Abstract
The co-inhibitory receptor lymphocyte activation gene 3 (LAG-3) is an immune checkpoint molecule that negatively regulates T cell activation, proliferation, and homeostasis. Blockade or deletion of LAG-3 in autoimmune-prone backgrounds or induced-disease models has been shown to exacerbate disease. We observed significantly fewer LAG-3+ CD4 and CD8 T cells from subjects with relapsing-remitting multiple sclerosis (RRMS) and type 1 diabetes. Low LAG-3 protein expression was linked to alterations in mRNA expression and not cell surface cleavage. Functional studies inhibiting LAG-3 suggest that in RRMS subjects, LAG-3 retains its ability to suppress T cell proliferation. However, LAG-3 expression was associated with the expression of markers of apoptosis indicating a role for low LAG-3 in T cell resistance to cell death. In T cells from RRMS subjects, we observed a global dysregulation of LAG-3 expression stemming from decreased transcription and persisting after T cell stimulation. These findings further support the potential clinical benefits of a LAG-3 agonist in the treatment of human autoimmunity.
Keywords: Autoimmunity, Cell surface molecules
Introduction
Autoimmune diseases are chronic and debilitating and affect more than 20 million people in the USA (1). Although immunotherapies have greatly improved treatment, we are still unable to prevent or cure autoimmune disease. An understanding of the molecular and cellular mechanisms underlying the loss of tolerance and progression to autoimmune disease is crucial to move this field forward. Here, we investigate the role of the inhibitory receptor, lymphocyte activation gene-3 (LAG-3) in two organ-specific autoimmune diseases: relapsing remitting multiple sclerosis (RRMS) and type 1 diabetes (T1D). Inhibitory receptors (IRs) counterbalance co-stimulatory signals and prevent excessive effector T cell activation contributing to autoimmunity (2). Consequently, the expression of IRs is necessary to promote appropriate self-tolerance, but overexpression can lead to an inability of T cells to mount effective immune responses against tumors or pathogens (3). This critical role in regulating immune responses has made IRs an attractive target for immunotherapy especially in the setting of cancer – where targeting CTLA-4 and PD-1 have been highly successful. More recently, clinical trial results from combination LAG-3/PD-1 immune checkpoint inhibition in patients with melanoma doubled the progression-free survival time compared to PD-1 blockade alone, underscoring the value of targeting LAG-3 in the clinic (4). Despite the clinical success of immune checkpoint inhibition in the treatment of cancer, these therapies are considered dangerous for the treatment of patients with underlying autoimmunity (5, 6) and have been associated with both disease relapse and death in patients with multiple sclerosis (MS) (7).
To date, almost all of the work investigating the role of LAG-3 in autoimmunity has been done in murine models. Experimental autoimmune encephalomyelitis (EAE) is a commonly used murine model for the inflammatory demyelinating disease, MS, and is considered a prototype for T cell-mediated autoimmune disease. In studies of the myelin oligodendrocyte glycoprotein (MOG)-specific TCR transgenic mouse EAE model, the upregulation of LAG-3 on myelin-specific CD4+ gut-induced intraepithelial lymphocytes that migrate to the CNS is required to reduce disease severity (8). The role of LAG-3 shedding in the context of EAE has also been explored as the inhibitory function of T cells is enhanced in non-cleavable LAG-3 (LAG-3NC) (9). Less severe EAE is seen in LAG-3NC CD4+ Foxp3- cells compared to LAG-3NC CD8 T cells or LAG-3 cleavable controls. Furthermore, LAG-3NC on CD4+ Foxp3- T cells reduced proliferation and increased cleaved caspase 3 (cCasp3) suggesting a selective role for LAG-3NC mediating T cell functionality and proliferation in EAE (10). In addition, studies where antigen-specific LAG-3+ Foxp3+ regulatory T cells are expanded through vaccination demonstrate enhanced suppression of antigen-specific autoreactive effector T cells and bystander immunosuppression (11). Thus, the LAG-3 inhibitory pathway may function to selectively immunosuppress activated T cells through a variety of pathways that may be clinically effective in the treatment of autoimmunity. While these studies looking at the impacts of Lag3 deletion or overexpression provide a foundation for hypothesizing a role for LAG-3 in autoimmunity, the source and extent of LAG-3 expression in human autoimmunity has not been investigated.
In this study, we address this gap in knowledge by investigating regulation of LAG-3 expression in primary T cells from patients with two organ-specific autoimmune diseases: RRMS and T1D. We explore the source of altered LAG-3 expression in these diseases and the functional implications of this altered expression.
Materials and Methods
Human subjects and samples
All samples used in this study were from the Benaroya Research Institute Registry and Repository. The study was approved by the Benaroya Research Institute’s IRB (protocol no. IRB07109) and all subjects gave written informed consent. Two patient cohorts and healthy controls were selected for these studies; both cohorts were heterogeneous with respect to disease duration, disease activity, and therapy for RRMS and T1D. The healthy control subjects were selected based on the absence of autoimmune disease or any family history of autoimmunity and were age- and sex-matched to the RRMS and T1D subjects. Individuals were diagnosed with RRMS based on the Revised McDonald Diagnostic Criteria for MS (12). All experiments were performed in a blinded manner. Characteristics of study participants are listed in Supplemental Table 1.
T cell isolation and culture
Thawed PBMCs were rested in serum-free X-VIVO 15 Medium (Lonza) for 1 h and washed with PBS. Total CD3 T cells, CD4 T cells, or CD8 T cells were purified by negative selection using magnetic-activated cell sorting (MACS) technology (Miltenyi). T cell stimulation was carried out in 96-well flat-bottom plates (Thermo Fisher Scientific) at 2 × 106 cells/ml in 200 μl of RPMI medium (supplemented with 10% human serum, 2 mM glutamine, 100 IU/ml penicillin, 0.1 mg/ml streptomycin) with plate-bound anti-CD3 (OKT3; 1 μg/ml) and soluble anti-CD28 (CD28.2; 2 μg/ml) for 24 h.
Flow cytometry
The relevant fluorochrome-conjugated anti-human surface antibodies used were specific for LAG-3 (R&D Systems, polyclonal goat IgG), ADAM10 (R&D Systems, clone 163003), ADAM17 (R&D Systems, clone 111633), CD25 (BioLegend, clone BC96), and CD95/Fas (BioLegend, clone DX2). Dead cells were discriminated by staining with Live/Dead™ fixable blue dead cell stain kit (Invitrogen). General T cell gating strategy, gating on LAG-3+ T cells, as well as ADAM10 and ADAM17 expression are shown in Supplemental Fig. 1A–D. For intracellular staining, cells were stained with surface markers (here a monoclonal LAG-3 antibody; eBioscience, clone 3DS223H), fixed in Fix/Perm buffer (eBioscience) for 30 min., washed in permeabilization buffer (eBioscience) twice, and stained for intracellular factors Ki67 (BD Biosciences, clone B56) and LAG-3 (R&D Systems, clone 874512) in permeabilization buffer for 30 min. on ice. To stain for cleaved caspase 3 (BD Biosciences, clone C92–605), cells were fixed and permeabilized using Fix Buffer I and Perm Buffer III (BD Biosciences), respectively. All cells were acquired on a LSR Fortessa (BD Biosciences) and data were analyzed using FlowJo version v10.6.2 (Tree Star).
LAG-3 shedding
Magnetically enriched CD3+ T cells were distributed to 96-well flat-bottom plates (Corning) at 2 × 106 cells/ml in 200 μl of supplemented RPMI medium (10% human serum) with plate-bound anti-CD3 (OKT3; 1 μg/ml), soluble anti-CD28 (CD28.2; 2 μg/ml), and either the pan-metalloproteinase inhibitor TAPI or the ADAM10 inhibitor GI254023X (20 μM; Selleck Chemicals). After 24 h, supernatants were collected and sLAG3 concentrations were determined using the LAG-3 Human ELISA Kit (Invitrogen).
Real-time qRT-PCR
Magnetically-enriched CD4 or CD8 T cells were either rested or stimulated with plate-bound anti-CD3 (OKT3; 1 μg/ml), soluble anti-CD28 (CD28.2; 2 μg/ml) at 1 × 106 cells/ml in 200 μl medium per well for 48 h. RNA was extracted from 6 × 105 cells using RNAqueous™-micro total RNA isolation kit (Invitrogen) with on-column DNA digestion (Qiagen). SuperScript III (Life Technologies) was used to generate complementary DNA and gene expression was measured by multiplex real-time PCR performed on an ABI 7500 Fast Real-Time PCR System. TaqMan expression assays for LAG3 (Hs00158563_m1), PDCD1 (Hs01550088_m1), TIGIT (Hs00545087_m1) and HAVCR2 (Hs00958618_m1) were used in combination with RPL36AL (Hs00733231_m1) for normalization (ThermoFisher Scientific). Quantitative detection of LAG3 mRNA levels was determined by 2−ΔΔCt calculations and reported relative to a Jurkat cell line (Clone E-61; ATCC TIB-152).
LAG-3 inhibition and proliferation
PBMCs were plated at 3.75 × 106 cells/ml in RPMI medium (supplemented with 10% human serum, 2 mM glutamine, 100 IU/ml penicillin, 0.1 mg/ml streptomycin) and a LAG-3 antagonist antibody (αLAG-3, Abcam, 17B4, 10 μg/ml) was added to select wells for 30 min. before the addition of PepTivator CMV pp65 and Adv5 hexon (Miltenyi Biotec, 175 ng/ml) and incubated at 37°C for 6 days.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9 and JMP software from SAS. To assess statistical significance, a one-way ANOVA with Holm-Sidak’s multiple comparison test with single paired variance was carried out to correct for multiple testing. Results were expressed as medians and differences were considered statistically significant at p < 0.05. Multiple variable and simple linear regression was performed, and the coefficient of determination (r2) was reported stratified by patient group. Bivariate and multivariate (partial) Pearson correlation analyses were also performed. Outliers were removed from Figure 1 using the ROUT method (coefficient Q = 0.1%) (13).
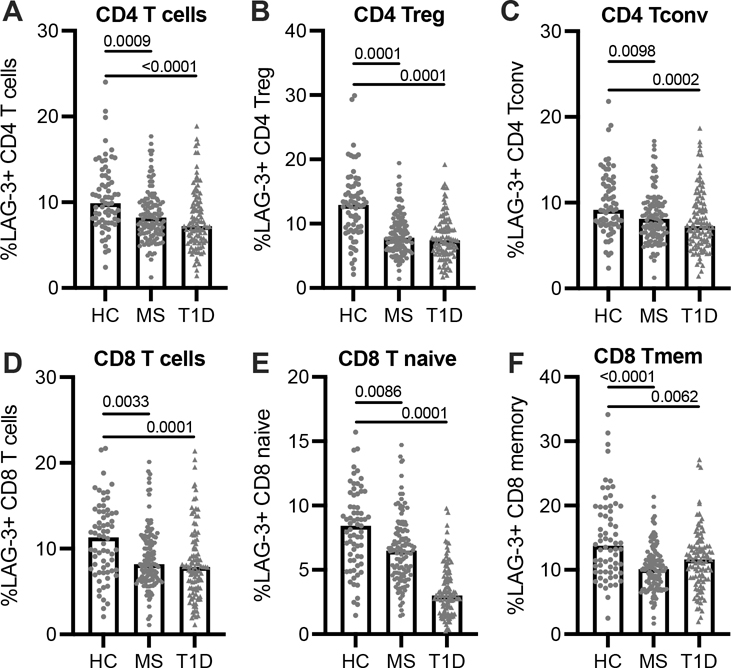
FIGURE 1.
Frequency of T cell LAG-3 surface expression. LAG-3+ frequency in: (A) CD4 T cells, (B) regulatory CD4 T cells (CD25+, CD127int), (C) conventional (non-Treg), (D) CD8 T cells, (E) naïve CD8 T cells (CD45RA+CD45RO-), and (F) memory CD8 T cells (CD45RA-CD45RO+) from HC (n = 69), T1D (n = 104), and RRMS (n = 121) subjects. Lines shown are median, and p values are shown after a one-way ANOVA. Results are shown from 25 independent experiments.
Results
Fewer LAG-3 positive T cells in RRMS and T1D
We measured LAG-3 cell surface expression on T cells from patients with RRMS (n = 121) and T1D (n = 104) compared to age- and sex-matched healthy control subjects (HC; n = 69) (Supplemental Table 1, Cohort 1). T1D subjects were not on disease modifying therapies (DMT), while 42% of RRMS subjects were treated with at least one DMT (natalizumab, glatiramer acetate, dimethyl fumarate, interferon beta-1a and 1b). We observed significantly fewer LAG-3+ CD4 T cells in both LAG-3 positive regulatory (CD4+ CD25hi CD127low) and conventional CD4 T cell subsets (Fig. 1A–C). Further gating of the CD4 population based on naïve and memory markers CD45RA and CD45RO showed fewer LAG-3+ CD4 memory cells in RRMS as compared to HC, but not in T1D (Supplemental Fig. 1E, 1F). Altered LAG-3 expression was not limited to the CD4 T cell compartment; there were also fewer LAG-3+ CD8 T cells in both RRMS and T1D compared to HC (Fig. 1D). While there is generally more LAG-3 expression on memory CD8 T cells than naïve, we observed fewer LAG-3+ cells in both memory and naïve CD8 T cells isolated from RRMS and T1D subjects (Fig. 1E, 1F). In RRMS subjects, the frequency of LAG-3 expression does not appear to be influenced by treatment with DMT (Supplemental Fig. 1G, 1H), disease flare (Supplemental Fig. 1I, 1J), age or sex (not shown). Together, these data suggest that T cell LAG-3 surface expression is globally dysregulated in subjects with RRMS and T1D.
Lower soluble LAG-3 levels in RRMS and T1D is primarily driven by the frequency of LAG-3+ T cells and not altered cleavage
Cell surface LAG-3 is regulated by proteolytic cleavage that results in the shedding of soluble LAG-3 (sLAG3) (14). This cleavage is mediated by the metalloproteinases ADAM10 and ADAM17 and is necessary for optimal T cell function (9). Since low LAG-3 cell surface expression on CD4 and CD8 T cells could be the result of enhanced proteolytic cleavage, we measured shedding of LAG-3 in T cell supernatants after activation. For these experiments, we selected a subset of subjects from Cohort 1 based on their resting surface LAG-3 expression. We selected HC with a LAG-3+ T cell frequency greater than the third quartile, and RRMS and T1D subjects with LAG-3+ frequency less than the first quartile. We found a significantly lower concentration of sLAG3 in supernatant taken from the RRMS and T1D cultures as compared to HC (Fig. 2A). In the same assay, we measured metalloproteinase expression at rest and upon activation. We observed significantly lower ADAM10 on both CD4 and CD8 resting T cells from RRMS and T1D subjects compared to HC at rest, while ADAM10 was lower on T1D CD8 cells (Supplemental Fig. 2A; Fig. 2B). Although ADAM10 expression increased upon activation, RRMS and T1D subjects had significantly lower ADAM10 in CD8 T cells (Fig. 2C) and RRMS subjects also had less ADAM10 expression on CD4 T cells after activation compared to HC (Supplemental Fig. 2B). ADAM17 was not differentially expressed in resting or stimulated T cells between cohorts (Supplemental Fig. 2C–F). To further address whether the altered frequency of LAG-3+ T cells could be influenced by shedding, we used metalloproteinase inhibitors to determine if blockade of ADAM10 and ADAM17 impacted cleavage to a greater degree in RRMS and T1D subjects. Both the pan-metalloproteinase inhibitor, TAPI-1, and the ADAM10 inhibitor, GI254023X, decreased the concentration of sLAG3 in conjunction with TCR stimulation (Fig. 2D). In cultures from RRMS and T1D subjects, the change in sLAG3 concentration after ADAM10 inhibition was comparable to that in HC (Fig. 2E). Together, these results indicate that while ADAM10 has reduced expression in RRMS and T1D, it is fully functional.
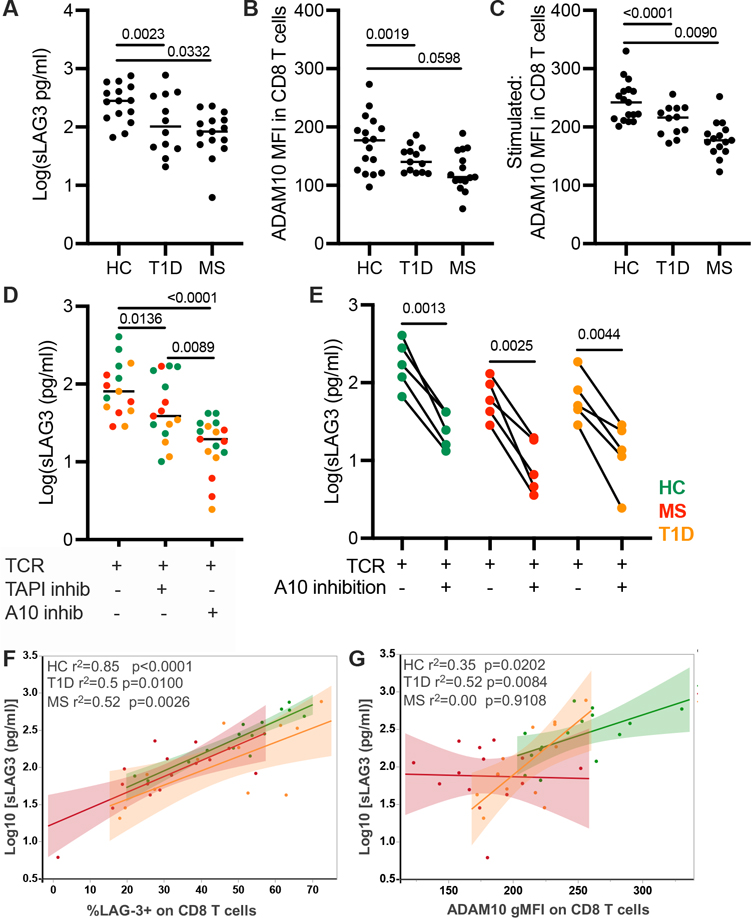
FIGURE 2.
The role of surface LAG-3 expression and ADAM10 expression in the generation of soluble LAG-3 (sLAG3). (A) The concentration of sLAG3 after 48-h TCR stimulation of CD3+ T cells. (B) The gMFI of ADAM10 in unstimulated CD8 T cells and (C) after a 48-h TCR stimulation. (D) Concentration of sLAG3 after 48-h TCR stimulation with or without the addition of the pan-metalloproteinase inhibitor, TAPI-1 or the ADAM10-inhibitor, GI254023X. (E) Concentration of sLAG3 after TCR stimulation and with ADAM10 inhibition in paired subjects. Results from one experiment, HC, n = 5; T1D, n = 4; RRMS, n = 4. (F) The frequency of CD8 T cell surface LAG-3 versus the concentration of sLAG3 after TCR stimulation in HC, RRMS, and T1D subjects (p < 0.0001, r2 = 0.6390 for all subjects). (G) The gMFI of ADAM10 on CD8 T cells versus the concentration of sLAG3 after TCR stimulation (p < 0.0001, r2 = 0.3450 for all subjects). Results from two independent experiments, HC (green), n = 17; RRMS (red), n = 15; and T1D (orange), n = 13. Lines shown are median and p values are shown after a (A-C) one-way ANOVA, (D, E) paired t test, and (F, G) Spearman correlation test.
After observing both decreased ADAM10 expression and fewer LAG-3+ T cells in RRMS subjects, we addressed which of these two factors most contributed to the concentration of sLAG3. We observed a positive correlation between the frequency of sLAG3 and surface LAG-3 expression on CD8 T cells when pooling all subjects together (p < 0.0001, r2 = 0.6390) (Fig. 2F) and similarly in CD4 T cells (p < 0.0001, r2 = 0.4150) (Supplemental Fig. 2G). In contrast to the correlation with LAG-3 frequency, the relationship between ADAM10 and sLAG3 was much weaker in CD8 and CD4 T cells (Fig. 2G, Supplemental Fig. 2H). In CD8 T cells from RRMS subjects, the weak relationship between ADAM10 expression and sLAG3 concentration suggests that altered expression of ADAM10 was not the primary cause of altered sLAG3. From this we conclude that alterations in LAG-3 cleavage are not the primary driver of altered LAG-3 expression on T cells in T1D and RRMS.
A linear regression analysis taking into account the surface expression of LAG-3 on CD8 T cells and cohort (HC, RRMS, T1D) found that the frequency of surface LAG-3 explains about 66% of the variability in the concentration of sLAG3 while ADAM10 expression and cohort type only explains 29%. Together, the frequency of surface LAG-3+ CD8 T cells and ADAM10 gMFI explain approximately 69% of the variability in the concentration of sLAG3, after controlling for cohort. The effect of adding cohort to this model had minimal impact (4%) on the total model r-squared value. Moreover, a multivariable model investigating interaction effects between surface LAG-3 and ADAM10 expression with cohort demonstrated that the relationship between these two factors and the concentration of sLAG3 did not differ between cohorts. This analysis implies that the expression of ADAM10 only modestly contributes to the cleavage of cell surface LAG-3, regardless of cohort.
Surface and intracellular LAG-3 protein is diminished after TCR stimulation in RRMS
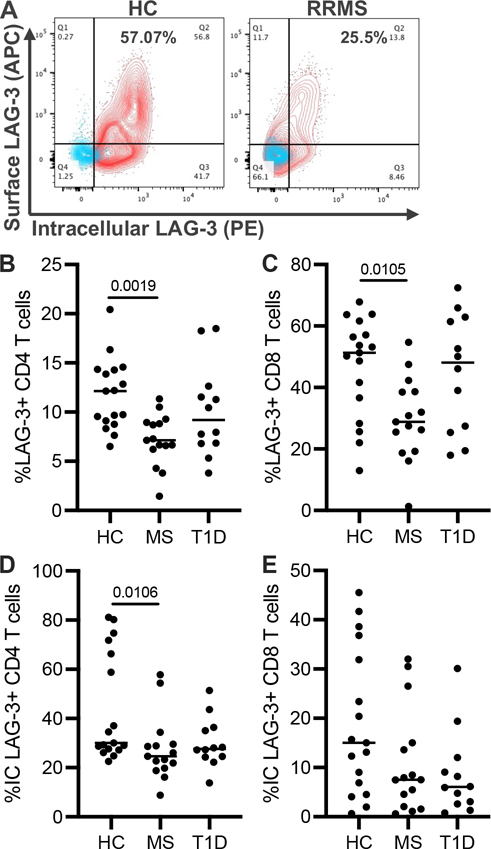
LAG-3 is stored in lysosomal compartments to facilitate rapid translocation to the cell surface following TCR stimulation; in the absence of stimulation, intracellular LAG-3 is degraded (15, 16). To determine whether intracellular LAG-3 is low after TCR stimulation, we used the same subset of subjects from Cohort 1 that were previously used for sLAG3 studies. To measure both surface and intracellular LAG-3, instead of using a polyclonal antibody, we used two monoclonal LAG-3 antibodies on separate colors (R&D Systems, 874512 and eBioscience, 3DS223H). Representative flow cytometry plots show LAG-3 expression in resting (blue) and stimulated (red) CD8 T cells from HC and RRMS subjects (Fig. 3A). Similar to our initial observations in resting T cells, following stimulation, surface LAG-3 in CD4 and CD8 T cells from RRMS subjects was significantly lower than in T cells from HC (Fig. 3B, 3C). Intracellular LAG-3 was also significantly lower in CD4 T cells and trended lower in CD8 T cells from RRMS subjects (Fig. 3D, 3E). While we did not observe statistically significant differences in surface LAG-3 expression on CD4 and CD8 T cells from T1D subjects, intracellular LAG-3 in CD8 T cells trended lower in these subjects. These data indicate that low LAG-3 protein production in RRMS contributes to the frequency of LAG-3+ T cells in RRMS, while in T1D there may be less intracellular LAG-3 available for cell surface expression.
FIGURE 3.
Frequency of intracellular and surface LAG-3 on T cells after stimulation. (A) Representative flow cytometry plots of surface and intracellular LAG-3 staining before (blue) and after (red) 48-h TCR stimulation in CD8 T cells from three independent experiments. After 48-h TCR stimulation of purified CD3+ T cells, the surface LAG-3 expression was measured in (B) CD4 and (C) CD8 T cells as well as the intracellular LAG-3 expression (D, E) from HC, RRMS, and T1D subjects. HC, n = 17; RRMS, n = 15; T1D, n = 12. Lines shown are median, and p values are shown after a one-way ANOVA.
Low LAG3 mRNA expression in RRMS
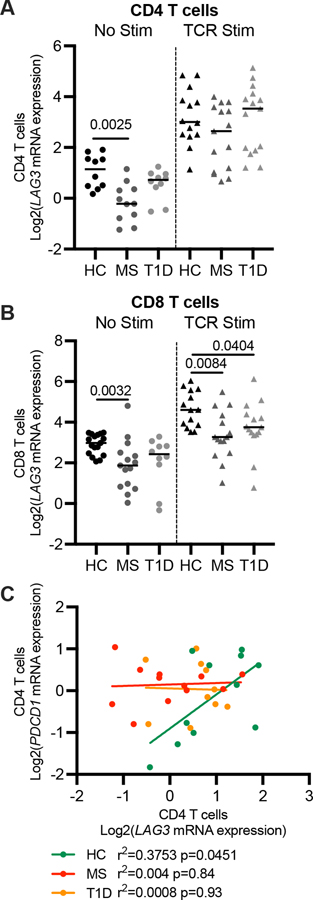
To determine whether low LAG-3 protein production and ultimately fewer LAG-3+ T cells from RRMS and T1D subjects was due to reduced transcription, we measured LAG3 mRNA levels using real-time quantitative RT-PCR. CD4 and CD8 T cells were magnetically enriched from frozen PBMCs isolated from a smaller cohort of RRMS (n = 20), T1D (n = 15) and HC (n = 19) subjects (Supplemental Table 1, Cohort 2). In T cells from subjects with RRMS, LAG3 mRNA expression was less than in resting CD4 (median 2.58-fold) and CD8 T cells (2.16-fold) from HC, while no difference was observed in T1D CD4 or CD8 T cells (Figure 4A, B). We next examined the expression of LAG3 after TCR stimulation and observed an increase in LAG3 transcript in all subjects. There were no differences in expression after stimulation within CD4 T cells (Fig. 4A); however, in CD8 T cells, both RRMS and T1D subjects expressed significantly less LAG3 after stimulation (Fig. 4B). The expression of LAG3 was not significantly different between RRMS subjects on and off DMT (Supplemental Fig. 3A). These findings suggest that low LAG3 mRNA levels contribute to less LAG-3 protein expression in RRMS subjects and may contribute in part to fewer LAG-3+ CD8 T cells in T1D.
FIGURE 4.
Transcriptional expression of LAG3 in T cells. LAG3 mRNA expression in resting (A) CD4 and (B) CD8 T cells. (C) PDCD1 mRNA levels correlated with LAG3 in HC but not RRMS and T1D subjects in resting CD4 T cells. qRT-PCR for LAG3 and PDCD1 was normalized to the ribosomal gene RPL36A and reported relative to a Jurkat cell line. HC, n = 19; T1D, n = 15; RRMS, n = 20, results shown are from three independent experiments. Lines shown are median, and p values are from a one-way ANOVA. Linear regressions are shown with Pearson correlation coefficient.
Knowing that IRs can function as a gene module in response to stimulation (3), we asked if altered LAG3 mRNA expression was part of a broader dysregulation of IRs in subjects with T1D and RRMS. Unlike our findings with LAG3, we found no differences in the mRNA expression of PDCD1 (PD-1), HAVCR2 (TIM3), and TIGIT in resting CD4 and CD8 T cells from individuals with RRMS and T1D when compared to HC (Supplemental Fig. 3B). Further, the positive correlation between LAG3 and PDCD1 transcript seen in HC was not present in resting CD4 T cells from RRMS and T1D subjects (Fig. 4C). After TCR stimulation, LAG3 mRNA levels were tightly correlated with TIGIT, PDCD1, and HAVCR2, indicating the relationship between LAG3 and other co-inhibitory molecules is retained upon activation, but not maintained in the resting state (Supplemental Fig. 3C).
LAG-3 inhibition enhances T cell proliferation in both HC and RRMS
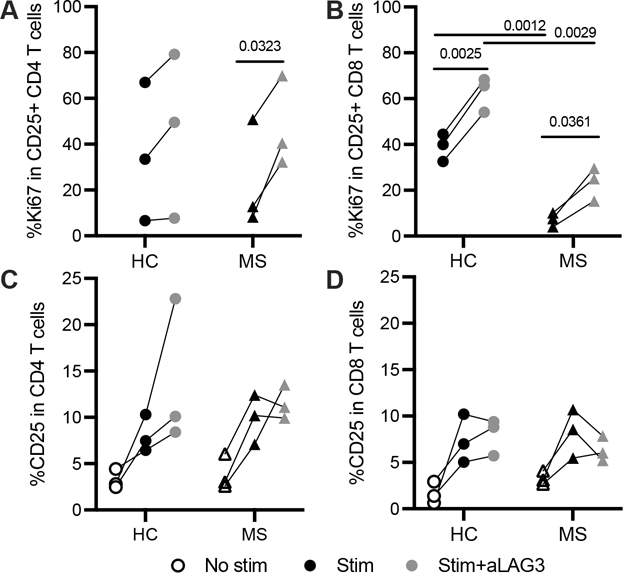
To address the functional implications of fewer LAG-3+ T cells in RRMS, we first investigated the effect of LAG-3 inhibition on T cell proliferation in samples from three HC and three off-therapy RRMS subjects selected from Cohort 1 on the basis of high and low surface LAG-3 expression, respectively. We measured proliferation after a six-day peptide stimulation in the presence or absence of a LAG-3 antagonistic antibody (αLAG-3) and observed more proliferation of LAG-3+ T cells when LAG-3 was blocked in both HC and RRMS T cells (Fig. 5A, B). Proliferation was comparable in activated CD4 T cells from HC and RRMS subjects (Fig. 5A); CD8 T cells from RRMS subjects were significantly less proliferative than those from HC (Fig. 5B). While T cell proliferation was higher after LAG-3 inhibition in both HC and RRMS, the CD8 T cells from RRMS subjects remained hypoproliferative relative to those from HC (Fig. 5B). While these data are gated on activated CD25+ T cells (the bulk of which are CD45RA- CCR7- effector memory), similar differences between groups were seen in total CD4 and CD8 T cells. Additionally, the frequency of CD25 was comparable between HC and RRMS subjects on both CD4 and CD8 T cells (Fig. 5C, 5D). LAG-3 inhibition comparably enhanced proliferation of T cells from RRMS and HC subjects suggesting T cell LAG-3 is functional in RRMS subjects.
FIGURE 5.
T cell proliferation after low-dose antigen stimulation and LAG-3 inhibition. Frequency of Ki67 after PBMC peptide stimulation (CMV/Adv5, 175 ng/ml, 6 days) in activated (A) CD4 and (B) CD8 T cells after subtracting the frequency of Ki67 in unstimulated T cells. Frequency of CD25 in (C) CD4 and (D) CD8 T cells. Unstimulated in blue; peptide-stimulated in black; peptide-stimulated with aLAG3 in light gray. ⚫ HC, ▲ RRMS. P values are paired or unpaired t tests. Results are from a single experiment, 3 HC and 3 RRMS subjects.
LAG-3 is associated with caspase 3 and Fas suggesting a link between LAG-3 and altered apoptosis in RRMS subjects
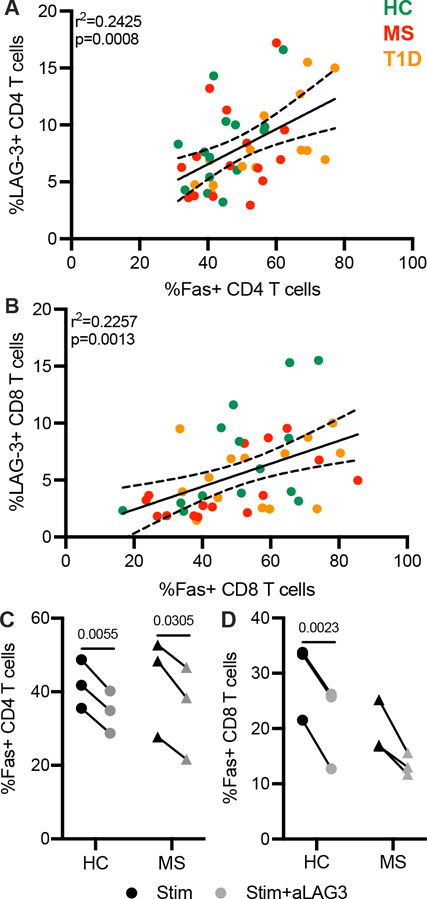
Earlier studies in LAG-3-deficient mice and non-cleavable LAG-3 mutants indicate a role for LAG-3 in promoting cell death. Furthermore, a resistance to activation-induced cell death has been implicated in the pathogenesis of MS and disease-modifying therapies have been shown to induce apoptosis (17–19). To determine the association of LAG-3 with markers of apoptosis, we used surface LAG-3 expression to select the same subset of patients from Cohort 1 as studied in Figure 2 (Supplemental Table 1) and then measured expression of Fas and caspase 3, both of which play a central role in apoptosis. Fas, once bound to its ligand (FasL), activates caspase 3 via caspase 8 leading to apoptosis (20). Here, we observed a positive correlation in the expression of LAG-3 and Fas in resting CD4 and CD8 T cells from HC, RRMS, and T1D subjects (Fig. 6A, 6B). LAG-3 inhibition decreased the frequency of Fas on CD4 and CD8 T cells from six HC and RRMS subjects after a six-day peptide stimulation (Fig. 6C, D).
FIGURE 6.
Fas expression and response to LAG-3 inhibition. Frequency of Fas positively correlates with LAG3 in (A) CD4 and (B) CD8 T cells in a pooled cohort of HC, RRMS, and T1D subjects. Results are from two independent experiments, HC, n = 14; RRMS, n = 15; T1D, n = 14. Frequency of Fas after peptide stimulation (CMV/Adv5, 175 ng/ml, 6 days; black) and aLAG3 (10 μg/ml; gray) in (C) CD4 and (D) CD8 T cells. Results are from a single experiment, 3 HC and 3 RRMS subjects (C, D). P values are shown after a Spearman correlation with 95% confidence interval shown (A, B) and a paired t-test (C, D).
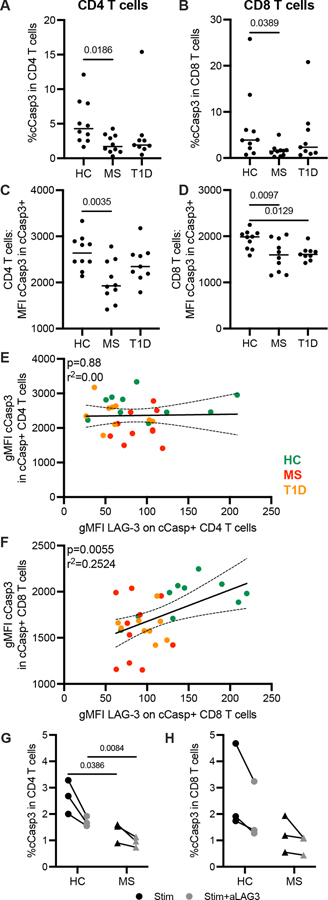
In addition, subjects with RRMS had fewer cleaved caspase 3 (cCasp3) positive CD4 and CD8 T cells as well as lower expression (gMFI) of cCasp3 on those cells; T cells from T1D subjects showed similar trends (Fig. 7A–D). Within cCasp3+ cells, there was a positive correlation between the expression of LAG-3 and cCasp3 in CD8 T cells, but not in CD4 T cells from pooled HC, RRMS, and T1D subjects (Fig. 7E, 7F). Blocking LAG-3 decreased the frequency of cCasp3+ CD4 T cells (p = 0.014) and CD8 T cells (p = 0.039) in all subjects (Fig. 7G, 7H). As we observed in resting T cells, there were fewer cCasp3+ T cells from RRMS subjects after stimulation and LAG-3 inhibition decreased cCasp3 to a lesser degree. LAG-3 expression is associated with the expression of both Fas and cCasp3 suggesting that low expression of LAG-3 may contribute to resistance to cell death.
FIGURE 7.
Cleaved caspase-3 expression and response to LAG-3 inhibition. Frequency of cleaved caspase 3 (cCasp3) in resting (A) CD4 and (B) CD8 T cells from HC, RRMS, and T1D subjects. Geometric MFI of cCasp3 within cCasp3+ (C) CD4 and (D) CD8 T cells. Correlation of geometric MFI of LAG-3 versus cCasp3 in cCasp3+ (E) CD4 and (F) CD8 T cells. Results are from two independent experiments, HC, n = 10; RRMS, n = 10; T1D, n = 10. Frequency of cCasp3 after peptide stimulation in (G) CD4 and (H) CD8 T cells. Results are from a single experiment, 3 HC and 3 RRMS subjects. Lines shown are median, and p values are shown after a one-way ANOVA (A-D), a Spearman correlation with 95% confidence interval shown (E, F) and an unpaired t-test (G, H).
Discussion
In this study, we investigated the source and extent of LAG-3 expression in two tissue-specific autoimmune diseases: RRMS and T1D. We found significantly lower protein expression of LAG-3 primarily due to alterations in mRNA expression and not cell surface cleavage in cells from subjects with RRMS and T1D. Our functional studies of the cleavage of LAG-3 in human autoimmunity corroborate murine findings on the generation of sLAG3 primarily by the ADAM10 sheddase and demonstrate the strong association of sLAG3 with LAG-3 surface expression.
We found fewer LAG-3+ T cells in individuals with RRMS and T1D compared to HC. Notably, this lower expression of LAG-3 was found in all major CD4 and CD8 T cell subsets. In T cells from RRMS, lower expression of LAG-3 was predominantly at the level of transcription, and not through post-transcriptional mechanisms such as extracellular transport or metalloprotease cleavage. In T1D, a single mechanism was not clearly identified, suggesting a more complex set of factors impacting LAG-3 expression in T cells from these subjects. Our findings are consistent with the literature where low LAG-3 expression has been linked to murine models of autoimmunity (21). While relevant patient-based studies are largely missing from the literature, fewer LAG-3+ CD4 T cells were recently reported in patients with active psoriatic arthritis (22). In an NOD mouse model, genetic deletion and LAG-3 inhibition have both been shown to accelerate the development of autoimmune diabetes (23). In the setting of EAE, upregulation of LAG-3 on myelin-specific CD4+ gut-induced intraepithelial lymphocytes limited disease severity (8). Prevention of LAG-3 cleavage on CD4 T cells reduced both disease severity and the number of pathogenic IL-17+IFN-γ+GM-CSF+ cells isolated from the brain (10).
We identified transcriptional dysregulation as the source of altered LAG-3 expression in T cells from RRMS subjects. This is supported by prior studies in MS where lower LAG3 mRNA expression in PBMCs is associated with a more severe MS outcome and increased likelihood of progression to secondary progressive MS, while high LAG3 expression correlates with persisting RRMS and lower disability score up to 10 years later (24). In T1D, the role of transcription in altered LAG-3 surface expression was less clear as LAG3 transcript was only significantly decreased in CD8 T cells after activation. Altered expression of LAG3 in T cells has been shown to be the result of epigenetic dysregulation in some cancer patients, suggesting a possible explanation for the lower frequency of LAG-3+ T cells (25–27). In addition, although LAG3 is part of an inhibitory receptor gene module that is co-regulated under certain conditions (3); we did not observe altered PDCD1, HAVCR2, or TIGIT mRNA expression in resting T cells suggesting that LAG3 is uniquely dysregulated in T1D and RRMS subjects. While we did not observe a decrease in the mRNA expression of HAVCR2 in MS subjects, a functional defect in TIM-3 immunoregulation has previously been reported to be reversed by treatment with glatiramer acetate or interferon beta (28, 29). It is possible that our findings are influenced by the makeup of our cohort wherein 80% of the subjects were taking DMT at the time of sample collection. Moreover, the altered relationship between PDCD1 and LAG3 in CD4 T cells may be indicative of epigenetic modifications causing a targeted change in LAG3 regulation.
LAG-3 cleavage is mediated by ADAM10 and ADAM17 and is associated with the inhibitory function of T cells (9). We examined the possibility that altered LAG-3 cleavage may contribute to the decrease in LAG-3+ T cells from subjects with RRMS and T1D. Differences in expression of these sheddases were limited to ADAM10, which was lower in T cells from RRMS and T1D subjects. Furthermore, T cells from RRMS and T1D subjects shed significantly less soluble LAG-3 after activation, which appeared to be driven by the frequency of LAG-3+ T cells and not by altered cleavage. We concluded that ADAM10 is functional in its ability to cleave surface LAG-3 in these subjects, as inhibition of ADAM10 and ADAM17 decreased the concentration of sLAG3 similarly in all subjects. Interestingly, inhibiting ADAM10 reduced the concentration of sLAG3 significantly more than the pan-metalloproteinase inhibitor, which is less effective at blocking ADAM10; this suggests ADAM10 is primarily responsible for the cleavage of sLAG3 from T cells in response to TCR stimulation. This observation corroborates recent findings that LAG-3 surface expression on CD4+ Foxp3- T cells was modulated by ADAM10-mediated cell surface shedding (10). We were surprised to observe decreased but functional ADAM10 in T cells from subjects with low LAG-3 where cleavage was not perturbed; future studies may explore the possibility of co-regulation between these two surface molecules.
Our functional studies implicate LAG-3 in the regulation of T cell proliferation and apoptosis in autoimmune disease. T cells from individuals with T1D and RRMS have been shown to have altered responses to activation and regulation. There is broad consensus that LAG-3 plays a role in regulating the expansion of activated T cells (30–32). Early research posited that LAG-3 was a negative regulator of T cell activation and function with the observation that blocking LAG-3 on CD4 T cell clones enhanced both proliferation and Th1 cytokine production (33). In assessing whether T cell LAG-3 was functional in RRMS subjects and how lower LAG-3 may alter the response to activation, we demonstrated that LAG-3 blockade resulted in enhanced proliferation in a similar respect in HC and RRMS subjects. Although we used only a small cohort for these experiments, it is interesting that antigen-stimulated CD8 T cells from RRMS subjects not prescribed DMT proliferated less than those from HC, both before and after LAG-3 inhibition. This may be due to underlying or acquired defects from chronic in vivo stimulation and independent of LAG-3 expression. Future studies may explore if dysregulated LAG-3 expression is also present in autoreactive cells that recognize CNS antigen. Our functional assays explored apoptosis in addition to proliferation as they relate to LAG-3 expression and inhibition. RRMS subjects exhibited both fewer cCasp3+ T cells and lower expression of cCasp3 on those cCasp3+ T cells that correlated with LAG-3; cCasp3 was further decreased after LAG-3 blockade. Taken together these data suggest low LAG-3 expression may be linked to the decrease in cell death observed in T cells from RRMS subjects and possibly also T1D subjects. Certain DMTs for RRMS have been shown to induce activation-induced cell death (18, 34) and the resistance of T cells to Fas-mediated apoptosis has been implicated in both the pathogenesis and prognosis of MS (17, 35, 36). We observed a modest correlation between Fas and LAG-3 expression in resting T cells. Blocking LAG-3 decreased the expression of Fas on T cells from both healthy and off-therapy RRMS subjects, further underscoring the association between LAG-3 expression and Fas-induced cell death. Our studies were generally not powered to examine the role of therapy in RRMS subjects and include subjects both on and off therapy. Future studies could explore the role of LAG-3 expression in the context of immuno-suppressive therapies, particularly in light of a recent pre-print reporting the upregulation of LAG-3 in the presence of IFN-β, which is also a first-line treatment in MS (37).
In human autoimmunity, targeting LAG-3 therapeutically could function to selectively downmodulate activated T cells (32). Yet the efficacy of such approaches will depend in part on the function and expression of LAG-3 on autoreactive T cells being targeted by these therapies. Our findings indicate that LAG-3 is dysregulated in RRMS and T1D. This LAG-3 dysregulation may contribute to the escape of autoreactive T cells in autoimmunity by evading apoptosis. We find that the dominant source of this dysregulation is a decrease in transcription rather than cleavage of the expressed protein. Thus, targeting LAG-3 therapeutically in RRMS and T1D should take into account the source of dysregulation. This study further endorses the need to study human autoimmune samples in order to develop an in depth understanding of the regulation and expression of therapeutic targets in the context of disease.
Supplementary Material
Key Points.
There are fewer LAG-3+ CD4 and CD8 T cells in subjects with RRMS and T1D.
LAG-3 protein expression is linked to alterations in mRNA expression.
Low LAG-3 may be associated with decreased in T cell death in RRMS subjects.
Acknowledgements
We thank the BRI Clinical Core Laboratory, in particular T. Nguyen and acknowledge the efforts of research assistants David Kook, Jenna Snavely, and Kim Varner in the Translational Research Program and the Diabetes Clinical Research Program in addition to Sylvia Posso for sample selection. We also thank A. Hocking and V. Green for assistance in editing and all participants in the BRI Registry and Repository.
Funding Acknowledgements
This work was supported by the National Institutes of Health (NIAID R01 AI132774; J.H.B.).
Abbreviations used in this article:
- LAG-3
lymphocyte activation gene-3
- RRMS
relapsing remitting multiple sclerosis
- T1D
type 1 diabetes
- IRs
inhibitory receptors
- MS
multiple sclerosis
- EAE
experimental autoimmune encephalomyelitis
- MOG
myelin oligodendrocyte glycoprotein
- LAG-3NC
non-cleavable LAG-3
- cCasp3
cleaved caspase 3
- sLAG3
soluble LAG-3
- HC
healthy control subjects
- DMT
disease modifying therapies
- PD-1
PDCD1
- TIM3
HAVCR2
- NS
not significant
Footnotes
Disclosures
The authors declare that no conflicts of interest exist pertaining to the contents of this manuscript.
References
- 1.National Institute of Environmental Health Sciences. 2020. NIEHS Autoimmune Diseases Fact Sheet: Autoimmune Diseases and Your Environment. In https://www.niehs.nih.gov/health/materials/autoimmune_diseases_and_your_environment_508.pdf
- 2.Joller N, and Kuchroo VK 2017. Tim-3, Lag-3, and TIGIT. Curr. Top. Microbiol. Immunol.: Emerging Concepts Targeting Immune Checkpoints in Cancer and Autoimmunity 410: 127–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, Nyman J, Marjanovic ND, Kowalczyk MS, and Wang C 2018. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558: 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bristol Myers Squibb. 2021. Bristol Myers Squibb Announces LAG-3-Blocking Antibody Relatlimab and Nivolumab Fixed-Dose Combination Significantly Improves Progression-Free Survival vs. Opdivo (nivolumab) in Patients with Previously Untreated Metastatic or Unresectable Melanoma. https://news.bms.com/news/details/2021/Bristol-Myers-Squibb-Announces-LAG-3-Blocking-Antibody-Relatlimab-and-Nivolumab-Fixed-Dose-Combination-Significantly-Improves-Progression-Free-Survival-vs.-Opdivo-nivolumab-in-Patients-with-Previously-Untreated-Metastatic-or-Unresectable-Melanoma/default.aspx 5/19/2021
- 5.Huang C, Zhu H-X, Yao Y, Bian Z-H, Zheng Y-J, Li L, Moutsopoulos HM, Gershwin ME, and Lian Z-X 2019. Immune checkpoint molecules. Possible future therapeutic implications in autoimmune diseases. J. Autoimmun. 104: 102333. [DOI] [PubMed] [Google Scholar]
- 6.Dougan M, and Pietropaolo M 2020. Time to dissect the autoimmune etiology of cancer antibody immunotherapy. J. Clin. Invest. 130: 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia CR, Jayswal R, Adams V, Anthony LB, and Villano JL 2019. Multiple sclerosis outcomes after cancer immunotherapy. Clin. Transl. Oncol. 21: 1336–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadowaki A, Miyake S, Saga R, Chiba A, Mochizuki H, and Yamamura T 2016. Gut environment-induced intraepithelial autoreactive CD4+ T cells suppress central nervous system autoimmunity via LAG-3. Nat. Commun. 7: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li N, Wang Y, Forbes K, Vignali KM, Heale BS, Saftig P, Hartmann D, Black RA, Rossi JJ, and Blobel CP 2007. Metalloproteases regulate T-cell proliferation and effector function via LAG-3. EMBO J. 26: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews LP, Somasundaram A, Moskovitz JM, Szymczak-Workman AL, Liu C, Cillo AR, Lin H, Normolle DP, Moynihan KD, and Taniuchi I 2020. Resistance to PD1 blockade in the absence of metalloprotease-mediated LAG3 shedding. Sci. Immunol 5: eabc2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krienke C, Kolb L, Diken E, Streuber M, Kirchhoff S, Bukur T, Akilli-Öztürk Ö, Kranz LM, Berger H, and Petschenka J 2021. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science 371: 145–153. [DOI] [PubMed] [Google Scholar]
- 12.Polman CH, Reingold SC, Edan G, Filippi M, Hartung HP, Kappos L, Lublin FD, Metz LM, McFarland HF, and O’Connor PW 2005. Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria”. Ann. Neurol. 58: 840–846. [DOI] [PubMed] [Google Scholar]
- 13.Motulsky HJ, and Brown RE 2006. Detecting outliers when fitting data with nonlinear regression–a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 7: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li N, Workman CJ, Martin SM, and Vignali DA 2004. Biochemical analysis of the regulatory T cell protein lymphocyte activation gene-3 (LAG-3; CD223). J. Immunol. 173: 6806–6812. [DOI] [PubMed] [Google Scholar]
- 15.Woo SR, Li N, Bruno TC, Forbes K, Brown S, Workman C, Drake CG, and Vignali DA 2010. Differential subcellular localization of the regulatory T-cell protein LAG-3 and the coreceptor CD4. Eur. J. Immunol. 40: 1768–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae J, Lee SJ, Park C-G, Lee YS, and Chun T 2014. Trafficking of LAG-3 to the surface on activated T cells via its cytoplasmic domain and protein kinase C signaling. J. Immunol. 193: 3101–3112. [DOI] [PubMed] [Google Scholar]
- 17.Durelli L, Conti L, Clerico M, Boselli D, Contessa G, Ripellino P, Ferrero B, Eid P, and Novelli F 2009. T-helper 17 cells expand in multiple sclerosis and are inhibited by interferon-β. Ann. Neurol. 65: 499–509. [DOI] [PubMed] [Google Scholar]
- 18.Boziki M, Lagoudaki R, Melo P, Kanidou F, Bakirtzis C, Nikolaidis I, Grigoriadou E, Afrantou T, Tatsi T, and Matsi S 2019. Induction of apoptosis in CD4 (+) T-cells is linked with optimal treatment response in patients with relapsing-remitting multiple sclerosis treated with Glatiramer acetate. J. Neurol. Sci. 401: 43–50. [DOI] [PubMed] [Google Scholar]
- 19.Petelin Z, Brinar V, Petravic D, Zurak N, Dubravcic K, and Batinic D 2004. CD95/Fas expression on peripheral blood T lymphocytes in patients with multiple sclerosis: effect of high-dose methylprednisolone therapy. Clin. Neurol. Neursurg. 106: 259–262. [DOI] [PubMed] [Google Scholar]
- 20.Elmore S 2007. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35: 495–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grebinoski S, and Vignali DA 2020. Inhibitory receptor agonists: the future of autoimmune disease therapeutics? Curr. Opin. Immunol. 67: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gertel S, Polachek A, Furer V, Levartovsky D, and Elkayam O 2021. CD4+ LAG-3+ T Cells are Decreased in Active Psoriatic Arthritis Patients and Their Restoration In Vitro is Mediated by TNF Inhibitors. Clin. Exp. Immunol. 206: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettini M, Szymczak-Workman AL, Forbes K, Castellaw AH, Selby M, Pan X, Drake CG, Korman AJ, and Vignali DA 2011. Cutting edge: accelerated autoimmune diabetes in the absence of LAG-3. J. Immunol. 187: 3493–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavon I, Heli C, Brill L, Charbit H, and Vaknin-Dembinsky A 2019. Blood Levels of Co-inhibitory-Receptors: A Biomarker of Disease Prognosis in Multiple Sclerosis. Front. Immunol. 10: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klümper N, Ralser DJ, Bawden EG, Landsberg J, Zarbl R, Kristiansen G, Toma M, Ritter M, Hölzel M, and Ellinger J 2020. LAG3 (LAG-3, CD223) DNA methylation correlates with LAG3 expression by tumor and immune cells, immune cell infiltration, and overall survival in clear cell renal cell carcinoma. J. Immunother. Cancer 8: e000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair VS, El Salhat H, Taha RZ, John A, Ali BR, and Elkord E 2018. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin. Epigenetics 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fröhlich A, Sirokay J, Fietz S, Vogt TJ, Dietrich J, Zarbl R, Florin M, Kuster P, Saavedra G, and Valladolid SR 2020. Molecular, clinicopathological, and immune correlates of LAG3 promoter DNA methylation in melanoma. EBioMedicine 59: 102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koguchi K, Anderson DE, Yang L, O’Connor KC, Kuchroo VK, and Hafler DA 2006. Dysregulated T cell expression of TIM3 in multiple sclerosis. J. Exp. Med. 203: 1413–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Anderson DE, Kuchroo J, and Hafler DA 2008. Lack of TIM-3 immunoregulation in multiple sclerosis. J. Immunol. 180: 4409–4414. [DOI] [PubMed] [Google Scholar]
- 30.Maçon-Lemaître L, and Triebel F 2005. The negative regulatory function of the lymphocyte-activation gene-3 co-receptor (CD223) on human T cells. Immunology 115: 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichtenegger FS, Rothe M, Schnorfeil FM, Deiser K, Krupka C, Augsberger C, Schlüter M, Neitz J, and Subklewe M 2018. Targeting LAG-3 and PD-1 to enhance T cell activation by antigen-presenting cells. Front. Immunol. 9: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angin M, Brignone C, and Triebel F 2020. A LAG-3–Specific Agonist Antibody for the Treatment of T Cell–Induced Autoimmune Diseases. J. Immunol. 204: 810–818. [DOI] [PubMed] [Google Scholar]
- 33.Huard B, Tournier M, Hercend T, Triebel F, and Faure F 1994. Lymphocyte-activation gene 3/major histocompatibility complex class II interaction modulates the antigenic response of CD4+ T lymphocytes. Eur. J. Immunol. 24: 3216–3221. [DOI] [PubMed] [Google Scholar]
- 34.Lopatinskaya L, Zwemmer J, Uitdehaag B, Lucas K, Polman C, and Nagelkerken L 2006. Mediators of apoptosis Fas and FasL predict disability progression in multiple sclerosis over a period of 10 years. Mult. Scler. 12: 704–709. [DOI] [PubMed] [Google Scholar]
- 35.Comi C, Leone M, Bonissoni S, DeFranco S, Bottarel F, Mezzatesta C, Chiocchetti A, Perla F, Monaco F, and Dianzani U 2000. Defective T cell fas function in patients with multiple sclerosis. Neurology 55: 921–927. [DOI] [PubMed] [Google Scholar]
- 36.Okuda Y, Apatoff BR, and Posnett DN 2006. Apoptosis of T cells in peripheral blood and cerebrospinal fluid is associated with disease activity of multiple sclerosis. J. Neuroimmunol. 171: 163–170. [DOI] [PubMed] [Google Scholar]
- 37.Sumida TS, Dulberg S, Schupp J, Stillwell HA, Axisa P-P, Comi M, Lincoln M, Unterman A, Kaminski N, and Madi A et al. 2020. Type I Interferon Transcriptional Network Regulates Expression of Coinhibitory Receptors in Human T cells. bioRxiv Preprint. Oct 31: doi: 10.1101/2020.1110.1130.362947 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.