the degranulation process, during which hatching enzymes would have been secreted.
Figure 7: spint2 is required for hatching gland cell survival before hatching.
A-B. DIC images of the head and yolk region of 48 hpf old wild-type (A) and spint2fr49/fr49 mutant (B) embryos. Inlets show a magnified view of the hatching gland cell region, in which granule-containing cells are observed in wild-type but not in mutant embryos. C-D. Overlay of DIC and fluorescent images of 48 hpf embryos transgenic for Tg(gsc:Cre)fr44Tg and Tg(ubi:Zebrabow-M). In wild type (C) gfp-expressing hatching gland cells are visible, whereas they are absent in spint2 morphant (D). E-K. WISH of ctslb in wild-type siblings (E,G,I) and spint2 mutants (F,H,J), revealing similar numbers of hatching gland cells in mutants and wild-type siblings at 28 hpf, but progressively fewer hatching gland cells in mutants compared to wild types at 40 hpf and 48 hpf. K. Quantification of numbers of ctslb-positive hatching gland cells at 18 hpf, 28 hpf, 40 hpf, 48 hpf in wild-type and spint2 mutant embryos, and at 48 hpf in spint2 morphants and siblings injected with 5-mismatch (5mm) control MO. Squares represent single embryos; significances were determined via a one-way ANOVA and Tukey’s post hoc test. Different letters indicate statistically significant differences (p < 0.0001). L-N. TUNEL staining of wild-type (L), spint2 mutant (M) and spint2 morphant (N) embryos at 36 hpf. spint2-deficient embryos display cell death in the hatching gland cells region. O-S. Wild-type hatching gland cells die after hatching. O-Q. Overlays of DIC and fluorescent images of embryos transgenic for Tg(gsc:Cre)fr44Tg and Tg(ubi:Zebrabow-M), showing GFP-positive hatching gland cells shortly after embryos have hatched at 54 hpf (O) but their subsequent loss over the next two days (P,Q). R,S. TUNEL staining of wild-type embryos showing no positive TUNEL cells in the hatching gland tissue in a 54 hpf embryo (R) but few TUNEL-positive hatching gland cells at 78 hpf (S).
Thus far, the fate of hatching gland cells in wild-type embryos after hatching had not been studied. Since upon the secretion of hatching enzymes, cells lose their characteristic granules, they are difficult to detect by DIC optics afterwards. Therefore, we applied the aforementioned lineage tracing approach to follow cells during the further course of development. Shortly after hatching (54 hpf), the hatching gland tissue is still densely packed and negative for TUNEL staining (Figure 7O,R). However, during the next two days, TUNEL-positive cells appear and the number of hatching gland cells drops dramatically (Figure 7P,Q,S), similar to the formerly described disappearance of hatching gland cells only markedly after hatching during Xenopus development (Drysdale and Elinson, 1991).
Together, this suggests that Spint2 is required to postpone the death of fully differentiated hatching gland cells for several days, thereby allowing the cells to de-granulate and the embryos to hatch.
spint2 is required in hatching gland cells themselves to promote their cohesive properties, collective migratory behavior and survival
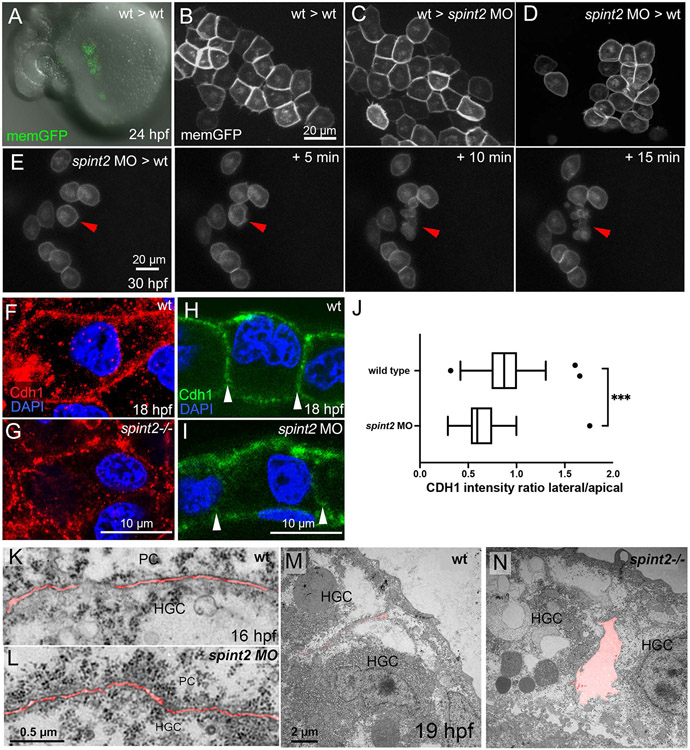
To get first insights into the molecular and cellular mechanisms via which Spint2 regulates hatching gland morphogenesis and hatching gland cell survival, we carried out tissue autonomy studies. spint2 is expressed both in hatching gland cells themselves as well as in the periderm (see above; Figures 1E-K), the directly adjacent tissue and potential adhesion and migration substrate (Figure 5C-E) and could therefore in principle be required in either of the two tissues. To distinguish between these possibilities, we generated chimeric embryos with a spint2 morphant hatching gland anlage in a wild-type environment or vice versa, transplanting labeled polster cells (the hatching gland cell precursors) into host embryos in which the endogenous mesendoderm had been chemically suppressed (Richardson et al., 2016; Shih and Fraser, 1996). Strikingly, upon transplantation of wild-type polster cells into spint2 morphant hosts (wt -> spint2 MO), wild-type hatching gland cells displayed wild-type-like polygonal cell shapes (Figure 8C) indistinguishable from those of wild-type cells in wt -> wt control transplants (Figure 8B), in addition to a wild-type-like spatial organization of the hatching gland itself (Figure 8A and data not shown). In contrast, upon transplantation of spint2 morphants polsters into wild-type hosts (spint2 MO -> wt), morphant hatching gland cells displayed mutant-like roundish cell shapes (Figure 8D), indistinguishable from those of non-chimeric spint2 morphants (see above; Figure 6M), in addition to a mutant-like spatial organization of the hatching gland itself (data not shown). Furthermore, as in non-chimeric mutants and morphants (see above; Figure 7L-N), spint2 morphant hatching cells in such spint2 MO -> wt chimeras underwent precocious cell death (Figure 8E; 6/6 chimeras), whereas wild-type cells in wt -> spint2 MO chimeras did not (data not shown; 0/7 chimeras). Together, this indicates that spint2 is required in hatching gland cells themselves to promote their cohesiveness, collective migration and survival, rather than in surrounding tissues such as the overlying layer of peridermal cells.
Figure 8: spint2 acts cell-autonomously to promote cohesiveness and survival of hatching gland cells.
A. Overlay of DIC and fluorescent image of live 24 hpf wild-type host, in which endogenous mesendoderm was chemically suppressed, and which was transplanted with polster cells (precursors of hatching gland cells) of a wild-type donor labeled with membrane-bound GFP (via transgene Tg(Ola.Actb:Hsa.hras-egfp)). B. Magnified view of fluorescently labeled hatching gland cells in A. C. Magnified view of fluorescently labeled wild-type hatching gland cells whose precursors had been transplanted into a spint2 morphant host, as in A. D. Magnified view of fluorescently labeled spint2 morphant hatching gland cells whose precursors had been transplanted into a wild-type host, as in A. E. Stills of a time-lapse recording of GFP-labeled spint2 morphant donor cells in a wild-type host at 30 hpf in 5 min intervals. Red arrowhead points to a cell undergoing cell death. F-I. Immunofluorescence of Cdh1 (E-cadherin) in hatching gland cells at 18 hpf. In whole mounts, cell contacts between wild-type hatching gland cells show continuous Cdh1 (red) localization (F), whereas in spint2fr49/fr49 mutants, Cdh1 localization is disrupted (G). Immunofluorescence on sections showing comparable Cdh1 levels (green) at apical and basal membranes of wild-type (H) and spint2 morphant (I) embryos but reduced Cdh1 levels at lateral membranes (indicated by arrowheads) between hatching gland cells of spint2 morphant. J. Tukey box and whiskers plot showing a decreased ratio of the mean intensity of CDH1 on lateral versus apical cell junctions of HGCs in spint2 morphant embryos compared to wild-type controls; N= 3 embryos, n= 36-38; *** indicates p value of 0.0002 determined via Student’s t test. K-N. Transmission electron microscopy (TEM) sections of wild-type (K) and spint2 morphant (L) embryos at 16 hpf, and of wild-type (M) and spint2fr49/fr49 mutant (N) embryos at 19 hpf, with intercellular spaces false-colored in red. Interspaces between hatching gland cells (HGC) and peridermal cells (PC) of spint2 morphant and wild-type control are unaltered (K,L), whereas interspaces between adjacent hatching gland cells (HGC) are increased in spint2fr49/fr49 embryo compared to wild-type sibling (M,N).
Spint2 acts independently of all tested potential protease targets and other downstream mediators
Spint2 has been reported to inhibit various serine proteases. Well-described and important in vivo targets are the TTSPs Matriptase/St14 (Kawaguchi et al., 2019; Szabo et al., 2019; Szabo et al., 2009; Tsai et al., 2014) and Hepsin (Nakamura et al., 2009), the GPI-anchored protease Prostasin (Faller et al., 2014; Szabo and Bugge, 2018), and membrane-associated Plasmin (Wu et al., 2019) (see Introduction). We attempted to identify the relevant protease whose deregulation is causative of the hatching gland phenotype obtained upon loss of Spint2 function in zebrafish embryos. Of note, in other systems, such membrane-tagged target proteases have been shown to be present in and on the same cells like the Spint1/2 proteins themselves, with physical binding between Spint1/2 and the proteases in the endoplasmatic reticulum, in secretory vesicles, and on the plasma membrane to regulate their trafficking, activity and shedding (Larsen et al., 2013; Murray et al., 2017; Nonboe et al., 2017; Oberst et al., 2005; Szabo et al., 2008). Therefore, and since we found Spint2 to be required in hatching gland cells themselves (see above; Figure 8A-E), potential Spint2 targets were first tested for hatching gland expression, followed by loss-of-function studies in case of positive expression results (data not shown, but summarized in Table 1). Applying different phylogeny and BLAST analyses, we found four zebrafish St14 orthologs. Of those, only st14c was weakly expressed in hatching gland cells. However, morpholino knockdown of spint2 together with three different morpholinos against st14c still resulted in scattered hatching gland cells. Additionally, the spint2 phenotype was not rescued upon concomitant genetic loss of st14c (mutant generated via TALEN technology; see Materials & Methods). Therefore, we conclude that the relevant target is not an St14 ortholog. The same applies to the single Hepsin orthologue hpn (Khandekar and Jagadeeswaran, 2014), which was not expressed in hatching gland cells. In contrast, most of the identified six zebrafish prostasin-related zebrafish genes showed hatching gland expression. Yet, single or combinatorial knockdown of them by morpholinos alleviated neither the scattered hatching gland morphology, nor the hatching gland cell death and the hatching deficiency phenotype of spint2 mutants or morphants. Similar negative results were obtained upon morpholino-mediated knockdown of plasminogen (pln) (Cheng et al., 2006), encoding the single zebrafish Plasmin precursor, and upon knockdown or chemical inhibition of multiple growth factor receptors and signaling pathways known as crucial targets or mediators of Matriptase function in mammalian systems (Tanabe and List, 2017). In conclusion, we can rule out the most likely candidates as being responsible for the hatching gland defects of zebrafish spint2 mutants, while the relevant target protease(s) remain unknown. Of note, also in the mouse, only some of the phenotypic traits of Spint2 mutants are Matriptase-dependent, while the proteases responsible for the development of the other traits are not known to date (Kawaguchi et al., 2019; Szabo et al., 2019; Szabo et al., 2009).
Table 1:
Summary of tested potential targets / mediators of Spint2.
| Gene | Expression in hatching gland cells |
lof method | Phenotype | Rescue of spint2 lof hatching gland phenotype |
|---|---|---|---|---|
| Zebrafish orthologs of mammalian St14 (Matriptase) | ||||
| st14a | no | |||
| st14b | no | |||
| zmp:0000001114 (st14c) | yes | 2 ATG MO, st14cfr50 TALEN mut. | no | |
| CR352265.1 (tmprss7) | no | |||
| Zebrafish orthologs of mammalian Prostasin | ||||
| zgc:92313 | weak | ATG MO | Shorter axis | no |
| si_dkey-16l2.17 | no | |||
| zgc:100868 | yes | ATG MO | Mild blistering phenotype 48 hpf | no |
| prss60.1 | n.d. | ATG MO | Developmental delay, blistering phenotype | no |
| prss60.2 | yes | 2 ATG MO | no | |
| prss60.3 | n.d. | splice MO | Curved axis, blistering phenotype | no |
| Other serine proteases | ||||
| hepsin | no | |||
| plasminogen | n.d. | ATG MO | no | |
| Serine protease signaling targets | ||||
| c-met | n.d. | ATG MO | no | |
| mst1ra | n.d. | ATG MO | no | |
| mstr1b | n.d. | ATG MO | no | |
| par2b | yes | ATG MO | Rescue of hai1a epidermal phenotype | no |
| EGFR signaling | Inhibitor PD168393 | Rescue of hai1a epidermal phenotype | no | |
| Pi3K signaling | Inhibitor LY294002 | Mild HGC migration phenotype | no | |
Expression was determined by WISH at 16.5 −24 hpf. The general phenotype obtained upon loss of function (lof) of the candidates in otherwise wild-type embryos is indicated when observed. The potential rescue of the spint2 hatching gland phenotype upon loss of function of the candidates in spint2 morphants was assayed for hatching gland morphogenesis, hatching gland cell survival, and hatching.
Hatching gland cells of spint2 mutants display altered E-cadherin localization
Even though lacking information about the relevant direct Spint2 target proteases involved in hatching gland morphogenesis and hatching gland cell survival, we continued to study possible downstream effectors. Cell-cell adhesion molecules possibly involved in hatching gland cell cohesion that had been formerly reported in the context of Spint1/2 proteins and in the context of cell survival are E-cadherin, also called Cdh1 (Carney et al., 2007; Szabo and Bugge, 2018; Wu et al., 2019) and the epithelial cell adhesion molecule EpCAM (Huang et al., 2018; Kawaguchi et al., 2019; Szabo et al., 2019; Wu et al., 2017). Both of them are expressed in zebrafish hatching gland cells, cdh1 from early prechordal plate stages onwards and known to be indispensable for prechordal plate cell migration (Dumortier et al., 2012; Montero and Heisenberg, 2004), and epcam only during later stages of hatching gland morphogenesis (Slanchev et al., 2009).
Using whole mount and section immunofluorescence analysis, we found E-cadherin rather evenly distributed along the cell membranes of mono-layered hatching gland cells of wild-type embryos. In comparison, in spint2 mutants and morphants of 18 hpf, E-cadherin levels were significantly reduced at lateral membranes of hatching gland cells, but rather normal at their apical and basal membranes facing peridermal and basal keratinocytes, respectively (Figure 8F-J). Consistently, TEM analysis revealed unaltered distances between hatching gland and peridermal cells of spint2 morphants both at 16 hpf (Figure 8K,L) and 19 hpf (data not shown), whereas gaps between adjacent hatching gland cells within the hatching gland monolayer were markedly increased (Figure 8M,N). These differential alterations in E-cadherin distribution and hatching gland cell adhesiveness caused by loss of Spint2 function are in line with the observed reduction in hatching gland cohesion (see above; Figures 6J-O and 8A-D) on one hand, but the unaltered velocity of their migration (see above; Figure 6E), which most likely occurs at the inner surface of the periderm, on the other hand. Together, this suggests that Spint2 might regulate cohesion and thereby collective migration of hatching gland cells via E-cadherin.
Spint2 and E-cadherin genetically interact to promote hatching gland morphogenesis and survival and E-cadherin can partially compensate for the loss of Spint2 to promote hatching gland cell cohesion
Mutants and morphants in cdh1 have been shown to display defects in the cohesion and collective migration of prechordal plate cells, the progenitors of the hatching gland cells (Dumortier et al., 2012; Montero and Heisenberg, 2004) long before defects are seen in spint2 deficient embryos - which precludes direct studies on potential Spint2-dependent essential roles of E-cadherin during such later stages of hatching gland morphogenesis. Instead, we performed genetic interactions studies, partially knocking down cdh1 gene function by injecting low amounts of a cdh1 morpholino (Montero and Heisenberg, 2004) in combination with low amounts of spint2 morpholino to study whether the two manipulations synergistically enhance each other’s effect. Indeed, whereas the injection of each of the morpholinos alone at this low concentration did not result in a phenotype, the combination of both caused a scattering of hatching gland cells (Figure 9A-C) similar to what was observed upon full knock down of spint2 (Figure 9D). Furthermore, the combined treatment had a synergistically enhancing effect on the death of hatching gland cells, indicated by increased numbers of TUNEL-positive hatching gland cells at 36 hpf (Figure 9E-I), reduced hatching gland cells numbers at 44 hpf (Figure 9J), and reduced hatching rates at 72 hpf (Figure 9K). In contrast to the hatching gland cell morphogenesis itself (Figure 9D), however, all of these later, cell death-related traits of spint2, cdh1 double hypomorphs were significantly weaker than in embryos upon complete loss of Spint2 function (Figure 9J,K).
Figure 9: spint2 and cdh1 genetically interact to promote hatching gland morphogenesis and hatching gland cell survival.
A-C. ctlsb WISH displays normal distribution of hatching gland cells in 34 hpf embryos injected with low amounts of spint2 MO (A) and low amounts of cdh1 MO (B), but a scattered hatching gland phenotype in embryos injected with both (C). Wild-type and full spint2 MO controls as used for quantification (D) are not shown, but compare with Figures 3H,I and 7E,F for 28 hpf. D. Quantification of the scattered hatching gland phenotype analyzed at 34 hpf; n=7-31. E-I. TUNEL staining of 36 hpf embryos shows no TUNEL-positive cells on yolk sac in wild type (E) and in embryos injected with low amounts of spint2 MO (F) or low amounts of cdh1 MO (G), but multiple in embryo injected with low amounts of both (H), and even more in full spint2 morphant (I). J. Quantification of hatching gland cell (HGC) numbers scored by ctslb WISH of 44 hpf embryos injected with different MOs. Squares represent individual embryos; significances were determined via a one-way ANOVA and Tukey’s post hoc test. Different letters indicate statistically significant differences (p < 0.0001). K. Quantification of percentages of embryos hatched after 3 dpf, injected with different MOs; N=3, n=25-94. Significances were determined via a one-way ANOVA and Tukey’s post hoc test. Different letters indicate statistically significant differences (p < 0.0001). L-O. Forced expression of spint2 or cdh1 restores polygonal shapes of spint2 morphant hatching gland cells. L,M. Confocal images of 24 hpf spint2 morphant embryos transgenic for Tg(bact:tdtomato-caax) with mosaic expression of eGFP together with spint2 (L) or cdh1 (M). N,O. Quantification of cell shapes as shown in panels L and M. Violin plots show that compared to regular spint2 morphant control hatching gland cells (GFP−), the circularity (N) as well as solidity (M) indices of spint2 morphant hatching gland cells expressing spint2 or cdh1 (GFP+) are decreased and more similar to wild-type values (compare with Figure 6N,O); N=5-8 embryos, n=38-82 cells; **** indicates p value < 0.0001 determined via one-way ANOVA and Tukey’s post hoc test.
Finally, and most strikingly, analyses of full spint2 morphant embryos injected with bi-cistronic plasmids driving heat-inducible expression of morpholino-insensitive spint2 or cdh1 together with the cell marker GFP to generate transgenic chimeras revealed that elevated levels of Cdh1 in spint2 morphant cells lead to a significant rescue of polygonal cell shapes and thus cohesion among hatching gland cells, similarly to, although more weakly than, the re-introduction of Spint2 itself (Figure 9L-O). Together, these data suggest that Spint2 promotes cohesion and thereby collective migration, as well as, at least to some extent, survival of hatching gland cells via E-cadherin.
In contrast, no genetic interaction in compromising hatching gland morphogenesis or hatching gland cell survival was observed when performing corresponding synergistic enhancement experiments with epcam and spint2 morpholinos, whereas the opposite, a genetic interaction with epcam, but not with cdh1, was revealed for Spint1 during the formerly described effects on the epithelial integrity of the embryonic epidermis (Carney et al., 2007) (Table 2). This provides further evidence for the importance and specificity of E-cadherin, rather than EpCAM, as a mediator of Spint2 function in the hatching gland, while revealing corresponding roles of EpCAM, rather than E-cadherin, downstream of Spint1 in other developmental contexts.
Table 2:
spint1a displays genetic interaction with epcam, but not with cdh1 (E-cadherin), in the developing epidermis, and spint2 with cdh1, but not with epcam, in the developing hatching gland.
| MO knock down | epidermal aggregates | scattered HGCs |
HGCs loss |
hatching defect |
|---|---|---|---|---|
| cdh1 low | − | − | − | − |
| epcam low | − | − | − | − |
| spint1a full | +++ | − | − | − |
| spint1a low | − | − | − | − |
| spint1a low + cdh1 low | − | − | − | − |
| spint1a low + epcam low | +++ | − | − | − |
| spint2 full | − | +++ | +++ | +++ |
| spint2 low | − | − | − | − |
| spint2 low + cdh1 low | − | +++ | + | ++ |
| spint2 low + epcam low | − | − | − | − |
Full or low amounts of morpholino oligonucleotides (MOs) were injected for complete or partial gene knock down. Embryos were assayed for the development of epidermal aggregates at 24 hpf, applying live imaging, and the development of a scattered hatching gland cell (HGC) phenotype at 34 hpf and hatching gland cell loss at 44 hpf, applying ctslb WISH, as well as their hatching rate at 72 hpf. −, no defects; +, weak defects; ++, intermediate defects; +++, strong defects. For spint2 – cdh1 interaction, compare with Figure 9D,J,K.
Discussion
Hatching gland morphogenesis involves dynamic cell intercalations and coordinated sheet migration
Morphogenetic movements of cells and tissues are important developmental processes that shape entire embryos and specific organs. Here, we describe the morphogenesis of the zebrafish hatching gland. Hatching gland cells derive from the mesendodermal prechordal plate and are required for the secretion of enzymes to degrade the chorion and facilitate hatching.
Of note, the hatching gland cells of zebrafish, although of mesendodermal origin, share crucial characteristics with their counterparts of the clawed frog Xenopus laevis, which are supposed to be of ectodermal origin (Drysdale and Elinson, 1991). Thus, as in zebrafish, the specification of the frog cells is dependent on the Krüppel-like factor Neptune (Kurauchi et al., 2010), the Xenopus orthologue of zebrafish Klf17 (Suzuki et al., 2019). Furthermore, the frog hatching gland cells express the orthologous hatching enzymes of the Astacin-family (Katagiri et al., 1997), and, most strikingly, Spint2 (Gawantka et al., 1998).
We show that zebrafish hatching gland morphogenesis involves the rearrangement of a multi-layered cluster, the polster, into a singled-layered epithelial sheet, accompanied by the progressive movement of this sheet between the two layers of the epidermis, the outer periderm and the basal keratinocyte layer, onto the yolk sac. Here, two different types of morphogenetic cell movements are involved, intercalations driving the multilayer-monolayer transition, and collective migration driving the anterior-wards displacement (Figure 4). Intercalation requires very dynamic changes of cell-cell contacts, and adhesiveness of cells has to be locally and temporally reduced but restored elsewhere and/or slightly later. We found similar dynamics to be at play during the subsequent collective migration of the cohesive monolayer. Over the course of this collective migration, the initially rather loosely organized hatching gland cells become more tightly attached to each other, lose neighbors less frequently, and display a change in cell shape towards more regular, epithelial-like polygonal characteristics (Figure 5). In line with this notion, they express epithelial markers as E-cadherin (cdh1; this study), Epithelial cell adhesion molecule (epcam; Slanchev et al., 2009), and the Claudins encoded by cld7b (Li et al., 2017) and cldnb (de la Paz et al., 2017). Together, this suggests that hatching gland cells need to acquire epithelial properties as a prerequisite of collective sheet migration.
Due to the employment of similar cellular mechanisms and, most likely, the underlying molecular control systems, and due to their large temporal overlap (see Figures 4M and 5F / Supplementary Movie 4), we were thus far unable to functionally dissect the impacts of sheet migration (at the front of the anlage) versus radial cell intercalations (at the rear of the anlage) on the movement of the hatching gland onto the yolk sac. However, while at later stages of hatching gland morphogenesis (20 hpf; Figure 5B), sheet migration seems to be the sole driver of hatching gland movement, as intercalations have ceased, we assume that at earlier stages both participate in force generation and are even interconnected, as is the case for several other morphogenetic movements of development and regeneration, such as convergent extension during zebrafish gastrulation (Tada and Heisenberg, 2012), or cutaneous wound healing in adult zebrafish (Richardson et al., 2016).
We also present evidence that the collective migration of zebrafish hatching gland cells strongly depends on E-cadherin-mediated cell cohesion among hatching gland cells, and that this requirement becomes evident even before cells display a corresponding loss of their polygonal shapes and epithelial integrity. A similar, but even earlier role of E-cadherin to promote collective cell migrations has been formerly demonstrated for the progenitors of the hatching cells, the prechordal plate cells, during zebrafish gastrulation (Arboleda-Estudillo et al., 2010; Dumortier et al., 2012; Montero and Heisenberg, 2004). During these early morphogenetic processes, prechordal plate cells use the overlying ectoderm as migration substrate (Montero and Heisenberg, 2004). We do not know whether after their out-wards transit into positions directly underneath the superficial enveloping layer / periderm (Figure 4A-E), the hatching gland cell precursors continue to use the new overlying cell layer, the periderm, as their migration substrate, or the underlying basal keratinocyte layer, or both. However, they are generally more closely attached to the periderm (Figure 5C-E), pointing to the inner surface of the periderm as the likely migration substrate. Notably, already at these early stages, peridermal cells and basal keratinocytes are tightly attached to each other via E-cadherin (Arora et al., 2020), and such attachments have to be released, so that hatching gland cells on their way onto the yolk sac can get in between. We do not know how this opening of the two epidermal layers is accomplished and whether it is regulated by the hatching gland cells or the epidermal cells themselves. However, it seems quite unlikely that the same, thus far unidentified Spint2 target proteases regulating hatching gland cohesiveness and collective migration are involved, as the distance between the two epidermal layers in front of the hatching gland cells (see Figure 4I for wild type) remains unaltered upon loss of Spint2 function (J.H., W.B. and M.H., unpublished results).
Spint2 is required for proper E-cadherin localization and cohesion among migrating hatching gland cells
We identified the Kunitz-type serine protease inhibitor Spint2 as a novel regulator of the cohesiveness of hatching gland cells. The loss of Spint2 function becomes primarily obvious when hatching gland cells have left the multi-layered polster and migrate collectively as a mono-layered sheet. Our time-lapse recording revealed that during regular collective migration of hatching gland cells in wild-type embryos, cells frequently lose and re-establish cell-cell contacts with each other (Figure 5), while in spint2 mutants, cell contacts are lost more frequently and more persistently (Figure 6), so that cells become distributed in a scattered, rather than a sheet-like fashion (Figures 3 and 4). This suggests that the integrity of the monolayer during sheet migration is achieved by proper balancing between proteolysis-mediated (direct or indirect) loss of cell-cell adhesion proteins involved in cohesion among hatching gland cells and its Spint2-mediated inhibition. Yet, we thus far failed to identify the involved proteases that are inhibited by Spint2 (Table 1).
However, we obtained several lines of evidence that they act – most likely indirectly - via the cell-cell adhesion protein E-cadherin. Thus, Spint2-deficient hatching gland cells display reduced E-cadherin levels at their lateral sides, the contact zones with other hatching gland cells. In contrast, E-cadherin levels at the apical and basal sides, where contacts with the overlying periderm and the underlying basal keratinocytes are formed, are unaffected in spint2 mutants (Figure 8). This is in line with the unaltered distances between hatching gland cells and the two epidermal cell layers (Figure 8), and with the unaltered overall migration speed of hatching gland cells - which most likely occurs on the surface of such epidermal cells (Figure 6). Finally, we provide evidence for the functional relevance of the effect of Spint2 on E-cadherin levels, demonstrating that the effects of partial loss of Spint2 function on hatching gland morphogenesis are synergistically enhanced by concomitant partial loss of E-cadherin function and that the defects in the cohesion of hatching gland cells caused by complete loss of Spint2 function can be significantly attenuated upon forced E-cadherin expression (Figure 9). Together, these data identify E-cadherin as a crucial (indirect) target and mediator of Spint2 function.
That E-cadherin levels in spint2-deficient hatching gland cells are only reduced in lateral, but not in apical and basal cell membrane domains further suggests that Spint2 affects membranous E-cadherin levels in a polarized manner, pointing to a potential connection of Spint2 function to the epithelial cell polarity system. The molecular basis of this connection, if existent, is totally unclear. One possibility could be that Spint2 affects secretory vesicular trafficking or endocytosis, both of which can occur in a polarized manner, distinguishing between different cell plasma domains (Román-Fernández and Bryant, 2016; Shafaq-Zadah et al., 2020; Zeng et al., 2017). In line with this notion, mammalian Spint2 has been reported to localize to cytoplasmic structures (Kataoka et al., 2000; Lee et al., 2018), and has been discussed to be involved in secretory pathways to maintain epithelial integrity (Kawaguchi et al., 2019).
Stabilizing effects of Spint2 proteins on E-cadherin levels have also been reported in multiple other contexts, although also here, the underlying molecular mechanisms have not been elucidated. For example, loss of Spint2 in mice with decreased prostasin activity leads to diminished E-cadherin levels in the intestine (Szabo and Bugge, 2018), and decreased levels of E-cadherin as a result of decreased SPINT2 levels were observed in non-small cell lung cancer cells (Ma et al., 2019) and in endometrial cancer cell lines (Nakamura et al., 2011). Furthermore, various serine proteases with the capacity of being inhibited by Spint2 have been shown to promote the disassembly of cell-cell junctions by modulating E-cadherin levels. For example, overexpression of Tmprss4 in lung and colon cancer cells (Jung et al., 2008) results in a loss of E-cadherin, and Matriptase-1 overexpression reduces E-cadherin localization at cell-cell contacts in a breast cancer cell line (Welman et al., 2012) and in the epidermis of zebrafish embryos (Carney et al., 2007).
Matriptase-1 can also act via the cell-cell adhesion molecule EpCAM, in this case via direct proteolytic cleavage, which in turn leads to the additional degradation of Claudins, crucial components of tight junctions and regulators of epithelial cell polarity (Wu et al., 2017). Of note, in the mouse and human intestine, this EpCAM-degrading activity of Matriptase-1 is under the control of Spint2 (Kawaguchi et al., 2019; Szabo et al., 2019; Wu et al., 2017), whereas according to our own synergistic enhancement studies for zebrafish hatching gland development, Spint2 does not interact with EpCAM, but with E-cadherin (Table 2). Rather, we identified Spint1a as the potential regulator of EpCAM during epidermal development (Table 2). A likely explanation for these differential effects and the differences between fish and mammals could be differences in the presence of protease targets of the Spint1/2 inhibitors. Thus, in both the mammalian intestine (see above) and the zebrafish epidermis (Carney et al., 2007), it is Matriptase-1 that mediates the effects of Spint2 or Spint1a on EpCAM, respectively. In contrast, for the role of Spint2 during zebrafish hatching gland development, Matriptase-1 can be ruled out as the relevant protease, as it is not made by hatching gland cells and its global loss fails to rescue or revert the defects (Table 1).
Spint2 is required for persistent directionality of collective hatching gland migration
Of note, diminished E-cadherin-dependent cohesion caused by the loss of Spint2 function does not affect the overall migration speed of hatching gland cells but impairs the directional persistence of their migration, with cells that do not follow one direct route but frequently change their direction and even turn around (Figure 6). This behavior is very similar to what has been formerly described for the loss of E-cadherin in other cell migration systems, including the collective migration of cell monolayers in scrape-wound assays (Desai et al., 2009) and the collective migration of the prechordal progenitors of hatching gland cells during zebrafish gastrulation (Arboleda-Estudillo et al., 2010; Dumortier et al., 2012), as well as the single cell migration of zebrafish primordial germ cells, which migrate along the surfaces of neighboring somatic cells (Grimaldi et al., 2020). In all of these systems, cells establish a front-rear polarity and form protrusions or blebs involved in migration at the side with the weakest E-cadherin-mediated cell-cell contacts. During prechordal plate migration, this further assures that front-rear polarities of migrating cells particular in the back of the group are oriented in parallel, adding to the robustness of the migration and its directionality against environmental challenges (Dumortier et al., 2012). Compared to the rather loosely organized interspace between the ectoderm and the yolk syncytial layer cells have been migrating through during gastrulation, such challenges should be much more pronounced during the later phases of hatching gland morphogenesis, when cells have to migrate between tightly attached epidermal cell layers (Figure 2). This might be one of the reasons why the Spint2 system has only been set up during such later phases of hatching gland morphogenesis, but not during the earlier phases of polster formation.
The identified roles of Spint2 during hatching gland morphogenesis are in line with described pro-invasive effects of Spint2 during cancer metastasis
Cancer metastasis has been shown to often involve collective cell migration (Friedl and Mayor, 2017), and cancer cells that migrate in clusters have a higher potential to metastasize (Janiszewska et al., 2020; Nagai et al., 2020). Interestingly, Spint2 has been suggested to have important functions in cancer cell invasion. In many cases, a downregulation of Spint2 has been implicated with increased cell motility and invasiveness, pointing to a tumor-suppressive role of Spint2. Many of these studies suggest an inhibitory role of Spint2 towards Matriptase and, therefore, hyperactive Matriptase as the cause of carcinogenesis (see for example, Tsai et al., 2014). Interestingly, however, few cases have been reported in which Spint2 promotes tumorigenesis. For example, the human SPINT2 gene has been shown to be aberrantly highly expressed in some non-small lung cell, colon, breast and prostate cancers (Roversi et al., 2018) and has been suggested to have a tumor-promoting role in ovarian cancer (Graumann et al., 2019). Moreover, Spint2 exhibits a positive role in the progression of oral squamous cell carcinomas (OSCC), with loss of Spint2 reducing matrigel invasion of two OSCC lines (Yamamoto et al., 2018). The molecular and cellular mechanism underlying these pro-invasive effects of Spint2 are not fully understood. However, in light of the data presented here, one could speculate that a pro-invasive function of Spint2 is especially important in situations that require a collective migration of metastasizing cells, with Spint2 promoting cohesion and migration robustness and directionality of invading cancer cells.
Spint2 is required for hatching gland cell survival
As another potential tumor-promoting effect of Spint2, and in addition to its function to promote cell cohesiveness and collective cell migration of hatching gland cells, Spint2 is also required for their survival. Whereas wild-type hatching gland cells only die several days after having released the hatching enzymes, spint2-deficient hatching gland cells undergo precocious cell death before they have completely matured and have released the hatching enzymes, resulting in the failure of embryos to hatch. Like during its effect on cohesion, Spint2 does so by acting within hatching gland cells themselves, rather than in adjacent tissues (Figure 8), suggesting that it is required to inhibit / delay an intrinsic cell death program. Increased and/or precocious cell death was also observed in intestinal crypts of adult zebrafish spint2 mutants (Figure 2) and in the atrophic intestinal epithelium of conditional Spint2 knockout mice (Szabo and Bugge, 2018), indicating that this cell survival-promoting role of Spint2 is not restricted to hatching gland cells or zebrafish embryos.
Future studies have to reveal the exact type of this cell death. However, remarkably, as for hatching gland cohesiveness, it seems to involve, at least to some extent, E-cadherin. Thus, upon combinatorial partial loss of spint2 and cdh1 function, the two display a similar synergistic enhancement in causing hatching gland cell death, although this interaction is not as strong as during their effect on cohesiveness and coordinated cell migration (Figure 9). A similar survival-promoting function of E-cadherin-mediated adhesion has also been observed in other systems. For example, loss of E-cadherin junctions in intestinal enterocytes and several other cell types has been shown to lead to anoikis (Bergin et al., 2000; Fouquet et al., 2004; Kantak and Kramer, 1998; Lugo-Martínez et al., 2009; Orford et al., 1999), a special type of programmed cell death that occurs upon the detachment of cells from their adhesion substrates - which can either be the extracellular matrix or other cell surfaces (reviewed in Orford et al., 1999; Paoli et al., 2013). Future experiments have to reveal whether Spint2 affects cell survival solely via such E-cadherin-mediated mechanisms also employed to promote cohesiveness, or whether parallel pathways are also involved, and, if so, at which level downstream of Spint2 they branch off.
Supplementary Material
In vivo time-lapse recording of DIC images of the top layer of hatching gland cells next to the head region, showing intercalation events. A wild-type embryo was mounted at 16 hpf and images were taken in 3 min intervals for 39 min. Hatching gland cells intercalating from lower layers are false-colored.
In vivo time-lapse recording of an embryo transgenic for Tg(−1.8gsc:gfp) and Tg(krt4:tdtomatocaax) starting at 11 hpf. Hatching gland cells labeled with GFP move relatively to the overlaying periderm labeled with tdTomato-caax (compare with Figure 5A for stills). Z-stacks of fluorescent images were recorded in 10 min intervals. Maximum intensity projections were converted into the movie.
In vivo time-lapse recording of an embryo transgenic for Tg(−1.8gsc:gfp) and Tg(krt4:tdtomatocaax) starting at 20 hpf. Hatching gland cells labeled with GFP move relatively to the overlaying periderm labeled with tdTomato-caax (compare with Figure 5B for stills). Z-stacks of fluorescent images were recorded in 15 min intervals. Maximum intensity projections were converted into the movie.
In vivo time-lapse recording of a mosaic wild-type embryo with transplanted cells transgenic for Tg(Ola.Actb:Hsa.hras-egfp), starting at 16.5 hpf, showing two initially neighboring hatching gland cells that become separated, with much higher protrusive activity of the anterior-wards departing cell. Two hatching gland cells intercalating from lower layers are false-colored in yellow and blue. Note that intercalations (left / anterior side) and cell migrations (right / posterior side) take place back to back, and that both most likely contribute to the anterior-wards net movement of the tissue. Bright-field and fluorescent images were recorded in 3 min intervals. Compare with Figure 5F for stills.
In vivo time-lapse recording of wild-type embryos injected with a 1.8gsc:LifeAct-Ruby plasmid, starting at 16.5 hpf, labeling active actin cytoskeletal rearrangements and dis- and reassociation events in hatching gland cells. Z-stacks of fluorescent images were recorded in 3 min intervals. Maximum intensity projections were converted into the movie. Compare with Figure 5G for stills.
In vivo time-lapse recording of wild-type embryos injected with a 1.8gsc:LifeAct-Ruby plasmid, starting at 20 hpf, displaying active actin cytoskeletal rearrangements but no dis- and reassociation events in hatching gland cells. Z-stacks of fluorescent images were recorded in 3 min intervals. Maximum intensity projections were converted into the movie. Compare with Figure 5H for stills.
In vivo time-lapse recording of a wild-type embryo transgenic for Tg(−1.8gsc:gfp) starting at 16.5 hpf, displaying dynamic, but cohesive migration of the hatching gland tissue. Z-stacks of fluorescent images were recorded in 5 min intervals. Maximum intensity projections were converted into the movie. Compare with Figure 6A for stills.
In vivo time-lapse recording of a spint2 morphant embryo transgenic for Tg(−1.8gsc:gfp) starting at 16.5 hpf, showing less-organized migration of the hatching gland cells compared to the wild type (Supplementary Movie 7) . Z-stacks of fluorescent images were recorded in 5 min intervals. Maximum intensity projections were converted into the movie.
In vivo time-lapse recording of a spint2fr49/fr49 embryo transgenic for Tg(−1.8gsc:gfp) starting at 16.5 hpf, showing less-organized migration of the hatching gland cells compared to the wild type. Z-stacks of fluorescent images were recorded in 5 min intervals. Maximum intensity projections were converted into the movie.
Acknowledgements
The authors wish to thank Christel Schenkel and Beatrix Martiny for excellent technical help, Zhiyuan Gong, Dirk Meyer, Alex Schier, and Jacek Topczewski for zebrafish lines and Anna Huttenlocher for providing the LifeAct-Ruby plasmid. Work in the laboratory of M.H. was supported by the German Research Foundation (DFG; SFB 829 and its Z2 project; FOR 2722) and the US National Institute of General Medical Sciences (GM63904).
References
- Arboleda-Estudillo Y, Krieg M, Stühmer J, Licata NA, Muller DJ, Heisenberg CP, 2010. Movement directionality in collective migration of germ layer progenitors. Curr Biol 20, 161–169. [DOI] [PubMed] [Google Scholar]
- Armistead J, Hatzold J, van Roye A, Fahle E, Hammerschmidt M, 2020. Entosis and apical cell extrusion constitute a tumor-suppressive mechanism downstream of Matriptase. J Cell Biol 219, e201905190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P, Dongre S, Raman R, Sonawane M, 2020. Stepwise polarisation of developing bilayered epidermis is mediated by aPKC and E-cadherin in zebrafish. eLife 9, e49064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajoghli B, Aghaallaei N, Heimbucher T and Czerny T (2004). An artificial promoter contruct for heat-inducible misexpression during zebrafish embryogenesis. Dev. Biol 271,416–430. [DOI] [PubMed] [Google Scholar]
- Bergin E, Levine JS, Koh JS, Lieberthal W, 2000. Mouse proximal tubular cell-cell adhesion inhibits apoptosis by a cadherin-dependent mechanism. American J Physiol Renal Physiol 278, F758–768. [DOI] [PubMed] [Google Scholar]
- Carney TJ, Feitosa NM, Sonntag C, Slanchev K, Kluger J, Kiyozumi D, Gebauer JM, Coffin Talbot J, Kimmel CB, Sekiguchi K, Wagener R, Schwarz H, Ingham PW, Hammerschmidt M, 2010. Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. PLoS Genet 6, e1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney TJ, von der Hardt S, Sonntag C, Amsterdam A, Topczewski J, Hopkins N, Hammerschmidt M, 2007. Inactivation of serine protease Matriptase1a by its inhibitor Hai1 is required for epithelial integrity of the zebrafish epidermis. Development 134, 3461–3471. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF, 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Guo L, Zhang Z, Soo HM, Wen C, Wu W, Peng J, 2006. HNF factors form a network to regulate liver-enriched genes in zebrafish. Dev Biol 294, 482–496. [DOI] [PubMed] [Google Scholar]
- Cooper MS, Szeto DP, Sommers-Herivel G, Topczewski J, Solnica-Krezel L, Kang HC, Johnson I, Kimelman D, 2005. Visualizing morphogenesis in transgenic zebrafish embryos using BODIPY TR methyl ester dye as a vital counterstain for GFP. Dev Dyn 232, 359–368. [DOI] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ, 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet 8, e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Paz JF, Beiza N, Paredes-Zuniga S, Hoare MS, Allende ML, 2017. Triazole fungicides inhibit zebrafish hatching by blocking the secretory function of hatching gland cells. Int J Mol Sci 18, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie J-M, Blanc G, 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evolutionary Biology 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai RA, Gao L, Raghavan S, Liu WF, Chen CS, 2009. Cell polarity triggered by cell-cell adhesion via E-cadherin. J Cell Sci 122, 905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Reichman-Fried M, Stebler J, Köprunner M, Dörries J, Meyer D, Esguerra CV, Leung T, Raz E, 2002. Guidance of Primordial Germ Cell Migration by the Chemokine SDF-1. Cell 111, 647–659. [DOI] [PubMed] [Google Scholar]
- Dumortier JG, Martin S, Meyer D, Rosa FM, David NB, 2012. Collective mesendoderm migration relies on an intrinsic directionality signal transmitted through cell contacts. Proc Natl Acad Sci U S A 109, 16945–16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale TA and Elison RP, 1991. Development of the Xenopus laevis hatching gland and its relationship of surface ectoderm patterning. Development 111, 469–478. [DOI] [PubMed] [Google Scholar]
- Faller N, Gautschi I, Schild L, 2014. Functional analysis of a missense mutation in the serine protease inhibitor SPINT2 associated with congenital sodium diarrhea. PLoS One 9, e94267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Brennan J, Grant D, Peale F, Rangell L, Kirchhofer D, 2007. Hepatocyte growth factor activator inhibitor-1 (HAI-1) is essential for the integrity of basement membranes in the developing placental labyrinth. Dev Biol 303, 222–230. [DOI] [PubMed] [Google Scholar]
- Feitosa NM, Zhang J, Carney TJ, Metzger M, Korzh V, Bloch W, Hammerschmidt M, 2012. Hemicentin 2 and Fibulin 1 are required for epidermal-dermal junction formation and fin mesenchymal cell migration during zebrafish development. Dev Biol 369, 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet S, Lugo-Martínez VH, Faussat AM, Renaud F, Cardot P, Chambaz J, Pinçon-Raymond M, Thenet S, 2004. Early loss of E-cadherin from cell-cell contacts is involved in the onset of Anoikis in enterocytes. J Biol Chem 279, 43061–43069. [DOI] [PubMed] [Google Scholar]
- Friedl P, Mayor R, 2017. Tuning Collective Cell Migration by Cell–Cell Junction Regulation. Cold Spring Harb Perspect Biol 9, a029199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, Schier AF, 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9, e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner MR, Daggett DF, Zon LI, Perkins AC, 2005. Zebrafish KLF4 is essential for anterior mesendoderm/pre-polster differentiation and hatching. Dev Dyn 234, 992–996. [DOI] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock C, Niehrs C, 1998. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev 77, 95–141. [DOI] [PubMed] [Google Scholar]
- Gong Z, Ju B, Wang X, He J, Wan H, Sudha PM, Yan T, 2002. Green fluorescent protein expression in germ-line trasmitted transgenic zebrafish under a stratified epithelial promoter from keratin8. Dev Dyn 223, 204–215. [DOI] [PubMed] [Google Scholar]
- Graumann J, Finkernagel F, Reinartz S, Stief T, Brodje D, Renz H, Jansen JM, Wagner U, Worzfeld T, Pogge von Strandmann E, Muller R, 2019. Multi-platform Affinity Proteomics Identify Proteins Linked to Metastasis and Immune Suppression in Ovarian Cancer Plasma. Front Oncol 9, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi C, Schumacher I, Boquet-Pujadas A, Tarbashevich K, Vos BE, Bandemer J, Schick J, Aalto A, Olivo-Marin J-C, Betz T, Raz E, 2020. E-cadherin focuses protrusion formation at the front of migrating cells by impeding actin flow. Nat Commun 11, 5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines L, Neyt C, Gautier P, Keenan DG, Bryson-Richardson RJ, Hollway GE, Cole NJ, Currie PD, 2004. Met and Hgf signaling controls hypaxial muscle and lateral line development in the zebrafish. Development 131, 4857–4869. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, van Eeden FJ, Granato M, Brand M, Furutani-Seiki M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nüsslein-Volhard C, 1996. dino and mercedes, two genes regulating dorsal development in the zebrafish embryo. Development 123, 95–102. [DOI] [PubMed] [Google Scholar]
- Heinz-Erian P, Müller T, Krabichler B, Schranz M, Becker C, Rüschendorf F, Nürnberg P, Rossier B, Vujic M, Booth IW, Holmberg C, Wijmenga C, Grigelioniene G, Kneepkens CMF, Rosipal S, Mistrik M, Kappler M, Michaud L, Dóczy L-C, Siu VM, Krantz M, Zoller H, Utermann G, Janecke AR, 2009. Mutations in SPINT2 Cause a Syndromic Form of Congenital Sodium Diarrhea. Am J Hum Genet 84, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Nowakowski M, Spronk C, Petersen SV, Andreasen PA, Kozminski W, Mulder FAA, Jensen JK, 2015. The solution structure of the MANEC-type domain from hepatocyte growth factor activator inhibitor-1 reveals an unexpected PAN/apple domain-type fold. Biochem J 466, 299–309. [DOI] [PubMed] [Google Scholar]
- Holt-Danborg L, Vodopiutz J, Nonboe AW, De Laffolie J, Skovbjerg S, Wolters VM, Müller T, Hetzer B, Querfurt A, Zimmer KP, Jensen JK, Entenmann A, Heinz-Erian P, Vogel LK, Janecke AR, 2019. SPINT2 (HAI-2) missense variants identified in congenital sodium diarrhea/tufting enteropathy affect the ability of HAI-2 to inhibit prostasin but not matriptase. Hum Mol Genet 28, 828–841. [DOI] [PubMed] [Google Scholar]
- Huang L, Yang Y, Yang F, Liu S, Zhu Z, Lei Z, Guo J, 2018. Functions of EpCAM in physiological processes and diseases (Review). Int J Mol Med 42, 1771–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohaya K, Yasumasu S, Araki K, Naruse K, Yamazaki K, Yasumasu I, Iuchi I, Yamagami K, 1997. Species-dependent migration of fish hatching gland cells that express astacin-like proteases in common. Dev Growth Differ 39, 191–197. [DOI] [PubMed] [Google Scholar]
- Janiszewska M, Primi MC, Izard T, 2020. Cell adhesion in cancer: Beyond the migration of single cells. J Biol Chem 295, 2495–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lee KP, Park SJ, Park JH, Jang YS, Choi SY, Jung JG, Jo K, Park DY, Yoon JH, Park JH, Lim DS, Hong GR, Choi C, Park YK, Lee JW, Hong HJ, Kim S, Park YW, 2008. TMPRSS4 promotes invasion, migration and metastasis of human tumor cells by facilitating an epithelial-mesenchymal transition. Oncogene 27, 2635–2647. [DOI] [PubMed] [Google Scholar]
- Kane DA, McFarland KN, Warga RM, 2005. Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development 132, 1105–1116. [DOI] [PubMed] [Google Scholar]
- Kantak SS, Kramer RH, 1998. E-cadherin regulates anchorage-independent growth and survival in oral squamous cell carcinoma cells. J Biol Chem 273, 16953–16961. [DOI] [PubMed] [Google Scholar]
- Katagiri C, Maeda R, Yamashika C, Mita K, Sargent TD, Yasumasu S,1997. Molecular cloning of Xenopus hatching enzyme and its specific expression in hatching enzyme and its specific expression in hatching gland cells. Int J Dev Biol 41, 19–25. [PubMed] [Google Scholar]
- Kataoka H, Itoh H, Uchino H, Hamasuna R, Kitamura N, Nabeshima K, Koono M, 2000. Conserved expression of hepatocyte growth factor activator inhibitor type-2/placental bikunin in human colorectal carcinomas. Cancer Lett 148, 127–134. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Kawaguchi M, Fukushima T, Shimomura T, 2018. Hepatocyte growth factor activator inhibitors (HAI-1 and HAI-2): Emerging key players in epithelial integrity and cancer. Path Int 68, 145–158. [DOI] [PubMed] [Google Scholar]
- Kawaguchi M, Kanemaru A, Sawaguchi A, Yamamoto K, Baba T, Lin CY, Johnson MD, Fukushima T, Kataoka H, 2015. Hepatocyte growth factor activator inhibitor type 1 maintains the assembly of keratin into desmosomes in keratinocytes by regulating protease-activated receptor 2-dependent p38 signaling. Am J Pathol 185, 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M, Yamamoto K, Takeda N, Fukushima T, Yamashita F, Sato K, Kitamura K, Hippo Y, Janetka JW, Kataoka H, 2019. Hepatocyte growth factor activator inhibitor-2 stabilizes Epcam and maintains epithelial organization in the mouse intestine. Commun Biol 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Qin L, Shimomura T, Kondo J, Matsumoto K, Denda K, Kitamura N, 1997. Purification and cloning of hepatocyte growth factor activator inhibitor type 2, a Kunitz-type serine protease inhibitor. J Biol Chem 272, 27558–27564. [DOI] [PubMed] [Google Scholar]
- Khandekar G, Jagadeeswaran P, 2014. Role of hepsin in factor VII activation in zebrafish. Blood Cells Mol Dis 52, 76–81. [DOI] [PubMed] [Google Scholar]
- Kurauchi T, Izutsu Y, Maeno M, 2010. Involvement of Neptune in induction of the hatching gland and neural crest of the Xenopus embryo. Differentiation 79, 251–259. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien C-B, 2007. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- Larsen BR, Steffensen SD, Nielsen NV, Friis S, Godiksen S, Bornholdt J, Soendergaard C, Nonboe AW, Andersen MN, Poulsen SS, Szabo R, Bugge TH, Lin CY, Skovbjerg H, Jensen JK, Vogel LK, 2013. Hepatocyte growth factor activator inhibitor-2 prevents shedding of matriptase. Exp Cell Res 319, 918–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kimelman D, 2002. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell 2, 607–616. [DOI] [PubMed] [Google Scholar]
- Lee SP, Kao CY, Chang SC, Chiu YL, Chen YJ, Chen MG, Chang CC, Lin YW, Chiang CP, Wang JK, Lin CY, Johnson MD, 2018. Tissue distribution and subcellular localizations determine in vivo functional relationship among prostasin, matriptase, HAI-1, and HAI-2 in human skin. PLoS One 13, e0192632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Song G, Zhao Y, Zhao F, Liu C, Liu D, Li Q, Cui Z, 2017, Claudin7b is required for the formation and function of inner ear in zebrafish. J Cell Physiol 233, 3195–3206. [DOI] [PubMed] [Google Scholar]
- List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH, 2006. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am J Pathol 168, 1513–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo-Martínez VH, Petit CS, Fouquet S, Le Beyec J, Chambaz J, Pinçon-Raymond M, Cardot P, Thenet S, 2009. Epidermal growth factor receptor is involved in enterocyte anoikis through the dismantling of E-cadherin-mediated junctions. Am J Physiol Gastrointest Liver Physiol 296, G235–244. [DOI] [PubMed] [Google Scholar]
- Ma Z, Liu D, Li W, Di S, Zhang Z, Zhang J, Xu L, Guo K, Zhu Y, Han J, Li X, Yan X, 2019. STYK1 promotes tumor growth and metastasis by reducing SPINT2/HAI-2 expression in non-small cell lung cancer. Cell Death Dis 10, 435–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor CW, Delaria KA, Davis G, Muller DK, Greve JM, Tamburini PP, 1997. Identification and cloning of human placental bikunin, a novel serine protease inhibitor containing two Kunitz domains. J Biol Chem 272, 12202–12208. [DOI] [PubMed] [Google Scholar]
- Melby AE, Warga RM, Kimmel CB, 1996. Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development 122, 2225–2237. [DOI] [PubMed] [Google Scholar]
- Montero J-A, Carvalho L, Wilsch-Bräuninger M, Kilian B, Mustafa C, Heisenberg C-P, 2005. Shield formation at the onset of zebrafish gastrulation. Development 132, 1187–1198. [DOI] [PubMed] [Google Scholar]
- Montero JA, Heisenberg CP, 2004. Gastrulation dynamics: cells move into focus. Trends Cell Biol 14, 620–627. [DOI] [PubMed] [Google Scholar]
- Murray AS, Varela FA, Hyland TE, Schoenbeck AJ, White JM, Tanabe LM, Todi SV, List K, 2017. Phosphorylation of the type II transmembrane serine protease, TMPRSS13, in hepatocyte growth factor activator inhibitor-1 and −2–mediated cell-surface localization. J Biol Chem 292, 14867–14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T, Ishikawa T, Minami Y, Nishita M, 2020. Tactics of cancer invasion: solitary and collective invasion. J Biochem. 167, 347–355. [DOI] [PubMed] [Google Scholar]
- Nagaike K, Kawaguchi M, Takeda N, Fukushima T, Sawaguchi A, Kohama K, Setoyama M, Kataoka H, 2008. Defect of hepatocyte growth factor activator inhibitor type 1/serine protease inhibitor, Kunitz type 1 (Hai-1/Spint1) leads to ichthyosis-like condition and abnormal hair development in mice. Am J Pathol 173, 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Abarzua F, Hongo A, Kodama J, Nasu Y, Kumon H, Hiramatsu Y, 2009. Hepatocyte growth factor activator inhibitor-2 (HAI-2) is a favorable prognosis marker and inhibits cell growth through the apoptotic pathway in cervical cancer. Ann Oncol 20, 63–70. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Hongo A, Kodama J, Hiramatsu Y, 2011. The role of hepatocyte growth factor activator inhibitor (HAI)-1 and HAI-2 in endometrial cancer. Int J Cancer 128, 2613–2624. [DOI] [PubMed] [Google Scholar]
- Netzel-Arnett S, Currie BM, Szabo R, Lin CY, Chen LM, Chai KX, Antalis TM, Bugge TH, List K, 2006. Evidence for a matriptase-prostasin proteolytic cascade regulating terminal epidermal differentiation. J Biol Chem 281, 32941–32945. [DOI] [PubMed] [Google Scholar]
- Nonboe AW, Krigslund O, Soendergaard C, Skovbjerg S, Friis S, Andersen MN, Ellis V, Kawaguchi M, Kataoka H, Bugge TH, Vogel LK, 2017. HAI-2 stabilizes, inhibits and regulates SEA-cleavage-dependent secretory transport of matriptase. Traffic 18, 378–391. [DOI] [PubMed] [Google Scholar]
- Oberst MD, Chen LY, Kiyomiya K, Williams CA, Lee MS, Johnson MD, Dickson RB, Lin CY, 2005. HAI-1 regulates activation and expression of matriptase, a membrane-bound serine protease. Am J Physiol Cell Physiol 289, C462–470. [DOI] [PubMed] [Google Scholar]
- Orford K, Orford CC, Byers SW, 1999. Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J Cell Biol 146, 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, Schier AF, 2013. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoli P, Giannoni E, Chiarugi P, 2013. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta 1833, 3481–3498. [DOI] [PubMed] [Google Scholar]
- Pogoda H-M, Riedl-Quinkertz I, Löhr H, Waxman JS, Dale RM, Topczewski J, Schulte-Merker S, Hammerschmidt M, 2018. Direct activation of chordoblasts by retinoic acid is required for segmented centra mineralization during zebrafish spine development. Development 145, dev159418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson R, Metzger M, Knyphausen P, Ramezani T, Slanchev K, Kraus C, Schmelzer E, Hammerschmidt M, 2016. Re-epithelialization of cutaneous wounds in adult zebrafish combines mechanisms of wound closure in embryonic and adult mammals. Development 143, 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román-Fernández A, Bryant DM, 2016. Complex Polarity: Building Multicellular Tissues Through Apical Membrane Traffic. Traffic 17, 1244–1261. [DOI] [PubMed] [Google Scholar]
- Roversi FM, Olalla Saad ST, Machado-Neto JA, 2018. Serine peptidase inhibitor Kunitz type 2 (SPINT2) in cancer development and progression. Biomed Pharmacother 101, 278–286. [DOI] [PubMed] [Google Scholar]
- Salomon J, Goulet O, Canioni D, Brousse N, Lemale J, Tounian P, Coulomb A, Marinier E, Hugot J-P, Ruemmele F, Dufier J-L, Roche O, Bodemer C, Colomb V, Talbotec C, Lacaille F, Campeotto F, Cerf-Bensussan N, Janecke AR, Mueller T, Koletzko S, Bonnefont J-P, Lyonnet S, Munnich A, Poirier F, Smahi A, 2014. Genetic characterization of congenital tufting enteropathy: epcam associated phenotype and involvement of SPINT2 in the syndromic form. Human Genet 133, 299–310. [DOI] [PubMed] [Google Scholar]
- Sano K, Inohaya K, Kawaguchi M, Yoshizaki N, Iuchi I, Yasumasu S, 2008. Purification and characterization of zebrafish hatching enzyme - an evolutionary aspect of the mechanism of egg envelope digestion. FEBS J 275, 5934–5946. [DOI] [PubMed] [Google Scholar]
- Schepis A, Barker A, Srinivasan Y, Balouch E, Zheng Y, Lam I, Clay H, Hsiao CD, Coughlin SR, 2018. Protease signaling regulates apical cell extrusion, cell contacts, and proliferation in epithelia. J Cell Biol 217, 1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W, 1997. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development 124, 327–342. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Strähle U, Scholpp S, 2013. Neurogenesis in zebrafish – from embryo to adult. Neural Dev 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaq-Zadah M, Dransart E, Johannes L, 2020. Clathrin-independent endocytosis, retrograde trafficking, and cell polarity. Curr Opin Cell Biol 65, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J, Fraser SE, 1996. Characterizing the zebrafish organizer: microsurgical analysis at the early-shield stage. Development 122, 1313–1322. [DOI] [PubMed] [Google Scholar]
- Shimomura T, Denda K, Kitamura A, Kawaguchi T, Kito M, Kondo J, Kagaya S, Qin L, Takata H, Miyazawa K, Kitamura N, 1997. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J Biol Chem 272, 6370–6376. [DOI] [PubMed] [Google Scholar]
- Sivagnanam M, Mueller JL, Lee H, Chen Z, Nelson SF, Turner D, Zlotkin SH, Pencharz PB, Ngan BY, Libiger O, Schork NJ, Lavine JE, Taylor S, Newbury RO, Kolodner RD, Hoffman HM, 2008. Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterology 135, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slanchev K, Carney TJ, Stemmler MP, Koschorz B, Amsterdam A, Schwarz H, Hammerschmidt M, 2009. The epithelial cell adhesion molecule EpCAM is required for epithelial morphogenesis and integrity during zebrafish epiboly and skin development. PLoS Genet 5, e1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ishizaka T, Yanagi K, Sone R, Sunaga Y, Ohga R, Kawahara A, 2019. Characterization of biklf/klf17-deficient zebrafish in posterior lateral line neuromast and hatching gland development. Sci Rep 9, 13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo R, Bugge TH, 2018. Loss of HAI-2 in mice with decreased prostasin activity leads to an early-onset intestinal failure resembling congenital tufting enteropathy. PLOS ONE 13, e0194660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo R, Callies LK, Bugge TH, 2019. Matriptase drives early-onset intestinal failure in a mouse model of congenital tufting enteropathy. Development 146, dev183392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo R, Hobson JP, Christoph K, Kosa P, List K, Bugge TH, 2009. Regulation of cell surface protease matriptase by HAI2 is essential for placental development, neural tube closure and embryonic survival in mice. Development 136, 2653–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo R, Hobson JP, List K, Molinolo A, Lin CY, Bugge TH, 2008. Potent inhibition and global co-localization implicate the transmembrane Kunitz-type serine protease inhibitor hepatocyte growth factor activator inhibitor-2 in the regulation of epithelial matriptase activity. J Biol Chem 283, 29495–29504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo R, Molinolo A, List K, Bugge TH, 2007. Matriptase inhibition by hepatocyte growth factor activator inhibitor-1 is essential for placental development. Oncogene 26, 1546–1556. [DOI] [PubMed] [Google Scholar]
- Tada M, Heisenberg CP, 2012. Convergent extension: using collective cell migration and cell intercalation to shape embryos. Development 139, 3897–3904. [DOI] [PubMed] [Google Scholar]
- Tanabe LM, List K, 2017. The role of type II transmembrane serine protease-mediated signaling in cancer. FEBS J 284, 1421–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Nagaike K, Takeda N, Itoh H, Kohama K, Fukushima T, Miyata S, Uchiyama S, Uchinokura S, Shimomura T, Miyazawa K, Kitamura N, Yamada G, Kataoka H, 2005. Hepatocyte growth factor activator inhibitor type 1 (HAI-1) is required for branching morphogenesis in the chorioallantoic placenta. Mol Cell Biol 25, 5687–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira Rosa J, Oralová V, Larionova D, Eisenhoffer GT, Eckhard Witten P, Huysseune A, 2019. Periderm invasion contributes to epithelial formation in the teleost pharynx. Sci Rep 9, 10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CH, Teng CH, Tu YT, Cheng TS, Wu SR, Ko CJ, Shyu HY, Lan SW, Huang HP, Tzeng SF, Johnson MD, Lin CY, Hsiao PW, Lee MS, 2014. HAI-2 suppresses the invasive growth and metastasis of prostate cancer through regulation of matriptase. Oncogene 33, 4643–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich F, Krieg M, Schötz EM, Link V, Castanon I, Schnabel V, Taubenberger A, Mueller D, Puech PH, Heisenberg CP, 2005. Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin. Dev Cell 9, 555–564. [DOI] [PubMed] [Google Scholar]
- Warga RM, Kimmel CB, 1990. Cell movements during epiboly and gastrulation in zebrafish. Development 108, 569–580. [DOI] [PubMed] [Google Scholar]
- Welman A, Sproul D, Mullen P, Muir M, Kinnaird AR, Harrison DJ, Faratian D, Brunton VG, Frame MC, 2012. Diversity of matriptase expression level and function in breast cancer. PLoS One 7, e34182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westcot SE, Hatzold J, Urban MD, Richetti SK, Skuster KJ, Harm RM, Lopez Cervera R, Umemoto N, McNulty MS, Clark KJ, Hammerschmidt M, Ekker SC, 2015. Protein-Trap Insertional Mutagenesis Uncovers New Genes Involved in Zebrafish Skin Development, Including a Neuregulin 2a-Based ErbB Signaling Pathway Required during Median Fin Fold Morphogenesis. PLoS One 10, e0130688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-J, Feng X, Lu M, Morimura S, Udey MC, 2017. Matriptase-mediated cleavage of EpCAM destabilizes claudins and dysregulates intestinal epithelial homeostasis. J Clin Invest 127, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SR, Lin CH, Shih HP, Ko CJ, Lin HY, Lan SW, Lin HH, Tu HF, Ho CC, Huang HP, Lee MS, 2019. HAI-2 as a novel inhibitor of plasmin represses lung cancer cell invasion and metastasis. Br J Cancer 120, 499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Kawaguchi M, Shimomura T, Izumi A, Konari K, Honda A, Lin CY, Johnson MD, Yamashita Y, Fukushima T, Kataoka H, 2018. Hepatocyte growth factor activator inhibitor type-2 (HAI-2)/SPINT2 contributes to invasive growth of oral squamous cell carcinoma cells. Oncotarget 9, 11691–11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A, 2010. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell 18, 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Feng S, Wu B, Guo W, 2017. Polarized Exocytosis. Cold Spring Harb Perspect Biol 9, a027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo time-lapse recording of DIC images of the top layer of hatching gland cells next to the head region, showing intercalation events. A wild-type embryo was mounted at 16 hpf and images were taken in 3 min intervals for 39 min. Hatching gland cells intercalating from lower layers are false-colored.
In vivo time-lapse recording of an embryo transgenic for Tg(−1.8gsc:gfp) and Tg(krt4:tdtomatocaax) starting at 11 hpf. Hatching gland cells labeled with GFP move relatively to the overlaying periderm labeled with tdTomato-caax (compare with Figure 5A for stills). Z-stacks of fluorescent images were recorded in 10 min intervals. Maximum intensity projections were converted into the movie.
In vivo time-lapse recording of an embryo transgenic for Tg(−1.8gsc:gfp) and Tg(krt4:tdtomatocaax) starting at 20 hpf. Hatching gland cells labeled with GFP move relatively to the overlaying periderm labeled with tdTomato-caax (compare with Figure 5B for stills). Z-stacks of fluorescent images were recorded in 15 min intervals. Maximum intensity projections were converted into the movie.
In vivo time-lapse recording of a mosaic wild-type embryo with transplanted cells transgenic for Tg(Ola.Actb:Hsa.hras-egfp), starting at 16.5 hpf, showing two initially neighboring hatching gland cells that become separated, with much higher protrusive activity of the anterior-wards departing cell. Two hatching gland cells intercalating from lower layers are false-colored in yellow and blue. Note that intercalations (left / anterior side) and cell migrations (right / posterior side) take place back to back, and that both most likely contribute to the anterior-wards net movement of the tissue. Bright-field and fluorescent images were recorded in 3 min intervals. Compare with Figure 5F for stills.
In vivo time-lapse recording of wild-type embryos injected with a 1.8gsc:LifeAct-Ruby plasmid, starting at 16.5 hpf, labeling active actin cytoskeletal rearrangements and dis- and reassociation events in hatching gland cells. Z-stacks of fluorescent images were recorded in 3 min intervals. Maximum intensity projections were converted into the movie. Compare with Figure 5G for stills.
In vivo time-lapse recording of wild-type embryos injected with a 1.8gsc:LifeAct-Ruby plasmid, starting at 20 hpf, displaying active actin cytoskeletal rearrangements but no dis- and reassociation events in hatching gland cells. Z-stacks of fluorescent images were recorded in 3 min intervals. Maximum intensity projections were converted into the movie. Compare with Figure 5H for stills.
In vivo time-lapse recording of a wild-type embryo transgenic for Tg(−1.8gsc:gfp) starting at 16.5 hpf, displaying dynamic, but cohesive migration of the hatching gland tissue. Z-stacks of fluorescent images were recorded in 5 min intervals. Maximum intensity projections were converted into the movie. Compare with Figure 6A for stills.
In vivo time-lapse recording of a spint2 morphant embryo transgenic for Tg(−1.8gsc:gfp) starting at 16.5 hpf, showing less-organized migration of the hatching gland cells compared to the wild type (Supplementary Movie 7) . Z-stacks of fluorescent images were recorded in 5 min intervals. Maximum intensity projections were converted into the movie.
In vivo time-lapse recording of a spint2fr49/fr49 embryo transgenic for Tg(−1.8gsc:gfp) starting at 16.5 hpf, showing less-organized migration of the hatching gland cells compared to the wild type. Z-stacks of fluorescent images were recorded in 5 min intervals. Maximum intensity projections were converted into the movie.