ABSTRACT
The domain of unknown function (DUF221 domain-containing) proteins regulates various aspects of plant growth, development, responses to abiotic stresses, and hormone transduction pathways. To understand the role of DDP proteins in tomato, a comprehensive genome-wide analysis was performed in the tomato genome. A total of 12 DDP genes were identified and distributed in 8 chromosomes in the tomato genome. Phylogenetically all SlDDPs were clustered into four clades, subsequently supported by their gene structure and conserved motifs distribution. The SlDDPs contained various cis-acting elements involved in plant responses to abiotic and various phytohormones stresses. The tissue-specific expression profile analysis revealed the constitutive expression of SlDDPs in roots, leaves, and developmental phases of fruit. It was found that SlDDP1, SlDDP3, SlDDP4, SlDDP9, SlDDP10, and SlDDP12 exhibited high expression levels in fruits at different development stages. Of these genes, SlDDP12 contained ethylene (ERE) responsive elements in their promoter regions, suggesting its role in ethylene-dependent fruit ripening. It was found that a single SlDDP induced by two or more abiotic and phytohormone stresses. These include, SlDDP1, SlDDP2, SlDDP3, SlDDP4, SlDDP7, SlDDP8, and SlDDP10 was induced under salt, drought, ABA, and IAA stresses. Moreover, tomato SlDDPs were targeted by multiple miRNA gene families as well. In conclusion, this study predicted that the putative DDP genes might help improve abiotic and phytohormone tolerance in plants, particularly tomato, rice, and other economically important crop plant species.
KEYWORDS: Gene family, miRNA target, expression profile, Duplications, Phylogeny
Introduction
Plants cope with abiotic and phytohormone stresses in several ways, including physiochemical, morphological, and ultrastructural changes in a cell at various molecular events. Therefore, uncovering the roles of distinct gene families against various stresses helps to identify their particular role.1 The availability of the plant genome enables us to identify and characterize various gene families under abiotic and biotic stresses. For instance, several gene families have been identified and characterized, such as NAC,2,3 bHLH,4 MAPK,5 bZIP,6–8 GRAS,9 Aux/IAA,10 TIFY,11 FKBP,12 PEPC,13 and HSF transcription factors.14 In addition, plants’ genomes contained plenty of stress-responsive proteins with highly conserved domain15,16 and play critical roles in various plant biological processes during stress conditions. Genes with such potential hypothetical domains are classified as a domain of unknown functions (DUF). In recent years, the rapid development of proteomics and genomics identified and sequenced plenty of species’ genomes having enormous DUF superfamilies.17 However, there have been some reports of many DUF gene families in plants including DUF221, DUF581, DUF668, DUF724, DUF810, DUF866, DUF936, DUF966, DUF1644, and DUF1618 in rice, Gossypium hirsutum, and Arabidopsis.18–21
The systematic study of DUF superfamily genes lays the foundation for analyzing these DUF family genes in regulating plant growth and development and tolerance to biotic and abiotic stresses. DUF proteins also function as a membrane protein associated with other related proteins, implying their roles as membrane integral proteins.22 The DUF-mediated stress resistance has been reported in only model plants, while comprehensive DUF gene family analysis in other plant species remains determined. For example, Brassica juncea dehydration-responsive gene (ERD4),22 rice drought-responsive gene AtCSC1 in Arabidopsis,23 and its homolog in rice (OsCA1) associated with osmotic regulation.23,24 DUF538 and DUF27 have the chlorophyll-binding ability25,26 and bind to ADP-ribose precisely,27 respectively. Arabidopsis DUF283 superfamily is essential for siRNA processing in gene silencing.28
DUF genes have also been related to phytohormone and abiotic stress responses, particularly drought and salinity. The OsSIDP366 and SIDP361 (DUF1644 superfamily) positively regulate the response to salinity and drought stress in rice.21,29 The OsSIDP366 overexpression exhibits more substantial salt tolerance and drought resistance.29 Similarly, SIDP361, OsDSR2, and OsDUF810 of DUF1644, DUF966, and OsDUF810 superfamily play a role in dehydration-mediated nutritional status regulation.20,21,29 Arabidopsis overexpressing the salt-inducing gene TaSRHP of DUF581 superfamily can enhance salinity tolerance and drought resistance.30 DUF221 domain-containing proteins (DDP) belong to the anoctamin/calcium-activated chloride channels/ TMEM16 family.31 The DDP proteins play an essential role in plant growth, development, phytohormone signaling, and responses to abiotic and biotic stresses.32 This suggesting that DUF domain-containing genes may play a direct or indirect role in pant tolerance.
Tomato (Solanum lycopersicum) is an important climacteric vegetable fruit crop and highly sensitive to abiotic stresses33,34 affecting plant growth, reduced photosynthesis rate, disrupted ions homeostasis, and tomato productivity.35,36 However, the status of this signature domain remains to be determined in tomato. Therefore, taking advantage of DDP putative role in various biological processes, we performed a comprehensive study of DDP gene family in the tomato genome. We predicted in-silico subcellular localization, generated an unrooted phylogeny, and analyzed all putative gene expression profiles in different organs/tissues. Additionally, we have performed salinity, drought (PEG), and phytohormone stresses and analyzed the temporal expression profile of SlDDPs. Thus, our data can have the potential to provide a foundation for functional validation of the tomato DUF221 genes and their role in tomato plant growth and development under stressful conditions.
Material and Method
Discovery of DDP Gene Family in the Tomato Genome
The tomato whole-genome sequence data were downloaded from the Solanaceae Genomics Network (SGN, https://www.solgenomics.net/).37 The Arabidopsis DDP protein sequences were retrieved from the TAIR database (https://www.arabidopsis.org/, Table S1).38 The DDP proteins of tomato (SlDDPs) were predicted using a hidden Markov model (HMM) profile retrieved from the Pfam database.39 The S. lycopersicum DDP protein sequences were searched by using the HMMSEARCH program.40 All redundant DDP sequences were excluded. The domains of putative sequences were verified with SMART program41 and NCBI CDD.42 Sequence Manipulation Suite (SMS)43 was used to predict the physicochemical properties of DDP peptide sequences, including molecular weight (MW, kDa), the grand average of hydropathy (GRAVY), and theoretical isoelectric point (pI). For DDP genes nomenclature in tomato, members of the gene family were named 1 to 12 in chronological order on the chromosomes. DDPs chromosomal location was obtained from SNG, and MAP2Chromomse program (v2) was used to visualize each gene on corresponding chromosome.
In-silico Subcellular Location, Conserved Motif, and Gene Structure of Tomato DDP Genes
The peptide sequences of deduced DDP proteins were submitted to WoLF PSORT program (https://wolfpsort.hgc.jp/)44 for in-silico protein cellular localization prediction. Tomato DDP protein sequences were scanned in the MEME program (https://meme-suite.org/meme/tools/meme)45 to identify conserved motifs with parameters used by Mondal et al.46 The gene intron/exon number and distribution were determined in the Gene Structure Display Server (GSDS, http://gsds.gao-lab.org/Gsds_about.php)47 by submitting corresponding CDS genomic sequences of SlDDPs.
Phylogeny and Gene Duplication of SlDDP Genes
The multiple sequence alignment of tomato DDP proteins was performed using Clustal Omega.48 An unrooted phylogenetic tree was generated using MEGAX software49 by neighbor-joining (NJJ) method50 with bootstrap set at 1000 replicates. MCScanX program (https://github.com/wyp1125/MCScanX) was used to predict SlDDP gene duplication events in the tomato genome. The non-synonymous (Ka), synonymous (Ks) nucleotide substitution rates, and the Ka/Ks ratios were predicted using k-estimator (http://en.bio-soft.net/format/KEstimator.html).51 The divergence time (T, mya; millions year ago) was calculated as follows: T = Ks/2y (y = 6.56 × 10−9).52
Tomato DDP Genes Cis-regulatory Elements, and miRNAs Target Prediction
A 2000bp long 5`UTR nucleotide sequence from the start codon was extracted for each DDP gene from SNG and submitted to PlantCARE database (http://bioinformatics.psb.ugent.be/web tools/plantcare/html/)48as query sequence for putative cis-regulatory motif prediction. In addition, to predict miRNAs target the putative DDPs, the cDNA sequences of each SlDDPs were submitted to psRNATarget (http://plantgrn.noble.org/psRNATarget/)53 against all tomato miRNAs reported in miRbase.54
Plant Growth and Material Collection
The plants of tomato cultivar Micro-Tom have grown in a greenhouse under control conditions: 14 h light /12 h dark photoperiod, at 25°C/20°C day/night temperature with relative humidity between 70% and 80% and photon density of about 120 µmol photons m−2s−.1,55 When tomato seedlings were 6-week-old, different plant parts, including root, leaves, flower (in bud/fully opened), and different developmental phases of fruit (1/2/3 cm, MG; mature green, B; breaker, B10; 10 days breaker) were collected for tissue/organ-specific expression analysis.
In the sixth week, tomato seedlings were treated with 200 mM NaCl, 0.01 mM abscisic acid (ABA), gibberellins (GA3), indole-3-acetic acid (IAA, Auxin), and polyethylene glycol (PEG).12 Plants were harvested at 0 h, 3 h, 6 h, 12 h, and 24 h intervals after treatments. Three independent biological replicates were collected, and six seedlings were used for each treatment. All the samples were immediately frozen in liquid nitrogen and stored at −80 °C until further analysis.
RNA Extraction, cDNA Preparation, and RT-qPCR Analysis
Total RNA extracted from selected samples (tissue-specific/hormone treated) using TRIZOL reagent according to manufacturer’s protocol. The RNA was qualified using nanodrop (Thermo USA), and the quality was assessed through 2% (w/v) gel electrophoresis. The first complementary DNA (cDNA) strand was prepared using Prime Script ™ RT reagent Kit with gDNA Eraser (Takara, JAPAN). Next, SYBR-Premix Ex Taq-II (TliRNaseH Plus) was used to conduct qRT-PCR on CFX96 Touch ™ Real-Time PCR Detection System (BIO-RAD, USA). The housekeeping gene SlUBQ (Solyc01g056940) was used as an internal control. The relative expression was calculated following 2−ΔΔCt method.56 Finally, the heat map was generated using MeV 4.9 software package. All the primers used in this study are listed in Table S2.
Results
Identification of DDP Genes in the Tomato Genome
Total 18 DDP genes were identified in the tomato genome using Arabidopsis DDP as a query in the tomato SNG genome. To determine the reliability and validity of putative DDP genes, the protein sequences of identified tomato DDPs were submitted to NCBI and SMART. Finally, 12 unique SlDDP genes (protein sequences are provided in Table S1) were identified and designated as SlDDP1 to SlDDP12. Twelve SlDDP genes were unevenly distributed across the 12 tomato chromosomes. Chromosome 2 contained a comparatively high number of tomato DDPs (four genes). While chromosome 8 contained two genes (SlDDP9 and SlDDP10), chromosomes 1, 4, 6, 7, 9, and 12 had a single gene (Fig. 1). The protein sequences analysis of putative SlDDP genes revealed that the protein length varied from 684 aa (SlDDP2) to 977 aa (SlDDP12) with MW ranged from 77.07 kDa (SlDDP11) to 110.05 kDa (SlDDP12). The GRAVY ranged from 0.027 (SlDDP12) to 0.383 (SlDDP5), and pI varied from 6.7 (SlDDP4) to 9.6 (SlDDP2, SlDDP6, and SlDDP7) suggested that tomato SlDDPs working in a wide range of microenvironment. In-silico subcellular location prediction revealed that all the SlDDP associated with the plasma membrane (Table 1).
Figure 1.
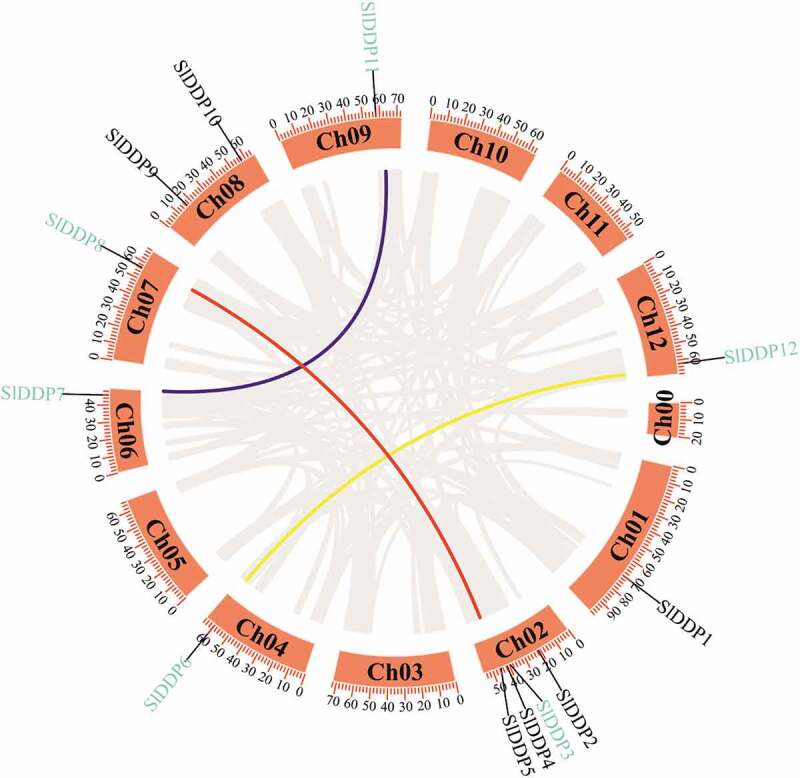
Circos plot showing the physical location of 12 DDP genes distribution in 12 tomato chromosomes, including chromosome 0 (for unallocated genes). The segmental duplication pairs are marked with blue color. The color lines in the circus plot indicate SlDDP segmental duplication between different chromosomes, including SlDDP3-SlDDP8, SlDDP7-SlDDP11, and SlDDP6-SlDDP12. The scale at the top of each chromosome indicates the size of the chromosome in MBs.
Table 1.
Characterization of protein sequences of 12 SlDDP gene family members in tomato genome
| Gene ID | Name | aa | MW | pI | GRAVY | Chromosome |
Subcellular location prediction | ||
|---|---|---|---|---|---|---|---|---|---|
| Number | Start | End | |||||||
| Solyc01g068500 | SlDDP1 | 705 | 80.8 | 8.91 | 0.264 | 1 | 70,131,982 | 70,142,492 | plas:9, E.R.:3, golg:2 |
| Solyc02g036260 | SlDDP2 | 684 | 78.31 | 9.6 | 0.082 | 2 | 21,172,510 | 21,179,750 | plas:13, vacu:1 |
| Solyc02g081030 | SlDDP3 | 686 | 78.35 | 8.3 | 0.093 | 2 | 39,662,050 | 39,674,092 | plas:8, vacu:2, E.R.:2, cyto:1, mito:1 |
| Solyc02g083430 | SlDDP4 | 831 | 93.93 | 6.7 | 0.127 | 2 | 41,421,699 | 41,424,194 | plas:10, E.R.:2, nucl:1, vacu:1 |
| Solyc02g088300 | SlDDP5 | 716 | 81.1 | 8.93 | 0.383 | 2 | 45,016,695 | 45,024,991 | plas:13, golg:1 |
| Solyc04g077400 | SlDDP6 | 719 | 81.45 | 9.64 | 0.309 | 4 | 59,898,021 | 59,905,315 | plas:10, vacu:2, E.R.:2 |
| Solyc06g084330 | SlDDP7 | 766 | 88.07 | 9.64 | 0.142 | 6 | 45,777,999 | 45,781,628 | plas:12, vacu:1, E.R.:1 |
| Solyc07g048110 | SlDDP8 | 796 | 91.05 | 9.14 | 0.181 | 7 | 56,620,600 | 56,630,095 | plas:11, vacu:2, E.R.:1 |
| Solyc08g023440 | SlDDP9 | 723 | 81.95 | 9.63 | 0.29 | 8 | 16,571,646 | 16,577,518 | plas:12, vacu:1, E.R.:1 |
| Solyc08g076310 | SlDDP10 | 815 | 94.03 | 9.46 | 0.057 | 8 | 57,437,506 | 57,445,845 | plas:12, chlo:1, E.R.:1 |
| Solyc09g064810 | SlDDP11 | 673 | 77.07 | 9.45 | 0.129 | 9 | 57,811,490 | 57,819,337 | plas:12, vacu:1, E.R.:1 |
| Solyc12g088230 | SlDDP12 | 977 | 110.05 | 9.57 | 0.027 | 12 | 62,114,857 | 62,122,490 | plas:8, vacu:3, E.R.:2, golg:1 |
aa; amino acid, MW; molecular weight, pI; isoelectric point, GRAVY; the grand average of hydropathy, nucl; nucleus, cyto; cytoplasm, chlo; chloroplast, plas; plasma membrane, vacu; vacuole, E.R; endoplasmic reticulum, golg; golgi apparatus
Phylogenetic Analysis, Gene Structure, and Conserved Motifs Analysis in SlDDPs
To find out peptide sequence conservation in DDPs, tomato DDP protein sequences were aligned. It was observed that DUF221 domain region was highly conserved across all SlDDPs (Fig. S1). To ascertain the phylogenetic relationship among tomato SlDDPs, an unrooted NJJ phylogenetic tree with 12 tomato SlDDPs (Fig. 2a) along with 14 AtDDPs from the Arabidopsis genome (Fig. 2b) was generated. The SlDDPs clustered into four groups (I, II, III, and IV). The SlDDPs pairwise similarity ranges from 26.13 (SlDDP10/ SlDDP4) to 84.94% (SlDDP3/ SlDDP2). It was found that SlDDP3 and SlDDP2 proteins have a similarity index of 89%, clustered together in group I. Similarly, SlDDP12 and SlDDP6 were grouped in group III with sequence similarity of 79.13% (Table S3, Fig. 2).
Figure 2.

Phylogeny of DDPs gene family. An unrooted neighbor-joining (NJJ) phylogenetic tree of (a) tomato SlDDPs and (b) with Arabidopsis DDPs were generated using MEGA 7 program. The bootstrap was set at 1000 replicates. All the DDPs were clustered into four clades named I, II, III, and IV.
To further gain insight into the structural diversity of SlDDPs genes, intron-exon organization, and conserved motifs numbers and their distribution were analyzed. It was found that the majority of SlDDPs exhibited similar gene structures sharing the same clusters. For instance, SlDDPs contained at least a single exon (SlDDP4) and a maximum of eleven exons (genes in phylogenetic cluster I) (Fig. 3a). A total of 10 conserved motifs (Table S4) were identified in SlDDPs consistent with their phylogenetic clustering. All the members of group-I contained 10 motifs; eight motifs are shared by group-II. Similarly, nine and four motifs are present in SlDDPs of group-III and group IV, respectively (Fig. 3b). Taken together, the members of tomato SlDDPs sharing similar gene structure and conserved motifs, implying functional similarly of the SlDDPs within the same group.
Figure 3.
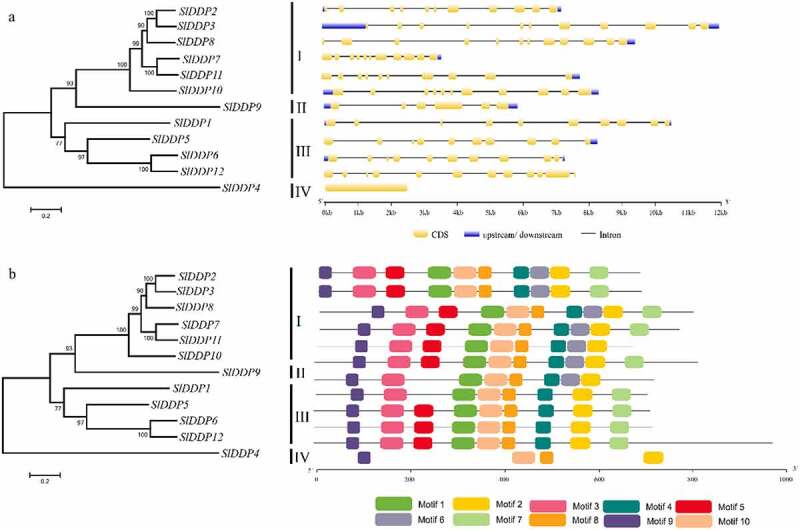
Gene structure analysis of tomato DDPs. (a) The number and distribution of exons and introns and (b) conserved motifs in SlDDPs identified using GSDS and MEME tools, respectively. The scale at the bottom is represented in Kb.
Gene Duplication of SlDDP Genes
Moreover, to elucidate the evolutionary relationship of SlDDP within the tomato genome, synteny analysis was performed. These results revealed that tomato SlDDP sharing three paralogous gene pairs displayed segmental duplication pairs in the whole genome while no tandem duplications were found (Fig. 1). These findings are consistent with the phylogenetic clustering of SlDDP gene. To assess the selection mode of the duplicated SlDDP genes, we estimated the average rate Ka vs. Ks by calculating the Ka/Ks ratio for each pair of duplicated SlDDP genes. In general, the Ka/Ks ratio < 1 suggests purifying selection; a ratio = 1 indicates neutral selection, while a ratio >1 indicates that these proteins may have been subject to positive selection. All the three segmental duplicated pairs in the tomato DDP family showed that the Ka/Ks ratios for these duplicated pairs were < 1. Based on the Ka/Ks analyses, we concluded that purifying selection may be primarily responsible for the function maintenance of SlDDP proteins. Based on a substitution rate of 6.5 × 10 − 9 substitutions per site per year, the duplication events for the three segmental duplications were estimated to have occurred approximately between 14.29 and 92.23 mya (Table S5).
Cis-regulatory Elements in Promoter Sequences of SlDDPs
To investigate the putative role of SlDDPs in plant development under abiotic and biotic stress, we analyzed the promoters of all putative SlDDPs. It was observed that the promoters of SlDDPs contained cis-regulatory elements related to plant development, phytohormone, and abiotic stress-responsive elements in their promoter regions. In addition, several phytohormone-related elements including, ABRE, TGA-element (auxin), ERE (ethylene), GARE-motif (gibberellic acid), TGACG-motif (methyl jasmonate), and SARE (salicylic acid), were detected. Furthermore, development-related elements, GCN4-motif, CAAT-box, and several abiotic stress-responsive elements such as MBS, HSE, ARE, TC-rich repeats were also observed (Fig. 4, Table S6).
Figure 4.
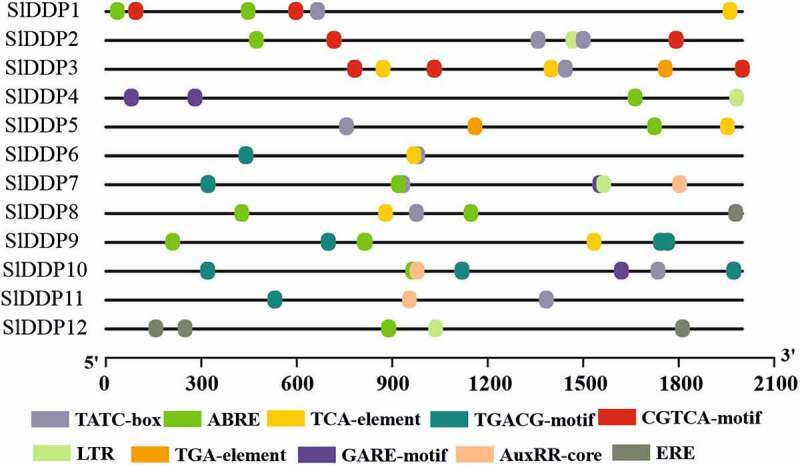
Predicted putative cis-regulatory elements in promoter regions of tomato DDP gene family members. TGA-element; Auxin-responsive element, ABRE; cis-acting element involved in the abscisic acid responsiveness, CGTCA/TGACG-motif; cis-acting regulatory element involved in the MeJA-responsiveness, ERE; Ethylene-responsive element, ABRE; involved in ABA responsiveness, LTR; cis-acting element involved in low-temperature responsiveness, AuxRR-core; cis-acting regulatory element involved in auxin responsiveness, TATC-box/GARE-motif; gibberellin-responsive element. A promoter sequence up to 2000 bp from 5`UTR was retrieved from SNG and submitted to PlantCARE database for cis-regulatory elements prediction.
miRNAs Targeting the DDP Family Members of the Tomato
To find out miRNAs targeting the SlDDPs, psRNATarget predicted that five SlDDPs gene family members were targeted by conserved miRNA. For instance, SlDDP3 or SlDDP5 was targeted by two different miRNAs gene families each. Sly-miRNA395 family (sly-miRNA395a and sly-miRNA395b) and sly-miRNA717 family (sly-miRNA717b), causing cleavage and inhibition of translation of SlDDP3. A single member from sly-miR319 family (sly-miR319b) and sly-miR6022 family member target to cleave of SlDDP5 gene. However, SlDDP7, SlDDP10, and SlDDP11 were of sly-miR6026, sly-miR6022, and sly-miR482a family members (Table S7).
Expression Patterns of SlDDPs in Different Parts of Tomato Plant
To predict the biological role of tomato SlDDPs genes, the expression of all the putative SlDDPs were investigated in various plant parts including, roots, leaves, flowers, and fruits at different development stages (Fig. 5). The results revealed that SlDDPs showed significant expression preference with expression higher in specific tissues. These indicated that these SlDDP genes play a role in the development of these tissues. However, few genes were expressed in an only single tissue or plant part. For example, SlDDP6 and SlDDP11 expressed with high levels in flower, SlDDP2 and SlDDP8 expressed with significant transcript abundance in the root.
Figure 5.
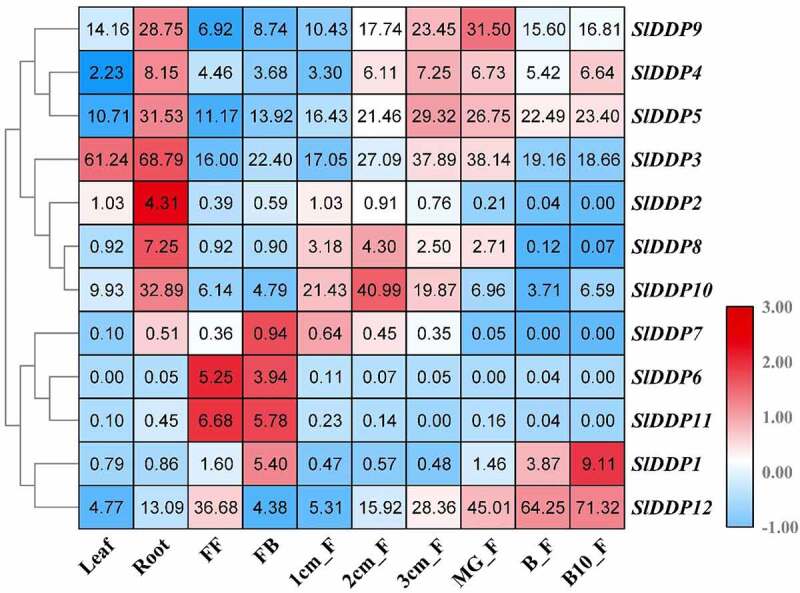
Tissue/organ-specific expression of SlDDPs in tomato. Expression profile of 12 SlDDPs in various plant parts, including root, leaf, flower, and fruits. FF; Fully opened flowers, FB; Flower bud, 1/2/3 cm_F; 1/2/3cm fruit, MG_F; Mature green fruit, B_F; Breaker fruit, and B10_F; ten days Breaker fruit. Heatmap was generated using log2 transformed RT-qPCR values.
Moreover, the genes with more significant transcript abundance include SlDDP9 in the root, mature green fruit, SlDDP5 in the root, 3 cm fruit, mature green fruit, SlDDP3 in leaves and root, SlDDP10 in root and 2 cm fruit, SlDDP1 in 10 days breaker fruit and SlDDP12 I breaker and ten-day breaker fruit. It was observed that some SlDDP genes such as SlDDP1, SlDDP3, SlDDP4, SlDDP7, SlDDP9, and SlDDP10 contained ethylene promoter (ERE, Fig. 4) in their promoter sequences. The expression profile of these genes during various stages of fruit development revealed their elevated transcript abundance except for SlDDP7. These suggested that these genes may play an essential role in ethylene-dependent tomato fruit ripening (Fig. 5).
Phytohormone and Abiotic Stress-inducible Expression Analysis of Putative Tomato DDPs
To further gain insight into the putative role of SlDDPs in tomatoes. We investigated the expression profiles of SlDDPs under two abiotic stresses and three phytohormones, including salt and drought, abscisic acid (ABA), gibberellins (GA3), and auxin (IAA).
For salt treatment, most SlDDP genes were upregulated over various time points, but few were downregulated upon exposure. SlDDP6, SlDDP11, and SlDDP12 were downregulated along with all-time points but, SlDDP1, SlDDP2, SlDDP3, SlDDP4, SlDDP7, SlDDP8, and SlDDP10 exhibited opposite trends and were peaked at 24 h interval. Similarly, SlDDP5 was peaked at 12 h, but SlDDP9 was peaked at 9 h (Fig. 6a). Under PEG stress, SlDDP2, SlDDP3, SlDDP6, SlDDP7, and SlDDP8 were upregulated at 24 h after stress. SlDDP11 was upregulated at 3 h time point and then downregulated in subsequent time intervals. SlDDP12 sharply upregulated till 6 h after treatment and then downregulated in later time points. Similarly, SlDDP3 and SlDDP4 were peaked at 12 h and then downregulated at 24 h. the elevated levels of SlDDP1 and SlDDP5 were found at 6 h and 24 h time points, but SlDDP9 was upregulated along with all-time intervals with maximum expressions at 24 h (Fig. 6b).
Figure 6.
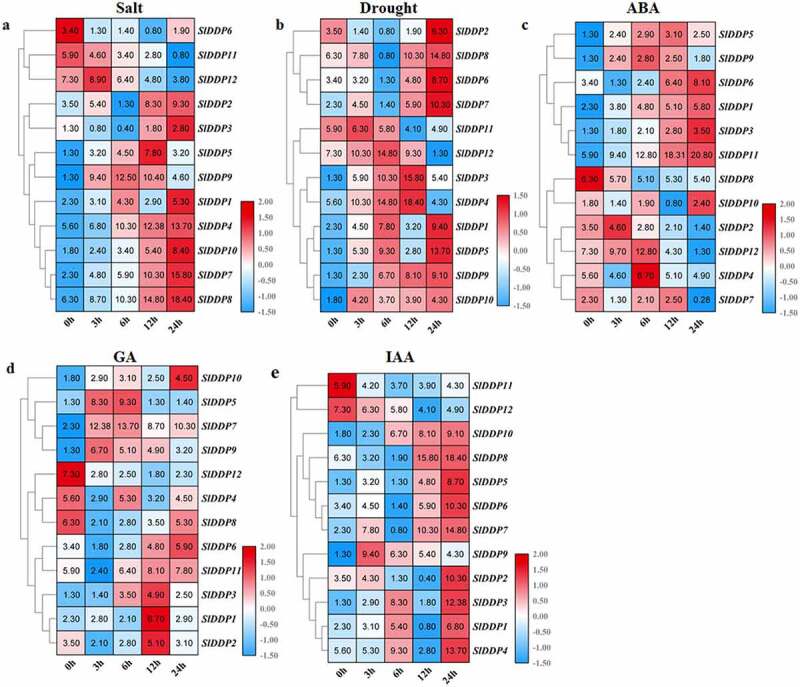
Expression analysis of SLDDPs under salinity, drought, and Phytohormones. Heatmap showing expression profile of 12 SlDDPs under (a) salt, (b) drought (PEG), (c) Abscisic acid (ABA), (d) Gibberellin (GA3), and (e) Auxin (IAA) at 0 h, 3 h, 6 h, 12 h, and 24 h time points. Plants at 0 h time interval were used as control. Heatmap was generated using log2 transformed RT-qPCR values.
Under ABA treatment, the transcript levels of SlDDP1. SlDDP3, SlDDP6, and SlDDP11 were increased sharply along with all-time intervals and detected maximum at 24 h. Similarly, the expression levels of SlDDP4, SlDDP9, and SlDDP12 were sharply increased till 6 h and then downregulated in later intervals. SlDDP10 was peaked at 24 h, but SlDDP2 showed opposite trends. Moreover, the expression of SlDDP5 and SlDDP7 was upregulated till 12 h and then suppressed (Fig. 6c). Under GA treatment, the transcript abundance of SlDDP6, SlDDP8, and SlDDP10 sharply upregulated and peaked at 24 h but, SlDDP4, SlDDP8, SlDDP9, and SlDDP12 was downregulated upon exposure to GA. The expression of SlDDP6 and SlDDP11 was significantly upregulated at 12 h and 24 h while SlDDP1, SlDDP2, and SlDDP3 were upregulated at 12 h interval only (Fig. 6d).
For auxin treatment, SlDDP11 and SlDDP12 were suppressed but, SlDDP2, SlDDP5, SlDDP6, SlDDP8, and SlDDP10 was peaked at 24 h after treatment. SlDDP9 was upregulated at 3 h, SlDDP1, SlDDP3, and SlDDP4 were peaked at 6 h and 24 h (Fig. 6e). In comparison, SlDDP1, SlDDP2, SlDDP3, SlDDP4, SlDDP7, SlDDP8, and SlDDP10 was peaked at 24 h under salt, drought, ABA, and IAA stresses. SlDDP3 was upregulated under drought and GA, SlDDP9 induced under salt and drought at 6 h and under GA and auxin at 3 h. SlDDP12 was induced under drought and ABA at 6 h but suppressed in GA and auxin upon exposure. Similarly, SlDDP8 was suppressed upon treatment, but SlDDP5 was induced under salt and drought at 12 h, and under drought and auxin at 24 h (Fig. 6a-e).
Discussion
The plant faces severe destruction from abiotic and biotic stresses during its life cycle, impacting its survival and productivity. Plants have developed tolerance mechanisms to alter their physiology and cellular biochemistry during stresses through changes in gene expression.57 Some of these expression products came from genes containing the hypothetical domain of unknown functions (DUF). One such domain is DUF221 and is a highly conserved membrane-associated protein and reported to regulate osmoregulation of calcium through plasma membrane.23 To date, the status of DUF221 genes family is mainly unexplored. In this study, DDP gene family was identified under two abiotic (salinity and drought) and three phytohormones (ABA, IAA, GA3) stress, according to previous studies in Arabidopsis,24 maize,58 and rice.8,59
The DDP proteins have been reported to be highly conserved across the plant lineage,57,60 which is also confirmed in our study (Fig S1). In this study, 12 DDP genes were identified in the tomato genome and were distributed on 8 chromosomes with a maximum number of genes located on chromosome 2 (Fig. 1). We found that tomato possesses a similar number of DDPs (Table 1) like maize.58 There is a loss of three DDPs in tomato compared to Arabidopsis, which possesses the highest number of DPPs 24, whilst one gene is compared to the rice genome.59 The number of DDPs in tomato, rice, and maize did not very much, indicating Arabidopsis genome undergo relatively conserved evolutionary history after their divergence. Tomato SlDDPs were clustered into four phylogenetic clades (Fig. 2) as reported previously in Arabidopsis, rice, and maize.24,58,59 To further gain insights into the structural changes in DDP genes that occur during the evolution, the gene structure and conserved motifs were analyzed. A high degree of variation in size and distribution of intron and exon was found. The SlDDPs with similar exon and intron were clustered together in a phylogenetic tree. Moreover, it was found in the distribution of conserved motifs (Fig. 3). Similar gene structural diversifications were observed in rice OsDDPs.8
Genome duplication plays a pivotal role in speciation and adaptation under various environmental conditions.61 The previous study has shown that segmental duplication was primarily responsible for the expansion of DDPs. For instance, four segmental duplications were found in Arabidopsis, and single pair was detected in rice.57 In our study, three pairs of SlDDP segmental duplication were found (Fig. 1). Like Arabidopsis, the segmental duplicates of SlDDPs were clustered together into a single clade (clade I and II) (Fig. 2). However, the duplicated pair of rice was found scattered in clade-I and clade-II.57 This outcome further substantiates the gene duplication in tomato and Arabidopsis during evolution, which might eventually allow the protein functional diversity by adaptive evolution.62 The approximate age of segmentally duplicated DDP (OsDDP3-OsDDP10) paralogues of rice is 64.2 MYA. In contrast, 29, 65, and 71 MYA of gene pairs AtDDP9-AtDDP13, AtDDP2-AtDDP14, and AtDDP12-AtDDP6 indicated that duplication of these pairs could have occurred before the appearance of Poaceae from the common ancestor ~55–70 mya63 or crucifers~24–40 mya,64 respectively. We found that tomato segmentally duplication DDPs (SlDDP3-SlDDP8, SlDDP6-SlDDP12, and SlDDP7-SlDDP11) indicated duplications of SlDDPs occurred before divergence Solanaceae from common ancestors about 50–52 mya.65
It was reported that rice OsDDPs were regulated by multiple miRNAs. For instance, OsDDP6 was targeted by multiple miRNA families belonging to the osa-miR818 family and osa-miR1436. Similarly, OsDDP10 is targeted by miRNA osa-miR6248.8 Barley miR81866 and rice osa-miR143666 and osa-miR624867 have been differentially regulated under drought, salinity, and arsenate stresses. It was found that tomato SlDDPs were also targeted by multi miRNA gene families (Table 73). For example, tomato SlDDP3 was targeted by sly-miRNA395a, sly-miRNA395, and sly-miRNA717b while sly-miR319b and sly-miR6022 target to cleavage tomato SlDDP5. These findings are suggesting that multiple miRNAs may regulate a single gene. Cis-regulatory elements play a pivotal role in controlling various aspects of plant growth and development under normal, abiotic, biotic, and phytochrome responses by regulating gene expression. Several cis-acting sequences related to phytohormone responses such as ERE, ABRE, GARE, AuxRR-core, TGA elements, abiotic stress-responsive such as HSE, MBS, and MYB were identified in promoter region of SlDDPs (Fig. 4, Table S6).
To our knowledge, although the relationship between AtDDP and ZmDDP proteins and stresses has been reported.58,68 The dynamic abiotic and phytohormone-responsive expression patterns of SlDDPs were still obscure. Expression pattern analysis of SlDDPs helped us to understand their possible functions and offer a thorough foundation for future functional studies. To provide the further foundation for functional characterization of tomato SlDDPs, expression profile analysis under drought, salinity, and phytohormone were evaluated at various time points. It was found that the majority of genes were induced under these stresses while few were downregulated. Ding et al.,58 showed that 12 of six ZmOSCAs were significantly upregulated, and expression of single ZmOSCA was down-regulated. The relative expression levels of OsOSCA1.1, −1.2, −2.1, −2.4, −2.5, and −4.1 were upregulated by PEG treatment.59 Interestingly, in this study, we found that SlDDP6, SlDDP11, and SlDDP12 were suppressed under salinity, but SlDDP6 was upregulated under drought stress (Fig. 6a-b), indicating that these genes might serve as key mediators of drought stress responses. SlDDP4, SlDDP8, and SlDDP12 were suppressed under GA and IAA stresses, but SlDDP4 was upregulated under IAA along with SlDDP2, SlDDP3, SlDDP4, SlDDP5, SlDDP6, SlDDP7, SlDDP8, and SlDDP10 (Fig. 6d-e). SlDDP1, SlDDP3, SlDDP6, and SlDDP11 were upregulated under ABA and IAA (Fig. 6c and Fig.6e). Moreover, SlDDP2 was upregulated under salinity, drought, GA, and IAA but suppressed under ABA, while SlDDP8 was suppressed under GA. This induced expression of SlDDPs under various stresses suggested that these genes may involve in multiple stress responses in the tomato plant.
Conclusion
In summary, a total of 12 tomato SlDDPs genes were identified in the whole genome. Gene structure, conserved motifs, and cis-regulatory elements prediction, and phylogeny were analyzed. The expression profile in various parts of the tomato plant was investigated to clue the possible biological and development role of these genes. However, differential abiotic (salinity and drought) and phytohormone inducible expression profile revealed their putative roles in abiotic and hormone transduction pathways. Furthermore, the prediction of miRNAs targets revealed that multiple-miRNAs regulate the expression of tomato SlDDPs. Together, our study will provide helpful information for further functional analysis of DDP genes in tomato and other related plant species.
Supplementary Material
Acknowledgments
Not applicable
Abbreviations
DDP, DUF221 domain-containing; MW, molecular weight; kDa, kilo Dalton; GRAVY, the grand average of hydropathy; pI, theoretical isoelectric point; ABA, abscisic acid; GA3, gibberellins; IAA, indole-3-acetic acid; PEG, polyethylene glycol; mM, Millimolar
Author’s contribution
MW designed and performed the whole experiment. MMA prepared material and performed RT-qPCR, and finally analyzed results. MW and IF performed all bioinformatics analyses. MW and MMA drafted the manuscript and revised the manuscript. All the author(s) have read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Sharoni AM, Nuruzzaman M, Satoh K, Moumeni A, Attia K, Venuprasad R.. Comparative transcriptome analysis of AP2/EREBP gene family under normal and hormone treatments, and under two drought stresses in NILs setup by aday selection and IR64. Molecular Genetics and Genomics. 2012;287(1):1–19. doi: 10.1007/s00438-011-0659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharjee P, Das R, Mandal A, Kundu P. Functional characterization of tomato membrane-bound NAC transcription factors. Plant Mol Biol. 2017;93(4–5):511–32. doi: 10.1007/s11103-016-0579-z. [DOI] [PubMed] [Google Scholar]
- 3.Zhu M, Chen G, Zhang J, Zhang Y, Xie Q, Zhao Z. The abiotic stress-responsive NAC-type transcription factor SlNAC4 regulates salt and drought tolerance and stress-related genes in tomato (solanum lycopersicum). Plant Cell Rep. 2014;33(11):1851–63. doi: 10.1007/s00299-014-1662-z. [DOI] [PubMed] [Google Scholar]
- 4.Sun H, Hj F, Hq L. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics. 2015;16(1):9. doi: 10.1186/s12864-014-1209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stulemeijer IJE, Stratmann JW, Joosten MHAJ. Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf-4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 2007;144(3):1481–94. doi: 10.1104/pp.107.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Fu F, Zhang H, Song F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (solanum lycopersicum L.). BMC Genomics. 2015;16(1):771. doi: 10.1186/s12864-015-1990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Y, Hu X, Li C, Xu X, Su C, Li J. SlbZIP38, a tomato bzip family gene downregulated by abscisic acid, is a negative regulator of drought and salt stress tolerance. Genes. 2017;8(12): 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proceedings of the National Academy of Sciences USA; 2017; 114:E897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W, Xian Z, Kang X, Tang N, Li Z. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant Biol. 2015;15(1):209. doi: 10.1186/s12870-015-0590-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waseem M, Ahmad F, Habib S, Li Z. Genome-wide identification of the auxin/indole-3-acetic acid (Aux/IAA) gene family in pepper, its characterisation, and comprehensive expression profiling under environmental and phytohormones stress. Sci Rep. 2018;8(1):12008. doi: 10.1038/s41598-018-30468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chini A, Ben-Romdhane W, Hassairi A, Aboul-Soud MAM. Identification of TIFY/JAZ family genes in solanum lycopersicum and their regulation in response to abiotic stresses. PloS One. 2017;12(6):e0177381–e. doi: 10.1371/journal.pone.0177381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waseem M, Ahmad F, Habib S, Gao Y, Li Z. Genome-wide identification of FK506-binding domain protein gene family, its characterization, and expression analysis in tomato (solanum lycopersicum L.). Gene. 2018;678:143–54. doi: 10.1016/j.gene.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 13.Waseem M, Ahmad F. The phosphoenolpyruvate carboxylase gene family identification and expression analysis under abiotic and phytohormone stresses in Solanum lycopersicum L. Gene. 2019;690:11–20. doi: 10.1016/j.gene.2018.12.033. [DOI] [PubMed] [Google Scholar]
- 14.Fragkostefanakis S, Mesihovic A, Simm S, Paupière MJ, Hu Y, Paul P. HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues. Plant Physiol. 2016;170(4):2461. doi: 10.1104/pp.15.01913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodacre NF, Gerloff DL, Uetz P. protein domains of unknown function are essential in bacteria. mBio. 2013 Dec 31;5(1):e00744-13. doi: 10.1128/mBio.00744-13. PMID: 24381303; PMCID: PMC3884060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauser R, Pech M, Kijek J, Yamamoto H, Titz B, Naeve F. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8(7):e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong H, Zhang H, Guo R, Wang Q, Huang X, Liao J. characterization and functional divergence of a novel DUF668 gene family in rice based on comprehensive expression patterns. Genes. 2019;10(12):980. doi: 10.3390/genes10120980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Wang P, Gao W, Long Y, Wang Y, Geng S. Genome-wide identification of the DUF668 gene family in cotton and expression profiling analysis of GhDUF668 in gossypium hirsutum under adverse stress. BMC Genomics. 2021;22(1):395. doi: 10.1186/s12864-021-07716-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li LH, Lv MM, Li X, Ye TZ, He X, Rong SH. The rice OsDUF810 family: osDUF810. 7 may be involved in the tolerance to salt and drought. Mol Biol. 2018;52:489–96. doi: 10.1134/S002689331804012X. [DOI] [PubMed] [Google Scholar]
- 20.Luo C, Guo C, Wang W, Wang L, Chen L. Overexpression of a new stress-repressive gene OsDSR2 encoding a protein with a DUF966 domain increases salt and simulated drought stress sensitivities and reduces ABA sensitivity in rice. Plant Cell Rep. 2014;33(2):323–36. doi: 10.1007/s00299-013-1532-0. [DOI] [PubMed] [Google Scholar]
- 21.Guo C, Luo C, Guo L, Li M, Guo X, Zhang Y. OsSIDP366, a DUF1644 gene, positively regulates responses to drought and salt stresses in rice. J Integr Plant Biol. 2016;58(5):492–502. doi: 10.1111/jipb.12376. [DOI] [PubMed] [Google Scholar]
- 22.Rai A, Suprasanna P, D’Souza SF, Kumar V. Membrane topology and predicted RNA-binding function of the ‘early responsive to dehydration (ERD4)’ plant protein. PLoS One. 2012;7(3):e32658. doi: 10.1371/journal.pone.0032658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hou C, Tian W, Kleist T, He K, Garcia V, Bai F. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014;24(5):632–35. doi: 10.1038/cr.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan F, Yang H, Xue Y, Kong D, Ye R, Li C. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in arabidopsis. Nature. 2014;514(7522):367–71. doi: 10.1038/nature13593. [DOI] [PubMed] [Google Scholar]
- 25.Gholizadeh A. DUF538 protein superfamily is predicted to be chlorophyll hydrolyzing enzymes in plants. Physiology and Molecular Biology of Plants. 2016;22(1):77–85. doi: 10.1007/s12298-015-0331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gholizadeh A Chlorophyll binding ability of non-chloroplastic DUF538 protein superfamily in plants. Proceedings of the National Academy of Sciences, India Section B: Biological Sciences; 2018; 88:967–76. [Google Scholar]
- 27.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24(11):1911–20. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin H, Chen F, Huan X, Machida S, Song J, Yuan YA. Structure of the arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein–protein interaction. Rna. 2010;16:474–81. doi: 10.1261/rna.1965310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Guo L, Guo C, Wang L, Chen L. Over-expression of a DUF1644 protein gene, SIDP361, enhances tolerance to salt stress in transgenic rice. Journal of Plant Biology. 2016;59(1):62–73. doi: 10.1007/s12374-016-0180-7. [DOI] [Google Scholar]
- 30.Hou X, Liang Y, He X, Shen Y, Huang Z. A novel ABA-responsive TaSRHP gene from wheat contributes to enhanced resistance to salt stress in arabidopsis thaliana. Plant Molecular Biology Reporter. 2013;31(4):791–801. doi: 10.1007/s11105-012-0549-9. [DOI] [Google Scholar]
- 31.Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134(6):1019–29. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaik R, Ramakrishna W. Bioinformatic analysis of epigenetic and microRNA mediated regulation of drought responsive genes in rice. PLoS One. 2012;7(11):e49331. doi: 10.1371/journal.pone.0049331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuartero J, Bolarin MC, Asins MJ, Moreno V. Increasing salt tolerance in the tomato. J Exp Bot. 2006;57(5):1045–58. doi: 10.1093/jxb/erj102. [DOI] [PubMed] [Google Scholar]
- 34.Giorgi F, Lionello P. Climate change projections for the Mediterranean region. Glob Planet Change. 2008;63(2–3):90–104. doi: 10.1016/j.gloplacha.2007.09.005. [DOI] [Google Scholar]
- 35.Julkowska MM, Testerink C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015;20(9):586–94. doi: 10.1016/j.tplants.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59(1):651–81. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 37.Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J. The SOL genomics network: a comparative resource for solanaceae biology and beyond. Plant Physiol. 2005;138(3):1310–17. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiser L, Subramaniam S, Li D, Huala E. Using the arabidopsis information resource (TAIR) to find information about arabidopsis genes. Current Protocols in Bioinformatics. 2017;60(1):1.11.1–1.45. doi: 10.1002/cpbi.36. [DOI] [PubMed] [Google Scholar]
- 39.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database):D281–D8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(suppl):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences U S A; 1998; 95:5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–d3. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stothard P. The sequence manipulation suite: javaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28(6):1102. 4. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 44.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–7. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings International Conference on Intelligent Systems for Molecular Biology; 1994; 2:28–36. AAAI Press. [PubMed] [Google Scholar]
- 46.Mondal TK, Ganie SA, Rana MK, Sharma TR. Genome-wide analysis of Zinc transporter genes of maize (zea mays). Plant Molecular Biology Reporter. 2014;32(2):605–16. doi: 10.1007/s11105-013-0664-2. [DOI] [Google Scholar]
- 47.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–97. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–27. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–49. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evoliution. 1987;4:406–25. [DOI] [PubMed] [Google Scholar]
- 51.Comeron JM. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15(9):763–64. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]
- 52.He Y, Liu X, Ye L, Pan C, Chen L, Zou T. genome-wide identification and expression analysis of two-component system genes in tomato. Int J Mol Sci. 2016;17:1204. doi: 10.3390/ijms17081204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(suppl_2):W155–9. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(D1):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waseem M, Li N, Su D, Chen J, Li Z. Overexpression of a basic helix–loop–helix transcription factor gene, SlbHLH22, promotes early flowering and accelerates fruit ripening in tomato (solanum lycopersicum L.). Planta. 2019;250(1):173–85. doi: 10.1007/s00425-019-03157-8. [DOI] [PubMed] [Google Scholar]
- 56.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods. 2001;25(4):402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 57.Ganie SA, Pani DR, Mondal TK. Genome-wide analysis of DUF221 domain-containing gene family in oryza species and identification of its salinity stress-responsive members in rice. PLOS ONE. 2017;12(8):e0182469. doi: 10.1371/journal.pone.0182469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding S, Feng X, Du H, Wang H. Genome-wide analysis of maize OSCA family members and their involvement in drought stress. PeerJ. 2019;7:e6765–e. doi: 10.7717/peerj.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Yuan F, Wen Z, Li Y, Wang F, Zhu T. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015;15:261. doi: 10.1186/s12870-015-0653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Häuser R, Pech M, Kijek J, Yamamoto H, Titz B, Naeve F. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:e1002815. doi: 10.1372/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytologist. 2009;183(3):557–64. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 62.Jia L, Clegg MT, Jiang T. Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 2004;134(2):575–85. doi: 10.1104/pp.103.027201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Shi X, Hao B, Ge S, Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytologist. 2005;165(3):937–46. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- 64.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–78. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schlueter JA, Dixon P, Granger C, Grant D, Clark L, Doyle JJ. Mining EST databases to resolve evolutionary events in major crop species. Genome. 2004;47:868–76. doi: 10.1139/g04-047. [DOI] [PubMed] [Google Scholar]
- 66.Deng P, Wang L, Cui L, Feng K, Liu F, Du X. Global identification of microRNAs and their targets in barley under salinity stress. PLoS One. 2015;10:e0137990. doi: 10.1371/journal.pone.0137990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Q. Novel miRNAs in the control of arsenite levels in rice. Funct Integr Genomics. 2012;12(4):649–58. doi: 10.1007/s10142-012-0282-3. [DOI] [PubMed] [Google Scholar]
- 68.Zhao X, Xu M, Wei R, Liu Y. Expression of OsCAS (calcium-sensing receptor) in an arabidopsis mutant increases drought tolerance. PLoS One. 2015;10(6):e0131272. doi: 10.1371/journal.pone.0131272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


